Abstract
We have designed the most efficient strategy to knock out genes in fission yeast Schizosaccharomyces pombe on a large scale. Our technique is based on knockout constructs that contain regions homologous to the target gene cloned into vectors carrying dominant drug-resistance markers. Most of the steps are carried out in a 96-well format, allowing simultaneous deletion of 96 genes in one batch. Based on our knockout technique, we designed a strategy for cloning knockout constructs for all predicted fission yeast genes, which is available in a form of a searchable database http://mendel.imp.ac.at/Pombe_deletion. We validated this technique in a screen where we identified novel genes required for chromosome segregation during meiosis. Here, we present our protocol with detailed instructions. Using this protocol, one person can knock out 96 S. pombe genes in 8 days.
INTRODUCTION
Large-scale knockout screens in fission yeast have been hindered so far by lack of an efficient knockout technique. A technique using long oligonucleotides (80 bp homology to the target sequence)1 was used in a pilot gene deletion project. As many as 20 out of 85 selected genes could not be deleted in a diploid strain2. Using the same approach, Martin-Castellanos et al.3 deleted 160 out of 184 selected meiotically upregulated genes. Recently, Korean BIONEER Corporation embarked on knocking out all fission yeast genes (http://pombe.bioneer.co.kr) using a long-oligonucleotide strategy. Although the long-oligonucleotide strategy1 has been widely used to knock out and modify genes in fission yeast, the low efficiency of this strategy suggests that it may not be optimal for large-scale screens.
We tested and compared different knockout strategies and decided that the most efficient strategy was to use constructs containing left and right homology regions 150–700 bp long cloned into a vector (Fig. 1). The vector was carrying dominant drug resistance markers conferring resistance to either nourseothricin or hygromycin B4-6. We chose the nourseothricin and hygromycin B resistance genes because these have so far not been widely used in fission yeast, which will facilitate the generation of deletions in most of the existing fission yeast strains. Most of the steps are carried out in a 96-well format, allowing simultaneous deletion of 96 genes in one batch.
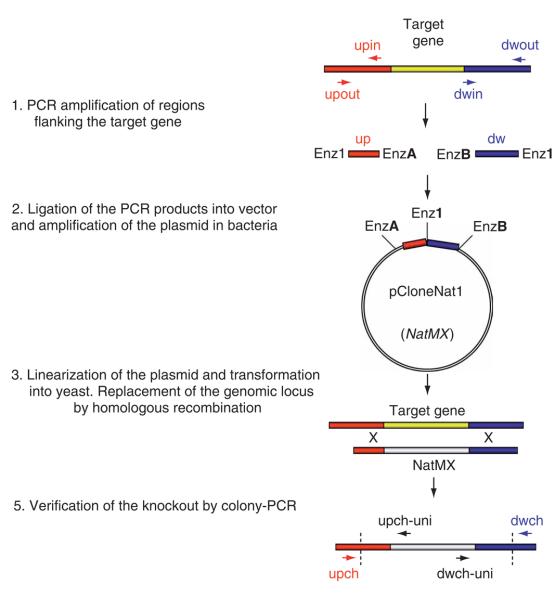
Figure 1.
Flowchart of the knockout strategy. Regions flanking the target gene upstream of the start codon and downstream of the stop codon are designated as “up” and “dw”, respectively. PCR primers were designed in silico so that 150–700 bp of up and dw regions are amplified. Combination of primers upout + upin and primers dwout + dwin is used to PCR amplify regions upstream and downstream of the target gene, respectively. These are the homology regions used to knock out the target gene. Primers encode restriction sites, which are used to clone the homology regions into the pCloneNat1 or pCloneHyg1 vector. The vector containing homology regions is linearized (linearization in the homology region increases efficiency of the targeting) and transformed into yeast. Homologous recombination is used to replace the target gene with the knockout construct. The knockout is verified in yeast transformants by colony PCR using combination of primers upch and upch-uni and primers dwch and dwch-uni.
We validated this technique in our initial screen where we knocked out 180 uncharacterized genes and identified proteins required for meiotic chromosome segregation, most notably the protector of centromeric cohesion Sgo1 (ref. 4). In this screen, we were able to knock out 180 genes out of 192 selected. The remaining genes, which resisted deletion, may be essential genes or possibly genes that are refractory to targeting by homologous recombination owing to a repressed state during vegetative growth7. Besides its superior knockout efficiency, our strategy has the added advantage that a library of knockout plasmids is created.
We further designed a strategy for cloning knockout constructs for all predicted fission yeast genes that is available in a form of a searchable database (http://mendel.imp.ac.at/Pombe_deletion). Apart from the cloning strategy, this webpage includes gel previews indicating mobility of the PCR-amplified homology regions, ligation products and checking PCR products as well as basic information about the protein, DNA sequence of the target gene and link to S. pombe genome database (GeneDB). Web-based tool that automatically suggests primer sequences for deletion, tagging and regulatable expression of S. pombe genes using long oligonucleotides is now also available8.
Apart from the fission yeast, this knockout technique should also be applicable to other yeast species (including pathogenic yeasts) where homologous recombination can be used to knock out genes.
MATERIALS
REAGENTS
PCR kit (Takara). PCR kits from other suppliers should also work. PCR premix will typically contain 1× PCR buffer (containing 1.5 mM MgCl2, 0.2 mM each dNTP mixture, 0.5 μM each primer, 2 U per 100 μl Taq DNA polymerase, template DNA)
Restriction enzymes (Roche)
Primers can be ordered in 96-well plates from Invitrogen. For each knockout, six primers are required (upout, upin, dwout, dwin, upch, dwch)
pCloneNat1 (Kim Nasmyth DNA collection number K4421) or pCloneHyg1 (Kim Nasmyth DNA collection number K4802) are the vectors used for cloning deletion constructs. They can be requested from Kim Nasmyth laboratory
S. pombe genomic DNA from which the homology regions can be amplified
For DNA purification from PCR, restriction digest or agarose gel, we used anion exchange columns from Qiagen. Products of any other supplier should work just as well
Salmon sperm DNA (Stratagene), for use as a carrier for yeast transformation
Qiaprep 96 turbo miniprep kit (Qiagen)
96-well plates for PCR (with cover stickers)
96-well tissue culture plates
EQUIPMENT
96-well plates for PCR (with cover stickers)
Adjustable 0.5–10 and 30–300 μl eight-channel pipettes (Eppendorf)
Adjustable electronic 1,200 μl eight-channel pipette (Eppendorf)
Multistep dispenser plus Combitips (Eppendorf)
96-well pinning tool (V&P Scientific)
electrophoresis units compatible with 96-well system (we used home-made, but it can be purchased from Bio-Rad)
Cutting tool for excising DNA bands from agarose gel (we used home-made). Alternatively, a scalpel can be used
PCR machine for 96-well plates
32 and 37 °C shaking incubators. We use incubators with sticky tape, which is convenient for shaking 96-well plates
REAGENT SETUP
Universal checking primers
For checking of bacterial inserts, two primers are required: upch-uni located approximately 90 bp upstream of the MCS and dwch-uni located approximately 60 bp downstream of the MCS. For checking of yeast transformants, the same primers are used, but there are two alternative primers, upch-uni2 located approximately 150 bp upstream of the MCS and dwch-uni2 located approximately 94 bp downstream of the MCS. upch-uni: GTCGTTAGAACGCGGCTACA; dwch-uni: TCTGGGCCTCCATGTCGCTGG; upch-uni2: GGCTGGCTTAACTATGCGGC; dwch-uni2: GCTGCGCACGTCA AGACTGTC.
Plasmid DNA
Prepare a midi-prep (Qiagen) of the cloning vector (pCloneNat1 or pCloneHyg1).
LiAc solution
Prepare 0.1 M lithium acetate in 1× Tris-EDTA (TE), pH 7.5.
PEG solution
Prepare 0.1 M lithium acetate and 40% (wt/vol) PEG 3350 in 1 × TE, pH 7.5
Competent Escherichia coli
Make transformation-competent Escherichia coli DH5α using CaCl2 method. Alternatively, use commercially available competent E. coli cells.
Media
Prepare standard 2× TY media for E. coli. For selection, add ampicillin (100 μg ml−1). For yeast cultivation, prepare standard YES medium supplemented with 0.15 g l−1 adenine and 0.1 g l−1 uracil, l-histidine, l-lysine and l-leucine. For selection, add clonat at 100 μgml−1 and hygromycin B at 200 μg ml−1.
Primers
Dissolve oligonucleotide primers in TE buffer to 100 μM concentration. To knock out 96 genes (one batch), you will need six 96-well plates with primers (upout, upin, dwout, dwin, upch, dwch).
Reagents required for deletion of one gene
Two 20-mer oligonucleotides, four 30-mer oligos, standard synthesis, desalted; 460 μl PCR reaction; 8.5 μlrestriction enzymes; 4 μl T4 DNA ligase; four DNA purification columns; competent bacteria and yeast cells plus reagents for transforming and growth media.
PROCEDURE
Day 1: preparing vector
1| Prepare vector pCloneNat1 or pCloneHyg1 (Fig. 2), which confer resistance to nourseothricin (clonat) and hygromycin B, respectively4-6. Per ligation you need 10 ng of vector digested with two restriction enzymes and dephosphorylated. We advise to prepare the vector very well: digest the vector with restriction enzyme, give it a long run on agarose gel electrophoresis, extract it from the gel using a Qiagen gel-extraction kit column and dephosphorylate it with shrimp alkaline phosphatase. Vector preparations digested with XhoI/BamHI can be ligated to inserts digested with XhoI/BglII, XhoI/BamHI and SalI/BglII. Vector preparations digested with HindIII/BamHI can be ligated to inserts digested with HindIII/BamHI and HindIII/BglII. Vector preparations digested with SspBI/BamHI can be ligated to inserts digested with SspBI/BamHI and SspBI/BglII. Inserts digested with HindIII/SspBI, XhoI/SspBI or HindIII/XhoI need to be ligated to vector preparations digested with the same enzymes. We used restriction enzymes from Roche with the following Roche buffers: buffer H for XhoI/BglII, SalI/BglII and XbaI digests; buffer B for HindIII/BamHI, XhoI/BamHI, SspBI/BamHI and XhoI/SspBI digests; buffer M for NheI; and buffer A for ApaI digests. Dephosphorylate the vector with shrimp alkaline phosphatase according to the manufacturer's instructions (we used phosphatase from USB).
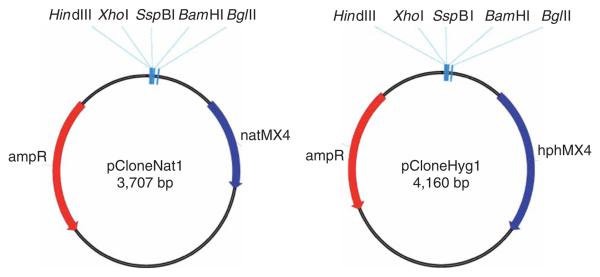
Figure 2.
Maps of cloning vectors pCloneNat1 and pCloneHyg1. Restriction sites used for cloning of the homology regions are indicated. Nucleotide sequence of pCloneNat1 (EF101285) and pCloneHyg1 (EF101286) can be found in GenBank.
▲ CRITICAL STEP Do not rush with preparing the vector. Vector that is not digested to completion and not prepared properly may result in high numbers of negative E. coli transformants containing empty vector without homology regions.
Preparing primers
2| Order primers (100 μM solution) in 96-well plates from the supplier of your choice. We chose Invitrogen. Per batch you will get six 96-well blocks containing required primers: upout, upin, dwout, dwin for cloning, and upch and dwch for yeast checking PCR (Fig. 1).
3| Upon receipt of the primers (100 μM solution), melt them at room temperature (approximately 23 °C) or in a microwave and add 5 μl of 100× TE to each well using a multistep dispenser (0.5 U with 1 U = 10 μl tip).
4| Dispense 90 μl of sterile water into four fresh PCR microtiter plates using a multistep dispenser (4.5 U with 1 U = 20 μl tip). To the first plate, add 5 μl of upout and 5 μl of upin primers (this is referred to as the up plate). To the second plate, add 5 μl of dwout and 5 μl of dwin primers (this is referred to as the dw plate), thus diluting the primers 1:20 and mixing the pairs together. To the third plate, add 5 μl of 100 μM upch primer (this is referred to as the upch plate) and to the fourth plate add 5 μl of 100 μM dwch oligo (this is referred to as the dwch plate), thus diluting the oligos approximately 1:20. Freeze the upch and dwch plates at −20 °C (they will be used in Step 37). The final concentration of primers will be 5 μM each (Fig. 3).
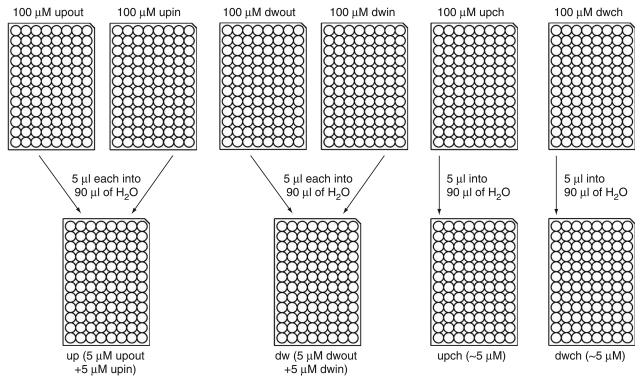
Figure 3.
Dilution of primers. Upon receipt of the primers, add 5 μl of 100× TE to each well. Dispense 90 μl of sterile water into four fresh PCR microtiter plates. To the first plate, add 5 μl of upout and 5 μl of upin primers (this is referred to as the up plate). To the second plate, add 5 μl of dwout and 5 μl of dwin primers (this is referred to as the dw plate). To the third plate, add 5 μl of 100 μM upch primer (this is referred to as the upch plate). To the fourth plate, add 5 μl of 100 μM dwch oligo (this is referred to as the dwch plate).
PCR amplification of homology regions
5| Make 10,000 μl of 1.1× PCR premix containing everything (DNA polymerase, dNTPs, 1.1× buffer, 5 μl of template (genomic) DNA), apart from primers. Dispense 50 μl per well into two PCR microtiter plates prechilled on ice using a multistep dispenser (2.5 U with 1 U = 20 μl tip). To the first plate, add using a multichannel pipette 5 μl from up plate (prepared in Step 4) containing 5 μM upout and 5 μM upin primers (this is referred to as the up PCR plate). To the second plate, add 5 μl from dw plate (prepared in Step 4) containing 5 μM dwout and 5 μM dwin primers (this is referred to as the dw PCR plate) (Fig. 4). Seal plates and run a PCR: 3 min 94 °C; 30 cycles of 40 s 94 °C, 50 s 60 °C, 90 s 72 °C; 5 min 72 °C.
6| Run an aliquot of the PCR on a gel: First dispense into two PCR microtiter plates 5 μl of 3× DNA loading buffer using a multistep dispenser (0.5 U with 1 U = 10 μl tip). From the PCR reaction, pipette 10 μl (using a 100 μl eight-way multichannel pipette) into microtiter plates with the loading buffer. Using the same pipette and tips, load 10 μl on a 1.5% agarose gel. Run the gel, take a picture and compare to lines 1 and 2 of the gel preview at http://mendel.imp.ac.at/Pombe_deletion.
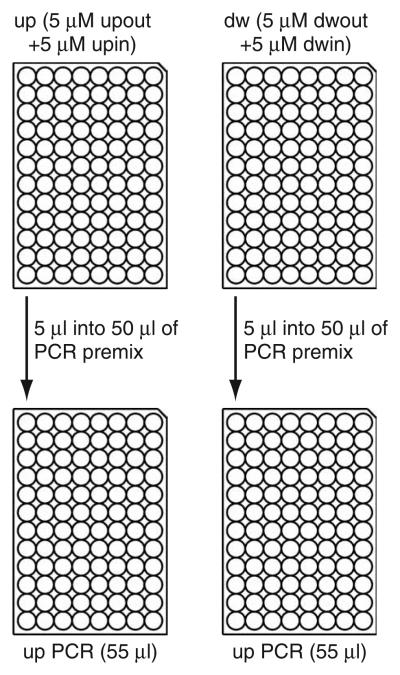
Figure 4.
Amplification of homology regions by PCR. Dispense 50 μl of the PCR premix per well into two PCR microtiter plates. Into the first plate, add 5 μl from up plate containing 5 μM upout and 5 μM upin primers (this is referred to as the up PCR plate). To the second plate, add 5 μl from dw plate containing 5 μM dwout and 5 μM dwin primers (this is referred to as the dw PCR plate).
? TROUBLESHOOTING
Cloning of homology regions into vector
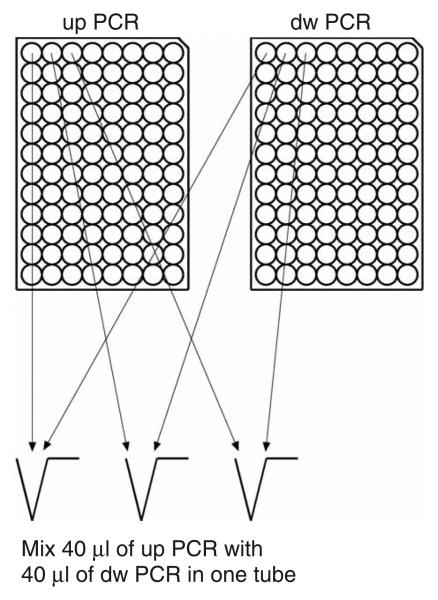
7| Mix 40 μl of the PCR reactions from the up plate (now containing upstream homology regions) with 40 μl of PCR reactions from the dw plate (now containing downstream homology regions) in a 1.5 ml microfuge tube (96 single tubes) (Fig. 5). Purify over a Qiagen gel-extraction kit column according to the manufacturer's instructions. Use 600 μl of buffer PB and elute with 30 μl of buffer EB.
8| Make the first restriction enzyme premixes: Per sample use 5.8 μl of 10× buffer, 2 μl of enzyme Enz1 and 22.2 μl of H2O. Dispense 30 μl into tubes using a multistep dispenser (3 U with 1 U = 10 μl tip). Mix by quick vortexing. Incubate for 2 h at 37 °C.
9| Purify over a Qiagen gel-extraction kit column. Use 300 μl of buffer PB and elute with 30 μl of buffer EB.
Figure 5.
Pooling of PCR-amplified homology regions—preparation for cloning. Mix 40 μl of the PCR reactions from the up plate with 40 μl of PCR reactions from the dw plate in a 1.5 ml microfuge tube.
■ PAUSE POINT Digested PCR products can be stored for months at −20 °C.
10| Make the first ligation premix: Per sample use 5.8 μl of 10× buffer, 2 μl of T4 DNA ligase and 22.2 μl of H2O. Dispense 30 μl into tubes using a multistep dispenser (3 U with 1 U = 10 μl tip). Mix by quick vortexing. Incubate overnight (16 h) at 4 °C.
Day 2
11| Purify over a Qiagen gel-extraction kit column. Use 300 μl of buffer PB and elute with 30 μl of buffer EB. Extraction of DNA from agarose gels is carried out in single tubes. If a gel extraction kit becomes available in a 96-well format, this may speed up the whole procedure.
12| Make the second restriction enzyme premix: Per sample use 5.8 μl of 10× buffer, 1.5 μl of EnzA, 1.5 μl of EnzB and 21.2 μl of H2O. Dispense 30 μl into tubes using the multistep dispenser (3 U with 1 U = 10 μl tip). Mix by quick vortexing. Incubate for 2 h at 37 °C.
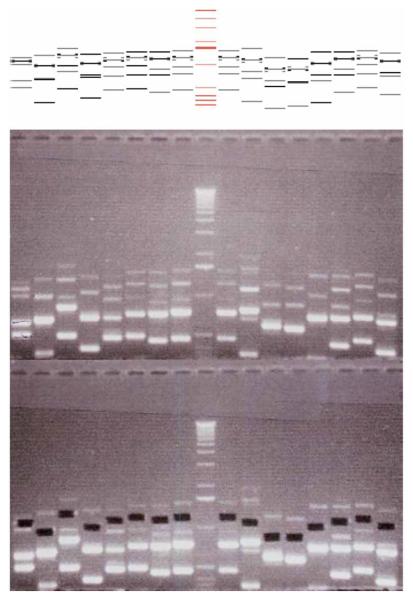
13| Add 12 μl of 6× DNA loading buffer and load on a 1% agarose gel. Use two gels for 96 samples and allow the samples to run twice as far as usual. PCR primers are designed such that two homology regions are of unequal size; therefore, five bands are expected to be seen after ligation. These correspond to the monomer of the smaller homology region, the monomer of the larger homology region, the homodimer of the smaller homology region, the desired heterodimer and the homodimer of the larger homology region. Using a cutting tool (alternatively, use a scalpel), cut out the band corresponding to correctly ligated heterodimer of two homology regions according to the gel preview at http://mendel.imp.ac.at/Pombe_deletion (the desired heterodimer is highlighted) (Fig. 6).
Figure 6.
Comparison of ligated homology regions with the gel preview at http://mendel.imp.ac.at/Pombe_deletion/. Bands highlighted on the gel preview represent correctly ligated homology regions. These bands are cut out of the gel and cloned into the vector.
▲ CRITICAL STEP Exposure to UV light can damage DNA. Work quickly and minimize the UV exposure.
14| Gel extract using a Qiagen gel-extraction kit column. Use 500 μl of buffer QG and elute with 30 μl of buffer EB.
■ PAUSE POINT Digested PCR products can be stored for months at −20 °C.
15| Transfer 10 μl of the eluted insert into a new PCR microtiter plate.
16| Ligate 10 μl of extracted insert with approximately 10 ng of digested vector (prepared in Step 1) using a commercial rapid ligation kit (we used Takara DNA Ligation Kit Version 2). Incubate the ligation according to the manufacturer's instructions (30 min for Takara DNA Ligation Kit Version 2). Do a control ligation without the insert. Alternatively, use T4 DNA ligase and incubate overnight at 4 °C. Per sample use 2 μl of 10× buffer, 1.5 μl of T4 DNA ligase, approximately 10 ng of cut and dephosphorylated vector and up to 20 μlH2O. Dispense 20 μl using the multistep dispenser (2 U with 1 U = 10 μl tip). Mix by quick vortexing. Incubate overnight (16 h) at 4 °C.
Transforming E. coli
17| Chill the microtiter plates with ligation reactions on ice. Add 150 μl of competent E. coli DH5α using a multistep dispenser (3 U with 1 U = 50 μl tip). Incubate for 30 min on ice and subject to heat shock for 90 s at 42 °C (in a thermocycler). Transfer the transformation mix to a 96-deep well block containing 1 ml of prewarmed 2× TY medium (LB will do) using a 300 μl multichannel pipette. Seal the plate and incubate for 30 min at 37 °C with shaking.
18| Spin down bacteria in a benchtop centrifuge (5 min, 2,000g, 23 °C). Pour off the supernatant, resuspend the pellet in each well in 100 μl of 2× TY medium (by vortexing with lid sticker on) and plate on 2× TY + ampicillin plates (100 μl per plate) (LB medium can also be used). Incubate overnight at 37 °C.
Day 3: checking bacteria by PCR followed by Enz1 digest 19
19| Prepare 4,000 μl PCR premix containing everything (DNA polymerase, dNTPs, buffer, primers) apart from the template. Use primers upch-uni and dwch-uni. If you use the TAKARA PCR kit, per 100 μl use 10 μl of 10× buffer, 8 μl of dNTPs, 6 μl of MgCl2, 0.5 μl of Taq polymerase, 0.5 μl of 100μM upch-uni primer, 0.5 μl of dwch-uni primer and 74.5 μl of H2O. Dispense 20 μl into two 96-well PCR microtiter plates prechilled on ice using a multistep dispenser (2 U with 1U = 10 μl tip).
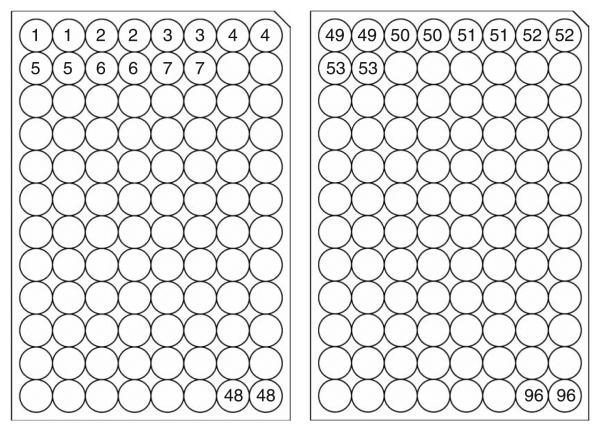
20| Prepare two new 96-well tissue culture microtiter plates. Dispense 100 μl of 2× TY + ampicillin medium into each well using a multistep dispenser (2 U with 1 U = 50 μl tip)(Fig. 7).
21| Check two E. coli colonies for each construct. Using a yellow tip, pick the first colony and place the tip on the eight-channel pipette. Then, pick a second colony and place the second tip on the pipette next to the first one. Continue with the next construct until all eight positions of the pipette are filled (this will contain four different constructs, two colonies for each) (Fig. 6). Dip five times into the plate containing the PCR premix (prepared in Step 19), then dip into the plate containing 2× TY + ampicillin medium (prepared in Step 20) and pipette up and down. Run the PCR: 3 min 94 °C; 30 cycles: 30 s 94 °C, 40 s 56 °C, 120 s 72 °C; 5 min 72 °C. Incubate the plates with 2× TY + ampicillin medium overnight at 37 °C.
22| Make restriction enzyme premix: Per sample use 0.5 μl of Enz1 and 4.5 μl H2O. Dispense 5 μl directly into each well of the 96-well microtiter plate containing PCR reactions, using a multistep dispenser (0.5 U with 1 U = 10 μl tip). Incubate for 2 h at 37 °C.
23| Run on a gel: First prepare 1.6× DNA loading buffer in a multichannel pipette reservoir. Pipette 15 μl into each well of the 96-well microtiter plates containing Enz1-digested PCR (from Step 22). Mix by pipetting up and down and load 15 μl on a 1.5% agarose gel.
24| Take a picture of the gel and compare it with the gel preview at http://mendel.imp.ac.at/Pombe_deletion. Identify positive clones (i.e., those with the insert of correct size that is also cleavable by the Enz1 enzyme) and sort them into one 96-deep-well plate containing 1.5 ml of 2× TY + ampicillin in each well. Alternatively, use two 48-deep-well plates containing 3 ml of 2× TY + ampicillin if more plasmid is needed.
Figure 7.
E. coli transformants ordered in 96-well plates. Prepare two 96-well tissue culture microtiter plates. Dispense 100 μl of 2× TY + ampicillin medium into each well. Inoculate two E. coli colonies for each construct.
▲ CRITICAL STEP Checking of the PCR product by Enz1 digestion is necessary. We observed and confirmed by sequencing that some of the ligated homology regions contained altered Enz1 recognition sequencethatcould notbecleaved by theEnz1.
? TROUBLESHOOTING
Day 4: preparing knockout plasmids
25| Prepare glycerol stock of E. coli transformants. Mix 50 μl of E. coli culture with 50 μl of 80% glycerol predispensed into a 96-well tissue culture plate and store at −80 °C.
26| Prepare plasmid DNA from the positive E. coli transformants. We used miniprep Qiagen columns. DNA purification columns in 96-well format are now available (e.g., Qiagen Turbo 96 kit), which should speed up the procedure.
■ PAUSE POINT E. coli transformants can be stored for years at −80 °C. Plasmid DNA can be stored for years at −80 °C.
Transforming yeast
27| Pipette 5 μl of prepared plasmid into a new 96-well PCR microtiter plate. Add 10 μl of restriction enzyme premix to linearize plasmids: Per sample use 1.5 μl of 10× buffer, 1.5 μl of Enz1 and 12 μl of H2O, dispensed using a multistep dispenser (1 U with 1 U = 10 μl tip). Incubate for 2 h at 37 °C.
28| Check the digests on a gel: First dispense 10 μl of 1.2× DNA loading buffer into a 96-well PCR microtiter plate using a multistep dispenser (1 U with 1 U = 10 μl tip)and add 2 μl of digested plasmid. Load 10 μl on a 1% agarose gel and run the gel.
29| Transfer the remaining digest into a 1.5 μl microfuge tube and transform your favorite yeast strain using your favorite protocol—we used the lithium acetate method. Plate cells on appropriate selective plates—YES + clonat or YES + hygromycin (Box 1).
30| Incubate plates after transformation at 32 °C until colonies appear (approximately 3 days).
BOX 1. S. pombe TRANSFORMATION.
Preparation of competent cells
(1) Grow yeast strain in 50 ml of YES in 250 ml Erlenmeyer flask at 32 °C overnight.
(2) Next morning, dilute the culture to OD600 = 0.2 (this is approximately 3 ml of the overnight culture into 100 ml of fresh YES) and incubate 100 ml of the culture in 500 ml Erlenmeyer flask for 4–5 h at 32 °C until the OD600 = 0.6.
(3) Spin for 3 min at 1,000g in two 50 ml Falcon tubes.
(4) Pour off the supernatant and resuspend in 20 ml of LiAc solution and pool the contents of two Falcon tubes.
(5) Spin for 3 min at 1,000g, pour off the supernatant and resuspend in 20 ml of LiAc solution.
(6) Spin for 3 min at 1,000g, pour off the supernatant and resuspend in 2 ml of LiAc solution.
(7) Incubate for 30 min at 32 °C.
Transformation
(8) Add to 1.5 ml microfuge tubes: The DNA to be transformed should be in a maximum 15 μl volume. Also include 150 μl of the suspension of competent yeast cells. Vortex briefly.
(9) Add 375 μl of PEG solution and 2 μl of single-stranded DNA (we used salmon sperm DNA 10 mg μl−1). Mix by inversion.
(10) Incubate for 45 min at 32 °C.
(11) Subject to heat shock by incubating for 5 min at 46 °C in a thermomixer. Let it cool down at room temperature for 10 min.
(12) Spin for 30 s at 2,000 r.p.m. and remove the supernatant using pipette.
(13) Resuspend the cells in 100 μl of YES and transfer to a 5 μl snapcap tube containing 1 ml of YES.
(14) Incubate overnight at 32 °C with shaking.
(15) The next morning, vortex the tubes and plate 200 μl on appropriate selective plates.
(16) Incubate for 3 days at 32 °C.
? TROUBLESHOOTING
Day 7: inoculating yeast transformants
31| Prepare four 96-well tissue culture plates containing 200 μl of YES + ClonNat in each well.
32| For each construct, pick up four colonies (using toothpick) and inoculate into 96-well tissue culture plates containing 200 μl of YES + ClonNat or YES + hygromycin. Incubate overnight at 32 °C with shaking at 250 r.p.m.
Day 8: checking yeast transformants
33| After overnight incubation, water may evaporate from the wells at the edge of the 96-well tissue culture plate. Fill them up with sterile H2O to the original volume (200 μl).
34| Using a 96-well pinning tool, transfer a drop from each well onto a 96-well compatible plate containing YES + ClonNat or YES + hygromycin agar medium and incubate at 32 °C. Cells from this plate should be used for the phenotypical analysis of the knockout strains.
35| Transfer 100 μl of the culture (resuspend properly by pipetting up and down) into a new 96-well PCR microtiter plate (repeat with all four plates). Spin down in a centrifuge (5 min, 1,800g) and pour off the supernatant. Resuspend in 50 μl of sterile H2O. A 7.5 μl portion of this suspension will be used as PCR template.
36| Prepare glycerol stock of the yeast transformants. Add 100 μl of 30% glycerol to the rest of the cultures. Freeze at −80 °C.
■ PAUSE POINT The transformants can be stored at −80 °C for years.
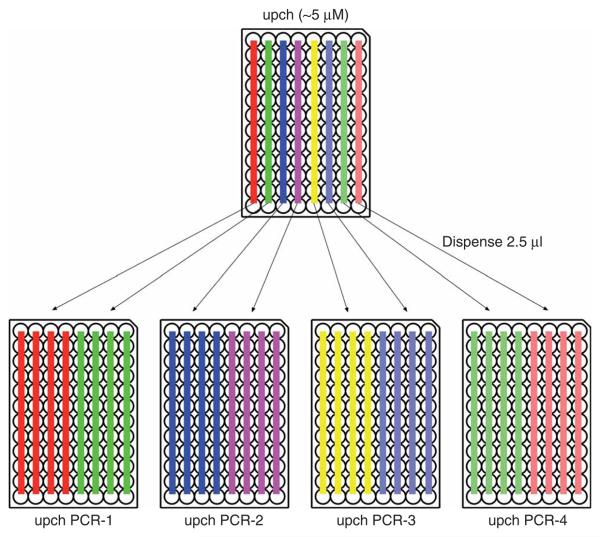
37| Dispense 2.5 μl of ~5 μM upch primer (from the upch plate prepared in Step 4) into four 96-well PCR plates on ice (referred to as upch PCR plates). Dispense 2.5 μl of ~5 μM dwch primer (from the dwch plate prepared in Step 4) into four 96-well PCR plates on ice (referred to as dwch PCR plates) (Fig. 8).
38| Prepare 6,000 μl of 1.6× PCR premix containing upch-uni primer. Prepare another 6,000 μl of 1.6× PCR premix containing dwch-uni primer. Dispense 15 μl of upch premix into four upch PCR plates and 15 μl of dwch premix into four dwch PCR plates. Keep the PCR plates on ice.
39| From the plates with cell suspensions (prepared in Step 35), pipette 7.5 μl into the corresponding upch PCR and dwch PCR plates. Make sure to properly resuspend sedimented cells before pipetting them. Keep PCR plates on ice and after adding cells transfer to 94 °C as quickly as possible. Run the PCR: 3 min 94 °C; 37 cycles: 50 s 94 °C, 60 s 60 °C, 180 s 72 °C; 5min 72 °C.
40| Run the PCR reactions on a gel: First prepare 1.6 DNA loading buffer in a multichannel pipette reservoir. Pipette 15 μl of the buffer into the PCR reactions, mix by pipetting up and down, and using the same tips load 15 μl on 1.5% agarose gel.
41| Run the gel and identify positive clones by the presence of the expected band according to the gel preview at http://mendel.imp.ac.at/Pombe_deletion.
Figure 8.
PCR checking of yeast transformants. Dispense 2.5 μl of upch primer into four 96-well PCR plates (referred to as upch PCR plates) and 2.5 μl of dwch primer into four 96-well PCR plates (referred to as dwch PCR plates). Dispense 15 μl of upch PCR premix into four upch PCR plates and 15 μl of dwch PCR premix into four dwch PCR plates and add the cell suspensions.
? TROUBLESHOOTING
● TIMING
Steps 1–10 (day one)
Steps 11–18 (day two)
Steps 19–24 (day three)
Steps 25–30 (day four)
Steps 31–32 (day seven)
Steps 33–41 (day eight)
Our suggestion is to start on Tuesday morning, transform yeast on Thursday and plate yeast transformants on Friday. Saturday and Sunday are free. On Monday, you should see colonies of yeast transformants on selective plates. Identify positive clones on Tuesday.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 6, 24, 41 | No PCR product | Reactions evaporated from the 96-well plates during the PCR |
Seal the cover sticker tightly |
| 6, 24, 41 | No PCR product | Annealing temperature was not optimal |
Repeat the PCR in single tubes using gradient annealing temperature |
| 22 | Ligated homology regions cannot be cleaved by Enz1 |
Enz1 recognition sequence has been altered |
Check more E. coli transformants |
| 24 | No positive clones (containing the insert) after transforming E. coli |
The cloning vector was not completely digested |
Do a control ligation with vector only (without the insert) to find out if the vector was properly digested |
| 30 | No colonies after transforming haploid yeast strain |
The gene is essential for cell growth |
Create heterozygous knockout in diploid strain |
ANTICIPATED RESULTS
Using this protocol, one person can knock out 96 S. pombe genes in 8 days. It is also possible that one person can do two batches (192 knockouts) simultaneously; however, if more than 192 knockouts are required, additional help is needed. We successfully used this protocol to knock out functionally uncharacterized genes whose expression is upregulated during meiosis9. In this screen, we were able to knock out 180 genes out of 192 selected.
Most of the steps are carried out in a 96-well format. However, single tubes are used for extraction of DNA from agarose gel and for plasmid purification. Recently, 96-well mini-prep kits became available (e.g., from Qiagen). In the future, gel extraction kits compatible with the 96-well system may also become available. Using these kits will eliminate single tubes and may speed up the whole procedure. This protocol can also be scaled down to knock out a single gene. We routinely use this protocol in our lab to knock out single genes. A diploid strain can be used to knock out essential genes. Knockout constructs can be verified by sequencing, which makes the whole procedure safe.
Knocking out your favorite gene may affect neighboring genes if they are in close proximity to the target gene. If this is the case, new primers should be designed to move the regions of homology. We have not considered this criterion when designing knockout constructs for all S. pombe genes (http://mendel.imp.ac.at/Pombe_deletion). To exclude the effect of neighboring genes on the knockout phenotype, we suggest to test complementation of the knockout phenotype by introducing the wild-type allele of the target gene.
ACKNOWLEDGMENTS
This work was supported by Boehringer Ingelheim and partly by Austrian Industrial Research Promotion Fund (FFF) and Austrian Science Fund (FWF) grants. We thank Georg Dietzl and Barry Dickson (IMP, Vienna) for help with setting up the cloning protocol and Stephen Kearsey (University of Oxford, UK) for helpful suggestions and reagents.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Rights and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.Decottignies A, Sanchez-Perez I, Nurse P. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Castellanos C, et al. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 2005;15:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabitsch KP, et al. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 6.Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- 7.Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penkett CJ, Birtle ZE, Bahler J. Simplified primer design for PCR-based gene targeting and microarray primer database: two web tools for fission yeast. Yeast. 2006;23:921–928. doi: 10.1002/yea.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregan J, et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]