Abstract
Ethanol-induced damage is largely attributed to its toxic metabolite, acetaldehyde. Clearance of acetaldehyde is achieved by its oxidation, primarily catalyzed by the mitochondrial class II aldehyde dehydrogenase (ALDH2). ALDH1B1 is another mitochondrial aldehyde dehydrogenase (ALDH) that shares 75% peptide sequence homology with ALDH2. Recent population studies in whites suggest a role for ALDH1B1 in ethanol metabolism. However, to date, no formal documentation of the biochemical properties of ALDH1B1 has been forthcoming. In this current study, we cloned and expressed human recombinant ALDH1B1 in Sf9 insect cells. The resultant enzyme was purified by affinity chromatography to homogeneity. The kinetic properties of purified human ALDH1B1 were assessed using a wide range of aldehyde substrates. Human ALDH1B1 had an exclusive preference for NAD+ as the cofactor and was catalytically active toward short- and medium-chain aliphatic aldehydes, aromatic aldehydes, and the products of lipid peroxidation, 4-hydroxynonenal and malondialdehyde. Most importantly, human ALDH1B1 exhibited an apparent Km of 55 μM for acetaldehyde, making it the second low Km ALDH for metabolism of this substrate. The dehydrogenase activity of ALDH1B1 was sensitive to disulfiram inhibition, a feature also shared with ALDH2. The tissue distribution of ALDH1B1 in C57BL/6J mice and humans was examined by quantitative polymerase chain reaction, Western blotting, and immunohistochemical analysis. The highest expression occurred in the liver, followed by the intestinal tract, implying a potential physiological role for ALDH1B1 in these tissues. The current study is the first report on the expression, purification, and biochemical characterization of human ALDH1B1 protein.
Introduction
Excessive alcohol consumption leads to a variety of psychological and pathological consequences. Chronic alcohol intake is often associated with damage to multiple organs, manifesting as hepatitis, liver cirrhosis, brain damage, and immune dysfunction. Many of the toxic effects of ethanol are attributable to ethanol metabolism. Ethanol is metabolized in the liver to acetaldehyde predominantly by class I alcohol dehydrogenase isoenzymes (ADH1A, ADH1B, and ADH1C) located in the cytosol of hepatocytes. Aldehyde dehydrogenases (ALDHs) then oxidize acetaldehyde to acetate and water (Crabb et al., 2004). Acetaldehyde, the intermediate metabolite of ethanol, is toxic and its accumulation contributes significantly to ethanol-induced tissue damage (Eriksson, 2001; Meier and Seitz, 2008). The toxicity of acetaldehyde is caused by it covalently binding biological macromolecules, such as proteins and nucleic acids and forming adducts that impair their functions (Niemelä, 2007). Acetaldehyde also induces lipid peroxidation, increased collagen synthesis, and glutathione depletion (Esterbauer et al., 1991). Moreover, protein adducts with acetaldehyde and products of lipid peroxidation trigger autoimmune responses that are associated with alcoholic liver disease (Viitala et al., 2000; Stewart et al., 2004). Therefore, metabolism and elimination of acetaldehyde is of great importance for cellular defense against ethanol-induced toxicities.
Among the 19 known human ALDHs, mitochondrial ALDH2, and, to a lesser extent, cytosolic ALDH1A1 has been shown to play a major role in acetaldehyde oxidation and elimination (Vasiliou and Pappa, 2000). Both enzymes, homotetramers with a 55-kDa subunit, exhibit low Km constants for acetaldehyde, namely 3.2 and 180 μM for human ALDH2 and ALDH1A1, respectively (Klyosov et al., 1996). The importance of ALDH2 in the clearance of acetaldehyde is exemplified in humans carrying the ALDH2*2 allele, which encodes a functional mutation (E487K) of ALDH2 and results in poor binding affinity to cofactor NAD+ and >90% loss of enzyme activity (Yoshida et al., 1984; Larson et al., 2005). Compared with wild-type individuals, blood acetaldehyde levels climb 20- and 6-fold higher in homozygous and heterozygous individuals carrying this polymorphism, respectively, for equivalent levels of alcohol consumption (Crabb et al., 1989). This causes the “alcohol flushing” syndrome characterized by the reddening of the face, neck, and, in some cases, the entire body due to dilation of capillaries (Eriksson, 2001). Similar elevations in acetaldehyde have been observed in the Aldh2-null mice (Yu et al., 2009). The second enzyme involved in acetaldehyde metabolism is ALDH1A1. In addition to its crucial role in retinoic acid biosynthesis (Duester, 2000), cytosolic ALDH1A1 has been shown to be important in acetaldehyde metabolism and alcohol preference in rodent studies (Little and Petersen, 1983; Bond and Singh, 1990; Bond et al., 1991). Human studies also report the association of ALDH1A1 variants with enzyme deficiency in white “flushers” (Eriksson, 2001). Nevertheless, the contribution of ALDH1A1 to acetaldehyde metabolism seems to be less important in humans than in rodents, probably because rodent ALDH1A1 has a much lower Km for acetaldehyde (15 μM) (Klyosov et al., 1996).
ALDH1B1, previously known as ALDHX and ALDH5, represents another mitochondrial ALDH isoenzyme. The human ALDH1B1 gene located on chromosome 9 spans a 5957-base pair region and is the only known ALDH gene with an intronless coding region (Hsu and Chang, 1991). The ALDH1B1 gene is composed of two exons and one intron and has only one known transcript variant (Black et al., 2009). ALDH1B1 enzyme, like ALDH2 and ALDH1A1, is predicted to be a homotetramer; the subunit contains 517 amino acids with an N-terminal 19-residue mitochondrial lead signal. Based on peptide sequence alignment, human ALDH1B1 shares 65 and 75% homology with human ALDH1A1 and ALDH2, respectively. Northern blot analysis in human tissues has shown high levels of ALDH1B1 mRNA in the liver and testis and relatively low levels in other tissues (Hsu and Chang, 1991; Stewart et al., 1996). ALDH1B1 has also been found to be expressed abundantly in mouse and bovine corneas (Stagos et al., 2010), in which cells are constantly exposed to UV-induced lipid peroxidation. To date, little is known regarding the biochemical properties and physiological roles of ALDH1B1. A study using crude lysate from HuH7 hepatoma cells reported that mitochondrial ALDH1B1 contributed to the oxidation of short-chain aldehydes including acetaldehyde and propionaldehyde, implying a role for ALDH1B1 in ethanol metabolism (Stewart et al., 1995). In agreement with this report, two recent large population-based studies identified two ALDH1B1 variants that were associated with drinking behavior (A69V) and alcohol-induced hypersensitivity (A86V) in whites (Husemoen et al., 2008; Linneberg et al., 2010). These findings strongly suggest that ALDH1B1 enzyme may be involved in ethanol detoxification and modifications in this enzyme may contribute to alcohol-related diseases. To expand our current knowledge on the catalytic properties of ALDH1B1, we cloned and purified human recombinant ALDH1B1. Different aldehydes, including acetaldehyde, were tested as substrates. In addition, we surveyed the expression profile of ALDH1B1 in multiple mouse and human tissues. This is the first report on the expression and biochemical characterization of human ALDH1B1 enzyme.
Materials and Methods
Chemicals.
4-Hydroxynonenal (4-HNE) was obtained from Cayman Chemical (Ann Arbor, MI). Malondialdehyde (MDA) was synthesized according to a method described previously (Esterbauer et al., 1991). All other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified,
Animals.
Male C57BL/6J wild-type mice (∼12 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, Maine). Aldh2(−/−) knockout mice were obtained from Dr. T. K. Kawamoto's laboratory (Department of Environmental Health, School of Medicine, University of Occupational and Environmental Health, Yahatanishi-ku, Kitakyushu, Japan) (Yu et al., 2009) and maintained in the C57BL/6J background. Mice were euthanized by CO2 inhalation followed by cervical dislocation. All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the University of Colorado Denver.
Construction, Expression, and Purification of Recombinant Human ALDH1B1.
The pCMV-XL4-ALDH1B1 plasmid harboring the full length of human ALDH1B1 cDNA (NM_000692.3) was purchased from Origene (Rockville, MD). The open reading frame (ORF) of ALDH1B1 was amplified by polymerase chain reaction (PCR) using the primer pair 5′-CAAGGTACCTACAGGAAAGCCCACCATGCTGCGCTTCCTGGCA-3′ (forward) and 5′-GTGAAGCTTTTACGAGTTCTTCTGAGGAACCTTGATGGTG-3′ (reverse). The forward primer was designed to contain a KpnI site (GGTACC) for subcloning and a sequence motif (CCACC) 5′-ward of the start codon ATG (in bold) to ensure correct initiation of translation in eukaryotic cells (Ding and Nam Ong, 2003). The reverse primer was designed to contain a HindIII site (AAGCTT) 3′-ward of the stop codon TAA (in bold). The 1.6-kilobase PCR product was then digested with KpnI and HindIII and subcloned into the pBlueBac4.5 expression vector (Invitrogen, Carlsbad, CA). The correct sequence of the inserted ORF of ALDH1B1 was confirmed by DNA sequencing analysis.
The pBlueBac4.5-ALDH1B1 plasmid was used to generate recombinant baculoviruses, which were plaque-purified and amplified in Sf9 insect cells by the Tissue Culture Core Facility at the University of Colorado Denver as described previously (Manzer et al., 2003). Approximately 6 × 108 Sf9 cells were harvested 48 h after infection by centrifugation at 1000g for 5 min and washed with phosphate-buffered saline, pH 7.4. Cell pellets were resuspended in lysis buffer (100 mM phosphate buffer, 1 mM EDTA, 0.1 mM 2-mercaptoethanol, 0.01% Triton X-100, 0.5 μg/ml leupeptin, 1 μg/ml pepstatin, 0.5 μg/ml aprotinin, and 100 μg/ml phenylmethylsulfonyl fluoride, pH 7.5), and cell suspensions were homogenized by sonication on ice. The resulting crude lysate was then centrifuged at 35,000g for 1 h at 4°C, and the supernatant was subjected to affinity purification by fast protein liquid chromatography as described previously (Pappa et al., 2003). In brief, 1 to 2 ml of Sf9 cell lysate was applied to a 1.6 × 6 cm 5′-AMP-Sepharose 4B affinity column (GE Healthcare, Pittsburgh, PA) pre-equilibrated with the binding buffer (100 mM potassium phosphate, 1 mM EDTA, 0.1 mM 2-mercaptoethanol, and 0.01% Triton X-100, pH 7.4). The bound ALDH1B1 was eluted by applying a gradient of 0 to 0.25 mM NAD+ dissolved in the binding buffer (0.005 mM increment per minute). Elution fractions (5 ml) were collected and examined for the presence of ALDH1B1 protein by Coomassie Blue staining and Western blot analysis. Fractions containing ALDH1B1 were then pooled and concentrated in concentrating buffer (10 mM Tris-HCl, pH 7.4) at 4°C using a Amicon concentrator (Millipore Corporation, Billerica, MA). The identity and purity of concentrated ALDH1B1 protein were confirmed by Coomassie Blue staining, Western blot analysis, and matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry (MS) analysis (see below).
Western Immunoblot.
Tissues from wild-type or Aldh2(−/−) knockout mice were collected, flash-frozen in liquid nitrogen, and stored at −80°C until use. Frozen tissues were homogenized in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Triton X-100, 0.25% sodium deoxycholate, 0.1% SDS, 50 mM Tris, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin, pH 7.4) on ice with a Tissue-Tearor (BioSpec Products, Bartlesville, OK). Tissue homogenates were centrifuged at 10,000g at 4°C for 20 min, and the supernatant was collected and used for tissue lysates. Proteins in tissue lysates (16–40 μg) or purified recombinant human ALDH1B1 (25–200 ng) were resolved by 10% SDS-PAGE and immunoblotted using primary antibodies as specified under Results. Rabbit polyclonal α-ALDH1B1 was raised against the human ALDH1B1 peptide sequence ELDTQQGPQVDKEQ FERVLGYIQL GQKEGAKLLCGGERFGERGFFIKPTVFGGVQDDMR (amino acids 353–411) by GenWay Biotech Inc. (San Diego, CA). To minimize the cross-reactivity toward ALDH2 protein, the antibody was preabsorbed by a ALDH2-bound column. Primary antibodies were used at dilutions of 1:5000 for anti-ALDH1B1, 1:1000 for polyclonal anti-ALDH2 (Lassen et al., 2005), 1:5000 for mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (Ambion, Austin, TX), and 1:10,000 for mouse monoclonal anti-β-actin (Sigma-Aldrich, St. Louis, MO). Blocking reagents and corresponding secondary antibodies were purchased from LI-COR Biosciences (Lincoln, NE) or Calbiochem (San Diego, CA) and used according to the manufacturer's protocol. Protein bands were visualized using an Odyssey Infrared Imaging System (LI-COR) or chemiluminescence (PerkinElmer Life and Analytical Sciences, Waltham, MA).
MALDI-TOF MS.
Purified recombinant human ALDH1B1 protein (2 μg) was resolved by 15% SDS-PAGE. The two protein bands, visualized by Coomassie Blue staining, were excised and digested with trypsin (0.05 μg/μl in 25 mM ammonium bicarbonate) for 16 h at 37°C. Digested peptides were then analyzed by MALDI-TOF MS on a 4800 plus MALDI TOF/TOF spectrometer (Applied Biosystems, Foster City, CA) equipped with a 200-Hz solid-state laser using a matrix of α-cyano-4-hydroxycinnaminic acid at 10 mg/ml in 50% acetonitrile and 0.05% trifluoroacetic acid. The sample was calibrated externally using a five-point peptide calibration mixture from Sigma-Aldrich (ProteoMass Peptide Calibration Kit). Mass spectral data were collected in standard reflector mode over the mass range of 600 to 5000 m/z with a total of 400 shots collected at a laser intensity of 3300. MS/MS spectra were obtained using the standard 1-kV acquisition method with collision-induced dissociation turned on and air as the collision gas with a threshold of 1 × 10−6 Torr. The laser intensity was set at 4300, and spectra were recorded for 2000 shots or until the accumulated spectra reached an estimated signal/noise ratio of 75 (five peaks and eight subspectra minimum). Database searches were performed on the MASCOT search engine (www.matrixscience.com; Matrix Science, London, UK) using Applied Biosystems GPS software. Combined searches of both MS and tandem mass spectrometry data were performed using the msdb database.
Enzyme Kinetic Studies.
The enzymatic activity of purified recombinant human ALDH1B1 was determined by monitoring the formation of NADH at 340 nm during the oxidation of aldehyde substrates using a Beckman DU-640 spectrophotometer. A molar extinction coefficient of 6.22 × 10−3 was used to calculate the rate of NADH formation (Pappa et al., 2003). Before the enzyme assay, ALDH1B1 protein was first reactivated by incubation with 50 mM 2-mercaptoethanol at 25°C in the dark for 1 h. Aldehyde substrates were prepared either in double-distilled H2O or 20% ethanol (the final concentration of ethanol being <1%). Eight to 10 concentrations of individual aldehyde substrate representing 3 to 5 concentrations below the apparent Km and 5 concentrations above the apparent Km were used. The reaction was initiated by adding the substrate into the reaction mixture containing 0.1 M sodium pyrophosphate, 1 mM NAD+, 1 mM pyrazole, 1.0 mM 2-mercaptoethanol, and 5 μg of activated ALDH1B1 protein. Production of NADH was monitored for 3 min at 25°C. For the Km study for NAD+ and NADP+, propionaldehyde (10 mM) was used as the substrate to define the activity. The disulfiram inhibition study was performed using propionaldehyde (100, 500, and 1000 μM) as the substrate. In these studies, 100× stock solution of disulfiram dissolved in dimethyl sulfoxide was added to the reaction mixture and preincubated with activated ALDH1B1 protein for 5 min before the addition of substrate.
The esterase activity of human ALDH1B1 was determined by monitoring the formation of p-nitrophenolate at 400 nm using a Beckman DU-640 spectrophotometer. A molar extinction coefficient of 18.3 × 10−3 was used to calculate the rate of p-nitrophenolate formation (Wang and Weiner, 1995). In brief, activated ALDH1B1 protein (5 μg) was mixed with p-nitrophenyl acetate (p-NPA) at various concentrations (0, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, and 2.0 mM) in 0.1 M sodium phosphate buffer (pH 7.4); p-Nitrophenolate formation was then monitored for 3 min at 25°C.
For all enzyme kinetic analyses, apparent kinetic constants (Vmax and Km) were determined using the Sigma Plot Enzyme Kinetic Module (version 7.0, 2001; SPSS Inc., Chicago, IL) by means of the Michaelis-Menten model. Enzyme activity is reported as nanomoles per minute per milligram of protein. Data are reported as means ± S.E. from three independent experiments carried out in triplicate.
Reverse Transcription and Quantitative Real-Time PCR.
Total RNA was isolated from mouse tissues using an RNeasy Mini kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. cDNA was synthesized with a SuperScript III RT Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction, using 5 μg of total RNA in a 20-μl reaction volume. Q-PCR reactions were carried out using 0.03 μg of cDNA by the TaqMan gene expression assay (Applied Biosystems) for mouse Aldh1b1 gene (primer identification number: Mm00728303_s1) and mouse Aldh2 gene (primer identification number: Mm00477463_m1) according to the manufacturer's protocol. Expression of β-actin was used for normalization of CT data according to the ΔΔCT method (Livak and Schmittgen, 2001). The mRNA level of ALDH1B1 in the liver was set as the control (= 1), and relative mRNA levels were expressed as fold of control. Data were reported as the mean ± S.E. of three to four animals.
Immunohistochemistry and Immunofluorescence.
Formalin-fixed and paraffin-embedded normal human tissues were procured by IHCtech (Aurora, CO) in accordance with approvals and guidelines to use for immunohistochemical (IHC) staining. Tissues from C57BL/6J wild-type male mice were fixed in 4% paraformaldehyde, dehydrated in graded ethanol solutions, and embedded in paraffin. Tissue sections (4-μm) were then deparaffinized, rehydrated, and processed for hematoxylin and eosin staining or for IHC staining of ALDH1B1 was performed using the TSA Biotin System Kit (PerkinElmer, Waltham, MA) according to the manufacturer's protocol. Primary antibodies were used at a dilution of 1:200 for rabbit polyclonal anti-ALDH1B1 (see above) or for rabbit preimmune serum (serum control). Secondary antibody was used at a dilution of 1:250 for goat polyclonal anti-rabbit IgG (Sigma-Aldrich). Images were obtained using a Nikon upright microscope linked to a Nikon digital camera. For immunofluorescence staining, tissue sections were blocked with 3% bovine serum albumin in phosphate-buffered saline for 1 h at room temperature, followed by incubation with rabbit polyclonal anti-ALDH1B1 (1:200) overnight at 4°C. Samples were then washed and stained with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO). Tissue sections were costained with 4,6-diamidino-2-phenylindole (DAPI) to identify the nucleus. Immunofluorescence staining was evaluated using a fluorescence microscope (Nikon Eclipse E600).
Results
Baculovirus-Mediated Expression and Purification of Recombinant Human ALDH1B1.
The ORF of human ALDH1B1 was inserted into the pBlueBac4.5 vector for recombination with baculovirus DNA, and recombinant viruses were used to infect Sf9 insect cells. Eleven independently isolated plaques were amplified and screened for the expression of ALDH1B1 protein by Western blot analysis in crude extracts of Sf9 cells (data not shown). The recombinant virus with the highest expression of ALDH1B1 protein was used for subsequent infection of Sf9 cells at a multiplicity of infection of 1. Cell lysates from infected Sf9 cells were prepared at 48 h after infection (as described under Materials and Methods) and used for the purification of recombinant human ALDH1B1.
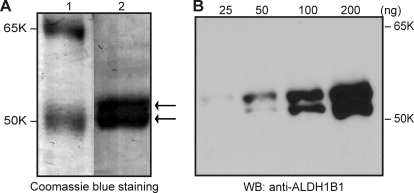
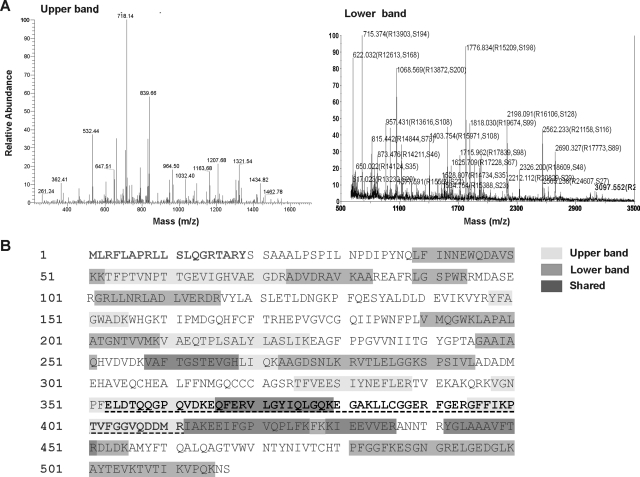
Purification of ALDH1B1 protein was performed by affinity chromatography using a 5′-AMP-Sepharose 4B column as described under Materials and Methods. This single-step process yielded approximately 200 to 300 μg of purified protein from 500 ml of Sf9 cell cultures. When visualized by Coomassie Blue staining on an SDS-PAGE gel (Fig. 1A), the purified proteins revealed two peptide bands running slightly above 50 kDa, in agreement with the predicted size of ALDH1B1 (≈55 kDa). By Western blot analysis (Fig. 1B), both peptides immunoreacted with a rabbit polyclonal antibody raised against the human ALDH1B1 peptide sequence (amino acids 353–411). To further verify the identity of the purified proteins, the two peptide bands were excised from a Coomassie Blue-stained SDS-PAGE gel and subjected to MALDI-TOF MS analysis after trypsin digestion (Fig. 2). The MALDI-TOF MS analysis showed 13 peaks for both the upper and lower band corresponding to a predicted amino acid sequence of ALDH1B1 protein (Fig. 2A), with a total sequence coverage of 30 and 37% for the higher and lower molecular weight band, respectively (Fig. 2B).
Fig. 1.
Purified recombinant human ALDH1B1 revealed by SDS-PAGE. A, purified human ALDH1B1 protein was visualized by Coomassie Blue staining after SDS-PAGE. Purified ALDH1B1 appeared as double protein bands running around 55 to 58 kDa. Lane 1, molecular weight marker; lane 2, recombinant human ALDH1B1 (2 μg). B, increasing amounts of ALDH1B1 protein (25–200 ng) were loaded and detected by Western blot (WB) using a rabbit polyclonal antibody (anti-ALDH1B1) against human ALDH1B1 peptide (see Materials and Methods). Both protein bands reacted with the antibody.
Fig. 2.
Confirmation of the identity of purified ALDH1B1. A, the two protein bands visualized by Coomassie Blue staining were excised and analyzed by MALDI-TOF MS. The two bands each showed 13 peaks corresponding to amino acid sequences of ALDH1B1 protein predicted from the ALDH1B1 cDNA. B, the predicted peptide sequence of human ALDH1B1. The N-terminal 19 residues (in bold) are the predicted mitochondrial lead signal. Distinct peptide sequences of upper and lower protein bands picked up by MALDI-TOF MS analysis are highlighted in light and dark gray, respectively. Peptide sequences positive for both protein bands are highlighted in darkest gray. The peptide used to generate the polyclonal antibody against ALDH1B1 is underlined and highlighted in bold.
Biochemical Characterization of Human ALDH1B1.
Our previous study demonstrated that recombinant mitochondrial ALDH2 enzyme is prone to inactivation, probably because of the oxidation of sulfhydryl residues that are essential for the catalytic activity of ALDHs (Lassen et al., 2005). Because ALDH1B1 shows ≈75% peptide sequence identity with ALDH2, we expected the same characteristic being shared by recombinant ALDH1B1. Indeed, after purification, recombinant human ALDH1B1 exhibited negligible aldehyde oxidation activity even at saturating concentrations of cofactor and substrate (data not shown). Under these same reaction conditions, recombinant human ALDH1B1, like recombinant ALDH2, was reactivated by preincubation with the reducing agent 2-mercaptoethanol at an optimal concentration at 50 mM (data not shown). Thus, for all the enzyme kinetic studies, recombinant human ALDH1B1 was preactivated by incubation with 50 mM 2-mercaptoethanol at 25°C for 1 h.
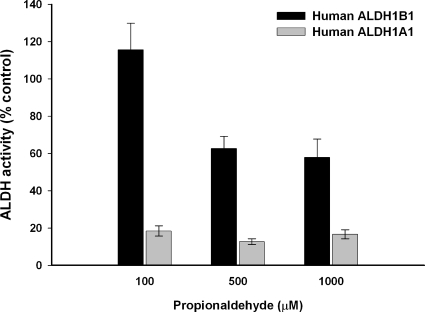
The kinetic properties of human ALDH1B1 enzyme were studied using a wide range of aldehydes as substrates and using either NAD+ or NADP+ as the cofactor. The apparent Km and Vmax values derived from these studies are summarized in Table 1. ALDH1B1 showed an absolute preference for NAD+ (Km = 3.6 μΜ) as the cofactor; no catalytic activity occurred when NADP+ was used. The recombinant human ALDH1B1 exhibited a high affinity for aliphatic aldehydes, including acetaldehyde, propionaldehyde, hexanal, and nonanal, as reflected by apparent Km values less than 100 μM; although there was a greater affinity toward medium-chain saturated aldehydes (Km = 0.4 and 0.8 μM for hexanal and nonanal, respectively) relative to that for short-chain aldehydes (Km = 55 and 14 μM for acetaldehyde and propionaldehyde, respectively). Human ALDH1B1 had an apparent Km of 50 μM for the aromatic aldehyde benzaldehyde. The order of the catalytic efficiency of human ALDH1B1 for these aldehydes, as reflected by Vmax/Km, was hexanal > nonanal > propionaldehyde > acetaldehyde > benzaldehyde. ALDH1B1 was also capable of metabolizing products of lipid peroxidation, namely 4-HNE and MDA, albeit with low affinity for substrate binding (Km > 400 μM) and poor catalytic efficiency (Vmax/Km < 1). On the other hand, ALDH1B1 exhibited no catalytic activity for oxidizing unsaturated aliphatic aldehydes, i.e., trans-2-hexenal, trans-2-nonenal, and trans-2-dodecenal. Because disulfiram is a potent inhibitor of both ALDH1A1 and, to a lesser extent, ALDH2 in vitro, we also tested whether ALDH1B1 was a target of disulfiram inhibition. As shown in Fig. 3, ALDH1B1 was less sensitive to inhibition by disulfiram than was ALDH1A1. At the lowest concentration of substrate tested (propionaldehyde = 100 μM), ALDH1B1 was not affected by disulfiram. Only at higher concentrations of propionaldehyde (500 and 1000 μM) was ALDH1B1 activity inhibited, with the extent of inhibition being <40%. On the other hand, human ALDH1A1 activity was inhibited >80% by the same concentration of disulfiram at all propionaldehyde concentrations.
TABLE 1.
Kinetic properties of recombinant human ALDH1B1
Apparent Km and Vmax values were determined by fitting the data to the Michaelis-Menten equation using SigmaPlot. Data represent mean ± S.E. from three independent experiments carried out in triplicate. The kinetic parameters for NAD+ and NADP+ were determined using propionaldehyde (10 mM) as the substrate. NAD+ (1 mM) was used for aldehyde substrate experiments.
| Substrate | Vmax | Km | Vmax/Km |
|---|---|---|---|
| nmol/(min · mg) | μM | ||
| NAD+ | 1277 ± 42 | 3.6 ± 0.5 | 359 |
| NADP+ | 0 | 0 | 0 |
| Acetaldehyde | 665 ± 55 | 55 ± 10 | 12 |
| Propionaldehyde | 789 ± 155 | 14 ± 3 | 56 |
| Hexanal | 1007 ± 30 | 0.4 ± 0.002 | 2398 |
| Nonanal | 686 ± 55 | 0.8 ± 0.1 | 879 |
| Benzaldehyde | 458 ± 36 | 50 ± 8 | 9.1 |
| 4-HNE | 2043 ± 163 | 3383 ± 304 | 0.6 |
| MDA | 348 ± 21 | 466 ± 13 | 0.7 |
| Unsaturated aldehydes (trans-2-hexenal, trans-2-nonenal, trans-2-dodecenal) | 0 | 0 | 0 |
| p-NPA | 1895 ± 91 | 288 ± 46 | 6.6 |
Fig. 3.
Inhibition of the dehydrogenase activity of ALDH1B1 and ALDH1A1 by disulfiram. Disulfiram (10 μM) was preincubated with recombinant human ALDH1B1 or ALDH1A1 for 5 min before the initiation of the reaction. Various concentrations of propionaldehyde were used as substrate. The ALDH activity of enzymes in presence of disulfiram was expressed as a percentage of the activity observed at the same concentration of substrate in the absence of disulfiram (control). Data represent mean ± S.E. from three independent experiments.
Mitochondrial ALDH2 has been reported to have esterase activity (Sheikh and Weiner, 1997). Given the 75% peptide homology shared by ALDH2 and ALDH1B1, we assessed whether ALDH1B1 also had esterase activity by monitoring the formation of p-NPA. Human ALDH1B1 exhibited maximal esterase activity of 1895 ± 91 nmol/(min · mg) and an apparent Km of 288 ± 46 μΜ for p-NPA. Compared with the reported Km of human ALDH2 for p-NPA (Sheikh and Weiner, 1997), this Km of ALDH1B1 (current study) was 40-fold higher.
Expression of ALDH1B1 in Mouse Tissues.
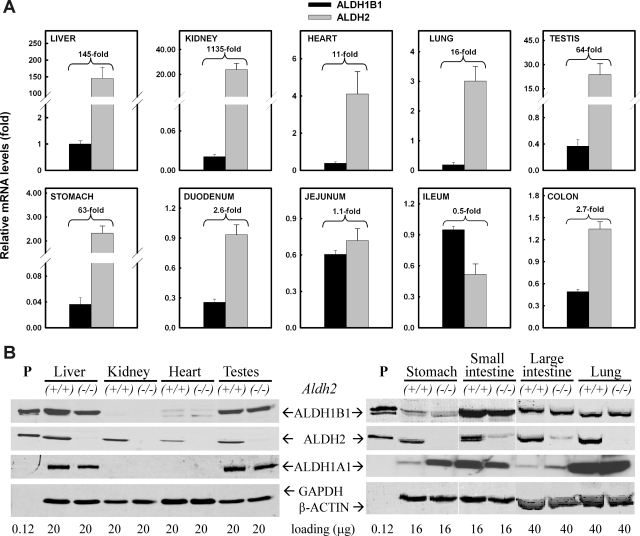
To understand the tissue distribution of ALDH1B1 expression, we first assessed the messenger level of the Aldh1b1 gene in various tissues from male C57BL/6J mice by Q-PCR and compared it with that of the Aldh2 gene (Fig. 4A). For easy comparison, the mRNA level of ALDH1B1 in the liver was set as the control (= 1), and relative mRNA levels were expressed as fold of control. The highest ALDH1B1 mRNA expression was detected in liver and part of the small intestine (ileum and jejunum); moderate levels (≈0.5-fold) were observed in the testis, distal colon, lung, heart, and duodenum and the lowest levels (≈0.03-fold) were found in the kidney and stomach. Compared with the Aldh1b1 gene, the Aldh2 gene was comparably expressed in intestinal tissues (<3-fold of ALDH1B1 mRNA level) but was more robustly expressed in other tissues examined, including the kidney, liver, testes, stomach, lung, and heart, namely 1135-, 145-, 64-, 63-, 16-, and 11-fold of ALDH1B1 mRNA, respectively.
Fig. 4.
Expression of ALDH1A1, ALDH1B1, and ALDH2 in mouse tissues. A, mRNA levels of ALDH1B1 and ALDH2 were measured by Q-PCR. The hepatic level of ALDH1B1 mRNA was set as the control (= 1) after normalization with β-actin. Relative levels of mRNA are reported as fold of control. The ratio of ALDH2/ALDH1B1 mRNA levels is indicated for each tissue examined. Data represent mean ± S.E. from three to four mice. B, protein levels of ALDH1A1, ALDH1B1, and ALDH2 were detected by Western blot in tissues from wild-type (+/+) and ALDH2-null (−/−) mice. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The protein levels of ALDH1B1, ALDH2, and ALDH1A1 were determined by Western blot analysis (Fig. 4B). To identify any potential compensatory interactions between these closely related ALDHs, we also investigated ALDH1B1 and ALDH1A1 expression in tissues from Aldh2(−/−) knockout mice (Fig. 4B). Partially in agreement with the mRNA expression profiles, ALDH1B1 protein was expressed abundantly in liver, small intestine, and testes. In these tissues, ALDH2 protein was also expressed at high levels. In large intestine and lung, ALDH1B1 and ALDH2 were expressed moderately and at comparable levels. Compared with ALDH1B1, ALDH2 seemed to be differentially expressed in certain tissues, including stomach, kidney, and heart. In a manner similar to that of the mitochondrial ALDHs, cytosolic ALDH1A1 showed high expression in liver, testes, and small intestine; interestingly, lung tissue had a relatively high abundance of ALDH1A1 expression. As expected, ALDH2 protein was undetectable in tissues from Aldh2(−/−) knockout mice. However, in these tissues ALDH1B1 and ALDH1A1 protein levels were no different from those obtained from wild-type mice, except that ALDH1A1 appeared to be up-regulated in stomach from Aldh2(−/−) knockout mice.
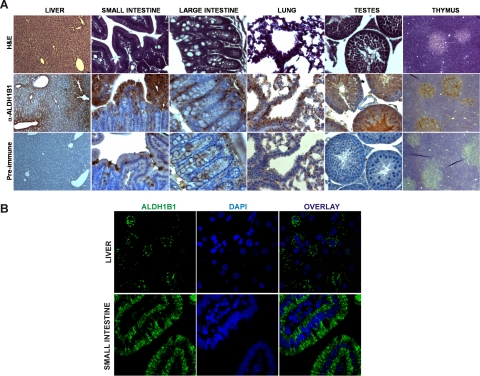
The distribution of ALDH1B1 in several mouse tissues was further investigated by immunohistochemical assay (Fig. 5A). In the liver, ALDH1B1 displayed a pericentral distribution and was detected in the cytoplasm of hepatocytes. In the small intestine, ALDH1B1 was intensely stained in the cytoplasm of absorptive epithelial cells. In the large intestine (colon), mild staining of ALDH1B1 was observed, mostly in locations where colon stem cells reside. In the lung, ALDH1B1 appeared to be expressed predominantly in the bronchiolar epithelium and Clara cells of small airways, especially in the terminal bronchioles. In the testes, intense ALDH1B1 immunoreactivity was observed in the epithelial cells lining the seminiferous tubules. In the thymus, ALDH1B1 seemed to be expressed specifically in the medulla, which contains fewer lymphocytes but more epithelial cells. To better visualize the cellular localization of ALDH1B1 protein, we performed confocal immunofluorescent staining for ALDH1B1 in the two tissues that showed the highest expression, i.e., liver and small intestine (Fig. 5B). The staining of ALDH1B1 exhibited a cytoplasmic punctate pattern in both tissues, supporting an organelle-associated localization of ALDH1B1.
Fig. 5.
Tissue distribution of ALDH1B1 protein by immunohistostaining. Mouse tissues were processed for immunohistochemical (A) or immunofluorescent (B) analysis of ALDH1B1 as described under Materials and Methods. A, representative images of tissue sections stained for general histology [hematoxylin and eosin (H&E)], ALDH1B1 immunoreactivity (α-ALDH1B1), and serum control (Pre-Immune). Liver displayed intense ALDH1B1 immunopositivity (brown) with a pericentral distribution. The absorptive epithelial cells of small intestine also showed high staining. Immunopositivity in the large intestine appeared to colocalize with stem cells. In the lung, ALDH1B1 was found to be expressed predominantly in the bronchiolar epithelium and Clara cells of small airways. In the testes, intense immunoreactivity was observed in the epithelial cells lining the seminiferous tubules. In the thymus, immunopositivity was detected specifically in the medulla. B, confocal immunofluorescent staining for ALDH1B1 (green) in the liver and small intestine exhibited a cytoplasmic punctate pattern, in agreement with an organelle-associated localization of ALDH1B1. Sections were costained with DAPI for nuclear staining. The overlay represents the ALDH1B1 and DAPI images combined. Original magnification, 400× (A); 1000× (B).
Expression of ALDH1B1 in Human Tissues.
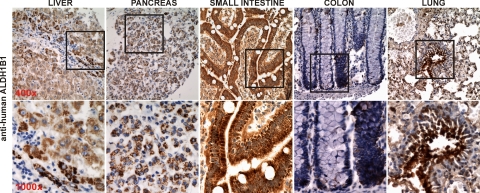
The expression of ALDH1B1 in human tissues, including liver, pancreas, small intestine, colon, and lung, was investigated by immunohistochemical staining (Fig. 6). The expression patterns of ALDH1B1, in terms of the distribution and intensity of immunopositivity, in these human tissues were quite consistent with what was seen in mouse tissues (Fig. 4A). Furthermore, the punctate positive-staining of human ALDH1B1 was clearly noted in the liver and pancreas.
Fig. 6.
Distribution of ALDH1B1 in human tissues by immunohistochemistry. Formalin-fixed and paraffin-embedded normal human tissues were processed in accordance with approvals and guidelines for IHC staining using polyclonal anti-human ALDH1B1. The absorptive epithelium of small intestine revealed the highest intensity of ALDH1B1 immunopositivity (brown). Hepatocytes and pancreatic acinar cells also showed strong positive staining for ALDH1B1, which displayed a cytoplasmic punctate pattern. The immunopositivity of ALDH1B1 in colon appeared to colocalize with stem cells. In the lung, ALDH1B1 seemed to be expressed predominantly in the bronchiolar epithelium and Clara cells of small airways. Original magnification, 400× (top panels); 1000× (bottom panels).
Discussion
The major enzyme that metabolizes acetaldehyde, the most toxic metabolite of ethanol, is mitochondrial ALDH2. The inactive variant (E487K) of human ALDH2 is responsible for ethanol-induced hypersensitivity, which serves as a deterrent against alcoholism in Asian populations (Yoshida et al., 1984). Although approximately 50% of Asians are carriers of this variant, it is nearly absent in whites. Recent studies have identified the genetic determinants of drinking behavior and alcohol hypersensitivity among whites to be polymorphisms in the human ALDH1B1 gene, which encodes another mitochondrial ALDH isoenzyme (Husemoen et al., 2008; Linneberg et al., 2010). These studies and others (Stewart et al., 1995) are suggestive of a potentially important role of ALDH1B1 in the metabolism of acetaldehyde, but more direct confirmation remains to be established. Herein, we have cloned, expressed, and purified recombinant human ALDH1B1 using a baculovirus expression system along with affinity chromatography. Human ALDH1B1 is catalytically active toward a wide range of aldehyde substrates and uses NAD+, but not NADP+, as the cofactor. The optimal substrates of human ALDH1B1 (Km < 1 μM) are medium-chain aldehydes, including hexanal and nonanal. Short-chain propionaldehyde and aromatic benzaldehyde are also good substrates for ALDH1B1. Most importantly, human ALDH1B1 exhibits an intermediate Km (55 μM) for acetaldehyde compared with human ALDH2 (Km = 3.2 μM) and human ALDH1A1 (Km = 180 μM) (Klyosov et al., 1996), suggesting that ALDH1B1 indeed represents a low Km ALDH for acetaldehyde. Thus, our study provides direct evidence to support the hypothesis that ALDH1B1 represents the second mitochondrial ALDH, aside from ALDH2, that actively oxidizes acetaldehyde and consequently may be important for the metabolism of ethanol.
Disulfiram is a drug used to treat chronic alcoholism by producing an undesirable acute sensitivity to alcohol. The effect of disulfiram is attributable to its action in elevating acetaldehyde levels by inhibiting ALDH1A1 and ALDH2 (Petersen, 1992). Recombinant human ALDH1A1 (Ki = 0.2 μM) is 60 times more sensitive to inhibition by disulfiram than recombinant human ALDH2 (Ki > 12 μM) (Lam et al., 1997). In the present study, recombinant human ALDH1B1 was inhibited by disulfiram but to a lesser extent than ALDH1A1 when propionaldehyde was used as substrate, a finding consistent with a previous study (Stewart et al., 1995). Such sensitivity of ALDH1B1 to disulfiram inhibition may have therapeutic ramifications for the use of disulfiram in vivo.
An interesting observation is that recombinant human ALDH1B1 expressed in Sf9 insect cells migrates as double bands by SDS-PAGE. Based on their locations relative to that of the molecular weight marker, the two protein bands correspond to ≈55 and ≈58 kDa peptides. MALDI-TOF MS and Western blot analysis confirmed the identity of both peptides to be ALDH1B1. We propose that the 58-kDa protein band represents the 517-residue intact translated peptide and the 55-kDa protein represents the mature form after the mitochondrial lead signal is excised. Alternatively, the 58-kDa protein band could be a post-translationally modified form of ALDH1B1, e.g., one that has been glycosylated or phosphorylated or both. Glycosylation has been documented previously for microsomal ALDH (Masaki et al., 1996). It has been determined that phosphorylation of ALDH2 occurs. One study reported that ALDH2 is phosphorylated and activated by the survival kinase protein kinase Cε and that activation of ALDH2 is cardioprotective (Chen et al., 2008). On the other hand, it has also been reported that carbon tetrachloride exposure inhibits ALDH2 activity through c-Jun NH2-terminal kinase-mediated phosphorylation; inactivation of ALDH2 contributes to carbon tetrachloride-induced liver damage (Moon et al., 2010). These studies suggest that the effect of phosphorylation of ALDH2 and probably of other ALDHs, is variable, leading to activation or inactivation of ALDH depending on the specific stress. Further studies are needed to determine whether phosphorylation is a mechanism regulating ALDH1B1.
ALDH1B1 messenger is actively transcribed in multiple human tissues (Hsu and Chang, 1991; Stewart et al., 1996). In the present study, we observed a similar tissue expression profile in mouse tissues. Liver had the highest level of ALDH1B1 and ALDH2 mRNAs relative to those of other tissues examined, supporting an important role of the two mitochondrial ALDHs in this organ. In most tissues, except in the intestine, ALDH2 messenger is predominant by up to 3 orders of magnitude. Much higher levels of ALDH2 mRNA relative to ALDH1B1 mRNA have also been found in human liver (Hsu and Chang, 1991). The predominance of hepatic ALDH2 over ALDH1B1 could possibly explain the inability of Asian carriers of the ALDH2*2 allele to efficiently metabolize acetaldehyde produced after alcohol consumption despite the presence of the ALDH1B1 enzyme. Along the mouse gastrointestinal tract, there seems to be a gradient of ALDH1B1 mRNA expression, which starts at baseline in the duodenum, peaks in ileum, and drops back to baseline in the colon. The levels of ALDH1B1 mRNA in these tissues are comparable to those of ALDH2 mRNA, suggesting a more important role of ALDH1B1 in mouse intestine. It is well established that the liver plays a major role in drug and xenobiotic biotransformation. However, metabolism during absorption across the intestinal wall also influences the bioavailability of orally administered compounds (van de Kerkhof et al., 2007). For instance, it has recently been reported that intestinal CYP1A1, but not hepatic CYP1A1, is the key enzyme in oral benzo[a]pyrene detoxification (Shi et al., 2010). Given that ethanol is usually administered via the oral route, ALDH1B1 in the intestinal wall may play an important role in the initial metabolism of ethanol or of orally ingested aldehydes, whereas ALDHs associated with the liver would be anticipated to process any aldehydes in the hepatic portal vein or in the systemic circulation. Interestingly, ADH1, the major alcohol-metabolizing enzyme converting ethanol to acetaldehyde, is predominantly expressed in the small intestine (Vaglenova et al., 2003). It should be noted that the tissue distribution of ALDH1B1 protein, in both human and mouse, agrees well with the pattern of ALDH1B1 messenger, indicating that the transcriptional regulation is probably the key mechanism controlling the expression of ALDH1B1 protein.
Genome-wise, the two mitochondrial ALDHs, namely ALDH2 and ALDH1B1, are conserved across taxa, including fungi, mammals, and plants (Liu and Schnable, 2002). Why would nature choose to have two closely related ALDHs present in the mitochondria? One simple explanation is that the two mitochondrial ALDHs have different biochemical functions and/or differential expression in tissues at different developmental stages. Several studies have associated ALDH1B1 with functions other than acetaldehyde metabolism. A recent microarray study showed that ALDH1B1 was up-regulated in medulloblastoma cell lines resistant to cyclophosphamide, suggesting a possible role of ALDH1B1 enzyme in cancer drug resistance (Bacolod et al., 2008). Another study reported that the dynamic expression of ALDH1B1 correlated with granulocytic development of hematopoietic stem cells; in this report, the author proposed a possible role of ALDH1B1 in retinoic acid metabolism mediating the effect (Luo et al., 2007). A transgenic mouse line having forced expression of the human ALDH2*2 variant exhibited distinct phenotypes, including small body size, reduced muscle mass, diminished fat content, and osteopenia (Endo et al., 2009), all of which are absent from ALDH2-null mice (Yu et al., 2009). One hypothesis to explain such a discrepancy is that the ALDH2*2 mutant subunit not only inactivates the ALDH2 wild-type subunit but also inactivates other ALDHs by forming heterotetramers, thereby blocking their functions. Given >75% homology in peptide sequence and the same mitochondrial localization, the ALDH1B1 subunit could be a potential binding partner of the ALDH2 subunit. Taken together, these observations indicate the involvement of ALDH1B1 in multiple physiological or pathological processes, the biochemical mechanism(s) of which merits further investigation.
In summary, the present study describes, for the first time, the expression, purification, and biochemical characterization of the human ALDH1B1 enzyme. Our results demonstrate that mitochondrial ALDH1B1 represents a low Km for ALDH for acetaldehyde metabolism and therefore is consistent with ALDH1B1 being actively involved in the metabolism of ethanol. The differential tissue distribution of ALDH1B1 argues in favor of a role for ALDH1B1 in intestinal tissues. The observed biochemical properties of ALDH1B1 are consistent with a physiological role(s) for this ALDH isoenzyme.
This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grant AA017754]; and the National Institutes of Health National Eye Institute [Grant EY17963].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.034678.
- ADH
- alcohol dehydrogenase
- ALDH
- aldehyde dehydrogenases
- 4-HNE
- 4-hydroxynonenal
- MDA
- malondialdehyde
- ORF
- open reading frame
- PCR
- polymerase chain reaction
- MALDI-TOF
- matrix-assisted laser desorption ionization/time of flight
- MS
- mass spectrometry
- PAGE
- polyacrylamide gel electrophoresis
- p-NPA
- p-nitrophenyl acetate
- Q-PCR
- quantitative real-time PCR
- IHC
- immunohistochemical
- DAPI
- 4,6-diamidino-2-phenylindole.
References
- Bacolod MD, Lin SM, Johnson SP, Bullock NS, Colvin M, Bigner DD, Friedman HS. (2008) The gene expression profiles of medulloblastoma cell lines resistant to preactivated cyclophosphamide. Curr Cancer Drug Targets 8:172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WJ, Stagos D, Marchitti SA, Nebert DW, Tipton KF, Bairoch A, Vasiliou V. (2009) Human aldehyde dehydrogenase genes: alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet Genomics 19:893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond SL, Singh SM. (1990) Studies with cDNA probes on the in vivo effect of ethanol on expression of the genes of alcohol metabolism. Alcohol Alcohol 25:385–394 [PubMed] [Google Scholar]
- Bond SL, Wigle MR, Singh SM. (1991) Acetaldehyde dehydrogenase (Ahd-2)-associated DNA polymorphisms in mouse strains with variable ethanol preferences. Alcohol Clin Exp Res 15:304–307 [DOI] [PubMed] [Google Scholar]
- Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. (2008) Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 321:1493–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Edenberg HJ, Bosron WF, Li TK. (1989) Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest 83:314–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Matsumoto M, Chang D, You M. (2004) Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc 63:49–63 [DOI] [PubMed] [Google Scholar]
- Ding WX, Nam Ong C. (2003) Role of oxidative stress and mitochondrial changes in cyanobacteria-induced apoptosis and hepatotoxicity. FEMS Microbiol Lett 220:1–7 [DOI] [PubMed] [Google Scholar]
- Duester G. (2000) Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem 267:4315–4324 [DOI] [PubMed] [Google Scholar]
- Endo J, Sano M, Katayama T, Hishiki T, Shinmura K, Morizane S, Matsuhashi T, Katsumata Y, Zhang Y, Ito H, et al. (2009) Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ Res 105:1118–1127 [DOI] [PubMed] [Google Scholar]
- Eriksson CJ. (2001) The role of acetaldehyde in the actions of alcohol (update 2000). Alcohol Clin Exp Res 25:15S–32S [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11:81–128 [DOI] [PubMed] [Google Scholar]
- Hsu LC, Chang WC. (1991) Cloning and characterization of a new functional human aldehyde dehydrogenase gene. J Biol Chem 266:12257–12265 [PubMed] [Google Scholar]
- Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, Beenfeldt Fredriksen S, Linneberg A. (2008) The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol Clin Exp Res 32:1984–1991 [DOI] [PubMed] [Google Scholar]
- Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. (1996) Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry 35:4445–4456 [DOI] [PubMed] [Google Scholar]
- Lam JP, Mays DC, Lipsky JJ. (1997) Inhibition of recombinant human mitochondrial and cytosolic aldehyde dehydrogenases by two candidates for the active metabolites of disulfiram. Biochemistry 36:13748–13754 [DOI] [PubMed] [Google Scholar]
- Larson HN, Weiner H, Hurley TD. (2005) Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J Biol Chem 280:30550–30556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen N, Estey T, Tanguay RL, Pappa A, Reimers MJ, Vasiliou V. (2005) Molecular cloning, baculovirus expression, and tissue distribution of the zebrafish aldehyde dehydrogenase 2. Drug Metab Dispos 33:649–656 [DOI] [PubMed] [Google Scholar]
- Linneberg A, Gonzalez-Quintela A, Vidal C, Jørgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL. (2010) Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy 40:123–130 [DOI] [PubMed] [Google Scholar]
- Little RG, 2nd, Petersen DR. (1983) Subcellular distribution and kinetic parameters of HS mouse liver aldehyde dehydrogenase. Comp Biochem Physiol C 74:271–279 [DOI] [PubMed] [Google Scholar]
- Liu F, Schnable PS. (2002) Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiol 130:1657–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Luo P, Wang A, Payne KJ, Peng H, Wang JG, Parrish YK, Rogerio JW, Triche TJ, He Q, Wu L. (2007) Intrinsic retinoic acid receptor α-cyclin-dependent kinase-activating kinase signaling involves coordination of the restricted proliferation and granulocytic differentiation of human hematopoietic stem cells. Stem Cells 25:2628–2637 [DOI] [PubMed] [Google Scholar]
- Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. (2003) Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol 22:329–338 [DOI] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. (1996) Membrane topology and retention of microsomal aldehyde dehydrogenase in the endoplasmic reticulum. J Biol Chem 271:16939–16944 [DOI] [PubMed] [Google Scholar]
- Meier P, Seitz HK. (2008) Age, alcohol metabolism and liver disease. Curr Opin Clin Nutr Metab Care 11:21–26 [DOI] [PubMed] [Google Scholar]
- Moon KH, Lee YM, Song BJ. (2010) Inhibition of hepatic mitochondrial aldehyde dehydrogenase by carbon tetrachloride through JNK-mediated phosphorylation. Free Radic Biol Med 48:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä O. (2007) Acetaldehyde adducts in circulation. Novartis Found Symp 285:183–192; discussion 193–197 [DOI] [PubMed] [Google Scholar]
- Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. (2003) Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J 376:615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen EN. (1992) The pharmacology and toxicology of disulfiram and its metabolites. Acta Psychiatr Scand Suppl 369:7–13 [DOI] [PubMed] [Google Scholar]
- Sheikh S, Weiner H. (1997) Allosteric inhibition of human liver aldehyde dehydrogenase by the isoflavone prunetin. Biochem Pharmacol 53:471–478 [DOI] [PubMed] [Google Scholar]
- Shi Z, Dragin N, Galvez-Peralta M, Jorge-Nebert LF, Miller ML, Wang B, Nebert DW. (2010) Organ-specific roles of CYP1A1 during detoxication of oral benzo[a]pyrene. Mol Pharmacol 78:46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagos D, Chen Y, Cantore M, Jester JV, Vasiliou V. (2010) Corneal aldehyde dehydrogenases: multiple functions and novel nuclear localization. Brain Res Bull 81:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MJ, Malek K, Crabb DW. (1996) Distribution of messenger RNAs for aldehyde dehydrogenase 1, aldehyde dehydrogenase 2, and aldehyde dehydrogenase 5 in human tissues. J Investig Med 44:42–46 [PubMed] [Google Scholar]
- Stewart MJ, Malek K, Xiao Q, Dipple KM, Crabb DW. (1995) The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem Biophys Res Commun 211:144–151 [DOI] [PubMed] [Google Scholar]
- Stewart SF, Vidali M, Day CP, Albano E, Jones DE. (2004) Oxidative stress as a trigger for cellular immune responses in patients with alcoholic liver disease. Hepatology 39:197–203 [DOI] [PubMed] [Google Scholar]
- Vaglenova J, Martínez SE, Porté S, Duester G, Farrés J, Parés X. (2003) Expression, localization and potential physiological significance of alcohol dehydrogenase in the gastrointestinal tract. Eur J Biochem 270:2652–2662 [DOI] [PubMed] [Google Scholar]
- van de Kerkhof EG, de Graaf IA, Groothuis GM. (2007) In vitro methods to study intestinal drug metabolism. Curr Drug Metab 8:658–675 [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A. (2000) Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology 61:192–198 [DOI] [PubMed] [Google Scholar]
- Viitala K, Makkonen K, Israel Y, Lehtimäki T, Jaakkola O, Koivula T, Blake JE, Niemelä O. (2000) Autoimmune responses against oxidant stress and acetaldehyde-derived epitopes in human alcohol consumers. Alcohol Clin Exp Res 24:1103–1109 [PubMed] [Google Scholar]
- Wang X, Weiner H. (1995) Involvement of glutamate 268 in the active site of human liver mitochondrial (class 2) aldehyde dehydrogenase as probed by site-directed mutagenesis. Biochemistry 34:237–243 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Huang IY, Ikawa M. (1984) Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA 81:258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HS, Oyama T, Isse T, Kitakawa K, Ogawa M, Pham TT, Kawamoto T. (2009) Characteristics of aldehyde dehydrogenase 2 (Aldh2) knockout mice. Toxicol Mech Methods 19:535–540 [DOI] [PubMed] [Google Scholar]