Abstract
Mammalian spermatogenesis is a continuum of cellular differentiation in a lineage that features three principal stages: 1) a mitotically active stage in spermatogonia, 2) a meiotic stage in spermatocytes, and 3) a postreplicative stage in spermatids. We used a microarray-based approach to identify changes in expression of cell-cycle genes that distinguish 1) mitotic type A spermatogonia from meiotic pachytene spermatocytes and 2) pachytene spermatocytes from postreplicative round spermatids. We detected expression of 550 genes related to cell-cycle function in one or more of these cell types. Although a majority of these genes were expressed during all three stages of spermatogenesis, we observed dramatic changes in levels of individual transcripts between mitotic spermatogonia and meiotic spermatocytes and between meiotic spermatocytes and postreplicative spermatids. Our results suggest that distinct cell-cycle gene regulatory networks or subnetworks are associated with each phase of the cell cycle in each spermatogenic cell type. In addition, we observed expression of different members of certain cell-cycle gene families in each of the three spermatogenic cell types investigated. Finally, we report expression of 221 cell-cycle genes that have not previously been annotated as part of the cell cycle network expressed during spermatogenesis, including eight novel genes that appear to be testis-specific.
Keywords: cell cycle, gametogenesis, gene regulation, meiosis, microarray, spermatogenesis, testis
Distinct networks of cell-cycle genes are associated with the mitotic, meiotic, and postreplicative stages of spermatogenesis.
INTRODUCTION
In the adult male mammal, spermatogenesis is a continuum of differentiation in a cell lineage for which three principal stages can be discerned: 1) a mitotic stage in spermatogonia, including cellular renewal and proliferation; 2) a meiotic stage in spermatocytes, including meiosis and genetic recombination; and 3) a postreplicative stage in spermatids, when haploid male germ cells undergo the differentiative process of spermiogenesis in the absence of any further replication [1–3]. The spermatogonial stem cell population replicates mitotically both to maintain itself and to give rise to mitotically active, differentiating spermatogonia. These differentiating cells then enter meiosis as primary spermatocytes that proceed through the first meiotic division to yield secondary spermatocytes and through the second meiotic division to produce postmeiotic spermatids. Spermatids then undergo extensive differentiation via the process of spermiogenesis to form spermatozoa.
A typical eukaryotic cell cycle progresses through mitosis (M phase) alternating with the much longer interphase, which is divided into three phases: 1) G1, 2) S, and 3) G2 [4]. A cell grows during the G1 phase, continues to grow and duplicates its chromosomes during the S phase, prepares for mitosis during the G2 phase, and divides during the M phase [5–7]. Cell-cycle genes are known to be differentially expressed during germ cell development in a wide range of organisms [8–11]. Spermatogonia are mitotic cells that can be expected to share mechanisms of cell-cycle regulation with other mitotic cells, including most somatic cell types. During meiosis I in primary spermatocytes, the DNA is replicated as in a mitotic cell cycle, but then homologous chromosomes pair and chiasmata form between homologues to facilitate genetic recombination and the subsequent proper segregation of chromosomal homologues to each secondary spermatocyte. This is followed by a second meiotic division without an intervening round of DNA replication that separates sister chromatids into haploid spermatids [12–14]. Because meiosis occurs uniquely in germline cells, spermatocytes can be expected to utilize at least some cell-cycle control mechanisms that are not used in mitotic somatic cells or in premeiotic spermatogonia. Spermiogenesis occurs in the absence of cytokinesis [15] and, therefore, might be expected to be characterized by cell-cycle control distinct from that in either spermatogonia or spermatocytes.
Cellular differentiation is mediated by differential gene expression, and spermatogenesis is no exception [16]. Microarrays provide a platform to simultaneously evaluate expression of thousands of genes [17–20]. Indeed, studies based on microarray or expressed sequence tag analyses have revealed dynamic changes in gene expression during spermatogenesis [18–22]. In the present study, our objective was to derive a comprehensive characterization of the expression of genes that contribute to regulation of the cell cycle throughout spermatogenesis, with a particular emphasis on changes in expression of these genes as spermatogenic cells progress from mitotic spermatogonia to meiotic spermatocytes and then to postreplicative spermatids. We addressed this objective by mining recently acquired microarray data published by Namekawa et al. [23] describing gene expression in type A spermatogonia representing the mitotically active stage, pachytene spermatocytes representing the meiotic stage, and round spermatids representing the postreplicative stage of spermatogenesis. We used this approach to specifically address the following questions regarding regulation of the cell cycle during spermatogenesis: 1) Are entire cell-cycle networks differentially expressed in mitotic, meiotic, and postreplicative spermatogenic cell types, respectively? 2) Are specific members of common cell-cycle networks differentially expressed in each spermatogenic cell type? 3) Are similar cell-cycle networks expressed in spermatogenic and somatic cell types, but with unique, spermatogenesis-specific members included in the former? 4) Are pathways that support or impact cell-cycle regulation differentially expressed during spermatogenesis?
Our results demonstrate that many of the same cell-cycle genes are expressed during all three stages of spermatogenesis in the mouse. However, striking changes were found in the expression levels of these genes as a function of the transition from mitotic spermatogonia to meiotic spermatocytes, followed by a general down-regulation of cell-cycle gene expression in postreplicative spermatids. We also observed remarkable changes in the relative expression levels of genes involved in different parts of the cell cycle in each spermatogenic cell type, and this appears to be associated with differential expression of cell-cycle phase-specific networks during each stage of spermatogenesis. Finally, we observed differential expression of members of specific gene families in mitotic, meiotic, and postreplicative spermatogenic cell types.
MATERIALS AND METHODS
Microarray Data Mining
Acquisition of microarray data.
We examined recently acquired microarray data from the Griswold laboratory derived from purified populations of spermatogenic cells isolated in the McCarrey laboratory as previously reported by Namekawa et al. [23] and others [24–27] (GEO accession no. GSE4193) to compare cell-cycle gene expression in mitotic type A spermatogonia, meiotic pachytene spermatocytes, and postreplicative round spermatids. Feature-level data (CEL) files were downloaded and imported into GeneSpring 7.2 software (Silicon Genetics) for further analysis.
Processing of microarray data in GeneSpring.
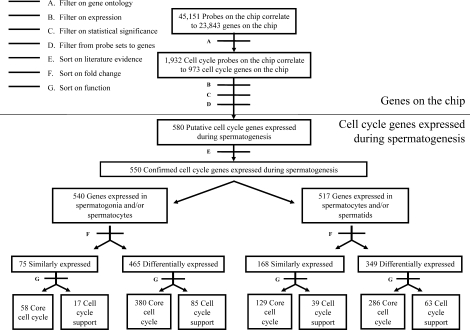
Figure 1 depicts a flow chart of the data analysis procedures used in the present study. Expression data were normalized within GeneSpring using normalization methods as previously described by Shima et al. [21]. To identify cell-cycle genes present on the chip, we examined the controlled vocabulary for the description of cellular components, molecular functions, and biological processes provided by Gene Ontology (GO) using the primary annotation term GO:0007049 and its subheadings as described previously [28, 29]. Of 45 101 probe sets present on the chip, 1923 were annotated by GO to be involved in cell-cycle control (Fig. 1, Filter A). These represent 973 different putative cell-cycle genes.
FIG. 1.
Pipeline for the analysis of cell-cycle genes. This flow chart summarizes the analyses performed based on bioinformatics filters applied. Filter A used GO terms to identify all cell-cycle gene probes represented on the Affymetrix GeneChip mouse genome 430 2.0 microarrays. Filter B was used to identify all probes that were expressed in one or more spermatogenic cell types with a raw intensity of 50 units or greater. Filter C used a one-way ANOVA to identify replicate probe sets expressed at statistically significant consistent levels. Filter D was used to correlate probe sets to individual genes. Filter E involved a thorough literature search to screen genes for prior evidence of involvement in the cell cycle. Filter F was used to identify genes that were differentially expressed by a fold-change of 1.5 or more. Filter G was used to sort “phase-specific, core cell-cycle genes” and “cell cycle-support genes” differentially expressed in spermatogonia and spermatocytes or spermatocytes and spermatids.
Analysis of putative cell-cycle genes expressed during spermatogenesis.
Affymetrix GeneChip microarray technology uses gene-specific probe sets to detect specific mRNAs in each sample. Each probe set is a collection of up to 11 probes designed to interrogate a given transcript, and each probe is 25 nucleotides in length. To eliminate poorly expressed probe sets, only those showing expression values with raw signal intensities of greater than 50 arbitrary fluorescence units were included in our analysis (Fig. 1, Filter B). Of the 1923 cell-cycle probe sets, 1489 were expressed with a raw intensity signal of 50 or more in one or more of the spermatogenic cell types examined. Our microarray expression datasets were derived from two different biological replicates for each cell type. Because this is less than the standard of three replicates commonly used to ensure consistency among samples, we performed a one-way ANOVA on the data for each individual probe set. A minimum P value of 0.05 was required to designate statistically significant, consistent expression between each of two replicates (Fig. 1, Filter C). Based on this criterion, expression of 816 cell-cycle probe sets was significantly consistent. Most genes were represented by only a single probe set on the microarray. Of those that were represented by more than one probe set, only expression data for those that showed concordant expression among all probe sets, or expression of a single probe set that could be considered more gene-specific because of positioning in the untranslated region, were included in subsequent analyses. The UniGene database and NetAffx portal supported by Affymetrix were then used to correlate the probe sets to specific genes (Fig. 1, Filter D). The 816 probe sets corresponded to 580 putative cell-cycle genes, all of which had unique UniGene identifiers.
Analysis of confirmed or predicted cell-cycle and cell cycle-support genes expressed during spermatogenesis.
Each of the 580 putative cell-cycle genes was then subjected to a thorough literature search based on key words and annotated manually for independent evidence of function in, or relationship to, the cell cycle (Fig. 1, Filter E). The starting point in each gene annotation was the ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics database [30]. Information about expression of each gene in mouse tissues and the role of each gene in cell-cycle regulation was obtained through literature analysis of articles extracted from PubMed. Databases including UniGene [31] and GermOnline [32–34] were used as additional sources of information regarding previously reported gene expression patterns.
Of the 580 putative cell-cycle genes expressed during spermatogenesis, 550 were confirmed as genes that impact the cell cycle based on independent reports in the literature showing that ablation or inhibition of the individual cell-cycle gene or the relevant cell cycle-support pathway causes disruption or aberrant progression of the cell cycle (Supplemental Table S1; all supplemental data are available online at www.biolreprod.org).
Analysis of patterns of core cell-cycle and cell cycle-support gene expression during spermatogenesis.
We first sorted the expressed core cell-cycle and cell cycle-support genes into two groups: 1) genes that were constitutively expressed during spermatogenesis (no change of ≥1.5-fold during spermatogenesis) and 2) genes that were differentially expressed during spermatogenesis (one or more changes of ≥1.5-fold during spermatogenesis) (Fig. 1, Filter F). We chose 1.5-fold as the minimum fold-change indicative of differential expression both because this has been used in previous microarray studies [35] and because we wanted to comprehensively identify changes in expression levels of cell-cycle genes during spermatogenesis. Importantly, this fold-change was considered to be valid only when applied to differences in expression levels obtained from duplicate samples that were determined by ANOVA to be significantly consistent (P < 0.05) as described above. Using this criterion, we found that 536 of the 550 cell-cycle genes showed differential expression among the mitotic, meiotic, and/or postreplicative stages of spermatogenesis. The remaining 14 cell-cycle genes were expressed constitutively throughout spermatogenesis. Using the standard k-means algorithm provided in GeneSpring, we clustered cell-cycle genes according to their expression patterns during spermatogenesis. We used “trajectory clustering,” a nonparametric method of clustering gene expression data from time-course experiments [36, 37]. The trajectories used in our clustering method were defined by the direction of change between adjacent cell types in a series. The direction of change can take on one of three possible values: 1) increasing, 2) decreasing, or 3) unchanged. For our analysis, we had three (N) cell types, so there could be N − 1 changes, and 3N−1, or nine, possible patterns. GeneSpring's k-means clustering algorithm divides genes into predetermined groups, and clusters are constructed so that the average behavior in each group is distinct from that of any other group [38]. The resulting patterns included genes that showed constitutive expression throughout spermatogenesis (one possible pattern), genes that showed differential expression at one of two transitions (mitotic to meiotic or meiotic to postmeiotic/postreplicative) during spermatogenesis (four possible patterns), or genes that showed differential expression at both transitions (mitotic to meiotic and meiotic to postmeiotic or postreplicative) during spermatogenesis (four possible patterns).
Classification of core cell-cycle and cell cycle-support genes.
We further categorized the core cell-cycle and cell cycle-support genes through literature searches based on functional annotation (Fig. 1, Filter G). We divided the core cell-cycle genes into two subcategories: 1) phase-specific genes (328 genes) for which published reports of ablation or inhibition showing disruption in the progression of the cell cycle at one or more specific phases were found (Supplemental Table S1) and 2) non-phase-specific genes (94 genes) for which either no reports of ablation or inhibition were found or published studies of ablation or inhibition did not indicate disruption of the cell cycle in a phase-specific manner (Supplemental Table S1). We further subdivided the phase-specific cell-cycle genes into 1) inducers (215 genes), defined as phase-specific cell-cycle genes for which functional analyses have demonstrated a role in promoting the frequency or extent of cell-cycle progression, and 2) inhibitors (113 genes), defined as phase-specific cell-cycle genes for which functional analyses have demonstrated a role in decreasing the frequency or extent of cell-cycle progression (Supplemental Table S1). Genes annotated as inducers or inhibitors of specific parts of the cell cycle were grouped separately, and heat maps were created to facilitate visualization of the expression data. This was done by hierarchical cluster analysis using the GeneSpring software [37]. Hierarchical clustering is an iteratively agglomerative clustering method used to produce gene trees. The resulting trees group the genes based on the similarity of their expression profiles [39].
Pathway analysis.
To characterize the unique gene networks associated with progression of the cell cycle during each stage of spermatogenesis, we used the pathway analysis tool available in GeneSpring GX 10 to identify genes encoding proteins that interact with any of the 343 core cell-cycle phase-specific genes expressed in one or more spermatogenic cells based on evidence from the literature. The pathway analysis algorithm in GeneSpring GX 10 is a proprietary Natural Language Processing algorithm that maps each expressed gene identifier to its corresponding gene entity in a knowledge base derived from published literature abstracts obtained through PubMed and maintained by the company supplying the GeneSpring GX 10 software (Agilent). This approach was used to create a list of “focus genes” as described in the GeneSpring GX 10 manual (available from www.chem.agilent.com). The software then overlaid this list of focus genes onto a global molecular network developed from information contained in the knowledge base to derive networks in which the focus genes are projected as nodes and interacting partners are clustered around these nodes based on their reported connectivity. This allowed us to group subsets of these focus genes into functional networks associated with each phase or subphase of the cell cycle. Because of the extreme complexity of these networks, we have posted them online as supplemental data (available at www.biolreprod.org and at http://darwin.cbi.utsa.edu/mccarreylab). For each of these cell-cycle phase- or subphase-specific networks, this online database includes an HTML map that allows the reader to click on any individual member of the network or any individual interaction between any two connected members of the network to link to additional information about each. The HTML maps also include relative expression of each member in each spermatogenic cell type as indicated by individual color-coded bars. This allowed us to correlate our expression data with previously reported functional data for each phase-specific, core cell-cycle gene.
Differential expression of genes encoding proteins that interact with the nodal core cell-cycle genes was assessed in our microarray data as described above. If a nodal focus gene plus one or more interacting partner genes were up-regulated by more than 1.5-fold in a particular spermatogenic cell type relative to either of the other two spermatogenic cell types investigated, we classified that subnetwork as up-regulated in that cell type (summarized in Table 1 and highlighted in red in the online expression maps).
TABLE 1.
Differential expression of cell cycle phase-specific gene networks.
Analysis of expression of members of cell-cycle gene families.
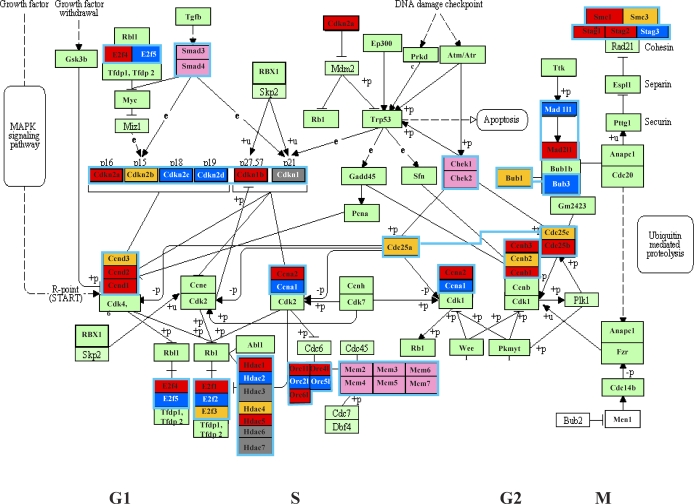
To characterize potential differential expression of specific members of families of core cell-cycle genes, we applied a color-coded scheme to the KEGG core cell-cycle network (http://www.genome.jp/kegg-bin/show_pathway?mmu04110). Thus, up-regulation of specific members of core cell-cycle gene families in each of the three spermatogenic cell types is represented by distinct colors in Figure 2.
FIG. 2.
KEGG cell-cycle pathway depicting differential expression of genes belonging to the same gene family. Core cell-cycle gene families containing members that were differentially expressed in spermatogonia, spermatocytes, and/or spermatids are color-coded red, dark blue, and gold, respectively. Genes colored light green were either the only member of that gene family to be expressed in spermatogenic cells or among multiple members of a gene family that were similarly expressed in spermatogenic cells. Boxes enclosed in light blue depict individual gene families. Genes colored gray were not expressed in any spermatogenic cell type analyzed. Genes colored white were not represented on the microarray and so could not be assessed for expression.
Validation of Microarray Results
Preparation of cells and RNA.
For quantitative RT-PCR (qRT-PCR) and RT-PCR experiments, populations of specific spermatogenic cell types enriched from mouse testes and distinct from those used for the microarray analysis were prepared using a StaPut 2–4% bovine serum albumin gradient as described previously [23]. The animals were housed in the University of Texas at San Antonio animal care facility, and the Institutional Animal Care and Use Committee of the University of Texas at San Antonio approved all of the animal experimentation protocols. Total RNA was recovered from each cell population using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Three replicate RNA samples were prepared from each purified cell type and used for each qRT-PCR or RT-PCR experiment. RNA quantity and quality were determined by ultraviolet (UV) absorption readings at 260 and 280 nm and agarose gel electrophoresis. An optical density ratio (OD260/280) of 1.8 along with evidence of intact ribosomal RNA bands following electrophoresis in agarose was considered to be ideal for subsequent use of the RNA in expression validation experiments.
Real-time qRT-PCR.
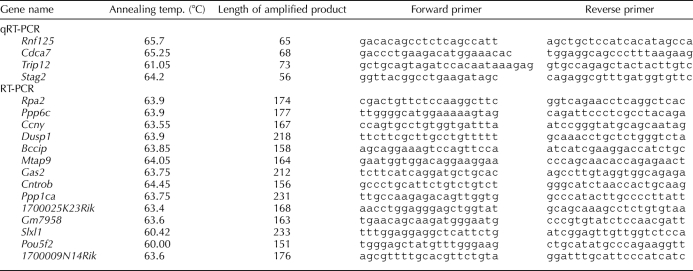
Primers for qRT-PCR experiments (Table 2, sets 1–4) were designed using RealTimeDesign software from Bioresearch Technologies. One microgram of total RNA from each purified cell type was reverse-transcribed in 20 μl of reaction medium using the iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer's instructions. PCR was performed using SYBR Premix Ex Taq (Takara) on an MJ Research PTC-100 Tetrad Thermal cycler as described previously [40]. All signals were normalized to that for somatic Gapdh, which was constitutively expressed throughout spermatogenesis (Supplemental Table S2) and was arbitrarily set to a value of 1.0. Relative quantification of gene expression was calculated using the comparative cycle threshold method as described previously by Livak and Schmittgen [41]. Each qRT-PCR data point shown in the present study is based on an average of results from three biological replicates.
TABLE 2.
Primers used for qRT-PCR and RT-PCR experiments.
Standard RT-PCR.
Primers (Table 2, sets 5–18) were designed to unique regions of exemplary transcripts representing 14 of the 221 cell-cycle genes not previously annotated as expressed during spermatogenesis. Following DNase treatment to remove any contaminating genomic DNA, 500 ng of total RNA from type A spermatogonia, 500 ng from pachytene spermatocytes, and 500 ng from round spermatids were reverse transcribed using an oligo(dT) primer. One microliter of the resulting cDNA was used for each 25-μl PCR reaction, involving an initial denaturation for 1 min at 94°C followed by 35 cycles of annealing at 55–62°C (exact temperature determined by primer sequence in each case; see Table 2), extension at 70°C for 15 sec, and denaturation for 1 min at 94°C. HotMaster Taq DNA Polymerase (Eppendorf) was used in all reactions. PCR products were visualized on the basis of ethidium bromide staining using a UV imaging system. In all cases, products from RT-PCR amplification of RNA from the somatic Gapdh gene served as a positive control, and no-template and no-RT controls (data not shown) were run with each primer set. Each reaction was conducted at least twice to confirm expression patterns. A 1-kb DNA ladder was used as a size marker.
RESULTS
Global Cell-Cycle Gene Expression Patterns During Spermatogenesis
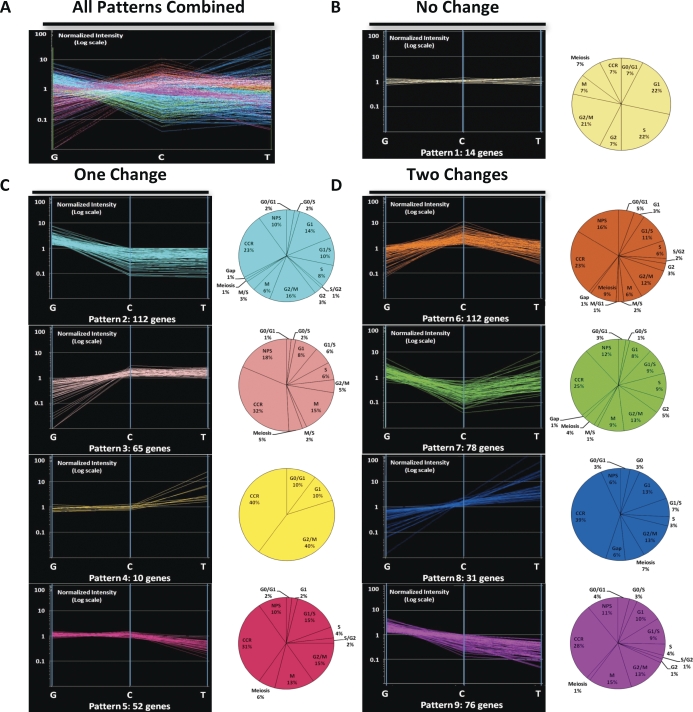
As with most microarray-based expression studies, our data describe levels of steady-state transcripts for each gene investigated and do not directly comment on the presence or absence of encoded proteins or any effects of posttranscriptional regulation. Among the 550 confirmed cell-cycle genes we found to be expressed during spermatogenesis, 520 were expressed in spermatogonia, 496 were expressed in spermatocytes, and 490 were expressed in spermatids. Of these, 453 were expressed in all three spermatogenic cell types examined, whereas 50 genes were expressed in two of the three cell types and the remaining 47 genes were expressed exclusively in only one cell type during spermatogenesis (see Supplemental Table S1 for designations of genes expressed in each cell type). Thus, although a great majority of these genes were expressed during all three stages of spermatogenesis, distinct sets of genes were expressed in spermatogonia, spermatocytes, and spermatids. The highly dynamic expression patterns during spermatogenesis of these genes are shown in Figure 3A.
FIG. 3.
Patterns of core cell-cycle gene expression during spermatogenesis. A) Cumulative cell-cycle gene expression profiles during spermatogenesis. The expression patterns of all 550 cell-cycle genes expressed during spermatogenesis are represented. This reveals dynamic changes in expression of these genes as spermatogenesis proceeds from mitotic spermatogonia (G) to meiotic spermatocytes (C) to postmeiotic spermatids (T). K-means clustering was used to group the 550 cell-cycle genes expressed during spermatogenesis into nine different expression patterns. B) Expression pattern 1 included those cell-cycle genes that showed no changes of 1.5-fold or more in level of expression between cell types. C) Expression patterns 2–5 included those genes that showed differential expression (≥1.5-fold change) between one pair of sequential spermatogenic cell types but not both. D) Expression patterns 6–9 included those genes that showed differential expression (≥1.5-fold change) between both pairs of sequential spermatogenic cell types. Each expression pattern is color-coded. In each case, the y-axis represents normalized intensity of expression of a gene in each of the three spermatogenic cell types represented on the x-axis. The pie charts adjacent to each expression pattern display the distribution of genes in each expression pattern annotated to be associated with specific phases or subphases of the cell cycle. Riken-predicted full-length transcripts and expressed sequence tags that are annotated by GO to be involved in cell-cycle function but with no available phase-specific annotations were categorized as “other” genes in each pattern.
Of the 550 genes expressed during spermatogenesis that were confirmed to impact the cell cycle, 439 were known (422 genes) or predicted (17 genes) to function in networks that directly regulate the cell cycle and, therefore, were termed “core cell-cycle genes.” The remaining 111 genes were known (103 genes) or predicted (eight genes) to function in distinct networks or pathways that support or interact with the cell cycle (e.g., pathways for mitogen-activated protein kinase [MAPK; 4 genes], apoptosis [21 genes], ubiquitination [55 genes], DNA repair [14 genes], circadian cycle regulation [1 gene], and cell signaling, such as Wnt signaling [2 genes], Notch signaling [4 genes], G-protein signaling [5 genes], Rho signaling [3 genes], calmodulin signaling [2 genes]) and were collectively termed “cell cycle-support genes.”
Genes with similar expression patterns likely encode proteins that function in related biological processes [42]. Therefore, we analyzed expression of the 550 cell-cycle genes during spermatogenesis and clustered those with similar patterns. We detected nine distinct expression patterns of cell-cycle genes during spermatogenesis, including one pattern shared by 14 genes showing constitutive expression throughout spermatogenesis (i.e., no changes of ≥1.5-fold in level of expression among the three spermatogenic cell types examined) (Fig. 3B). The other 536 cell-cycle genes showed differential (≥1.5-fold change) expression between mitotic spermatogonia and meiotic spermatocytes, between meiotic spermatocytes and postreplicative spermatids, or both. Specifically, we detected four patterns involving differential expression between two of the three cell types (Fig. 3C) and another four patterns involving differential expression among all three cell types (Fig. 3D).
We further classified the core cell-cycle genes represented within each expression pattern based on their phase-specific annotations to determine if specific phases of the cell cycle predominated in each (see pie charts in Fig. 3, but note that this analysis was performed only on core cell-cycle genes and not on cell cycle-support genes). We found only a single annotated G0 gene (RBL2/p130, up-regulated in spermatids) and a single annotated M/G1 gene (Ran, up-regulated in spermatocytes) to be expressed during spermatogenesis. However, multiple genes associated with each of the other phases of the cell cycle were represented among each of the expression patterns shown in Figure 3. Genes up-regulated in spermatogonia included those associated with the G1, G1/S, G1/G2, S, G2/M, and M phases of the cell cycle, indicative of the ongoing mitotic activity in these cells. A similar emphasis on expression of genes associated with ongoing replication was seen in spermatocytes, except that a marked increase was seen in expression of meiosis-specific genes, as expected in this meiotic cell type. Finally, proportionally greater expression of genes associated with the gap phases was observed in spermatids, which presumably reflects the postreplicative status of these cells.
Comparison of Spermatogonia and Spermatocytes
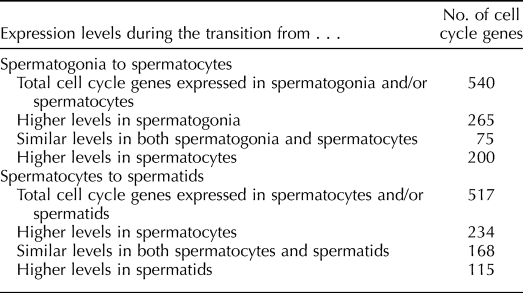
Because of the distinct transitions in cellular states (mitotic, meiotic, and postreplicative) associated with spermatogonia, spermatocytes, and spermatids, respectively, and the dynamic changes in expression of cell-cycle genes associated with these transitions (Fig. 3A), we next focused on the changes in cell-cycle gene expression that distinguish spermatogonia from spermatocytes and spermatocytes from spermatids. A total of 540 cell-cycle genes were expressed in spermatogonia, spermatocytes, or both (Table 3). Of these, 75 were expressed with similar intensity in both cell types (Fig. 3, patterns 1, 4, and 5). Approximately 36% (27/75) of the similarly expressed genes function primarily during the G1/S and G2/M phases of the cell cycle. The remaining 465 genes expressed in spermatogonia and/or spermatocytes were differentially expressed. Among these, 265 were up-regulated in spermatogonia relative to spermatocytes (Fig. 3, patterns 2, 7, and 9), with 69 having expression in spermatogonia 10-fold or greater than that in spermatocytes. The remaining 200 differentially expressed genes were up-regulated in spermatocytes relative to spermatogonia (Fig. 3, patterns 3, 6, and 8), with 40 having expression in spermatocytes 10-fold or greater than that in spermatogonia. Five of the cell-cycle genes up-regulated in spermatogonia and four of those up-regulated in spermatocytes appeared to be testis-specific (see below). All cell-cycle genes expressed in spermatogonia and/or spermatocytes are listed in Supplemental Table S3. Figure 4A shows a heat map summarizing expression patterns of cell-cycle phase-specific genes in spermatogonia and/or spermatocytes, with the genes grouped (where possible) according to their function in each phase. This heat map reveals that a majority of the phase-specific genes were expressed in antagonistic patterns in these two cell types. This suggests that genes associated with each phase of the cell cycle are expressed in both mitotic spermatogonia and meiotic spermatocytes, but that different networks of cell-cycle genes associated with each phase are preferentially expressed in each cell type.
TABLE 3.
Grouping of cell cycle genes based on expression.
FIG. 4.
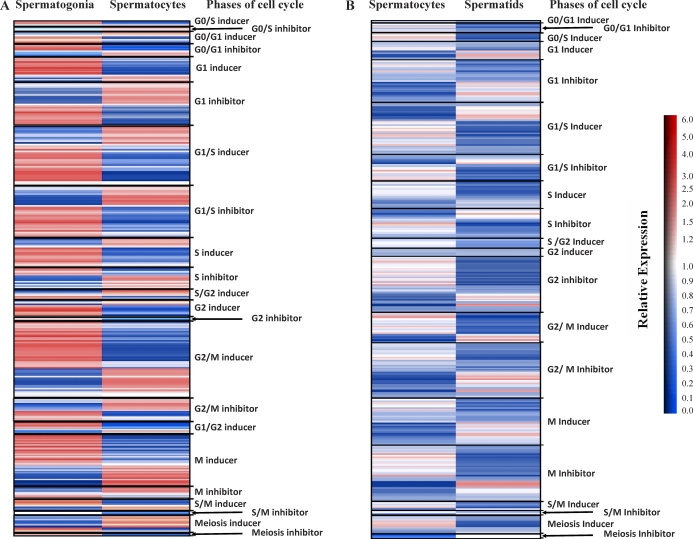
Differential expression of phase-specific cell-cycle genes during spermatogenesis. Heat maps depict the extent of differential expression of core cell-cycle genes during spermatogenesis. Cell-cycle genes belonging to phases including G0/S, G0/G1, G1, G1/S, S, S/G2, G2, G2/M, G1/G2, M, S/M, and Meiosis were further grouped as “inducers” or “inhibitors” of specific phases of the cell cycle based on reports of gene function derived from the literature. The color bar indicates the relative extent of differential expression between cell types (red, up-regulated expression; blue, down-regulated expression; white, little or no significant change in expression level). A) Transition from spermatogonia to spermatocytes. Of the 541 cell-cycle genes expressed in either spermatogonia, spermatocytes, or both, 307 were clustered according to cell-cycle phase annotations to produce this heat map. B) Transition from spermatocytes to spermatids. Of the 521 cell-cycle genes expressed in either spermatocytes, spermatids, or both, 162 were clustered as phase-specific genes according to cell-cycle phase annotations to produce this heat map.
Of the 163 cell-cycle genes with phase-specific annotations that were differentially expressed in spermatogonia and spermatocytes, 93 were annotated as G0-, G0/G1-, G0/S-, G1-, or G1/S-associated genes. Of these, 45 were inducers of the G0/G1, G0/S, G1, or G1/S phases, and 48 were inhibitors of these phases. A majority (32/45) of the inducers of these phases were up-regulated in spermatogonia, whereas a majority (29/48) of the inhibitors of these phases were up-regulated in spermatocytes. Among the S-phase genes expressed in these two cell types, we found that genes known to regulate mitotic DNA replication (e.g., Cdc23), minichromosome maintenance proteins (e.g., MCM), replication checkpoint (e.g., Prim1), and S-phase inhibitors (e.g., Gak and Clspn) were all down-regulated in spermatocytes. However, genes known to be essential for other aspects of DNA replication, including those encoding DNA polymerases and PCNA, were up-regulated in spermatocytes. Genes up-regulated in spermatocytes also included those involved with regulation of the first meiotic prophase (e.g., Siah1a), chromosomal condensation (e.g., Ppp1cb and Ppp2ca), and synaptonemal complex assembly (e.g., Sycp1, Syce1, and Syce2). Interestingly, some genes annotated as meiosis-specific genes in the literature were up-regulated in spermatogonia relative to spermatocytes [43]. These genes included Stra8, which is required for initiation of meiosis [44]. Overall, these results suggest that the transition from mitosis in spermatogonia to meiosis in spermatocytes is characterized by a switch from relatively high activity of parts of the cell-cycle network that promote initiation into, and progression through, the G1 phase in spermatogonia to relatively high activity of phases of the network that promote entry into a meiosis-specific cell cycle in spermatocytes. All cell-cycle genes expressed in spermatogonia and/or spermatocytes are listed in Supplemental Table S3.
Comparison of Spermatocytes and Spermatids
A total of 517 cell-cycle genes were expressed in spermatocytes and/or spermatids (Table 3), and 168 cell-cycle genes were expressed at similar levels in both cell types (Fig. 3, patterns 1–3). Forty-nine of these regulate the G1, G1/S, G2/M, or meiotic phases of the cell cycle. Among the 349 cell-cycle genes differentially expressed in spermatocytes and spermatids (Fig. 3, patterns 4–9), 234 were up-regulated in spermatocytes (Fig. 3, patterns 5, 6, and 9), whereas 115 were up-regulated in spermatids (Fig. 3, patterns 4, 7, and 8). Only 18 of these differentially expressed genes showed differences in expression levels of 10-fold or greater between the two cell types (3/234 genes up-regulated in spermatocytes and 15/115 genes up-regulated in spermatids). Thus, despite the large proportion of cell-cycle genes that showed differential expression between spermatocytes and spermatids, the magnitude of this differential expression was generally much lower than that observed between spermatogonia and spermatocytes. Thirty cell-cycle genes expressed in spermatocytes became completely silenced in spermatids (Fig. 3, patterns 5, 6, and 9). Of these, 33% (10/30) were annotated to regulate the G0/G1, G0/S, or G1/S phases of the cell cycle. Silencing of these genes in spermatids would appear to correlate with cessation of cellular proliferation during this stage of spermatogenesis.
Figure 4B shows a heat map summarizing expression patterns of cell-cycle phase-specific genes in spermatocytes and/or spermatids. A comparison of Figure 4A and Figure 4B reveals a general suppression of cell-cycle gene expression in spermatids. That this results from a cessation in replication activity in spermatids is suggested by our observation that genes known to induce the M or S phases of the cell cycle were generally down-regulated in spermatids relative to spermatocytes, whereas those known to inhibit these phases were generally up-regulated in spermatids. Not surprisingly, many cell-cycle genes annotated as meiosis-specific also showed significant down-regulation in spermatids relative to spermatocytes. Interestingly, no G0 genes were expressed during this transition, despite the fact that these cells were exiting the cell cycle. These results suggest that the transition from meiosis to postmeiosis during spermatogenesis is characterized by a switch from relatively high activity of phases of the cell-cycle network that promote meiosis-specific cell-cycle progression in spermatocytes to relatively high activity of phases of the network that promote exit from the G2 phase and entry into the postreplicative differentiation process in spermatids. All cell-cycle genes expressed in spermatocytes and/or spermatids are listed in Supplemental Table S4.
Pathway Analysis
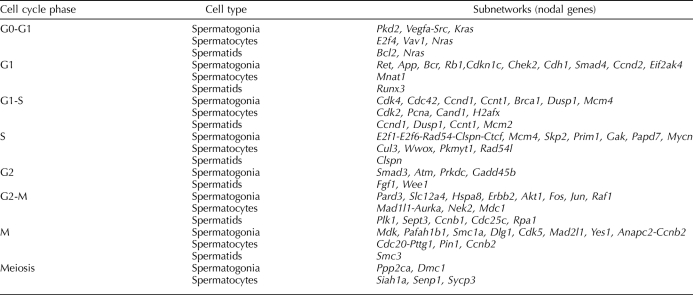
The observation that different subsets of core cell-cycle genes associated with each phase of the cell cycle were expressed in each spermatogenic cell type led us to wonder if these subsets might represent distinct, identifiable networks or subnetworks of genes, the differential expression of which might be related to the distinct mitotic, meiotic, and postreplicative states of the three spermatogenic cell types we studied. To test this hypothesis, we used the pathway analysis tool available in GeneSpring GX 10 to search for groups of genes encoding proteins that have previously been reported to interact and that are coordinately expressed in spermatogonia, spermatocytes, or spermatids. The detailed results of this analysis can be seen in the HTML and expression maps available online (www.biolreprod.org and http://darwin.cbi.utsa.edu/mccarreylab). This approach yielded novel networks of interacting genes associated with five phases (G1, S, G2, M, and meiosis) and three phase-transitions (G0 to G1, G1 to S, and G2 to M) during the cell cycle. Within each of these networks, we identified subnetworks centered around nodal core cell-cycle genes that were differentially expressed in each spermatogenic cell type (Table 1). Thus, 48 subnetworks associated with specific cell-cycle nodal genes were up-regulated in spermatogonia relative to spermatocytes or spermatids, whereas 18 subnetworks associated with other cell-cycle nodal genes and three meiosis-specific subnetworks were up-regulated in spermatocytes relative to spermatogonia and spermatids. In spermatids, only 16 subnetworks associated with cell-cycle nodal genes were up-regulated relative to spermatogonia and spermatocytes (Table 1 and Supplemental Data).
To determine if specific members of cell-cycle gene families are differentially expressed during spermatogenesis, we used the KEGG cell-cycle pathway to depict families of core cell-cycle genes and, where appropriate, used a color scheme to show differential expression of specific family members in different spermatogenic cell types (Fig. 2). Thus, genes up-regulated in spermatogonia, spermatocytes, and spermatids are colored red, blue, and gold, respectively, in Figure 2. As reported previously, members of the cyclin A (A1, A2, and A3), cyclin B (B1, B2, and B3), and cyclin D (D1, D2, and D3) families all showed differential patterns of gene expression during spermatogenesis [45]. However, we also found differential expression of members of other core cell-cycle gene families, including cyclin inhibitory kinase genes (Cdkn2a [p16], p17, Cdkn2c [p18], and Cdkn1b [p27]), G2/M checkpoint genes (Bub1, Bub3, Mad1, Mad2, Smc1, and Smc3), ORC (origin recognition complex) genes (Orc1l, Orc2l, Orc3l, Orc4l, Orc5l, and Orc6l), histone deacetylase genes (Hdac1, Hdac2, Hdac3, Hdac4, Hdac5, Hdac6, and Hdac7), and E2f transcription family genes (E2f1, E2f2, E2f3, E2f4, E2f5, and E2f6). Similarly, we noted differential expression of members of the septin and NIMA (never in mitosis gene a)-related expressed kinase 2 gene families (Supplemental Table S1), although these are not included in the KEGG pathway shown in Figure 2.
Validation of Cell-Cycle Gene Expression During Spermatogenesis
We used three approaches to confirm the validity of the expression patterns of cell-cycle genes indicated by our microarray-based studies of spermatogenic cells.
Validation from the literature.
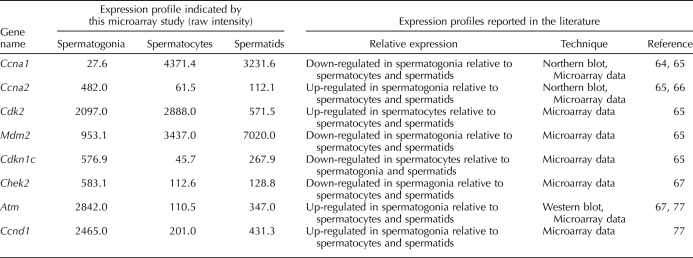
Table 4 lists eight well-known cell-cycle genes for which the expression patterns obtained from our microarray dataset matched those previously reported in the literature, confirming our microarray results for these particular genes.
TABLE 4.
Expression patterns of exemplary cell cycle genes during spermatogenesis.
Validation by RT-PCR.
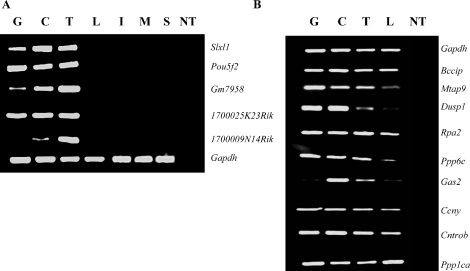
Our microarray results revealed expression in spermatogenic cells of 221 cell-cycle genes, which although included in previously reported microarray-based expression databases have not previously been specifically annotated as expressed during spermatogenesis in the mouse. To validate this expression, we selected nine exemplary genes previously reported to be important in regulating cell-cycle phase-specific function in other tissues. These included Bccip, Dusp1, and Cntron (centrobin), involved in G1/S-phase regulation [46–51]; Gas2, involved in the regulation of apoptosis [52, 53]; Rpa2, involved in DNA replication and damage repair [54]; Mtap9, involved in organization of the bipolar mitotic spindle [55]; Ppp6c and Ppp1ca, involved in regulation of the G2/M transition [56, 57]; and Ccny (cyclin Y), a RIKEN-predicted gene similar to the cyclin fold protein 1 gene [58]. (While preparing this manuscript, Cntron and Mtap9 were reported to be expressed in the testis by another group [48, 59].) Our microarray results also corroborated expression of eight meiosis-specific cell-cycle genes not previously reported to be expressed in spermatogenic cells by any other study. We validated spermatogenic expression of five of these genes, including 1700025K23Rik, involved in chromosome segregation [60], and 1700009N14Rik, Gm7958, Slxl1, and Pou5f2, which are RIKEN-predicted genes showing structural similarities to known cell-cycle genes [61]. Protein-based BLASTP analysis [62] indicates that three of these genes, Slxl1, Pou5f2, and Gm7958, show more than 90% similarity to Xlr-related, meiosis regulated-like genes (Xlr gene family), which are known to be involved in meiosis [61], whereas 1700009N14Rik shows 84% homology to guanosine triphosphate-binding protein (mammalian Ranp homologue), which is involved in the maintenance of nuclear organization, characteristic of genes known to regulate meiotic prophase [63].
We designed primers specific to each transcript (Table 2, sets 5–18) and validated the expression pattern of each gene by RT-PCR analysis of RNA samples from purified populations of type A spermatogonia, pachytene spermatocytes, and round spermatids as well as from exemplary somatic tissues, including liver, intestine, muscle, and spleen (Fig. 5). The results suggest testis-specific expression of the five cell-cycle genes annotated as meiosis-specific (Fig. 5A) and, as expected, confirmed expression in both spermatogenic cells and somatic tissues of the remaining nine genes previously reported to be expressed in somatic tissues (Fig. 5B). Slxl1, Pou5f2, and 1700025K23Rik appeared to be constitutively expressed in all three spermatogenic cell types, whereas Gm7958 and 1700009N14Rik appeared to be differentially expressed among the three spermatogenic cell types. Similarly, Bccip, Rpa2, Ccny, Cntrob, and Ppp1ca appeared to be constitutively expressed in all spermatogenic cell types as well as in somatic liver cells, whereas Mtap9, Dusp1, and Ppp6c appeared to be robustly expressed in spermatogenic cells but only faintly expressed in liver cells. Finally, Gas2 appeared to be robustly expressed in spermatocytes and spermatids but faintly expressed in spermatogonia and liver. These results confirm spermatogenic cell expression of these cell-cycle genes previously reported to be expressed in somatic tissues.
FIG. 5.
RT-PCR validation of expression of cell-cycle genes during spermatogenesis. A) Validation of spermatogenesis-specific expression of meiosis-specific cell-cycle genes. Expression of five cell-cycle genes annotated as meiosis-specific (Slxl1, Pou5f2, Gm7958, 1700025K23Rik, and 1700009N14Rik) was examined in spermatogonia (G), spermatocytes (C), and spermatids (T), along with liver (L), intestine (I), muscle (M), and spleen (S) as somatic controls to confirm tissue-specific expression of these genes. B) Validation of spermatogenic cell expression of ubiquitous cell-cycle genes. Expression of nine cell-cycle genes annotated as ubiquitously expressed (Bccip, Mtap9, Dusp1, Rpa2, Ppp6c, Gas2, Ccny, Cntrob, and Ppp1ca) was examined in spermatogonia (G), spermatocytes (C), and spermatids (T), along with liver (L) to confirm ubiquitous expression in spermatogenic cells and in a somatic tissue. Gapdh expression was used to normalize efficiency of cDNA synthesis and as a positive control for ubiquitous expression. NT, no template control.
Validation by qRT-PCR.
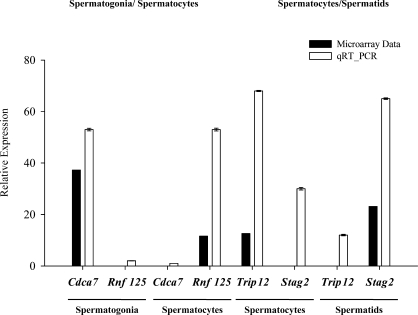
Validation of differential gene expression detected in our microarray experiments was accomplished by performing qRT-PCR on selected genes. We examined our microarray data for expression of the 221 cell-cycle genes that had not previously been annotated as expressed during spermatogenesis and selected four examples of apparent differential expression for further validation. We selected two genes, ring finger protein 125 (Rnf125) and cell division cycle-associated protein 7 (Cdca7), that were among the five most differentially expressed genes in spermatogonia and spermatocytes, with Cdca7 expression being 33-fold higher in spermatogonia than in spermatocytes and Rnf125 expression being 15-fold higher in spermatocytes than in spermatogonia. We selected two more genes, thyroid hormone receptor interactor 12 (Trip12) and stromal antigen 2 (Stag2), that were differentially expressed in spermatocytes and spermatids, with Trip12 expression being 20-fold higher in spermatocytes than in spermatids and Stag2 expression being 25-fold higher in spermatids compared to spermatocytes in our microarray data. Figure 6 shows results of qRT-PCR analyses of the expression of these four genes in RNA samples recovered from purified populations of type A spermatogonia, pachytene spermatocytes, and round spermatids isolated independently from the samples used for microarray analysis. The cell type-specific expression levels were normalized to that for the constitutively expressed Gapdh gene in each of these same cell types. For all four genes, the qRT-PCR results confirmed differential expression consistent with that indicated by our microarray results. In each case, the fold-difference in expression between the two spermatogenic cell types examined by qRT-PCR exceeded that detected by our microarray analysis, indicating that if anything, our microarray results represent an underestimate of the extent of differential expression of cell-cycle genes during spermatogenesis.
FIG. 6.
Validation of differential expression of cell-cycle genes by qRT-PCR. Four cell-cycle genes for which microarray analysis indicated significant differential expression during spermatogenesis were selected for validation by qRT-PCR. These included two cell-cycle genes differentially expressed in spermatogonia and spermatocytes (Rnf125 and Cdca7) and two cell-cycle genes differentially expressed in spermatocytes and spermatids (Trip12 and Stag2). Results are expressed as the mean ± SD of the relative expression of each gene in the different spermatogenic cell types and are representative of three independent experiments. In each case, the extent of differential expression indicted by the microarray analysis is compared to that indicated by qRT-PCR analysis. All values were normalized to that of Gapdh expression in each corresponding cell type.
DISCUSSION
Spermatogenesis is a developmentally dynamic process that involves significant changes in cell-cycle activity, including a mitotic stage in spermatogonia, a meiotic stage in spermatocytes, and a postreplicative stage in spermatids. However, relatively little is known about changes in the expression patterns of specific cell-cycle genes that distinguish these stages. In this context, we have used cDNA microarray technology to profile the expression of 550 cell-cycle or cell cycle-support genes expressed during the various stages of spermatogenesis. We found that a majority of genes known to be involved in regulating the cell cycle (550/973 cell-cycle genes represented on the Affymetrix gene chip) were expressed at some point during spermatogenesis, and that most of these genes were expressed throughout spermatogenesis. However, as would be predicted by the significant transitions in cell-cycle activity during spermatogenesis, we observed differential expression of most of these genes among the different spermatogenic cell types investigated.
The present study represents the first detailed description of spermatogenic cell expression for 221 of the 550 cell-cycle genes we found to be expressed during spermatogenesis, thus significantly expanding our knowledge of the gene network(s) responsible for regulating the cell cycle in male germ cells. Included among these 221 genes are eight genes that appear to be testis-specific. These were previously predicted to be cell-cycle genes [61], but their expression patterns had not been determined. Together with six previously described testis-specific cell-cycle genes [64–66], this brings the total number of apparent testis-specific cell-cycle genes to 15, of which eight are meiosis-specific and seven are associated with the G1/S, G2/M, or S phases of the cell cycle.
As expected, the profile of cell-cycle genes expressed in mitotic spermatogonia was similar to that reported previously for many mitotically active somatic cell types [67–70]. This included predominant expression of genes associated with the G1, G1/S, G2/M, and M phases of the cell cycle. However, this was followed by a shift in expression patterns of cell-cycle genes that accompanies the transition from mitotic spermatogonia to meiotic spermatocytes. Thus, in spermatocytes, we observed enhanced expression of genes associated with the S and S/M phases of the cell cycle, along with up-regulation of many cell-cycle genes previously annotated as meiosis-specific. In postreplicative spermatids, we observed a general down-regulation of cell-cycle gene expression, which was offset by elevated expression of certain genes associated with inhibition of the G1, G1/S, S, G2/M, and meiosis phases. Nevertheless, some genes annotated as inducers of the G1, G1/S, and M phases were still up-regulated in spermatids. These genes may contribute to functions other than inducing proliferation in these nonproliferating cells, such as contributing to DNA repair activities.
Despite the significant differences in expression patterns of cell-cycle genes that distinguished spermatogonia from spermatocytes and spermatocytes from spermatids, we were struck by the observation that at least some genes associated with each phase of the cell cycle were expressed in all three cell types, including in spermatogonia and spermatocytes and, to a lesser extent, in spermatids. What was equally striking, however, were the major changes in expression levels of specific genes associated with each phase of the cell cycle in each spermatogenic cell type. For instance, as can be seen in the heat maps presented in Figure 4, very few cell-cycle genes were expressed at similar levels in consecutive cell types. Rather, most cell-cycle genes that were expressed at relatively high levels in one spermatogenic cell type were down-regulated in the preceding or subsequent cell type.
Thus, whereas these results seem to suggest that some expression of genes is associated with each phase of the cell cycle during each stage of spermatogenesis, they also reveal a great deal of diversity among specific networks of cell-cycle genes expressed in each cell type. This was borne out by our pathway analysis, which allowed us to identify genes encoding proteins that interact with differentially expressed core cell-cycle genes and led us to the discovery that distinct subnetworks of genes associated with specific phases or subphases of the cell cycle are, indeed, differentially regulated as a function of progression through spermatogenesis (Table 1 and Supplemental Data). This in turn suggests that these distinct cell-cycle phase-specific gene networks directly regulate the different replication states during each stage of spermatogenesis (mitotic, meiotic, and postreplicative).
As reported previously for other cell types, we found the cell-cycle network to be biologically robust in spermatogenic cells, with many different genes encoding interchangeable components (i.e., redundancy encoded by multiple members of specific gene families) [71]. Thus, it appears that the different stages of spermatogenesis are also distinguished by the utilization of different specific members of families of cell-cycle genes, as shown in Figure 2. Such differences may represent specializations that facilitate optimal regulation of the cell cycle in each spermatogenic cell type in the context of the significant differences in chromatin structure and composition as well as chromosome status (e.g., 2C, 4C, and 1C) at each stage of spermatogenesis. In yeast, it has been shown that whereas both the mitotic and meiotic S phases depend on the cyclins (cylcin B1 and cyclin B2), mitotic and meiotic cells use different mechanisms to activate these cyclins based on the availability of cyclin-dependent kinase 2 [72–76]. Although the spermatogonial population we investigated included cells in all phases of the mitotic cell cycle but our pachytene spermatocyte population included only cells that had undergone the S phase approximately 1 wk or more before their isolation, our data appear to be consistent with the idea that fundamental differences exist between regulation of the mitotic and meiotic S phases as described in yeast [72]. This is suggested by our observation that Cdk2 and its regulators (Cdc25a, Chek1, and Chek2) are differentially expressed in mouse spermatogonia and spermatocytes, respectively.
In the Introduction, we posed four questions regarding expression of cell-cycle genes during spermatogenesis. Our microarray analysis now positions us to offer answers to these questions. First, are entire cell cycle networks differentially expressed in mitotic, meiotic, and postreplicative spermatogenic cell types? Our results demonstrate that genes associated with many, if not all, phases of the cell cycle are expressed at each stage during spermatogenesis. However, our pathway analysis revealed distinct networks of genes associated with specific phases or subphases of the cell cycle that are differentially expressed during different stages of spermatogenesis.
Second, are specific members of common cell-cycle gene networks differentially expressed in each spermatogenic cell type? We observed extensive differential expression of specific genes associated with each phase of the cell cycle in spermatogonia, spermatocytes, and spermatids, respectively. Thus, we confirmed that many members of cell-cycle gene families are differentially expressed at each stage of spermatogenesis to the extent that it appears unique gene networks regulate the cell cycle in each of the three spermatogenic cell types we investigated.
Third, are similar cell-cycle networks expressed in spermatogenic and somatic cell types, but with unique, spermatogenesis-specific members included in the former? Our results revealed expression of apparent testis-specific genes during all stages of spermatogenesis, although a majority of these genes were preferentially expressed in meiotic spermatocytes. In general, cell-cycle pathways expressed in spermatogenic cells appear to be similar to those expressed in somatic cells, albeit with the involvement of a small fraction of testis-specific genes in the former and with dynamic changes in expression of different phase-specific subnetworks of cell-cycle genes in mitotic, meiotic, and postreplicative spermatogenic cell types.
Fourth, are pathways that support or impact cell-cycle regulation differentially expressed during spermatogenesis? Our data indicate that cell cycle-support genes in the MAPK and apoptotic pathways were up-regulated in spermatogonia relative to spermatocytes, whereas those in the DNA repair and ubiquitination pathways were up-regulated in spermatocytes relative to spermatogonia. Relative to spermatocytes, the MAPK, DNA repair, and ubiquitination pathways were up-regulated in spermatids, whereas the apoptosis pathway was down-regulated.
In summary, we have used a microarray-based analysis to examine expression of cell-cycle genes during spermatogenesis in the mouse. Our data significantly expand the catalogue of cell-cycle genes known to be expressed in these cells and demonstrate dramatic transitions in the expression of most of these genes that coincide with transitions in the state of the cell cycle in each spermatogenic cell type. These results indicate that transitions during spermatogenesis from mitotic to meiotic to postreplicative cell types are accompanied by equally dynamic transitions in the expression of different networks of cell-cycle genes, and they suggest that at least some of this differential gene expression may play key roles in regulating progression of spermatogenic cells through the process of spermatogenesis.
Supplementary Material
Acknowledgments
We thank the Computational Biology Initiative (CBI) of the University of Texas at San Antonio for technical support and members of the McCarrey laboratory for assistance with qRT-PCR and RT-PCR methods.
Footnotes
This work was supported in part by NIH grant HD46637 to J.R.M.
REFERENCES
- McCarrey JR.Cell and Molecular Biology of the Testis. New York:Oxford University Press;1993: 58–89. [Google Scholar]
- Zhao GQ, Garbers DL.Male germ cell specification and differentiation. Dev Cell 2002; 5: 537–547. [DOI] [PubMed] [Google Scholar]
- Eddy EM.Male germ cell gene expression. Recent Prog Horm Res 2002; 57: 103–128. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Tanaka TU.The chromosome cycle: coordinating replication and segregation. Second in the cycles review series. EMBO Rep 2005; 6: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JW, Dolbeare F, Pallavicini MG, Beisker W, Waldman F.Cell cycle analysis using flow cytometry. Int J Radiat Biol Relat Stud Phys Chem Med 1986; 49: 237–255. [DOI] [PubMed] [Google Scholar]
- Norbury C, Nurse P.Controls of cell proliferation in yeast and animals. Ciba Found Symp 1990; 150: 168–177. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Kaufman DG.Cell cycle control, DNA repair and initiation of carcinogenesis. FASEB J 1993; 7: 1188–1191. [DOI] [PubMed] [Google Scholar]
- Lehner CF.Pulling the string: cell cycle regulation during Drosophila development. Semin Cell Biol 1991; 2: 223–231. [PubMed] [Google Scholar]
- Kimble J, Crittenden SL.Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 2007; 23: 405–433. [DOI] [PubMed] [Google Scholar]
- Hansen D, Schedl T.The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germline. Curr Top Dev Biol 2006; 76: 185–215. [DOI] [PubMed] [Google Scholar]
- Rajesh C, Pittman DL.Cell cycle regulation in mammalian germ cells. Results Probl Cell Differ 2006; 42: 343–367. [DOI] [PubMed] [Google Scholar]
- Lee B, Amon A.Meiosis: how to create a specialized cell cycle. Curr Opin Cell Biol 2001; 13: 770–777. [DOI] [PubMed] [Google Scholar]
- Wolgemuth DJ, Laurion E, Lele KM.Regulation of the mitotic and meiotic cell cycles in the male germline. Recent Prog Horm Res 2002; 57: 75–101. [DOI] [PubMed] [Google Scholar]
- Wolgemuth DJ.Insights into regulation of the mammalian cell cycle from studies on spermatogenesis using genetic approaches in animal models. Cytogenet Genome Res 2003; 103: 256–266. [DOI] [PubMed] [Google Scholar]
- Abou-Haila A, Tulsiani DR.Mammalian sperm acrosome: formation, contents, and function. Arch Biochem Biophys 2000; 379: 173–182. [DOI] [PubMed] [Google Scholar]
- McCarrey JR.Spermatogenesis as a model system for developmental analysis of regulatory mechanisms associated with tissue-specific gene expression. Semin Cell Dev Biol 1998; 9: 459–466. [DOI] [PubMed] [Google Scholar]
- Rossi P, Dolci S, Sette C, Capolunghi F, Pellegrini M, Loiarro M, Di Agostino S, Paronetto MP, Grimaldi P, Merico D, Martegani E, Geremia R.Analysis of the gene expression profile of mouse male meiotic germ cells. Gene Expr Patterns 2004; 4: 267–281. [DOI] [PubMed] [Google Scholar]
- Almstrup K, Nielsen JE, Hansen MA, Tanaka M, Skakkebaek NE, Leffers H.Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol Reprod 2004; 70: 1751–1761. [DOI] [PubMed] [Google Scholar]
- Schlecht U, Demougin P, Koch R, Hermida L, Wiederkehr C, Descombes P, Pineau C, Jegou B, Primig M.Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol Biol Cell 2004; 15: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jegou B, Primig M.The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci U S A 2007; 104: 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD.The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319–330. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Yan MSC, Klysik M, Matzuk M.Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril 2001; 76: 550–554. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT.Postmeiotic sex chromatin in the male germline of mice. Curr Biol 2006; 16: 660–667. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Berg WM, Paragioudakis SJ, Zhang PL, Dilworth DD, Arnold BL, Rossi JJ.Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol 1992; 154: 160–168. [DOI] [PubMed] [Google Scholar]
- Romrell LJ, Bellvé AR, Fawcett DW.Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol 1976; 49: 119–131. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Millette CF, Bhatnagar YM, O'Brien DA.Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem 1977; 25: 480–494. [DOI] [PubMed] [Google Scholar]
- Bellve AR.Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 1993; 225: 84–113. [DOI] [PubMed] [Google Scholar]
- Jiang X, Nariai N, Steffen M, Kasif S, Kolaczyk ED.Integration of relational and hierarchical network information for protein function prediction. BMC Bioinformatics 2008; 9: 350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Harris MA.The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr Protoc Bioinformatics 2008; 23: 7.2.1–7.2.9. [DOI] [PubMed] [Google Scholar]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A.UniProtKB/Swiss-Prot. Methods Mol Biol 2007; 406: 89–112. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, Wagner L.Database resources of the National Center for Biotechnology. Nucleic Acids Res 2003; 31: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A, Niederhauser-Wiederkehr C, Moore J, Hermida L, Primig M.The GermOnline cross-species systems browser provides comprehensive information on genes and gene products relevant for sexual reproduction. Nucleic Acids Res 2007; 35: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr C, Basavaraj R, Sarrauste de Menthiere C, Koch R, Schlecht U, Hermida L, Masdoua B, Ishii R, Cassen V, Yamamoto M, Lane C, Cherry M, et al. Database model and specification of GermOnline Release 2.0, a cross-species community annotation knowledgebase on germ cell differentiation. Bioinformatics 2004; 20: 808–811. [DOI] [PubMed] [Google Scholar]
- Primig M, Wiederkehr C, Basavaraj R, Sarrauste de Menthiere C, Hermida L, Koch R, Schlecht U, Dickinson HG, Fellous M, Grootegoed JA, Hawley RS, Jegou B, et al. GermOnline, a new cross-species community annotation database on germ-line development and gametogenesis. Nat Genet 2003; 35: 291–292. [DOI] [PubMed] [Google Scholar]
- Imai H, Honda S, Kondo N, Ishibashi K, Tsukahara Y, Negi A.The up-regulation of angiogenic gene expression in cultured retinal pigment epithelial cells grown on type I collagen. Curr Eye Res 2007; 32: 903–910. [DOI] [PubMed] [Google Scholar]
- Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC.Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia 2003; 8: 287–307. [DOI] [PubMed] [Google Scholar]
- Do JH, Choi DK.Clustering approaches to identifying gene expression patterns from DNA microarray data. Mol Cells 2008; 25: 279–288. [PubMed] [Google Scholar]
- Almon RR, DuBois DC, Pearson KE, Stephan DA, Jusko WJ.Gene arrays and temporal patterns of drug response: corticosteroid effects on rat liver. Funct Integr Genomics 2003; 3: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flight RM, Wentzell PD.Preliminary exploration of time course DNA microarray data with correlation maps. OMICS 2010; 14: 99–107. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Geyer CB, Hornecker JL, Patel KT, McCarrey JR.In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis. Mol Cell Biol 2007; 27: 7871–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Vidal M.Protein interaction maps for model organisms. Nat Rev Mol Cell Biol 2001; 2: 55–62. [DOI] [PubMed] [Google Scholar]
- Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Féret B, Vernet N, Codreanu CA, Chambon P, Ghyselinck NB.STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J Cell Sci 2008; 121: 3233–3242. [DOI] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC.Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A 2006; 103: 2474–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Wu J.Involvement of cyclins in mammalian spermatogenesis. Mol Cell Biochem 2008; 315: 17–24. [DOI] [PubMed] [Google Scholar]
- Meng X, Yue J, Liu Z, Shen Z.Abrogation of the transactivation activity of p53 by BCCIP down-regulation. J Biol Chem 2007; 282: 1570–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham SM, Clark AR.Dual-specificity phosphatase 1: a critical regulator of innate immune responses. Biochem Soc Trans 2006; 34: 1018–1023. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim S, Jeong Y, Rhee K.Centrobin/Nip2 expression in vivo suggests its involvement in cell proliferation. Mol Cells 2009; 28: 31–36. [DOI] [PubMed] [Google Scholar]
- Qin L, Li X, Ko JK, Partridge NC.Parathyroid hormone uses multiple mechanisms to arrest the cell cycle progression of osteoblastic cells from G1 to S phase. J Biol Chem 2005; 280: 3104–3111. [DOI] [PubMed] [Google Scholar]
- Meng X, Liu J, Shen Z.Inhibition of G1 to S cell cycle progression by BCCIP beta. Cell Cycle 2004; 3: 343–348. [PubMed] [Google Scholar]
- Sun H, Tonks NK, Bar-Sagi D.Inhibition of Ras-induced DNA synthesis by expression of the phosphatase MKP-1. Science 1994; 266: 285–288. [DOI] [PubMed] [Google Scholar]
- Lee KK, Tang MK, Yew DT, Chow PH, Yee SP, Schneider C, Brancolini C.Gas2 is a multifunctional gene involved in the regulation of apoptosis and chondrogenesis in the developing mouse limb. Dev Biol 1999; 207: 14–25. [DOI] [PubMed] [Google Scholar]
- Brancolini C, Schneider C.Phosphorylation of the growth arrest-specific protein Gas2 is coupled to actin rearrangements during G0→G1 transition in NIH 3T3 cells. J Cell Biol 1994; 124: 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantha RW, Vassin VM, Borowiec JA.Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem 2007; 282: 35910–35923. [DOI] [PubMed] [Google Scholar]
- Saffin JM, Venoux M, Prigent C, Espeut J, Poulat F, Giorgi D, Abrieu A, Rouquier S.ASAP, a human microtubule-associated protein required for bipolar spindle assembly and cytokinesis. Proc Natl Acad Sci U S A 2005; 102: 11302–11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai A, Tada M, Furuuchi K, Ishikawa S, Makiyama K, Hamada J, Okada F, Kobayashi I, Fukuda H, Moriuchi T.Expression of AIE-75 PDZ-domain protein induces G2/M cell cycle arrest in human colorectal adenocarcinoma SW480 cells. Cancer Lett 2004; 211: 209–218. [DOI] [PubMed] [Google Scholar]
- Sun Y, Tan M, Duan H, Swaroop M.SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal 2001; 3: 635–650. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, et al. Analysis of the mouse transcriptome based on functional annotation of 60 770 full-length cDNAs. Nature 2002; 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Venoux M, Delmouly K, Milhavet O, Vidal-Eychenie S, Giorgi D, Rouquier S.Gene organization, evolution and expression of the microtubule-associated protein ASAP (MAP9). BMC Genomics 2008; 9: 406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill AL, Herring BE, Fox AP.Stable silencing of SNAP-25 in PC12 cells by RNA interference. BMC Neurosci 2006; 7: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalier D, Eloy L, Garchon HJ.Sex-specific gene expression during meiotic prophase I: Xlr (X linked, lymphocyte regulated), not its male homologue Xmr (Xlr related, meiosis regulated), is expressed in mouse oocytes. Biol Reprod 2002; 67: 1646–1652. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.Basic local alignment search tool. J Mol Biol 1990; 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, McKay MJ.A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep 2004; 5: 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Matzuk MM, Sung WK, Guo Q, Wang P, Wolgemuth DJ.Cyclin A1 is required for meiosis in the male mouse. Nat Genet 1998; 20: 377–380. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Wright WW, Dicandeloro P, Wilson E, Kopf GS, Jelinsky SA.Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc Natl Acad Sci U S A 2008; 105: 8315–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnik SE, Wolgemuth DJ.The developmentally restricted pattern of expression in the male germline of a murine cyclin A, cyclin A2, suggests roles in both mitotic and meiotic cell cycles. Dev Biol 1996; 173: 69–78. [DOI] [PubMed] [Google Scholar]
- Rossi P, Dolci S, Sette C, Capolunghi F, Pellegrini M, Loiarro M, Di Agostino S, Paronetto MP, Grimaldi P, Merico D, Martegani E, Geremia R.Analysis of the gene expression profile of mouse male meiotic germ cells. Gene Expr Patterns 2004; 4: 2672–2681. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Bäumer N, Schultz N, Hamra FK, Schrader MG, Sandstede ML, Berdel WE, Serve H, Müller-Tidow C.Expression patterns of mitotic and meiotic cell cycle regulators in testicular cancer and development. Int J Cancer 2005; 116: 207–217. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H.The division of Drosophila germline stem cells and their precursors requires a specific cyclin. Curr Biol 2005; 15: 328–333. [DOI] [PubMed] [Google Scholar]
- Furuta T, Tuck S, Kirchner J, Koch B, Auty R, Kitagawa R, Rose AM, Greenstein D.EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol Biol Cell 2000; 11: 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H.Biological robustness. Nat Rev Genet 2004; 11: 826–837. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Hodson JA.Mitotic replication initiation proteins are not required for premeiotic S phase. Nat Genet 2000; 3: 263–268. [DOI] [PubMed] [Google Scholar]
- Dirick L, Goetsch L, Ammerer G, Byers B.Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 1998; 281: 1854–1857. [DOI] [PubMed] [Google Scholar]
- Weinberg RA.The retinoblastoma protein and cell cycle control. Cell 1995; 81: 323–330. [DOI] [PubMed] [Google Scholar]
- Aleem E, Kiyokawa H, Kaldis P.Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol 2005; 7: 831–836. [DOI] [PubMed] [Google Scholar]
- Katich SC, Zerfass-Thome K, Hoffmann I.Regulation of the Cdc25A gene by the human papillomavirus type 16 E7 oncogene. Oncogene 2001; 20: 543–550. [DOI] [PubMed] [Google Scholar]
- Hamer G, Kal HB, Westphal CH, Ashley T, de Rooij DG.Ataxia telangiectasia mutated expression and activation in the testis. Biol Reprod 2004; 70: 1206–1212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.