Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2
Mimitou and Symington show that reducing association of Ku proteins with DNA double-strand breaks is an important function of resection initiation factors, paralleling recently described similar functions for the Fanconi anaemia pathway in antagonizing NHEJ in favour of homologous recombination.
Keywords: end joining, homologous recombination, Ku, Mre11, Sae2
Abstract
In this study, we investigate the interplay between Ku, a central non-homologous end-joining component, and the Mre11–Rad50–Xrs2 (MRX) complex and Sae2, end-processing factors crucial for initiating 5′-3′ resection of double-strand break (DSB) ends. We show that in the absence of end protection by Ku, the requirement for the MRX complex is bypassed and resection is executed by Exo1. In contrast, both the Exo1 and Sgs1 resection pathways contribute to DSB processing in the absence of Ku and Sae2 or when the MRX complex is intact, but functionally compromised by elimination of the Mre11 nuclease activity. The ionizing radiation sensitivity of a mutant defective for extensive resection (exo1Δ sgs1Δ) cannot be suppressed by the yku70Δ mutation, indicating that Ku suppression is specific to the initiation of resection. We provide evidence that replication-associated DSBs need to be processed by Sae2 for repair by homologous recombination unless Ku is absent. Finally, we show that the presence of Ku exacerbates DNA end-processing defects established in the sae2Δ sgs1Δ mutant, leading to its lethality.
Introduction
DNA lesions arise spontaneously during normal cell metabolism or after treatment with DNA-damaging agents. Among these lesions, DNA double-strand breaks (DSBs) are considered the most deleterious and if unrepaired or repaired inappropriately, they can lead to mutagenic events, such as chromosome loss, deletions, duplications or translocations. DSBs are repaired through non-homologous end joining (NHEJ), which directly rejoins DNA ends with no or limited homology, or by homologous recombination (HR), which requires a homologous template for repair and generally preserves genetic information at the break site. In Saccharomyces cerevisiae, both pathways require the Mre11–Rad50–Xrs2 (MRX) complex, which is rapidly recruited to DSBs and signals checkpoint activation through the Tel1/ATM kinase, tethers DNA ends and regulates the initiation of 5′-3′ resection (Stracker et al, 2004; Mimitou and Symington, 2009). In addition, NHEJ requires Ku (a heterodimer encoded by the YKU70 and YKU80 genes in S. cerevisiae), Lif1, Nej1 and Dnl4 (DNA ligase IV), whereas HR requires proteins encoded by the RAD52 epistasis group genes (Krogh and Symington, 2004; Daley et al, 2005).
The choice of the repair pathway used to repair DSBs is highly regulated to ensure that the cell engages the most appropriate one, thus optimizing genome stability. This is corroborated by the finding that certain types of DSB repair such as V(D)J recombination and meiotic recombination are linked to specific repair pathways (Keeney, 2001; Lee et al, 2004). The types of ends generated and the cell cycle stage are critical determinants governing the choice between repair pathways. NHEJ is the predominant pathway in G1, whereas HR is activated during S/G2 (Moore and Haber, 1996; Karathanasis and Wilson, 2002; Aylon et al, 2004; Ira et al, 2004; Barlow et al, 2008). One step where cell cycle control is exerted by cyclin-dependent kinases (CDK) is the 5′-3′ nucleolytic degradation of DNA ends, which generates 3′ single-stranded DNA (ssDNA) tails, the substrate for binding by the Rad51 protein to initiate HR (Aylon et al, 2004; Ira et al, 2004; Zierhut and Diffley, 2008). In S. cerevisiae, end resection takes place by a two-step mechanism. Initially, the MRX complex with Sae2 endonuclease catalyse the removal of a short oligonucleotide(s) from the 5′ ends of the break. In the second step, the short 3′ overhangs created are further processed by two alternative pathways, one dependent on the 5′-3′ exonuclease Exo1 and the other dependent on the Sgs1 helicase and Dna2 helicase/endonuclease (Gravel et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008). Sae2 is directly phosphorylated by CDK activating the initiation of end processing; in addition, nuclear entry of Dna2 during S-phase is regulated by CDK (Huertas et al, 2008; Kosugi et al, 2009).
The MRX complex and Ku rapidly, and almost simultaneously, bind independently to DNA ends after DSB formation (Wu et al, 2008). Mre11 exhibits exo- and endonuclease activities that are required for processing of meiotic DSBs, a subset of ionizing radiation (IR)-induced DSBs and DNA hairpins, but are dispensable for NHEJ, telomere maintenance and processive 5′-3′ resection of DNA ends generated by HO endonuclease (Bressan et al, 1998; Moreau et al, 1999; Rattray et al, 2001; Lobachev et al, 2002; Llorente and Symington, 2004). Ku requires a free DSB end for binding and once bound protects ends and mediates recruitment of downstream NHEJ factors (Daley et al, 2005). The dissociation of Ku from DSB ends in vivo is dependent on MRX and the timing correlates with bulk resection in preparation of HR (Wu et al, 2008). Several lines of evidence suggested that Ku dissociation is not merely a de facto result of resection, but instead is required to allow resection to occur. Deletion of YKU70 was shown to increase resection initiation both at DSBs and telomeres (Lee et al, 1998; Maringele and Lydall, 2002; Clerici et al, 2008), partially rescue the IR and methylmethane sulphonate (MMS) hypersensitivity observed in mre11Δ and rad50Δ mutants (Bressan et al, 1999; Wasko et al, 2009) and increase Rfa1 foci formation in response to I-SceI-induced DSBs during G1 (Barlow et al, 2008). Similarly, sae2Δ and mre11 nuclease-defective mutants exhibit persistent Mre11 and Sae2 foci at DSBs, supporting a more general mechanism by which MRX-Sae2 regulate protein turnover at the DNA ends (Lisby et al, 2004).
These observations suggest that the first step of end resection executed by MRX-Sae2 serves to create a substrate less suitable for Ku binding thus committing cells to extensive resection and HR. To test this hypothesis, we combined genetic and physical assays to determine whether the loss of the first step in DSB resection can be rescued by concomitant loss of Ku. Indeed, we show that the DNA damage sensitivity of mutants defective for resection initiation, but not bulk resection, is suppressed in the absence of Ku. Exo1 and Sgs1, which are required for extensive resection, are responsible for this suppression. Finally, we show that the lethality of the sae2Δ sgs1Δ mutant can be bypassed by the yku70Δ mutation or by high-copy expression of EXO1, but not by the dnl4Δ mutation. These findings suggest that Ku inhibits growth by blocking access to Exo1 preventing resection in strains lacking Sae2 and Sgs1, and not by promoting lethal end-joining events.
Results
Suppression of the radiation sensitivity of mre11 mutants by deletion of YKU70
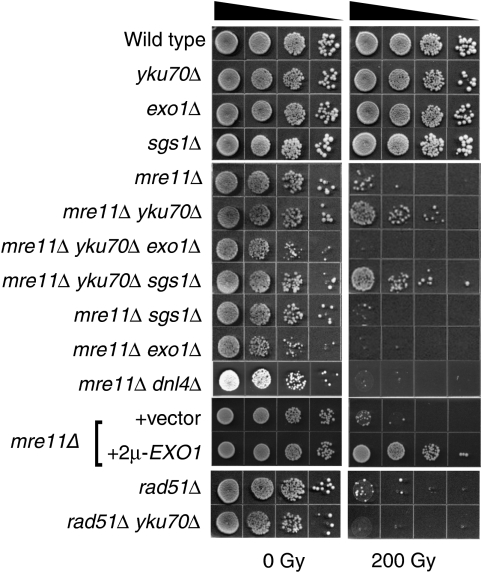
Null mutation of any of the three genes encoding members of the MRX complex renders the cells highly sensitive to IR (Ivanov et al, 1992; Tsubouchi and Ogawa, 1998; Bressan et al, 1999; Moreau et al, 2001). Notably, the mre11Δ IR sensitivity was shown to be suppressed by concomitant deletion of YKU70 (Bressan et al, 1999). The increased IR resistance of mre11Δ yku70Δ mutants is thought to originate from the loss of end protection by Ku allowing DSB ends to be processed even in the absence of Mre11. Given the redundancy of DSB resection pathways, we asked whether the yku70Δ suppression of the mre11Δ IR sensitivity is dependent on SGS1 and/or EXO1 by determining the plating efficiency of various mutant strains after IR exposure. In agreement with previous studies, we found that mre11Δ mutants exhibit high IR sensitivity (100-fold decrease in survival at 200 Gy), which is suppressed by deletion of YKU70 (Figure 1). The suppression does not apply to all HR mutants, as rad51Δ cannot be suppressed by yku70Δ, supporting the link between increased end processing and loss of DSB end protection. The IR sensitivity of the mre11Δ yku70Δ sgs1Δ mutant is comparable with the sensitivity of the mre11Δ yku70Δ double mutant, indicating that the suppression is independent of Sgs1. Conversely, exo1Δ negated the suppression, suggesting that in the absence of the MRX complex, Ku blocks access to Exo1 (Figure 1). In agreement with this hypothesis, EXO1 over-expression also suppressed the mre11Δ IR sensitivity (Figure 1) (Chamankhah et al, 2000; Tsubouchi and Ogawa, 2000; Moreau et al, 2001; Lewis et al, 2002). Deletion of DNL4 did not suppress the mre11Δ IR sensitivity, supporting the hypothesis that it is the loss of end protection by Ku that allows increased 5′-3′ end processing (Figure 1). Analogous findings were reported in Schizosaccharomyces pombe in which deletion of pku70 suppressed the IR and MMS sensitivity of rad50 or rad32 mutants in an exo1+-dependent manner (Tomita et al, 2003; Williams et al, 2008).
Figure 1.
Suppression of the mre11Δ IR sensitivity by YKU70 deletion. Exponentially growing cells of the indicated genotypes were 1:10 serially diluted, spotted onto YPD or selective plates and exposed to the indicated IR dose.
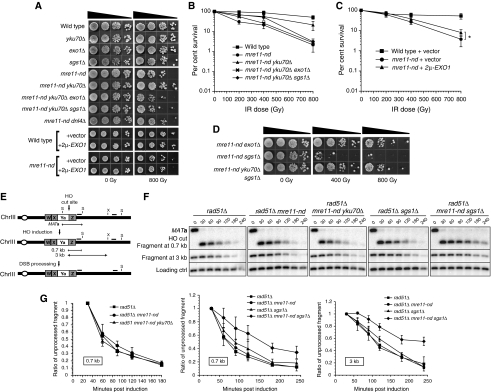
To determine whether the presence of Ku at DSB ends interferes with end processing in the presence of a structurally but not functionally competent MRX complex, we used an allele of MRE11 (mre11-H125N) encoding a protein lacking endo- and exonuclease activities (Moreau et al, 1999; Krogh et al, 2005). For simplicity, this nuclease-defective allele is referred to as mre11-nd. In agreement with the previous studies, the mre11-nd mutant exhibited IR sensitivity only at high doses, with a 17-fold decrease in survival at 800 Gy (Figure 2A and B) (Moreau et al, 1999). Deletion of YKU70 in the mre11-nd mutant increased the IR resistance at 800 Gy by seven-fold (P=0.01) (Figure 2A and B). Interestingly, this increased resistance is dependent on both Exo1 and Sgs1. High-copy expression of EXO1 increased the mre11-nd resistance at 800 Gy by only two-fold (P=0.03) (Figure 2C), consistent with a previous study (Moreau et al, 2001). Our findings suggest that in the presence of a defective MRX complex, Ku provides a partial block to processing DSB ends by Exo1 and Sgs1.
Figure 2.
Phenotype of mre11-nd and mre11-nd sgs1Δ mutants. (A) Suppression of the mre11-nd IR sensitivity by the yku70Δ mutation (quantitation in (B)) or high-copy expression of EXO1 (quantitation in (C), *P=0.03, unpaired t-test). (D) Radiation sensitivity of mre11-nd mutants in conjunction with sgs1Δ or exo1Δ mutations. (E) Schematic representation of the chromosome III MAT locus used in the physical assay to assess resection of an HO-induced DSB. The 5′-3′ degradation destroys the StyI (S) and XbaI (X) recognition sites, which translates into the disappearance of the StyI/XbaI digestion fragments. (F) Southern blot analysis and (G) cut fragment intensity plots showing the kinetics of the cut fragment intensity disappearance as a ratio of the intensity 30 min after induction. The means from four experiments are presented, error bars indicate s.d.
The mre11-nd defect was further characterized using a physical assay that monitors resection of an HO-induced DSB at the MAT locus in strains with an integrated PGAL1–HO fusion. The assay was performed in rad51Δ mutants in which the processed ends do not engage in repair facilitating their detection. Following synchronous HO cleavage by addition of galactose, resection at different distances from the break can be monitored by detecting restriction enzyme fragments with probes specific for that region. As 5′-3′ resection proceeds, a StyI site at 0.7 kb distal to the break and an XbaI site at 3 kb distal to the break are sequentially rendered ss; therefore, resistant to digestion (Figure 2E). As a result, after digesting genomic DNA with StyI/XbaI, the intensity of the bands corresponding to the DNA fragments diminishes over time. This analysis revealed that the disappearance of the fragment indicative of resection past the StyI site at 0.7 kb (Figure 2F and G) or the XbaI site at 3 kb (Figure 2F) is not altered in the mre11-nd strain, consistent with a previous study (Llorente and Symington, 2004). Moreover, deletion of YKU70 in the mre11-nd background did not increase processing of the cut fragment. Although the increased requirement for the Mre11 nuclease in response to high IR doses could reflect a dosage effect, we have previously shown that the mre11-nd mutant is proficient for resection of multiple HO-induced DSBs (Llorente and Symington, 2004). Thus, we favour the hypothesis that the differential phenotype and suppression of mre11-nd mutants by deletion of YKU70 in IR sensitivity and resection assays reflects the different requirements for processing IR (‘dirty') versus endonuclease (‘clean')-induced DSBs. For IR-induced breaks, the requirement for the Mre11 nuclease to process some ends is increased, making the suppression by the loss of Ku more obvious.
Sgs1 becomes important in the absence of the nuclease activity of Mre11
Given that the suppression of the sensitivity to IR of mre11-nd by yku70Δ requires Sgs1, we hypothesized that in the presence of a defective MRX complex the helicase and nuclease activities of Sgs1-Dna2 provide some redundant activity to initiate end processing. Consistent with this idea, the mre11-nd sgs1Δ double mutant exhibited a synergistic defect in IR resistance, with a 1700-fold decrease in survival at 800 Gy compared with the wild-type strain (Figure 2D). A similar finding was reported by Budd and Campbell (2009). The mre11-nd exo1Δ mutant exhibited higher resistance to IR than the mre11-nd sgs1Δ mutant, suggesting that Exo1 is less important for the initial processing of ends than Sgs1-Dna2 when Ku is present (Figure 2D). The yku70Δ mutation conferred a significant increase in the IR resistance of the mre11-nd sgs1Δ mutant, again consistent with the view that Ku prevents access of ends to Exo1. The exo1Δ mre11-nd sgs1Δ triple mutant is inviable preventing analysis of end processing in the absence of these overlapping functions (Mimitou and Symington, 2008).
For a more quantitative measure of DSB end processing, the disappearance of restriction fragments indicative of resection 0.7 and 3 kb from the HO-cut site was monitored in rad51Δ mre11-nd, rad51Δ sgs1Δ and rad51Δ mre11-nd sgs1Δ strains (Figure 2F). As noted above, the rad51Δ and rad51Δ mre11-nd mutants exhibited similar kinetics in the disappearance of the cut fragments over time. The rad51Δ sgs1Δ appeared slightly more defective in resection at both 0.7 and 3 kb, but the difference is not statistically significant. We consistently found a resection defect at both 0.7 and 3 kb in the rad51Δ mre11-nd sgs1Δ mutant (Figure 2F and G). At 120 min after HO induction, there is a two-fold decrease in the amount of ends resected at 0.7 kb (P=0.006) and a 2.5-fold decrease at 3 kb (P=0.0003) compared with rad51Δ. These results suggest that Sgs1 provides some redundant activity to initiate end resection in the absence of the Mre11 nuclease activity. This requirement for Sgs1 is more pronounced for IR-induced breaks than endonuclease-induced ends as evidenced by the high IR sensitivity of the mre11-nd sgs1Δ mutants compared with the subtle HO end-resection defect.
sae2Δ mutants exhibit high IR sensitivity that is suppressed by deletion of YKU70
MRE11 and SAE2 belong to the same epistasis group with respect to DSB resection and sae2Δ mutants exhibit a similar phenotype to mre11-nd mutants (Rattray et al, 2001; Lobachev et al, 2002; Clerici et al, 2005). When tested for IR sensitivity, the sae2Δ mutant showed a 255-fold decrease in survival at 800 Gy. The IR sensitivity of the sae2Δ mutant is higher than mre11-nd and is exhibited at lower IR doses (Figure 3A and B). At 800 Gy, the mre11-nd mutant is 15-fold more resistant than the sae2Δ mutant (P=0.0026) (Figures 2B and 3B).
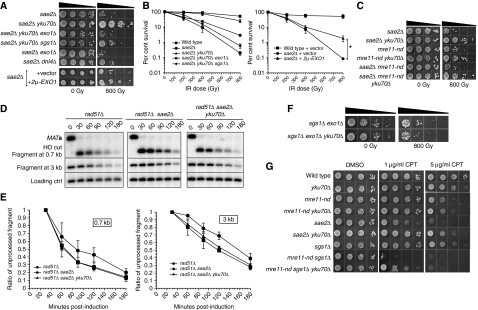
Figure 3.
Suppression of the sae2Δ mutant phenotype by YKU70 deletion. Radiation sensitivity of sae2Δ mutants: (A) spot assays and (B) survival plots as described in Figure 2C. *P=0.01 (unpaired t-test). (C) Epistatic relationship between sae2Δ and mre11-nd mutants, as shown by IR spot assays. Resection physical assay: (D) Southern blot analysis and (E) cut fragment intensity plots as described in Figure 2G. (F) Radiation sensitivity of sgs1Δ exo1Δ mutants, as indicated by spot assays. (G) CPT sensitivity of mre11-nd and sae2Δ mutants. Exponentially growing cells in SC minimal medium were 1:10 serially diluted and spotted on SC plates containing the indicated concentration of camptothecin in DMSO.
The IR sensitivity of the sae2Δ mutant is highly suppressed by the yku70Δ mutation, resulting in almost equivalent survival to wild type. Similarly to the mre11-nd mutant, this effect is dependent on both SGS1 and EXO1. Note that the sae2Δ sgs1Δ yku70Δ mutant is viable, whereas the sae2Δ sgs1Δ mutant is not (discussed later). This suggests that loss of end protection by Ku allows Exo1 and Sgs1 to initiate 5′-3′ resection of DSBs, which normally requires Sae2. The requirement for exo1+ in the suppression of ctp1 (the functional counterpart of SAE2) by pku70Δ was previously reported in S. pombe (Limbo et al, 2007). High-copy expression of EXO1 also resulted in a significant suppression of the IR sensitivity of the sae2Δ mutant (24-fold increase in survival at 800 Gy, P=0.01) (Figure 3A and B), supporting the model that Exo1 competes with Ku at ends to initiate resection of a subset of breaks. The suppression conferred by yku70Δ in sae2Δ mutants could also be attributed to defects in NHEJ that allow time for redundant resection factors to act. Indeed, we found that sae2Δ dnl4Δ mutants are slightly more resistant to IR than sae2Δ (10-fold increase in survival at 800 Gy, P=0.0061), but still more sensitive than sae2Δ yku70Δ (Figure 3A). This suggests that both the end protection and NHEJ functions of Ku contribute to compromise initiation of DSB resection in sae2Δ mutants, in agreement with studies of resection of HO-induced DSBs in G1 cells (Clerici et al, 2008). However, we cannot exclude the possibility that the slight suppression rendered by dnl4Δ is due to the decreased stability of Ku binding at DSBs (Zhang et al, 2007). Notably, this suppression could not be detected in mre11-nd cells that are more resection proficient than sae2Δ (Figure 2A).
The differential IR sensitivity of sae2Δ and mre11-nd mutants prompted us to test their epistatic relationship (Figure 3C). In the presence of Ku, the more severe phenotype conferred by sae2Δ is observed for the double mutant, but in the absence of Ku, the more severe phenotype conferred by mre11-nd is evident. This suggests that once the inhibitory function of Ku is bypassed, Sae2 is no longer required and a functional MRX complex is able to initiate resection, whereas the MRX complex remains compromised in the sae2Δ mre11-nd yku70Δ mutant.
The resection of sequences 0.7 and 3 kb distal to the HO DSB was monitored by the disappearance of restriction fragments in sae2Δ strains as described above (Figure 2E). Unlike mre11-nd, the sae2Δ mutant exhibited a slight delay in the disappearance of both fragments (Figure 3D and E). More specifically, at 0.7 kb from the DSB, 43% of the fragment remains unprocessed 120 min after HO induction compared with 27% in rad51Δ (P=0.02). A similar difference was observed at 3 kb from the break in which 69% remained unprocessed after 120 min compared with 49% observed in rad51Δ (P=0.0003). Deletion of YKU70 in the rad51Δ sae2Δ mutant allowed increased processing of both fragments with kinetics similar to those observed for rad51Δ control cells (Figure 3E). The sae2Δ resection defect is less pronounced than the IR sensitivity, which is likely due to the increased requirement for Sae2-dependent cleavage of dirty ends. However, we cannot rule out the possibility that sae2Δ cells can repair one or two DSBs, but not the large number of DSBs created by the doses of IR used to observe sensitivity.
To further validate the hypothesis that the suppression observed by the loss of Ku is due to increased initiation of resection, we tested whether mutants defective for extensive resection can be suppressed by yku70Δ. End resection is initiated in the exo1Δ sgs1Δ mutant, but only proceeds for about 100–700 nt (Mimitou and Symington, 2008; Zhu et al, 2008). The exo1Δ sgs1Δ mutant is sensitive to IR, but the sensitivity cannot be suppressed by yku70Δ (Figure 3F), suggesting that the block to resection established by Ku takes place at the first step of end processing.
To ensure that the phenotypes observed for resection-defective mutants are not specific to IR-induced breaks, we also used camptothecin (CPT), which creates replication-associated DSBs. Similar to the findings with IR, the mre11-nd mutant was sensitive only at high CPT doses and was slightly suppressed by yku70Δ (Figure 3G). The sae2Δ mutant exhibited a higher CPT sensitivity than mre11-nd, and this was suppressed in the absence of Ku (Figure 3G). The synergistic sensitivity of combining mre11-nd with sgs1Δ was also observed for CPT, and, similar to IR, could be suppressed by yku70Δ. The phenotypes described are reminiscent of those observed after treating cells with IR, consistent with the idea that Ku protects DSB ends from degradation and this becomes limiting to resection when the MRX-Sae2 initial processing is compromised.
The suppression of the sae2Δ IR sensitivity by yku70Δ led us to test whether the sporulation defect of sae2Δ/sae2Δ diploids might be similarly suppressed. We made a diploid homozygous null for SAE2 and YKU70 (sae2Δ/sae2Δ yku70Δ/yku70Δ) and compared its sporulation efficiency to SAE2/SAE2 and sae2Δ/sae2Δ diploids. As previously reported for W303, wild-type sporulation efficiency was approximately 50%, whereas sae2Δ/sae2Δ and sae2Δ/sae2Δ yku70Δ/yku70Δ failed to sporulate (data not shown). This indicates that during meiotic DSB processing, the absence of Ku is not enough to allow processing of the Spo11-bound ends by Sgs1 and/or Exo1.
Increased levels of Ku sensitize mre11-nd and sae2Δ mutants to IR
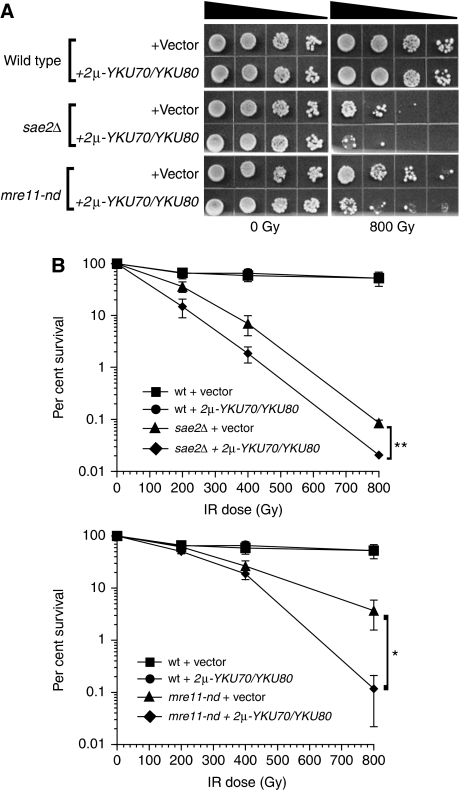
If Ku blocks resection in mutants compromised for the initial clipping of DNA ends, then one would predict that increased expression of Ku would render mre11-nd and sae2Δ mutants more sensitive to IR. This was tested by measuring the IR resistance of wild type, mre11-nd and sae2Δ mutants transformed with a high-copy number (2 μ) plasmid expressing YKU70 and YKU80. There was no effect on the survival of wild-type cells over-expressing Ku, suggesting that in the presence of functional MRX-Sae2, the initial cleavage is not compromised (Figure 4A and B). However, high-copy expression of YKU70-YKU80 sensitized the mre11-nd and sae2Δ mutants to 800 Gy by 30-fold (P=0.01) and four-fold (P=0.009), respectively (Figure 4B). It is notable that the survival of mre11-nd mutants over-expressing Ku at 800 Gy dropped to a level comparable with that seen by the sae2Δ mutant transformed with the empty vector, 0.11 and 0.08%, respectively. We conclude that, under conditions where the initial processing of the DSB ends cannot take place, increased/prolonged presence of Ku at DNA ends further impedes initiation of resection.
Figure 4.
YKU70/YKU80 over-expression sensitizes sae2Δ and mre11-nd mutants to IR. (A) Spot assays and (B) survival plots of wild type, sae2Δ and mre11-nd mutants transformed with empty or YKU70/YKU80 over-expressing vectors. Exponentially growing cells in SC-Ura to maintain selection of the plasmids were 1:10 serially diluted, spotted onto SC-Ura plates and exposed to IR. The means from at least three experiments are presented, error bars indicate s.d.; *P=0.01, **P=0.009.
Suppression of the synthetic lethality of rad27Δ sae2Δ and sgs1Δ sae2Δ mutants by yku70Δ
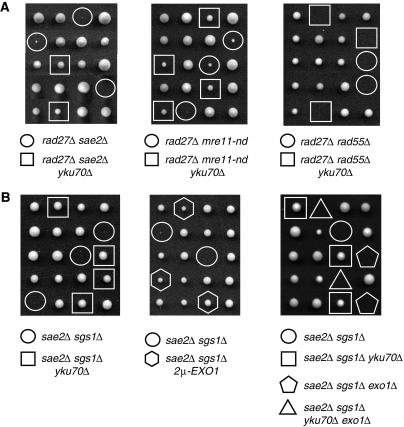
The RAD27 gene encodes a nuclease that functions to process Okazaki fragments during lagging strand DNA synthesis (Tishkoff et al, 1997). Deletion of RAD27 is lethal in combination with mutation of any one of the RAD52 group genes, including mre11Δ, sae2Δ and mre11-H125N, suggesting that MRX-Sae2 are required to process lesions generated in a rad27Δ strain (Symington, 1998; Moreau et al, 1999; Debrauwere et al, 2001). To determine whether deletion of YKU70 can suppress the rad27Δ synthetic lethality with sae2Δ or mre11-nd, diploids heterozygous for RAD27 and MRE11 or SAE2 were generated, and after sporulation, tetrads were dissected to determine whether viable rad27Δ mre11-nd yku70Δ or rad27Δ sae2Δ yku70Δ spores could be obtained (Figure 5A). After dissection, the plates were incubated at 23°C because the rad27Δ growth defect is suppressed at lower temperature. Even at 23°C, the rad27Δ sae2Δ double mutant was found to be lethal, but the lethality was suppressed by yku70Δ (Figure 5A). Some viable rad27Δ mre11-nd spore colonies were recovered that grew very slowly. Nevertheless, yku70Δ suppressed this growth defect to some extent, as rad27Δ mre11-nd yku70Δ mutants consistently survived after dissection (Figure 5A). To ensure that the suppression of the synthetic lethality is related to DNA end processing, we also made diploids heterozygous for YKU70, RAD27 and RAD55, and after sporulation found that no viable rad27Δ rad55Δ yku70Δ spores could be recovered (Figure 5A). Finally, deletion of YKU70 could not suppress the synthetic lethality of rad27Δ mre11Δ (data not shown). Although elimination of Ku partially suppresses the IR sensitivity of the mre11Δ mutant, the mre11Δ yku70Δ double mutant is still highly sensitive to IR with no survivors at 500 Gy (data not shown). Thus, it seems likely that the large number of lesions generated in the mre11Δ rad27Δ mutant cannot be repaired even in the absence of Ku.
Figure 5.
Genetic interactions between rad27Δ, mre11-nd, sae2Δ and sgs1Δ mutants. Viability and genotypes of spores derived from diploids heterozygous for the indicated mutations. (A) Deletion of YKU70 suppresses the synthetic lethality/growth defect of rad27Δ mutants with sae2Δ and mre11-nd, but not with rad55Δ. (B) Loss of Yku70 or EXO1 over-expression rescues the sae2Δ sgs1Δ synthetic lethality.
As yku70Δ completely suppresses the rad27Δ sae2Δ growth defect, we tested whether yku70Δ can also suppress the sae2Δ sgs1Δ synthetic lethality (Tong et al, 2001; Ooi et al, 2003). The majority of the sae2Δ sgs1Δ mutants were inviable, but some small spore colonies formed (<20% were viable). However, the sae2Δ sgs1Δ yku70Δ triple mutant grew remarkably well (Figure 5B). We were unable to derive the sae2Δ sgs1Δ yku70Δ exo1Δ quadruple mutant from crosses, indicating that the survival of the triple mutant is dependent on Exo1 (Figure 5B). Furthermore, by introducing a high-copy EXO1 plasmid into a diploid heterozygous for SAE2 and SGS1 viable sae2Δ sgs1Δ spores that inherited the plasmid were recovered (Figure 5B). These data suggest that lethality of the sae2Δ sgs1Δ mutants is caused by their inability to process some physiological DNA intermediates that are also substrates for Ku, most likely DSBs.
Telomere phenotypes associated with sae2Δ sgs1Δ
Telomeres are specialized DNA–protein complexes that define the physical ends of linear chromosomes and protect them from end degradation. The protruding ssDNA 3′ overhang, the G tail, has a central function in modulating telomere length, as it serves as a substrate for extension by telomerase and it is formed by active resection of the C-strand after completion of DNA synthesis (Gilson and Geli, 2007). Ku is one of the factors that bind to telomeres protecting them from degradation (Polotnianka et al, 1998). As telomeres are physiological DNA structures that share important similarities with DSBs, it seemed possible that an alteration of the natural chromosome ends might be responsible for the sae2Δ sgs1Δ lethality.
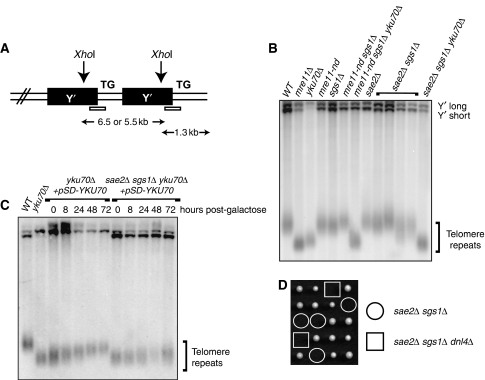
The sae2Δ sgs1Δ double mutant was recently shown to have short telomeres, a defect in telomere addition in G2-arrested cells and no detectable G tails (Bonetti et al, 2009). As described above, the sae2Δ sgs1Δ double mutant shows poor viability in the W303 strain background, but some rare viable spore colonies could be recovered. Analysis of the telomeres in these rare survivors indicated that their telomeres had normal length with occasional wider distribution (Figure 6A and B). Analysis of the mre11-nd sgs1Δ double mutant also revealed normal telomere length (Figure 6B). The yku70Δ mutant is known to exhibit short telomeres (Porter et al, 1996), and, as expected, the sae2Δ sgs1Δ yku70Δ strain was also shown to have short telomeres. The normal telomere length observed for the slow growing sae2Δ sgs1Δ double mutants could mean that they have already escaped from senescence and elongated their telomeres (Bonetti et al, 2009).
Figure 6.
Telomere-associated phenotypes of sae2Δ sgs1Δ yku70Δ mutants. (A) Schematic representation of the telomeric Y′ elements and TG repeats. XhoI liberates a wide band of ∼1.3 kb in wild-type cells, which is used to evaluate telomere repeat length in other genetic backgrounds. (B) Southern blot analysis of XhoI-digested genomic DNA as detected with a Y′ probe. (C) Telomere length analysis after induction of YKU70 expression with galactose in the yku70Δ and sae2Δ sgs1Δ yku70Δ mutant. (D) Viability and genotypes of spores derived from diploids heterozygous for the indicated mutations.
As the survival of the sae2Δ sgs1Δ mutant depends on the absence of YKU70, and the telomeres have no detectable ssDNA tails (Bonetti et al, 2009), it seemed possible that Ku-dependent telomere fusions might contribute to the lethality. To test this, and in order to bypass the difficulties imposed by the sae2Δ sgs1Δ synthetic lethality, an indirect approach was used. The viable sae2Δ sgs1Δ yku70Δ mutant was transformed with a plasmid expressing YKU70 under a galactose-inducible promoter. The strain was maintained in glucose-containing medium to retain viability. Addition of galactose to the medium to induce YKU70 expression led to a gradual telomere lengthening as shown by Southern blot analysis (Figure 6C). Notably, the sae2Δ sgs1Δ mutant exhibited slower and less efficient elongation of its telomeres after YKU70 expression was restored, suggesting that the telomeres in sae2Δ sgs1Δ mutants are less accessible to telomerase. To detect whether telomere–telomere fusions (T-TF) are formed in the sae2Δ sgs1Δ mutant, a PCR-based assay was used (Mieczkowski et al, 2003). A mec1Δ tel1Δ sml1Δ mutant, which was previously shown to form T-TF, was used as a positive control (Mieczkowski et al, 2003). T-TF were readily detected in the mec1Δ tel1Δ sml1Δ mutant, but not in wild type or sae2Δ sgs1Δ yku70Δ after induction of YKU70 with galactose (data not shown). Moreover, we noticed that the cells maintained their ability to divide when YKU70 was over-expressed, albeit slowly. These data suggest that T-TF are probably not the reason for the sae2Δ sgs1Δ lethality. This is supported by the observation that deletion of DNL4, the ligase responsible for T-TF formation, does not suppress the sae2Δ sgs1Δ lethality (Figure 6D).
Discussion
The 5′-3′ processing of DSB ends leads to the formation of 3′ ss tails that serve as precursors for HR and as sensors for the DNA damage checkpoint (Zou and Elledge, 2003; Krogh and Symington, 2004). Initiation of DSB processing is tightly regulated and provides a critical step in which the choice between NHEJ and HR repair pathways is made. The 5′-3′ resection of DSBs is under the control of CDK1 and to date, Sae2 and Dna2 are the only resection factors known to be cell cycle regulated (Huertas et al, 2008; Kosugi et al, 2009). CDK1-dependent phosphorylation of Dna2 is required for its nuclear localization during S-phase and phosphorylation of Sae2 at a conserved CDK site (Ser 267) was shown to correlate with initiation of resection and commitment to HR (Huertas et al, 2008; Kosugi et al, 2009). Similar findings were reported for the human and S. pombe Sae2 counterparts, CtIP and Ctp1, respectively, with the latter being controlled at the transcriptional level (Limbo et al, 2007; Huertas and Jackson, 2009; Yun and Hiom, 2009).
The initiation of end processing requires the MRX complex and Sae2, but as both Mre11 and Sae2 have nuclease activity and mutation in either results in a similar phenotype, it is not clear which nuclease is responsible for clipping 5′ ends (Lengsfeld et al, 2007; Mimitou and Symington, 2008; Zhu et al, 2008). Unfortunately, there are no recognizable nuclease motifs present in Sae2, and an sae2 nuclease-defective mutant awaits identification. During meiosis, the initial endonucleolytic processing by MRX and Sae2 is obligatory to remove Spo11, which is covalently bound to the 5′ ends of DSBs at hotspots (Neale et al, 2005; Mimitou and Symington, 2009). However, it is unclear whether this two-step mechanism is needed to resect DSB ends formed by different means. Here, we sought to investigate what purpose this initial processing might serve by testing whether competition is established by NHEJ when the MRX-Sae2-cleavage step is compromised. We found that the IR hypersensitivity of mre11Δ mutants is partially suppressed by yku70Δ in an Exo1-dependent manner, suggesting that when ends are not bound by MRX, Ku acts as a block to resection by Exo1. It is also possible that MRX bound to ends is inhibitory to Exo1-initiated resection (Farah et al, 2009). The observation that Sgs1 is not required for the suppression suggests that the MRX complex recruits Sgs1 to ends, consistent with the finding that Sgs1 associates with Mre11 upon DNA damage-induced checkpoint activation (Chiolo et al, 2005).
Unlike mre11Δ, the yku70Δ suppression of mre11-nd and sae2Δ was dependent on Exo1 and Sgs1, suggesting that in the presence of a structurally competent but functionally compromised MRX complex, binding of Ku limits access to both proteins. These results are also consistent with the MRX recruiting Sgs1 to DSBs. Surprisingly, the extent of the mre11-nd and sae2Δ IR sensitivity, as well as their suppression by yku70Δ, differs significantly (Figures 2 and 3). More specifically, after 800 Gy, sae2Δ survival was 0.2%, whereas mre11-nd was 3%, and yku70Δ restored survival to 26% in sae2Δ and 21% in mre11-nd mutants. Furthermore, over-expression of YKU70-YKU80 resulted in greater IR sensitivity in the mre11-nd strain than in sae2Δ (Figure 4), and over-expression of EXO1 conferred higher resistance in the sae2Δ strain than in mre11-nd. Similarly, we could detect a subtle defect in endonuclease-induced DSB processing in sae2Δ, but not mre11-nd mutants.
Several hypotheses could account for this behaviour. First, Sae2 could have a dual function: as a regulator of Mre11 nuclease function and as a nuclease itself. In the presence of Sae2 and the nuclease-defective Mre11, some processing might take place, which would be more efficient for endonuclease than IR-induced DSBs (Bressan et al, 1998; Moreau et al, 1999; Llorente and Symington, 2004). On the other hand, complete absence of Sae2 would be expected to result in a more severe defect in initial processing because of loss of both nucleases. A second hypothesis is that the physical presence of Sae2 at the DNA ends is crucial to avoid NHEJ. In support of this, a previous study showed a 60-fold increase in NHEJ in sae2Δ mutants, compared with a seven-fold increase in mre11-nd mutants (Lee and Lee, 2007). It is possible that the initial processing step by MRX and Sae2 functions to remove Ku from DNA ends, analogous to the removal of Spo11 during meiosis. Thus, in the absence of clipping by the Mre11 nuclease and Sae2 Ku remains bound to DNA ends, preventing resection by Exo1 and promoting NHEJ. The third hypothesis is supported from studies in S. pombe, in which the Rad32Mre11 nuclease activity was shown to be involved in the removal of Top2 from 5′ DNA ends as well as Top1 from 3′ DNA ends, whereas Ctp1 is involved in the removal of covalently bound Top2, but inhibits Top1 removal (Hartsuiker et al, 2009). This suggests that Ctp1 protects the 3′ end from processing, and if conserved, could mean that Sae2 is important in protecting the integrity of the 3′ end for productive Rad51 filament formation.
It is also notable that even though deletion of YKU70 significantly suppressed the end-processing defect of sae2Δ, it failed to suppress its meiotic defect. As the IR suppression requires EXO1 and SGS1, it could mean that (i) neither Exo1 nor Sgs1-Dna2 can process Spo11 bound ends or (ii) one of them is inactive for processing during meiosis. A recent study showed that Exo1 has the major function in meiotic DSB processing after Spo11 removal with Sgs1-Dna2 providing some residual activity in exo1Δ cells, indicating that both activities are functional in meiotic cells (Manfrini et al, 2010). These results suggest that Exo1 and Sgs1-Dna2 can process DSBs subsequent to Spo11 removal, but are ineffective in removal of the Spo11 adduct from 5′ ends.
We found that viability of rad27Δ sae2Δ mutants depends on the absence of Ku, suggesting the accumulation of DNA intermediates in rad27Δ mutants that are substrates for both Sae2 and Ku. One hypothesis is that these substrates are replication intermediates, which need to be resected to allow repair by HR. These intermediates are probably collapsed replication forks caused by the presence of unrepaired lagging strand lesions from the preceding S-phase. HR was shown to be the major DSB repair pathway at collapsed replication forks (Saleh-Gohari et al, 2005). The SAE2 requirement in the rad27Δ background could reflect the need to initiate resection of replication-associated DSBs to channel repair by HR. Indeed, rad27Δ is lethal in combination with mutations in the genes of the RAD52 epistasis group. Moreover, the sensitivity of sae2Δ mutants to CTP is suppressed by elimination of Ku, suggesting that DSBs formed after replication fork collapse can be bound by Ku and the presence of Sae2 is important to antagonize this binding. In agreement with our results, two recent studies show that DNA repair defects observed in Caenorhabditis elegans, mammalian or chicken cells deficient for Fanconi anaemia components, can be suppressed by loss of Ku (Adamo et al, 2010; Pace et al, 2010). This suggests that during replication cells use specialized factors to antagonize Ku, whose engagement with DSB intermediates during S-phase may inhibit the processing of DSBs required for loading of HR factors, while also promoting illegitimate repair.
Similarly, the ability to restore resection and enable HR-mediated repair of replication-associated DSBs could be the basis of the yku70Δ suppression of the sae2Δ sgs1Δ mutant lethality. The sgs1 mutants exhibit aberrant DNA replication phenotypes, including increased frequency of replication fork collapse after fork stalling (Cobb et al, 2003, 2005; Liberi et al, 2005), and such events could increase the need for Sae2 to prevent illegitimate repair of DSBs by NHEJ. However, unlike rad27Δ, sgs1Δ mutations are not synthetically lethal with mutations in genes of the RAD52 epistasis group, suggesting that there is something more contributing to the sae2Δ sgs1Δ lethality. Telomeres are logical candidates because they are actively resected and bound by Ku, similar to DSBs. It was recently shown that sae2Δ sgs1Δ mutants fail to form the 3′ G tails implicated in recruiting telomerase to maintain normal telomere length (Bonetti et al, 2009). Elimination of Ku at telomeres allows access to Exo1 (Maringele and Lydall, 2002), and this could rescue the formation of 3′ G tails in the sae2Δ sgs1Δ mutant. Although this could explain the yku70Δ suppression of the sae2Δ sgs1Δ lethality, it is important to note that the mre11Δ mutant is equally defective in G-tail formation, but is viable with sgs1Δ (although growth is slow). In addition, the mre11Δ yku70Δ double mutant grows poorly because two independent pathways for telomerase recruitment are defective (Nugent et al, 1998). It is possible that MRX inhibits recruitment of Exo1 to DSBs and the sae2Δ sgs1Δ double mutant lethality is due to the block to Exo1 recruitment imposed by both Ku and MRX. In contrast, Exo1 might still be recruited, albeit inefficiently, in the mre11Δ sgs1Δ double mutant.
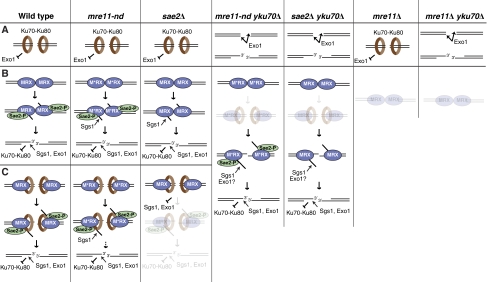
On the basis of our findings, we suggest the following models (Figure 7). DSBs can exist in three different dynamic states: Ku bound (Figure 7A), MRX bound (Figure 7B) and MRX-Ku bound (Figure 7C). The Ku-bound state, which is the only state present in mre11Δ mutants, blocks access to Exo1. Loss of Ku allows Exo1 to initiate resection at a subset of ends contributing to the Exo1-dependent suppression observed in mre11-nd yku70Δ, sae2Δ yku70Δ and mre11Δ yku70Δ mutants. The MRX-bound state is competent to initiate resection, unless Sae2 is absent or the nuclease activity of Mre11 is compromised. In this case, some redundancy from Sgs1 allows resection to initiate creating an intermediate committed to HR. The MRX-Ku-bound state presents the cells with a choice: HR or NHEJ? The fast and simultaneous recruitment of MRX and Ku at DSB ends presents an intermediate that can be readily used for NHEJ, and this is likely to be the default in G1. When cells transit through S/G2 and have a template for HR, Sae2 is recruited to ends in order to override the block to resection imposed by Ku. Mre11 with Sae2 catalyses the removal of oligonucleotides from the DSB ends resulting in short 3′ overhangs (Jazayeri et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008), a step that serves to create more favourable substrates for Sgs1 and Exo1 and remove Spo11-bound or chemically modified ends present at DSBs after IR. In the context of this work, the MRX-Sae2 cleavage may provide a substrate less suitable for Ku (and maybe MRX) binding; thus, preventing NHEJ and committing repair by HR. Ku preferentially binds double-stranded DNA ends over ssDNA and would be expected to show lower affinity for the clipped ends (Mimori and Hardin, 1986; Tuteja and Tuteja, 2000). Moreover, the short ssDNA tails formed after MRX-Sae2 cleavage could be bound by RPA, which in turn could promote DSB resection through its interaction with Dna2 (Bae et al, 2001). In the absence of Sae2, the cells fail to override the Ku barrier and instead use NHEJ, a pathway competent for repair of endonuclease induced, but less so for IR-induced DSBs. In mre11-nd cells, the physical/functional presence of Sae2 provides some line of defence against Ku and NHEJ, and even though the MRX-Sae2 cleavage is defective, Sgs1-Dna2 recruitment allows resection to initiate. The yku70Δ suppression of mre11-nd and sae2Δ is also dependent on Exo1, but at this point, we cannot distinguish between Exo1 degradation of ends unbound or bound by MRX.
Figure 7.
Models for the interplay between resection machinery and Ku at DSB ends. In wild-type cells, DSBs can exist in three dynamic states: (A) Ku-bound, which blocks access to Exo1, (B) MRX-bound, which can initiate end processing and (C) MRX-Ku-bound, which can initiate NHEJ and removal of Ku is required to allow resection initiation. Recruitment of Sae2 in G2 and clipping of the ends allows access to the processive resection machinery and creates an intermediate that can no longer convert states and commit to HR. In nuclease-defective mutants of Mre11, though compromised for initial processing, the presence of Sae2 channels ends to HR and redundant activity from Sgs1-Dna2 allows initiation of resection. In the absence of SAE2, the MRX-bound ends can still initiate resection, presumably with some assistance by Sgs1, whereas the Ku-bound and MRX-Ku-bound ends are blocked. When the end protection by Ku is lost, in mre11-nd yku70Δ and sae2Δ yku70Δ for example, MRX-naked ends can be resected by Exo1. For the MRX-bound ends even if compromised for the initial clipping, the absence of Ku allows Sgs1 (and maybe Exo1) to assist in initiating resection of the DSB. Finally, in the absence of Mre11, where the only state present is the Ku-bound state, access of Exo1 is blocked in a Ku-dependent manner.
Materials and methods
Media, growth conditions and genetic methods
Rich medium (yeast extract–peptone–dextrose, YPD), synthetic complete (SC) medium lacking the appropriate amino acids or nucleic acid bases, sporulation medium and genetic methods were as described previously (Sherman et al, 1986). YPLactate, rich medium containing 2% lactate (pH 5.5) and supplemented with adenine, was used for the galactose induction of HO in DSB end-resection assays. Sporulation medium contained 1% potassium acetate and the appropriate amino acids or nucleic acid bases at 1/5 of the concentration used in SC medium. Where indicated CPT dissolved in DMSO was added to liquid SC agar before pouring into plates. Hygromycin B (Sigma) to 300 μg/ml was used for selection of the hphMX4 cassette and G418 (Sigma) to 200 μg/ml was used for selection of the kanMX6 cassette. Transformation of yeast cells was performed by the lithium acetate method (Ito et al, 1983). Yeast cells were grown at 30°C, unless otherwise indicated.
Yeast strains and plasmids
The strains used were derived from W303 and are listed in the Supplementary Table 1. Most of the strains were constructed by crossing isogenic strains present in our laboratory collection to produce haploid progeny of the indicated genotypes. Strain LSY1091 was made by one-step gene replacement of W1588-4C with EcoRI/SphI-digested pMJ536 (a gift of M Lichten) to replace the SAE2 coding region with the kanMX6 cassette. Strains containing the integrated PGAL1–HO cassette were generated by crossing LSY1009-1 to strains within the laboratory collection. LSY1009-1 was constructed by transforming W1588-4A with the BstEII and PvuII linearized YIPade3HO as previously described (Sandell and Zakian, 1993).
The 2 μ-plasmid over-expressing YKU70/YKU80, pML550.46, was kindly provided by MP Longhese. The plasmid over-expressing EXO1, pEM-EXO1, was created by moving the EXO1 containing XhoI/SacII fragment from pSM502 (Moreau et al, 2001) to the corresponding sites of pRS426. pSD-YKU70 was obtained by cloning a BamHI/NheI PCR fragment (coordinates 838176–840023 on chromosome XIII) into the corresponding sites of pESC-URA (Stratagene). Plasmid YIPade3HO was kindly provided by V Zakian.
Gamma irradiation survival assays
Cells were grown in liquid YPD or SC-Ura medium to mid-log phase. The cultures were serially 1:10 diluted and spotted onto solid YPD or SC-Ura plates. The plates were irradiated in a Gammacell-220 irradiator containing 60Co for the designated dose. The plates were incubated for 3 days at 30°C before survivors were counted. For quantitations, the mean per cent survival from at least three independent experiments is presented.
Analysis of telomere lengths
Genomic DNA was isolated from 5 ml cultures and digested with XhoI. The products were examined by Southern blot analysis with pYT14 as a hybridization probe. Wild-type strains yield a terminal restriction fragment of 1.3 kb, which includes 400 bp of the G1–3T telomeric repeat (Porter et al, 1996).
Physical analysis of HO-induced DSB end resection
For the end-resection experiments, yeast cells were initially grown in 5 ml of YPD medium for 18 h and then transferred to YPLactate. Cultures were grown to an optical density at 600 nm (OD600) of 0.3–0.5 and then HO was induced by addition of galactose to a final concentration of 2%. Cell samples were removed before and after induction, harvested by centrifugation and the cell pellets were washed with H2O and stored at −80°C. DNA isolated by glass bead disruption and phenol extraction protocol was digested with XbaI and StyI and separated by electrophoresis through 1% agarose gels. DNA fragments were transferred to nylon membranes and hybridized with multiple radiolabelled DNA probes. For the detection of the HO-cut fragment next to the break site, the MAT-0.7 kb probe was used. This probe was generated by PCR amplification of MAT sequences distal to the HO-cut site (coordinates 201176–201580 on chromosome III sequence). For the detection of the fragment 3 kb distal to the HO-cut site, the MAT-3 kb probe was used (coordinates 204184–204893 on chromosome III). Quantities of DNA loaded at each time point were normalized using a DNL4 probe (coordinates 334672–335378 on chromosome XV). Intensities of bands on Southern blots corresponding to probed DNA fragments were analysed using ImageJ. DSB end resection for each time point was estimated as a ratio of the signal intensity corresponding to the fragment of interest 30 min after induction and represents the mean of two independent experiments in each of the two different inductions (four trials total).
Supplementary Material
Acknowledgments
We thank H Klein, M Lichten, M Longhese, R Rothstein and V Zakian for yeast strains and plasmids, and WK Holloman for comments on the paper. This project was supported by a grant from the National Institutes of Health (GM041784).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A (2010) Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Bae KH, Kim JA, Seo YS (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461 [DOI] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP (2009) Multiple pathways regulate 3′ overhang generation at S cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Bressan DA, Baxter BK, Petrini JH (1999) The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol 19: 7681–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL (2009) Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One 4: e4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamankhah M, Fontanie T, Xiao W (2000) The Saccharomyces cerevisiae mre11(ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics 155: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G (2005) Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol 25: 5738–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP (2008) The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280: 38631–38638 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39: 431–451 [DOI] [PubMed] [Google Scholar]
- Debrauwere H, Loeillet S, Lin W, Lopes J, Nicolas A (2001) Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc Natl Acad Sci USA 98: 8263–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah JA, Cromie GA, Smith GR (2009) Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc Natl Acad Sci USA 106: 9356–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM (2009) Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell 33: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Korolev VG, Fabre F (1992) XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132: 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V (2008) Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J 27: 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis E, Wilson TE (2002) Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S (2001) Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA 106: 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A, Symington LS (2005) Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB (2004) RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117: 171–184 [DOI] [PubMed] [Google Scholar]
- Lee K, Lee SE (2007) Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176: 2003–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA (2002) Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160: 49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Manfrini N, Guerini I, Citterio A, Lucchini G, Longhese MP (2010) Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J Biol Chem 285: 11628–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Mieczkowska JO, Dominska M, Petes TD (2003) Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci USA 100: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori T, Hardin JA (1986) Mechanism of interaction between Ku protein and DNA. J Biol Chem 261: 10375–10379 [PubMed] [Google Scholar]
- Moore JK, Haber JE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16: 2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS (2001) Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S (2005) Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol 8: 657–660 [DOI] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD (2003) DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet 35: 277–286 [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ (2010) Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 329: 219–223 [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol 8: 831–834 [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res 24: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray AJ, McGill CB, Shafer BK, Strathern JN (2001) Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics 158: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T (2005) Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol 25: 7158–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA (1993) Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75: 729–739 [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J (1986) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH (2004) The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 3: 845–854 [DOI] [PubMed] [Google Scholar]
- Symington LS (1998) Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res 26: 5589–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Filosi N, Gaida GM, Kolodner RD (1997) A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88: 253–263 [DOI] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H (1998) A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol 18: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H (2000) Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R, Tuteja N (2000) Ku autoantigen: a multifunctional DNA-binding protein. Crit Rev Biochem Mol Biol 35: 1–33 [DOI] [PubMed] [Google Scholar]
- Wasko BM, Holland CL, Resnick MA, Lewis LK (2009) Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair (Amst) 8: 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K (2009) CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459: 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE (2007) Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 14: 639–646 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF (2008) Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.