Abstract
Perturbation of DNA replication initiation arrests human cells in G1, pointing towards an origin activation checkpoint. We used RNAi against Cdc7 kinase to inhibit replication initiation and dissect this checkpoint in fibroblasts. We show that the checkpoint response is dependent on three axes coordinated through the transcription factor FoxO3a. In arrested cells, FoxO3a activates the ARF-∣Hdm2-∣p53 → p21 pathway and mediates p15INK4B upregulation; p53 in turn activates expression of the Wnt/β-catenin signalling antagonist Dkk3, leading to Myc and cyclin D1 downregulation. The resulting loss of CDK activity inactivates the Rb-E2F pathway and overrides the G1-S transcriptional programme. Fibroblasts concomitantly depleted of Cdc7/FoxO3a, Cdc7/p15, Cdc7/p53 or Cdc7/Dkk3 can bypass the arrest and proceed into an abortive S phase followed by apoptosis. The lack of redundancy between the checkpoint axes and reliance on several tumour suppressor proteins commonly inactivated in human tumours provides a mechanistic basis for the cancer-cell-specific killing observed with emerging Cdc7 inhibitors.
Keywords: cell cycle, checkpoint, Dkk3, DNA replication origin, FoxO3A
Introduction
An estimated 30 000 replication origins are spread along human chromosomes and it is understood that chromatin structure, adjacent sites of transcription and epigenetic parameters all affect origin selection (Mechali, 2001; Biamonti et al, 2003). Initiation of DNA replication is a two-step process. In early G1, the origin recognition complex (ORC) cooperates with Cdc6 and Cdt1 in loading the Mcm2–7 helicase to form a ‘licensed’ pre-replicative complex pre-RC). In late G1, the origin is ‘fired’ by CDKs and Cdc7/Dbf4 kinase. Cdc7 phosphorylates the Mcm2, 4 and 6 subunits, thereby inducing a conformational change that stimulates MCM helicase activity and exposes a domain of Mcm5 required for Cdk2-dependent loading of Cdc45 and the replisome containing RPA, PCNA and DNA polymerase α-primase (Sclafani and Holzen, 2007). In addition to its highly conserved function in origin firing, other, less well understood, functions have been reported for Cdc7 kinase. These include activation of the ATR-Chk1 pathway in response to DNA damage and DNA replication stress (Takeda et al, 1999; Costanzo et al, 2003; Dierov et al, 2004; Tenca et al, 2007; Kim et al, 2008), cohesin loading onto chromatin required for chromosomal segregation in mitosis (Takahashi et al, 2008), regulation of exit from mitosis (Miller et al, 2009) and double-strand break formation during meiotic recombination (Matos et al, 2008).
As the two-step-replication model excludes the formation of replication-competent origins once S phase has started, it has been argued on the basis of experimental evidence that a putative cell cycle checkpoint could delay progression from G1 into S phase if replication initiation is perturbed (Blow and Gillespie, 2008). In breast epithelial cells, for example, RNAi against ORC2 impairs DNA replication, causing G1 arrest with low cyclin E–Cdk2 activity (Machida et al, 2005). Inhibition of pre-RC assembly by overexpressing a stable form of geminin causes G1 arrest associated with low CDK activity in fibroblasts (Shreeram et al, 2002). Blocking activation of the MCM helicase through RNAi against CDC7 also causes G1 arrest in fibroblasts and leads to elevated p53 levels, p21 induction and hypo-phosphorylated Rb (Montagnoli et al, 2004). These findings, therefore, suggest that somatic cells can respond directly to impairment of the DNA replication initiation machinery by blocking S phase entry (Blow and Gillespie, 2008). In contrast, inhibition of origin licensing or firing has been shown to cause apoptotic cell death in a range of different cancer cell lines. This is thought to arise as a result of transformed cells entering S phase with inadequate numbers of competent origins to complete chromosomal replication, arguing for loss of the putative origin activation checkpoint in cancer. As only a limited number of replication forks can be established when replication initiation is perturbed, it is plausible that apoptosis is triggered as a result of fork stalling/collapse in cancer cells with active intra S phase checkpoint mechanisms or mitotic catastrophe arising from partially replicated chromosomes in more transformed cells (Blow and Gillespie, 2008).
The cancer-cell-specific killing reported for emerging pharmacological Cdc7 inhibitors, while normal cells undergo a non-genotoxic G1 arrest, has generated widespread interest in small molecule inhibitors of the DNA replication initiation machinery (Jackson, 2008; Montagnoli et al, 2008; Swords et al, 2010). However, very little is known about the molecular architecture and circuitry of the proposed origin activation checkpoint on which tumour specificity is dependent. Here, we have used RNAi against CDC7 to inhibit replication initiation and elucidate the molecular architecture of the checkpoint in human fibroblasts.
Results
Cdc7 depletion in IMR90 fibroblasts causes cell cycle arrest in G1
We set out to determine whether Cdc7 depletion can activate a checkpoint response to impaired DNA replication initiation by transfecting IMR90 cells with three different siRNAs with sequences corresponding to the CDC7 cDNA. Notably, two of the CDC7 siRNAs have been characterized in a previous study (Montagnoli et al, 2004), whereas the third has been validated by the manufacturer (Ambion, Warrington, UK) (Supplementary Table 1 and Supplementary Figure 1A–D). All three oligos efficiently reduced CDC7 mRNA levels (Supplementary Figure 1B). On the basis of the highest knock-down score and consistency in replicate experiments (Supplementary Figure 1B–D), oligo CDC7-A (referred to here as ‘CDC7-siRNA’) was used for all experiments shown, except those in which siRNA specificity was shown with an alternative siRNA (oligo CDC7-B).
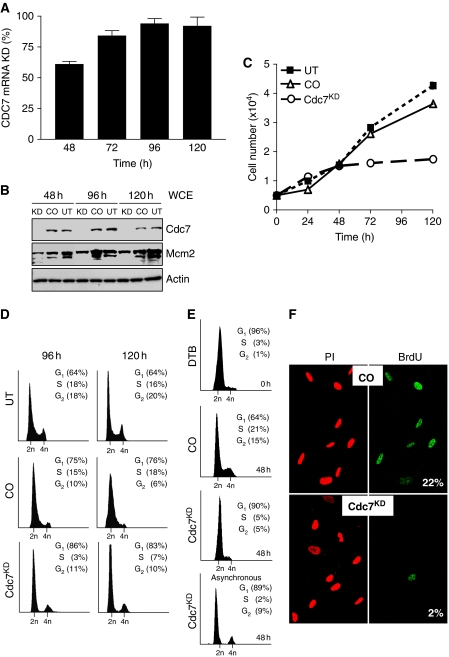
Relative to control-siRNA (CO), transfection of IMR90 cells with CDC7-siRNA reduced CDC7 mRNA levels by 65% 48 h post-transfection, by 85% at 72 h and by >95% at 96 h (Figure 1A). Correspondingly, in whole cell extracts (WCE), Cdc7 protein levels started to fall by 24 h and were undetectable from 48 h until 120 h post-transfection (Figure 1B). Consistent with efficient Cdc7 depletion, we noted a decrease in total Mcm2 protein levels and a shift from hyper-phosphorylated to slower migrating, hypo-phosphorylated Mcm2 isoforms (Montagnoli et al, 2004) (Figure 1B). Downregulation of Cdc7 expression caused a cessation of cell proliferation with cell numbers reaching a plateau 48 h post-transfection (Figure 1C). The majority of CDC7-siRNA-transfected cells accumulated with G1 DNA content. Although a small fraction of cells showed a G2/M DNA content, cells with less than 2C DNA content were not detected (Figure 1D), indicating that Cdc7-depleted cells remained viable. In cells that were synchronized by release from double thymidine block and directly transfected with CDC7-siRNA, the majority of cells again showed a G1 DNA content (90%), whereas the small G2/M peak noted for asynchronous cells (9%) was lost with remaining cells equally distributed between the S and G2/M fractions (5% each) (Figure 1E). To provide further evidence that Cdc7 depletion is causing a G1 arrest, cells were transfected with CDC7-siRNA and control-siRNA, pulsed with BrdU and immunostained with anti-BrdU antibodies. In keeping with the cell cycle profile, the percentage of BrdU-incorporating cells dropped from 22% in cells transfected with control-siRNA (23% in untreated cells) to 2% in Cdc7-depleted cells (Figure 1F). Interestingly, 6 days after transfection with CDC7-siRNA, IMR90 cells re-expressed Cdc7 and resumed cell proliferation, showing a degree of confluency seen in control cells (Supplementary Figure 1E–G).
Figure 1.
Cdc7 depletion in IMR90 fibroblasts causes cell cycle arrest in G1. (A) Time course of CDC7 mRNA knock-down (KD) in IMR90 cells relative to cells transfected with control-siRNA (CO). (B) Whole cell extracts (WCE) prepared from untreated (UT), CO and Cdc7KD cells were analysed by immunoblotting with the indicated antibodies (β-actin—loading control). (C) At the indicated time points, cell number was measured in UT, CO and Cdc7KD cell populations. (D) DNA content of UT, CO and Cdc7KD cells at 96 and 120 h post-transfection. (E) DNA content of double thymidine-arrested cells (DTB), CO and Cdc7KD cells 48 h after release from double thymidine block and transfection, and asynchronous Cdc7KD cells 48 h post-transfection. (F) At 96 h post-transfection, cells were pulsed for 2 h with BrdU, fixed and detected with an FITC-conjugated anti-BrdU antibody. DNA was stained with propidium iodide (PI). Numbers show the percentage of cells incorporating BrdU.
To control for siRNA specificity, we studied an alternative CDC7-siRNA (oligo CDC7-B; Supplementary Table 1 and Supplementary Figure 1B and C) targeting a different region of the transcript. Oligos CDC7-B and CDC7-A showed comparable gene silencing efficacy (mRNA and protein reduction) and induced similar phenotypic effects (accumulation of cells with G1 DNA content) (Figure 1; Supplementary Figure 2A and B). Moreover, rescue experiments were performed in which the RNAi effect was reversed through expression of a CDC7 gene variant refractory to silencing by oligo CDC7-A. Under these conditions, IMR90 cells were able to recover from the cell cycle arrest caused by Cdc7 depletion, as shown by flow cytometry and BrdU-incorporation data strongly resembling those of control cells (Supplementary Figure 3A–C). We, therefore, reasoned that CDC7 oligos A and B are equivalent and that the cell cycle arrest phenotype directly correlates with Cdc7 depletion and is unlikely to be due to concomitant non-specific downregulation of an unknown gene (off-target effects). Taken together, these data show that after Cdc7 depletion diploid human fibroblasts arrest cell cycle progression in G1, remaining in a non-proliferative state from which they can re-enter the cell cycle after Cdc7 levels have been restored.
CDK activity and replication initiation factors are downregulated in response to Cdc7 depletion
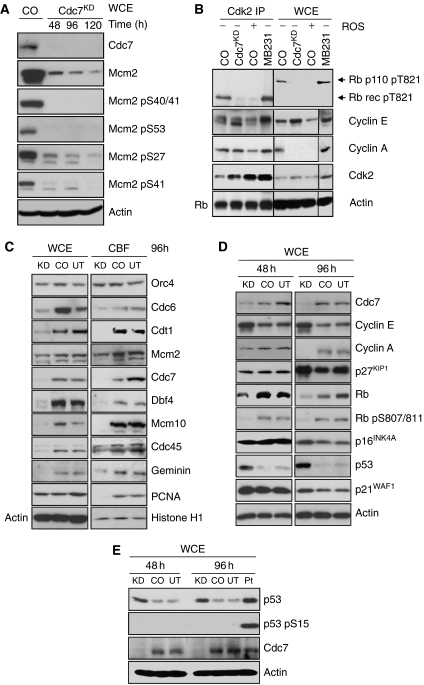
The Mcm2–7-replicative helicase is an important target for S phase-promoting kinases during origin activation, and the Mcm2 subunit, in particular, has been shown to be a substrate for both CDK and Cdc7. Phosphosites have been mapped in the N-terminal tail of Mcm2 for Cdc7 (Ser-40, Ser-53 and Ser-108) and Cdk2 (Ser-13, Ser-27 and Ser-41) in vitro and in vivo (Montagnoli et al, 2006). To test whether the Cdc7-depletion-induced checkpoint response involves downregulation of S phase-promoting CDK activity, we transfected IMR90 cells with CDC7-siRNA and studied Mcm2 phosphorylation at the mapped CDK and Cdc7 phosphosites. In keeping with a previous study (Montagnoli et al, 2004), Cdc7 depletion caused a decrease in Mcm2 levels and reduced the electrophoretic mobility of the protein in polyacrylamide gels, showing the presence of hypo-phosphorylated Mcm2 isoforms (Figure 2A). Although Mcm2 total protein levels consistently dropped in Cdc7-depleted cells, the extent of Mcm2 reduction varied between experiments. The average reduction in the intensity of Mcm2 protein bands (relative to control-siRNA-transfected cells) was 45% at 48 h, 62% at 96 h and 76% at 120 h post-transfection (Image J densitometry analysis). Mcm2 Ser-40/41 and Ser-53 phosphorylation was abolished when Cdc7 kinase was downregulated (48–120 h post-transfection), whereas phosphorylation at the CDK phosphosites Ser-27 and Ser-41 was significantly reduced after 96 h (Figure 2A). To confirm the loss of CDK activity in Cdc7-depleted cells, we carried out in vitro kinase assays with immunoprecipitated Cdk2 (Figure 2B). As expected, in vitro phosphorylated, recombinant truncated Rb protein was detected with an anti-Thr-821-phosphorylated Rb antibody in IMR90 and MDA-MB231 breast cancer cells (controls), but not in IMR90 cells transfected with CDC7-siRNA or treated with the CDK inhibitor roscovitine.
Figure 2.
CDK activity and replication initiation factors are downregulated in response to Cdc7 depletion. (A) WCE prepared from IMR90 cells transfected with control-siRNA (CO) and CDC7-siRNA (Cdc7KD) were analysed by immunoblotting with the indicated antibodies (β-actin—loading control). (B) WCE prepared from CO and Cdc7KD cell populations and from CO cells treated with 200 μM roscovitine (ROS) for 24 h (negative control) and MDA-MB231 breast cancer cells (positive control) were immunoprecipitated with anti-cyclin A and anti-cyclin E antibodies. Cdk2 immunoprecipitates (Cdk2 IP) were subjected to an in vitro kinase assay using recombinant truncated Rb protein (p56) as substrate. In vitro phosphorylation was detected with a specific anti-Rb-phospho-Thr-821 antibody. Note that lanes 1–8 were run on the same polyacrylamide gel and proteins transferred to the same PVDF membrane by semi-dry electroblotting. The membrane was subsequently cut as indicated for optimized immunodetection. (C) WCE and chromatin-bound protein fractions (CBF) prepared from untreated (UT), CO and Cdc7KD cells (96 h post-transfection) were analysed by immunoblotting with the indicated antibodies (β-actin and histone H1—loading controls). (D) WCE from UT, CO and Cdc7KD cells (48 and 96 h post-transfection) were analysed by western blotting with the indicated antibodies (β-actin—loading control). (E) WCE from UT, CO and Cdc7KD cells (48 and 96 h post-transfection) and from cells treated for 24 h with 17 μM cisplatin (Pt) were analysed by western blotting with the indicated antibodies (β-actin—loading control).
The decline in Mcm2 levels in Cdc7-depleted cells (Figure 2A) raises the possibility that perturbation of origin activation may result in downregulation of replication initiation factors and/or affect the stability on chromatin of pre-RCs already formed. To address this question, IMR90 cells transfected with CDC7- and control-siRNAs were biochemically fractionated into WCE and chromatin-bound fractions (CBF) and immunoblotted with antibodies against replication initiation factors (Figure 2C). Orc4 levels in untreated and siRNA-transfected cells did not vary over the course of the experiment. On the contrary, in Cdc7 depleted, but not control cells, protein levels of Cdc6, Cdt1, Mcm2, Dbf4, Mcm10 and Cdc45 were significantly downregulated, whereas levels of these replication initiation factors associated with chromatin were also reduced. Notably, the DNA polymerase processivity factor PCNA was undetectable in CBF from Cdc7-depleted cells (Figure 2C).
Next, we sought to establish whether the Cdc7-depletion-induced checkpoint response involves any known cell cycle regulators of the G1-S transition. Compared with control cells, at 96 h post-transfection Cdc7-depleted cells showed a marked increase in cyclin E levels, whereas cyclin A levels were reduced below the detection limit (Figure 2D). The loss of cyclin A (Figure 2D) and lack of chromatin-bound PCNA (Figure 2C) further support the notion of a late G1 arrest in Cdc7-depleted cells. The Cdc7-depleted cells also showed early loss of Rb phosphorylation at Ser-807/811, thought to be either Cdk4 or Cdk2 phosphorylation sites (Connell-Crowley et al, 1997; Chi et al, 2008), slightly raised p16 levels, p53 stabilization and increased levels of p21 (Figure 2D). Phosphorylation of p53 at Ser-15 (Figure 2E) and Chk1 at Ser-345 (Supplementary Figure 4) was not detected in Cdc7-depleted or control cells, indicating that the ATM/ATR checkpoint pathways were not activated. These results show that Cdc7 depletion results in downregulation of replication initiation factors and low CDK activity, consistent with the observed increase in p21 levels and the appearance of hypo-phosphorylated Rb.
Cdc7 depletion affects expression of genes required for cell cycle progression and proliferation
To investigate signalling pathways affected by CDC7 knock-down, we performed gene expression microarray (GEM) analysis on samples prepared from IMR90 cells transfected with CDC7-siRNA and control-siRNA (84 h post-transfection; see Supplementary Materials and Methods). Differentially regulated genes were analysed according to their membership of Kyoto Encyclopedia of Genes and Genomes human signalling pathways (Supplementary Table 2). The overall expression profile of genes in the cell cycle cluster was significantly altered between control cells and Cdc7-depleted cells (P<0.0001). Genes encoding Cdc6 and Mcm2–7, a number of mitosis regulatory factors, A-, B- and D-type cyclins, and Cdk1 and Cdk6 were all significantly downregulated in Cdc7-depleted cells, whereas CDKN2B (p15INK4B) was strongly upregulated (Supplementary Figure 5A). Genes in the p53 network cluster were also found to be differentially regulated (P<0.0001; Supplementary Figure 5B). The DNA damage checkpoint kinases ATM and ATR were strongly downregulated in Cdc7-depleted cells (Supplementary Figure 5B), consistent with the absence of p53 phosphorylation at Ser-15 (Figure 2E) and Chk1 phosphorylation at Ser-345 (Supplementary Figure 4). Although the pro-apoptotic genes BAX and FAS were upregulated, caspase 9 (CASP9) and caspase 3 (CASP3) were downregulated (Supplementary Figure 5B), suggesting that Cdc7 depletion may sensitize fibroblasts to pro-apoptotic signals, but does not activate the cell death effector machinery. Among p53-target genes, SIAH1, known to ubiquitinate β-catenin (Matsuzawa and Reed, 2001), and Dickkopf homolog 3 (DKK3), which blocks β-catenin accumulation in the nucleus (Wei et al, 2006; Lee et al, 2009), were strongly upregulated (Supplementary Figure 5B and C), pointing towards possible coupling between the p53 network and Wnt/β-catenin signalling pathway after CDC7 knock-down. This supposition was further supported by the notion that the overall expression profile of genes in the Wnt-signalling cluster was significantly altered between control and Cdc7-depleted cells (P=0.014; Supplementary Table 2; Supplementary Figure 5C). The GEM data indicate that the Cdc7-depletion-induced checkpoint overrides the transcriptional programme in a way that tilts the balance in favour of cell cycle arrest and reduced competency for cell proliferation. The GEM data used here may be found in the Array Express data repository (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-2115.
The Cdc7-depletion-induced checkpoint is p53 dependent
As the ATM/ATR checkpoint pathway does not appear to be active in Cdc7-depleted cells, we reasoned that the balance between Hdm2 and p14ARF may constitute the main mechanism for controlling p53 levels. Immunoblot analysis of nuclear extracts (NEs) prepared from Cdc7-depleted cells confirmed an increase in ARF levels, which correlated with loss of Hdm2 and p53 stabilization (Supplementary Figure 6A). Hdm2 protein was also not detectable in nucleolar subfractions prepared from Cdc7-depleted cells (data not shown). In keeping with the biochemical data, Cdc7-depleted cells showed strong ARF immunostaining compared with control cells (Supplementary Figure 6B). Hdm2 transcript levels increased two-fold in Cdc7-depleted cells relative to control-transfected cells 72 h post-transfection and were comparable at later time points, arguing against transcriptional downregulation of Hdm2 because of siRNA off-target effects (Supplementary Figure 7). A previous study in dermal fibroblasts showed that the Cdc7-depletion-induced checkpoint is dependent on p53 (Montagnoli et al, 2004). To confirm an active function for p53 in our experimental system, we used RNAi to downregulate p53 in IMR90 cells previously arrested by Cdc7 depletion (Supplementary Figure 8A). Notably, whereas Mcm2 phosphorylation at Ser-27 was abolished in Cdc7-depleted cells, phosphorylation at this mapped Cdk2 phosphosite was detectable in doubly depleted Cdc7/p53 cells. On the contrary, Mcm2 phosphorylation at the mapped Cdc7 phosphosite was strongly reduced in both Cdc7- and Cdc7/p53-depleted cells (Supplementary Figure 9). These data show that S phase-promoting CDK activity was restored in doubly depleted Cdc7/p53 cells. In keeping with the findings reported by Montagnoli et al, doubly depleted Cdc7/p53 cells failed to arrest cell cycle progression and instead progressed through S/G2 (Supplementary Figure 6C and D). Failure to elicit the Cdc7-depletion-induced checkpoint under CDC7 and P53 double knock-down conditions, however, eventually resulted in apoptotic cell death (Supplementary Figure 6E and F). Interestingly, cytoplasmatic protein fractions prepared from doubly depleted Cdc7/p53 cells revealed reduced levels of the β-catenin antagonist Dkk3 (Supplementary Figure 6G), supporting the notion of possible coupling between the p53 network and Wnt/β-catenin signalling pathway after Cdc7 depletion (Supplementary Table 2 and Supplementary Figure 5B and C). Note that in keeping with a previous report (Hsieh et al, 2004), peptide blocking and N-glycanase treatment identified the different bands detected with Dkk3 antibody as N-glycosylated Dkk3 isoforms (Supplementary Figure 10).
The Cdc7-depletion-induced checkpoint is dependent on p53 activity upstream of Wnt/β-catenin signalling antagonist Dkk3 to downregulate Myc and cyclin D1 expression
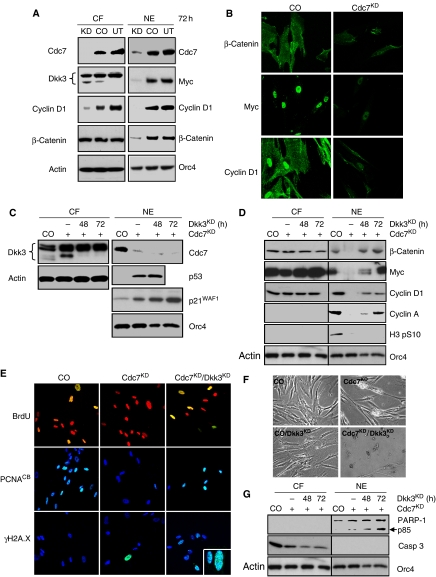
Dkk3 is known to block nuclear accumulation of β-catenin (Lee et al, 2009), resulting in downregulation of its downstream targets including CCND1 (cyclin D1) and MYC (Clevers, 2006). As DKK3 upregulation in Cdc7-depleted cells is dependent on p53 (Supplementary Figures 5C and 6G), we reasoned that p53 may affect cell cycle progression by indirectly blocking Wnt/β-catenin signalling. Indeed, immunoblot analysis of cytoplasmatic fraction (CF) and NE prepared from Cdc7-depleted cells showed reduced nuclear β-catenin and low Myc and cyclin D1 levels in conjunction with an increase in the inducible, faster migrating isoform of Dkk3 (Figure 3A). Consistent with the biochemical data, Cdc7-depleted cells showed only weak Myc and cyclin D1 immunostaining compared with control cells (Figure 3B). Notably, inducible Dkk3 expression and reduced nuclear β-catenin and cyclin D1 protein levels were not detected in IMR90 cells arrested through specific activation of the p53 pathway by low dose actinomycin D (Choong et al, 2009) (Supplementary Figure 11), further supporting a close relationship between p53-dependent Dkk3 upregulation and DNA replication control.
Figure 3.
p53-dependent upregulation of Wnt/β-catenin signalling antagonist Dkk3 is required for Cdc7-depletion-induced cell cycle arrest. (A) Cytoplasmatic protein fractions (CF) and crude nuclear extracts (NE) from untreated (UT), control-siRNA (CO) and CDC7-siRNA (Cdc7KD)-transfected IMR90 cells (72 h post-transfection) were analysed by immunoblotting with the indicated antibodies (β-actin and Orc4—loading controls). (B) At the same time point, CO and Cdc7KD cells were fixed and β-catenin, Myc and cyclin D1 detected by indirect immunofluorescence using a fluorescein-labelled secondary antibody. (C, D) CF and NE samples prepared from CO, Cdc7KD and doubly depleted Cdc7/Dkk3 (Cdc7KD/Dkk3KD) cells 48 and 72 h post-transfection were analysed by immunoblotting with the indicated antibodies. (E) 72 h post-transfection CO, Cdc7KD and Cdc7KD/Dkk3KD cells were pulsed for 2 h with BrdU, fixed and detected with an FITC-conjugated anti-BrdU antibody. Chromatin-bound PCNA and γH2A.X (inset: higher magnification) were detected by indirect immunofluorescence with anti-PCNA and anti-γH2A.X antibodies and a fluorescein-labelled secondary antibody. DNA was stained with propidium iodide (BrdU) or DAPI (PCNA and γH2A.X). Apoptotic cell death was detected in doubly depleted Cdc7KD/Dkk3KD cells by (F) phase contrast microscopy and by (G) immunoblot analysis of CF and NE with the indicated antibodies (β-actin and Orc4—loading controls).
To directly test whether the p53 → Dkk3-∣β-catenin axis is essential for the checkpoint response, we downregulated Dkk3 through RNAi in IMR90 cells previously arrested by Cdc7 depletion (Supplementary Figure 8B). Immunoblot analysis of CF and NE prepared at 48 and 72 h post-transfection shows loss of the inducible form of Dkk3 in the Cdc7-depleted background and confirms maintenance of elevated p53 and p21 levels (Figure 3C). Note that downregulation of the slower migrating, heavily glycosylated Dkk3 isoforms (upper bands) only occurs at later time points (Supplementary Figure 10B and C). Cdc7 depletion alone diminished the pool of nuclear β-catenin and reduced Myc and cyclin D1 levels (Figure 3D). As expected, this resulted in G1 arrest as shown by cells failing to incorporate BrdU (Figure 3E), loss of chromatin-bound PCNA (Figure 3E) and cyclin A and histone H3 Ser-10 phosphorylation becoming undetectable (Figure 3D). Importantly, in doubly depleted Cdc7/Dkk3 cells, nuclear levels of β-catenin, Myc and cyclin D1 were restored 72 h post-transfection (Figure 3D). In contrast to Cdc7-depleted cells, doubly depleted Cdc7/Dkk3 cells did not arrest in G1 and instead progressed into S phase, as shown by high levels of chromatin-bound PCNA (Figure 3E), BrdU incorporation (Figure 3E), cyclin A detection (Figure 3D) and flow cytometry (Supplementary Figure 12C). Notably, doubly depleted Cdc7/Dkk3 cells exhibited γH2A.X immunostaining indicative of double-strand breaks (Figure 3E) and did not appear to progress to G2/M as shown by the lack of histone H3 Ser-10 phosphorylation (Figure 3D). Induction of apoptosis in doubly depleted Cdc7/Dkk3 cells was confirmed morphologically (Figure 3F), through detection of caspase 3 activation and PARP-1 cleavage (Figure 3G), and by flow cytometric detection of cells with less than 2C DNA content (Supplementary Figure 12C). These results show that Dkk3-mediated downregulation of Myc and cyclin D1, critical components of the cell cycle engine required for progression through G1 phase, is essential for a functioning origin activation checkpoint.
The Forkhead transcription factor FoxO3a coordinates the ARF-∣Hdm2-∣p53 → p21, p53 → Dkk3-∣β-catenin and p15INK4B-∣cyclin D–CDK axes of the origin activation checkpoint
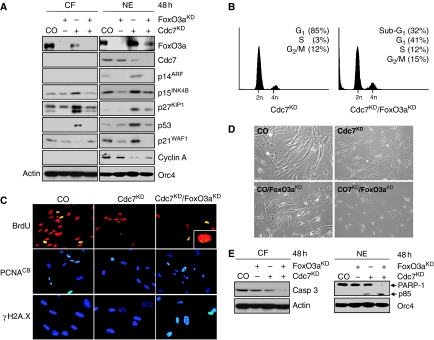
Cdc7-depleted IMR90 cells showed six-, seven- and four-fold increases in CDKN2A (ARF), CDKN2B (p15) and CDKN1B (p27) mRNA transcript levels (Supplementary Figure 13A). As FoxO3a can mediate the expression of all three of these cell cycle regulatory genes in response to different stimuli (Brunet et al, 1999; Bouchard et al, 2007; Katayama et al, 2008), we hypothesized that this transcription factor could represent an important node in the Cdc7-depletion-induced origin activation checkpoint. Immunoblot analysis showed a pool of FoxO3a present in the cytoplasm of untreated control cells and revealed nuclear accumulation of FoxO3a in Cdc7-depleted cells (Figure 4A). Concurrent with the nuclear accumulation of FoxO3a in Cdc7-depleted cells, we noted an increase in nuclear ARF, p53 and p21 levels and in nuclear and cytoplasmatic p15 and p27 levels (Figure 4A). Protein levels of these cell cycle regulators were not raised and Dkk3 expression was not induced in synchronized late G1 phase control-transfected IMR90 cells or in the G1 DNA content fraction collected by one-way sorting of propidium iodide (PI)-stained control cells (Supplementary Figure 14). Hence, the observed increase in checkpoint protein levels is not an indirect consequence of cell cycle position. Co-knock-down of CDC7 and FOXO3A (Supplementary Figure 8C) reduced CDKN2A mRNA levels by 89%, CDKN2B levels by 85% and CDKN1B levels by 50% compared with CDC7 knock-down alone (Supplementary Figure 13B). Concordantly, ARF became undetectable by western blotting and p53, p15, p21 and p27 were reduced to background levels seen in untreated control cells (Figure 4A). When expression of these proteins was compared in Cdc7-depleted and oxidatively stressed cells (note that reactive oxygen species are known regulators of FoxO family members), p53, p21, p15 and p27 upregulations were found in both cell populations, whereas increased ARF levels and inducible Dkk3 expression were restricted to Cdc7-depleted cells (Supplementary Figure 15). This suggests that inducible ARF and Dkk3 expression are more specific events associated with DNA replication control, whereas p15 and p27 upregulation in Cdc7-depleted-arrested cells overlaps with a common FoxO-induced cell cycle arrest pathway. Taken together, these expression profiles support the supposition that triggered by impaired origin activation FoxO3a may, either directly through transcriptional mechanisms or indirectly, mediate the expression of Dkk3, INK4 and CIP/KIP CDK inhibitors.
Figure 4.
FoxO3a-mediated upregulation of ARF and CDK inhibitors is an essential step for arresting cell cycle progression in Cdc7-depleted cells. (A) Cytoplasmatic protein fractions (CF) and crude nuclear extracts (NE) prepared from CO, FoxO3aKD, Cdc7KD and doubly depleted Cdc7/FoxO3a (Cdc7KD/FoxO3aKD) IMR90 cells 48 h post-transfection were analysed by immunoblotting with the indicated antibodies (β-actin and Orc4—loading controls). Note that the faster migrating band represents the hypo-phosphorylated, active form of p27 and the slower migrating band its hyper-phosphorylated inactive form (Rodier et al, 2001; Chopra et al, 2002) (B) DNA content of Cdc7KD and Cdc7KD/FoxO3aKD cells 48 h post-transfection. (C) 48 h post-transfection CO, Cdc7KD and Cdc7KD/FoxO3aKD cells were pulsed for 2 h with BrdU, fixed and detected with an FITC-conjugated anti-BrdU antibody (inset: higher magnification). Chromatin-bound PCNA and γH2A.X were detected as described in the legend to Figure 3. Apoptotic cell death was detected 48 h post-transfection in doubly depleted Cdc7KD/FoxO3aKD cells (D) by phase contrast microscopy and (E) by immunoblot analysis of CF and NE with the indicated antibodies (β-actin and Orc4—loading controls).
We reasoned that if the origin activation checkpoint is dependent on FoxO3a, doubly depleted Cdc7/FoxO3a cells should bypass the cell cycle blockade and progress into S phase. As predicted, flow cytometric analysis of doubly depleted Cdc7/FoxO3a cells 48 h post-transfection revealed a small S phase population, a marginally increased fraction of cells with G2/M DNA content and a large population of cells with less than 2C DNA content (Figure 4B). The notion that doubly depleted Cdc7/FoxO3a cells had progressed into S phase was further supported by positive immunostaining for chromatin-bound PCNA and detection of BrdU incorporation (Figure 4C). Notably, doubly depleted Cdc7/FoxO3a cells incorporated only low levels of BrdU into DNA (Figure 4C, insert) and showed strong immunostaining of the γH2A.X surrogate marker for DNA fragmentation (Figure 4C). Induction of apoptosis was confirmed by flow cytometry (Figure 4B), morphologically (Figure 4D), and through detection of caspase 3 activation and PARP-1 cleavage (Figure 4E).
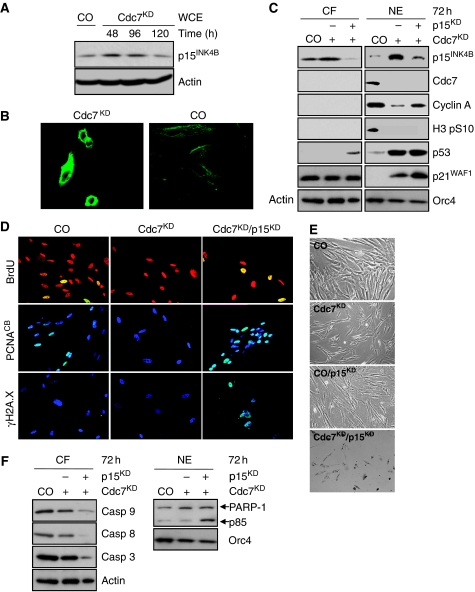
In Cdc7-depleted cells, p15 expression is strongly upregulated at both transcriptional (Supplementary Figure 13A) and protein level (Figure 5A and B). By binding to and inhibiting cyclin D–CDK4/6 complexes, p15 may inactivate the CDK-Rb-E2F pathway in Cdc7-depleted cells, which could be critical for shifting the balance between growth-promoting and anti-proliferative signals in favour of cell cycle arrest. To address this question, we downregulated p15 in IMR90 cells previously arrested by Cdc7 depletion (Supplementary Figure 8D). Immunoblot analysis shows knock-down of p15 expression in the Cdc7-depleted background (Figure 5C). In contrast to Cdc7-depleted cells arresting in G1, doubly depleted Cdc7/p15 cells entered S phase despite elevated p53 and p21 levels as shown by flow cytometry (Supplementary Figure 12D), chromatin-bound PCNA and BrdU incorporation (Figure 5D), and increased cyclin A detection (Figure 5C). Doubly depleted Cdc7/p15 cells also exhibited replication stress in the form of strong γH2A.X immunostaining (Figure 5D) and induced apoptosis as confirmed by flow cytometry (Supplementary Figure 12D), morphologically (Figure 5E), and through detection of caspase activation and PARP-1 cleavage (Figure 5F). Importantly, single knock-down control experiments showed that p15, FoxO3a, Dkk3 or p53 depletion alone does not cause strong S phase stimulation or apoptosis in this experimental system (Supplementary Figure 16).
Figure 5.
The Cdc7-depletion-induced cell cycle arrest is p15INK4B dependent. (A) Upregulation of p15 levels in Cdc7-depleted IMR90 cells was confirmed by immunoblotting WCE prepared from CO cells and Cdc7-depleted cells 48, 96 and 120 h post-transfection with antibodies against p15 and β-actin (loading control). (B) Cdc7KD and CO cells were fixed 96 h post-transfection and p15 detected by indirect immunofluorescence using a fluorescein-labelled secondary antibody. (C) CF and NE prepared from CO, Cdc7KD and doubly depleted Cdc7/p15 (Cdc7KD/p15KD) cells 72 h post-transfection were analysed by immunoblotting with the indicated antibodies (β-actin and Orc4—loading controls). (D) 72 h post-transfection CO, Cdc7KD and Cdc7KD/p15KD cells were pulsed for 2 h with BrdU, fixed and detected with an FITC-conjugated anti-BrdU antibody. Chromatin-bound PCNA and γH2A.X were detected as described in Figure 3 legend. Apoptotic cell death was detected 72 h post-transfection in doubly depleted Cdc7KD/p15KD cells by (E) phase contrast microscopy and by (F) immunoblot analysis of CF and NE with the indicated antibodies (β-actin and Orc4—loading controls).
Cdc7 depletion with an alternative CDC7-siRNA (oligo CDC7-B) triggered the same molecular changes in the described checkpoint pathways as CDC7 knock-down with oligo CDC7-A (Supplementary Figure 2C). The cell cycle arrest phenotype was reversed through expression of a CDC7 gene variant refractory to silencing by oligo CDC7-A (Supplementary Figure 3D and E). Moreover, the arrested phenotype was fully reproducible in a different fibroblast strain (Supplementary Figure 17). These control experiments further reinforce our findings and argue against RNAi off-target effects.
It can be postulated that if the checkpoint pathways elucidated here for Cdc7-depleted cells are manifested by impaired origin activation, these pathways should be similarly activated by depletion of other replication initiation proteins. To address this question, we compared the cellular response to Cdc7 depletion in IMR90 cells with the response caused by RNAi against ORC2, an origin licensing factor that acts upstream of Cdc7 in the DNA replication initiation pathway. Indeed, ORC2 knock-down led to an accumulation of cells with G1 DNA content (Supplementary Figure 18A and B). As expected, immunoblotting revealed changes in protein levels and subcellular localization of FoxO3a, ARF, p15, p21, p27, p53 and Dkk3 similar to those found in Cdc7-depleted cells (Supplementary Figure 18C). Moreover, CDK activity, determined indirectly through Mcm2 phosphorylation at the Cdk2 phosphosite Ser-27, was reduced and cyclin D1 downregulated in Orc2-depleted cells (Supplementary Figure 18C). Thus, at least partially overlapping checkpoint pathways appear to be activated by targeting either CDC7 or ORC2, reinforcing the conclusion that the Cdc7-depletion-induced checkpoint is triggered by impaired origin activation.
Discussion
Cells have evolved elaborate checkpoint mechanisms for maintenance of the genome. Checkpoints guard critical cell cycle transitions by preventing future events from happening if the prior event is not completed and error free. Previous studies have shown that inhibition of replication initiation proteins arrests the cell cycle in G1, pointing towards the existence of a checkpoint that prevents premature entry into S phase until a sufficient number of origins are replication competent (Blow and Gillespie, 2008). Here, we have used RNAi against CDC7 to reveal the activities of the origin activation checkpoint and to investigate its molecular architecture.
Our results show that Cdc7 depletion arrests cell cycle progression in diploid human fibroblasts. Immunoblotting showed that the DNA polymerase processivity factor PCNA was not associated with chromatin in Cdc7-depleted cells, whereas the proportion of BrdU-incorporating cells was reduced by >90% compared with control cells. Moreover, cyclin A, a marker for entry into S-G2-M phase, became undetectable in arrested cells. Phosphorylation of Chk1 on Ser-345, involved in activation of this checkpoint kinase in response to blocked DNA replication (Zhao and Piwnica-Worms, 2001), was also not detectable in Cdc7-depleted cells, arguing against the cell cycle arrest being triggered in early S phase by a signal generated from a few stalled replication forks that escaped the block. Downregulation of the DNA damage checkpoint controls kinases ATM and ATR and the lack of p53 phosphorylation on Ser-15 argues further against early S phase arrest in Cdc7-depleted cells. These results, therefore, suggest that fibroblasts depleted of Cdc7 arrest before entry into S phase. We found p53 and p21 levels to be elevated and Rb hypo-phosphorylated in Cdc7-depleted cells, whereas in vitro kinase assays confirmed a loss of CDK activity, which could mediate the G1 arrest.
Conceptually, the process of DNA checkpoint control is viewed as analogous to a signal transduction pathway. In this analogy, if replication initiation is impaired for any reason, a signal is detected by ‘sensor’ proteins and then sent by ‘transducer’ proteins to ‘effector’ proteins, which block the cell cycle until a sufficient number of origins are replication competent. Owing to experimental limitations of transient RNAi gene silencing, in particular the half-life of the CDC7 message and/or protein, our study has been restricted to elucidating the transducer and effector mechanisms of the origin activation checkpoint. Future investigations into how impaired replication initiation is sensed and signalled to transducer proteins, will be critically dependent on the availability of specific small molecule Cdc7 kinase inhibitors, which should allow activation of the checkpoint on a more rapid time scale.
As the FoxO subfamily has been previously implicated in cell cycle arrest (Huang and Tindall, 2007), we reasoned that this family of stress-activated transcription factors might serve as transducer proteins in the origin activation checkpoint. FoxO transcription factors have been shown to rapidly modulate the expression of genes involved in cell cycle transitions (Huang and Tindall, 2007) and themselves are regulated by subcellular localization, with cytoplasmatic sequestration preventing transactivation of target genes (Calnan and Brunet, 2008). Our results show that Cdc7-depletion causes nuclear accumulation of FoxO3a. Among the cohort of known FoxO3a gene targets, we found CDKN2A (ARF) (Bouchard et al, 2007), CDKN2B (p15) (Katayama et al, 2008) and CDKN1B (p27) (Chandramohan et al, 2008) to be upregulated both at transcriptional and protein level in Cdc7-depleted cells. We show that FoxO3a downregulation in cells previously arrested by Cdc7 depletion reduces expression of these critical cell cycle regulators to background levels and bypasses the G1 arrest with cells entering an abortive S phase followed by apoptosis. The same phenotype was also established by co-depletion of Cdc7 and p15, indicating that p15 is an effector of the G1 arrest, most likely through inhibiting cyclin D–CDK4/6 activity. Our results, therefore, show that FoxO3a activity is essential for a functioning origin activation checkpoint and highlight the importance of the FoxO3a → p15 axis (Figure 6). Previous work has established that degradation of the Xenopus homologue of p27 occurs at replication origins and is coupled with the initiation of DNA synthesis (Furstenthal et al, 2001; You et al, 2002), whereas in human breast epithelial cells, p27 is stabilized after Orc2 depletion and associated with the cyclin E–Cdk2 complex (Machida et al, 2005). Thus, it is possible that FoxO3a-dependent p27 upregulation forms a separate axis of the origin activation checkpoint that results in inhibition of the cyclin E–Cdk2 complex.
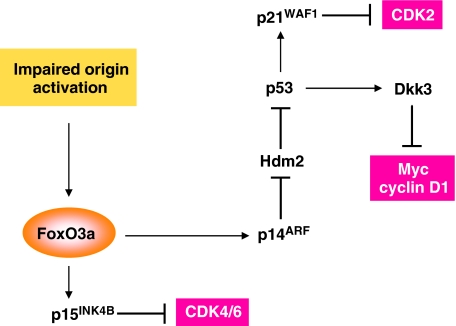
Figure 6.
A proposed model of the origin activation checkpoint.
We noted elevated levels of p53 and p21 in Cdc7-depleted fibroblasts. As the ATM/ATR checkpoint pathway (Abraham, 2001) does not appear to be active in these cells, FoxO3a-dependent upregulation of ARF provides a plausible explanation for p53 stabilization. By antagonizing the E3 ubiquitin ligase activity of Hdm2, ARF is known to stabilize p53 and increase its transcriptional activity (Pomerantz et al, 1998). One way in which ARF activates nucleoplasmic p53 is by sequestering Hdm2 in the nucleolus (Weber et al, 1999). Given the absence of any detectable Hdm2 either in crude NEs or nucleolar subfractions, however, we speculate that in Cdc7-depleted cells, ARF stabilizes p53 through Hdm2 proteolysis (Zhang et al, 1998) rather than nucleolar sequestration (Figure 6). We show that p21 induction in Cdc7-depleted cells is dependent on p53. The ability of p21 to shut down the activity of E-type and A-type cyclin–CDK complexes ensures that cell cycle progression is blocked in G1. If the cell has managed to escape the checkpoint and advance into S phase with an insufficient number of replication-competent origins, p21 can block progression through S, G2 and M phase by inhibiting A-type and B-type cyclin–CDK complexes and by interfering with PCNA function, thereby halting further advance of replication forks (Sherr and Roberts, 1999). We show that p53 downregulation in fibroblasts previously arrested in G1 by Cdc7 depletion bypasses the cell cycle blockade and results in cells proceeding through S phase to the G2/M boundary before inducing apoptosis. We, therefore, conclude that the FoxO3a → ARF-∣Hdm2-∣p53 → p21 axis is also essential for a functioning origin activation checkpoint (Figure 6).
GEMs revealed a second critical function for p53 in the checkpoint. Among known p53-target genes that were differentially expressed in cells arrested by CDC7 knock-down is DKK3, a negative regulator of the Wnt/β-catenin signalling pathway (Niehrs, 2006). Dkk3 is a glycoprotein that prevents nuclear accumulation of β-catenin, resulting in downregulation of its downstream targets MYC and CCND1 (Lee et al, 2009). As expected, immunoblotting showed an increase in the inducible, faster migrating Dkk3 isoform in Cdc7-depleted cells and revealed strongly diminished nuclear β-catenin levels as well as Myc and cyclin D1 downregulation. Dkk3 upregulation was reversed in cells co-depleted of Cdc7 and p53, whereas nuclear β-catenin, Myc and cyclin D1 levels were restored upon downregulation of Dkk3 in cells previously arrested by Cdc7 depletion. We speculate that Dkk3-mediated cyclin D1 downregulation halts progression through most of G1, whereas a decrease in transcription of the MYC gene prevents Myc-driven gene expression changes that normally drive the cell through G1 (Figure 6). It can be postulated, for example, that a sharp fall in Myc levels leads to downregulation of the cyclin D2 and CDK4 genes and a reduction in Cul1 levels, a protein required for p27 degradation (Eilers and Eisenman, 2008). Reduced Myc levels may also attenuate the expression of E2F1-3 and prevent Myc-Max-mediated repression of p15 and p21 expression (Eilers and Eisenman, 2008). As Myc has been implicated in stabilizing pre-RCs assembled at origins (Dominguez-Sola et al, 2007), Dkk3-mediated downregulation of MYC expression might also explain the decrease in chromatin-bound origin licensing factors seen in Cdc7-depleted cells. The ability of Myc to modulate the actions of a number of positive and negative regulators of cell cycle advance provides a firm rationale for the requirement to downregulate this critical driver of cell proliferation if cells sense inhibition of the DNA replication initiation pathway. Importantly, we show that Dkk3 downregulation in Cdc7-depleted-arrested fibroblasts also bypasses the cell cycle blockade, enabling cells to proceed into an abortive S phase followed by apoptosis. These data highlight the critical function of Dkk3 in linking the FoxO3a → ARF-∣Hdm2-∣p53 axis to the Wnt/β-catenin signalling pathway (Figure 6). We conclude that FoxO3a lies at the core of a complex molecular circuitry that resets the regulatory dials of the cell cycle clock in response to blocked-replication initiation and thereby shifts the balance of growth-promoting and growth-inhibiting mechanisms in a way that favours G1 arrest. Pathway specificity tests in fibroblasts arrested by oxidative stress (Chen et al, 2004) or through specific activation of the p53 pathway by low dose actinomycin D (Choong et al, 2009) show that while p15 and p27 upregulation in Cdc7-depleted cells overlap with the common FoxO-induced cell cycle arrest pathway (Huang and Tindall, 2007), inducible ARF and Dkk3 expression are more specific events associated with DNA replication control. Our microarray data indicate that the resulting loss of CDK activity inactivates the Rb-E2F signalling pathway, which in turn overrides the regular E2F-driven G1/S transcriptional programme.
The pathways elucidated in this study in response to impaired origin activation comply with two essential criteria of checkpoint control originally proposed by Hartwell and Weinert (1989). First, checkpoints are considered external control mechanisms that are not required for forward cell cycle progression. Conforming to this defining feature, FoxO3a-mediated activation of the three checkpoint axes does not occur during normal G1 progression when the potential for errors in the DNA replication initiation pathway is minimal. The second is the relief-of-dependence criterion, which stipulates that ‘the existence of a control mechanism is suggested when one finds chemicals, mutants or other conditions that … permit a late event to occur even when an early, normal prerequisite event is prevented’ (Hartwell and Weinert, 1989). In line with this hallmark of checkpoint control, double depletion of Cdc7 and any of the checkpoint proteins FoxO3a, p15, p53 or Dkk3 abrogates the checkpoint itself and allows S phase entry. Single knock-downs of these checkpoint components, on the contrary, do not cause strong S phase stimulation on their own.
Several different conditions must be met during G1 phase to ensure origin activation and DNA synthesis. These include origin recognition through ORC binding, origin licensing through the assembly of ORC, Cdc6, Cdt1 and the replicative MCM helicase into pre-RCs, origin activation through CDK and Cdc7-dependent phosphorylation events resulting in origin unwinding and the recruitment of the replisome required for initiation of DNA synthesis. Thus, as noted by Khodjakov and Rieder (2009), there is the possibility of multiple checkpoints, each detecting one of these conditions or, alternatively, that a range of abnormalities may cause a single condition detected by just one checkpoint. A G1 arrest phenotype, for example, has been reported in response to RNAi against ORC2 (Machida et al, 2005) and inhibition of MCM loading by overexpressing a stable form of geminin (Shreeram et al, 2002) or MCM helicase activity by RNAi against CDC7 (Montagnoli et al, 2004). Thus, the question arises whether these abnormalities are activating multiple different checkpoints or a single master checkpoint. It is of interest in this context that in our study the cellular effects of ORC2 knock-down were remarkably similar to those caused by CDC7 knock-down, suggesting that overlapping checkpoint pathways are governing at least origin licensing and origin firing. The activation of overlapping pathways by Orc2 and Cdc7 downregulation also argues against the possibility that the checkpoint described here is triggered by loss of other, less well understood, Cdc7 functions in, for example, mitotic chromosomal segregation (Takahashi et al, 2008) or DNA damage response (Takeda et al, 1999; Costanzo et al, 2003; Dierov et al, 2004; Tenca et al, 2007; Kim et al, 2008).
In separate studies, we found that RNAi against CDC7 in primary human breast and bronchial epithelial cells triggers a cell cycle blockade that phenotypically and at the molecular level strongly resembles the G1 arrest described here for diploid human fibroblasts (Rodriguez-Acebes et al, 2010; Kingsbury et al, in preparation). This suggests that the origin activation checkpoint is conserved in somatic cells of different embryological origin. The integrated nature of the underlying molecular circuitry explains how cells arrested by Cdc7 depletion can bypass the cell cycle arrest if the activity of important constituents of any of the three axes identified in this study is inhibited. Cells that bypass the checkpoint are able to support a low level of DNA synthesis and either arrest in S phase or proceed to the G2/M boundary if p53 function is lost. One explanation for FoxO3a-, p15-, p53- and Dkk3-depleted cells with reduced Cdc7 levels being able to synthesize DNA is the initiation of replication forks from isolated origins that escaped checkpoint function. Irrespective of p53 status, entry into S phase with an insufficient number of replication-competent origins is lethal, with cells experiencing replication stress and inducing apoptosis.
Recent studies have established that inhibition of the DNA replication initiation machinery causes cancer-cell-specific killing (Blow and Gillespie, 2008), validating replication initiation as a new target class for cancer cell chemotherapy (Jackson, 2008). Inhibiting an early step in replication initiation upstream of DNA polymerase should result in G1 arrest and, therefore, be non-genotoxic in normal cells with a functioning checkpoint. Given the reappearance of proliferative activity in fibroblasts several days after RNAi against CDC7, we speculate that the G1 arrest enforced in normal cells might also be reversible. The high levels of apoptosis reported for cancer cell lines subjected to inhibition of origin licensing or firing (Shreeram et al, 2002; Feng et al, 2003; Montagnoli et al, 2004) indicate that the origin activation checkpoint is most likely lost or impaired in transformed cells. This is in keeping with the dependency of the underlying molecular circuitry on a number of tumour suppressor proteins (p53, Hdm2, ARF, FoxO3a, Dkk3 and Rb), one or more of which are commonly inactivated during tumourigenesis. Thus, our findings support the concept of emerging pharmacological Cdc7 inhibitors (Montagnoli et al, 2008; Swords et al, 2010) as potentially powerful anti-cancer agents with broad tumour spectrum activity. Knowledge of the complex regulatory network underlying the checkpoint may help predict individual patient response to Cdc7 inhibitors in the future.
Materials and methods
Cell culture and cell synchronization
IMR90 (ATCC# CCL-186), a diploid human fibroblast adherent cell strain derived from foetal lung tissue, was obtained from LGC Standards (Middlesex, UK) at population doubling (PD) 12. All culture passages and PDs were recorded and all experiments were performed with IMR90 cells under a PD of 22. WI-38 (ATCC# CCL-75) diploid human fibroblasts were obtained from LGC standards at PD 6. All experiments with WI-38 cells were performed under a PD of 15. IMR90 and WI-38 cells were cultured at 37°C with 5% CO2 in DMEM (Invitrogen, Paisley, UK) supplemented with 10% defined FCS (Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin. For preparation of synchronous cell populations, IMR90 cells were synchronized in early S phase by two sequential 25 h blocks in 2.5 mM thymidine (Sigma, Gillingham, UK) separated by a 12 h interval without thymidine (Krude et al, 1997).
Cell treatments
IMR90 fibroblasts were oxidatively stressed through H2O2 treatment as described (Chen et al, 2004) with the following minor modifications. Cells were cultured in medium containing 600 μM H2O2 for 2 h. After 24 h, cells were treated again for 2 h with 600 μM H2O2 and collected 4 h after the second treatment. Specific activation of the p53 pathway in IMR90 cells was induced by 24 h low dose (1 nM) actinomycin D treatment as described (Choong et al, 2009).
Cell population growth assessment and cell cycle analysis
Phase contrast microscopy was performed with an inverted Axiovert 200 M (Carl Zeiss, Welwyn Garden City, UK) and Axiovision software. Flow cytometric cell cycle analyses were performed as described (Eward et al, 2004). The proportion of cells with less than 2C DNA content was calculated as an estimate of the apoptotic cell population.
Cell sorting
For preparation of G1 phase cell populations, 5 × 106 of each asynchronous control cells and Cdc7-depleted cells were collected and fixed for 3 h at −20°C in 80% methanol in PBS with added protease inhibitors (one complete EDTA-free protease inhibitor cocktail tablet (Roche Diagnostics, Burgess Hill, UK) per 25 ml of buffer). Cells were precipitated by centrifugation at 500 g for 5 min and resuspended in a mixture containing 50 μg/ml PI, 20 μg/ml RNase A and protease inhibitors. The cells were sorted on a DAKO/Beckman Coulter MoFlo High Speed Sorter (Beckman Coulter Inc., Orange County, CA). The forward scatter signal was used to trigger the detection of cells and the PI signal fluorescence was linearly quantified to rationalize DNA content after excitation at 488 nm in the orange/red channel (613/20 nm bandpass filter). Cells in G1 phase of the cell cycle were sorted through one-way sorting into round-bottomed Falcon tubes containing PBS with protease inhibitors (Roche Diagnostics). Cells were precipitated at 16 000 g for 5 min and lysed with modified RIPA buffer (300 mM NaCl).
Cell fractionation and immunoprecipitation
For western blot analysis, cells were harvested and WCE and subcellular fractions were prepared. For WCE, cells were lysed for 45 min on ice in modified RIPA lysis buffer (50 mM Tris–HCl, 300 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA and protease inhibitors) and sonicated for 10 s. For crude nuclear extraction, cells were lysed in buffer containing 10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1.0 mM DTT and protease inhibitors and gently homogenized. Nuclei were precipitated by centrifuging at 1000 g for 5 min at 4°C and the CF was removed. For preparation of NE, nuclear pellets were washed twice with the same buffer, lysed in modified RIPA buffer for 30 min, sonicated and centrifuged at 13 000 g. Nucleoli and CBF were isolated as described (Muramatsu and Onishi, 1978; Kingsbury et al, 2005). In vitro kinase assays were performed as described (Jinno et al, 2002) with minor modifications. Briefly, cell lysates were incubated at 4°C for 2 h with cyclin A and cyclin E antibodies. The kinase activity of immunoprecipitated Cdk2 in complex with cyclin A and cyclin E was assayed as described (Jinno et al, 1999). Phosphorylation of truncated Rb (Cdk2 substrate; QED Bioscience, San Diego, CA) was detected with anti-Rb phospho-Thr-821 antibody (Invitrogen).
Immunoblotting
Protein concentration was determined using the DC Bio-Rad protein assay kit (Bio-Rad Hemel Hempstead, UK); 60 μg of total protein was loaded in each lane and separated by 4–20% SDS–PAGE. Protein was transferred from polyacrylamide gels onto PVDF membranes (Bio-Rad) by semi-dry electroblotting. Blocking, antibody incubations and washing steps were performed as described (Kingsbury et al, 2005). Antibodies used for immunoblotting and immunofluorescence (see below) included caspase 3 from Novus (Littleton, CO); Chk1-pSer-345 (2341) from Cell Signaling Technology (Danvers, MA); caspase 9 (F-7), cyclin D1 (H-295), p16INK4A (H-156), c-Myc (N-262), PCNA (F-2), Cdc6 (180.2) and histone H1 (AE-4) from Santa Cruz (Santa Cruz, CA); FoxO3a, caspase 8, β-catenin and phospho-histone H3 (Ser-10) from Millipore (Billerica, MA); cyclin A (6E6) and cyclin E (Ab-4) (HE-172) from Thermo Fisher Scientific (Fremont, CA); p27KIP1, Orc4, Mcm2 (BM28), p21WAF1, cyclin B1, PARP-1, pRb and Orc2 from BD Biosciences (Oxford, UK); p15INK4B (15PO6), Cdt1, Hdm2 and Dkk3 from Abcam (Cambridge, UK); p53 (Ab-6) from Merck (Beeston, UK); Cdc7 from MBL International (Woburn, MA); p14ARF (DCS-240) and β-actin from Sigma; p53 phospho-Ser-15, Rb phospho-Ser-807/811, phospho-histone γH2A.X (Ser-139) from New England Biolabs (Hitchin, UK) and Mcm2 phospho-Ser-53, Mcm2 phospho-Ser-27, Mcm2 phospho-Ser-41, Mcm2 phospho-Ser-40/41 and Mcm2 phospho-Ser-108 from Bethyl Laboratories (Montgomery, TX). Affinity-purified rabbit polyclonal anti-geminin antibody G95 was generated as described (Wharton et al, 2004). Affinity-purified rabbit polyclonal antibodies to Mcm10, Cdc45 and Dbf4 were generated by Eurogentec (Seraing, Belgium) following the manufacturer's protocol. The positive control lysate for p53 phospho-Ser-15 was prepared from IMR90 cells treated with 17 μM cisplatin for 24 h (Pabla et al, 2008). Neuroblastoma SK-N-SH cell lysate from Insight Biotechnologies (Wembley, UK) was used as a positive control for Dkk3 protein. Immunodetection of Dkk3 was blocked by preincubation of Dkk3 antibody with recombinant human Dkk3 (1118-DK-050; R&D Systems Europe, Abingdon, UK) (1:1 w/w).
N-glycanase digestion
Cytoplasmatic protein fractions (125 μg or 50 μg total protein) were treated with N-glycanase (recombinant from Chriseobacterium (Flavobacterium) meningosepticum, expressed in Escherichia coli; ProZyme, Hayward, CA) as described (Krupnik et al, 1999).
Immunofluorescence
For detection of BrdU incorporation, cells pulsed for 2 h with 100 μM BrdU were fixed with 3.7% paraformaldehyde for 5 min and permeabilized with 0.2% Triton X-100 for 5 min. Coverslips were incubated with 2N HCl for 1 h, washed with PBS and incubated for 1 h at 37°C with primary anti-BrdU antibody (Alexis Biochemicals, Exeter, UK), diluted 1/10 vol/vol in 0.1% BSA in PBS containing 50 ng/ml PI and 50 ng/ml RNase A. Fluorescence confocal microscopy of random fields of cells was performed on a Leica TCS SP confocal microscope (Leica, Milton Keynes, UK). Images of the rhodamine (red) and fluorescein (green) channels were obtained using Leica TCS PowerScan software. At least 400 cells were routinely scored for each treatment and quantitated as percentages of the total number of cells. Indirect immunofluorescence of chromatin-bound PCNA was performed as described (Miura and Sasaki, 1999) with minor modifications. For detection of β-catenin, Myc, cyclin D1, p15INK4B and γ-H2A.X, after paraformaldehyde fixation and permeabilization, coverslips were saturated with blocking buffer (5% BSA in PBS) for 1 h at RT, incubated with primary antibody diluted in blocking buffer for 1 h at 37°C and incubated overnight at 4°C. FITC-conjugated anti-mouse antibody from Dako (Glostrup, Denmark) was used at dilution 1/1000. Slides were mounted in Vectashield mounting medium (Vector Laboratories, Peterborough, UK) with 1.5 μg/ml DAPI to visualize DNA. Coverslips for p14ARF detection were fixed with methanol/acetone (3:2 vol/vol) and stained as described above.
RNA interference
CDC7, p53 and CDKN2B (p15INK4B) expression was inhibited with double-stranded RNA oligos for CDC7 (A- and B- custom, and V—validated siRNAs), p53 (custom siRNA) and p15INK4B (silencer pre-designed and validated siRNAs) synthesized by Ambion (Supplementary Table 1). DKK3 was specifically inhibited with a cocktail of two ON-TARGET plus siRNAs (J-018352-11 and -12; Dharmacon, Fremont, CA) and FOXO3A was inhibited with siGENOME SMART pool (M-003007-02-0005, Dharmacon). Non-targeting siRNA was used as a negative control. Lipofectamine 2000 (Invitrogen) was used in all transfections according to the manufacturer's recommendations. Briefly, cells were seeded at a density to reach 50% confluence on the day of transfection. The transient transfections were performed using 10 nM of CDC7 siRNA duplex or 10 nM of ORC2 siRNA duplex. For double knock-downs, cells were first transfected with CDC7-siRNA and after 72 h (after replating at low density for CO cells) transfected with either CDC7 (10 nM) and control (10 nM) oligos, or CDC7 (10 nM) and p53 (10 nM) or FOXO3A (10 nM) oligos. DKK3 and p15INK4B cotransfections were performed with CDC7 oligo (10 nM) and a cocktail of two DKK3 oligos (each 10 nM) or a cocktail of p15INK4B oligos 1, 2 and 3 (each 5 nM). In this case, CDC7 oligo pulsing at 72 h was adjusted with 20 and 15 nM control (CO) oligo, respectively, to reach the same amount of oligos in both samples (single and double knock-down). For p53, Dkk3, FoxO3a and p15 single knock-downs, cells were first transfected with CO oligo and after 72 h replated at low density and transfected with CO, CO and p53-siRNAs (p53KD), or CO and DKK3 (Dkk3KD) or FOXO3A (FoxO3aKD) or CDKN2B (p15KD) oligos. siRNAs were complexed with transfection reagent in serum-free and antibiotic-free culture medium according to the manufacturer's instructions (Invitrogen). Cells were incubated from 48 to 120 h. All experiments were performed at least three times. The transfection efficiency was determined for fluorescein-conjugated non-specific siRNA-transfected cells (BLOCK-iT Transfection Optimization kit; Invitrogen) using a Leica TCS SP confocal fluorescence microscope. Selective silencing of the corresponding proteins was confirmed by western blotting. For rescue experiments, the full 1725 bp CDC7 cDNA sequence containing four silent, single base pair mutations in the 21 bp CDC7-siRNA (oligo-A) interaction region was inserted into pCMV6-AC expression vector (OriGene) to fully abolish the siRNA effect.
RNA extraction and qRT–PCR
To evaluate the efficiency of transfection with CDC7, p53, DKK3, p15INK4B and FOXO3A siRNAs, mRNA transcription levels of CDC7, p53, p15INK4B, DKK3 isoforms A and B and FOXO3A were detected by qRT–PCR. Total RNA was isolated using a PureLink Micro-to-Midi kit (Invitrogen) according to the manufacturer's instructions. Reverse transcription reactions using 40 ng of total RNA in a final reaction volume of 20 μl were performed in one step using SuperScript III Platinum SYBR Green One Step qRT–PCR Kit (Invitrogen). Relative quantitation data were obtained using the comparative Ct method with Realplex software according to the manufacturer's protocol (Eppendorf, Heidelberg, Germany). Glyceraldehyde-3-phosphate dehydrogenase was used to normalize each of the extracts for amplifiable human DNA. Primers (Supplementary Table 3) were provided by Eurofins MWG Operon (Ebersberg, Germany). Cycle conditions are available upon request.
Additional information on cRNA labelling and hybridization for microarray and Microarray data processing and analysis is available in the Supplementary data section.
Supplementary Material
Acknowledgments
We thank Hye-Kyung Hong and Richard Sainsbury for insightful discussion of the data and Arnold Pizzey and Tomas Adejumo for supporting the flow cytometric analyses with their expertise and advice. This study has been supported by Cancer Research UK scientific programme grant C428/A6263 (KS and GHW) and by an MRC Centre Grant (MWT).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Biamonti G, Paixao S, Montecucco A, Peverali FA, Riva S, Falaschi A (2003) Is DNA sequence sufficient to specify DNA replication origins in metazoan cells? Chromosome Res 11: 403–412 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ (2008) Replication licensing and cancer—a fatal entanglement? Nat Rev Cancer 8: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA (2007) FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev 21: 2775–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Calnan DR, Brunet A (2008) The FoxO code. Oncogene 27: 2276–2288 [DOI] [PubMed] [Google Scholar]
- Chandramohan V, Mineva ND, Burke B, Jeay S, Wu M, Shen J, Yang W, Hann SR, Sonenshein GE (2008) c-Myc represses FOXO3a-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor. J Cell Biochem 104: 2091–2106 [DOI] [PubMed] [Google Scholar]
- Chen JH, Stoeber K, Kingsbury S, Ozanne SE, Williams GH, Hales CN (2004) Loss of proliferative capacity and induction of senescence in oxidatively stressed human fibroblasts. J Biol Chem 279: 49439–49446 [DOI] [PubMed] [Google Scholar]
- Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE (2008) Identification of CDK2 substrates in human cell lysates. Genome Biol 9: R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong ML, Yang H, Lee MA, Lane DP (2009) Specific activation of the p53 pathway by low dose actinomycin D: a new route to p53 based cyclotherapy. Cell Cycle 8: 2810–2818 [DOI] [PubMed] [Google Scholar]
- Chopra S, Fernandez De MS, Lam EW, Mann DJ (2002) Jab1 co-activation of c-Jun is abrogated by the serine 10-phosphorylated form of p27Kip1. J Biol Chem 277: 32413–32416 [DOI] [PubMed] [Google Scholar]
- Clevers H (2006) Wnt/beta-catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L, Harper JW, Goodrich DW (1997) Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell 8: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J (2003) An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 11: 203–213 [DOI] [PubMed] [Google Scholar]
- Dierov J, Dierova R, Carroll M (2004) BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell 5: 275–285 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, la-Favera R (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN (2008) Myc's broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eward KL, Obermann EC, Shreeram S, Loddo M, Fanshawe T, Williams C, Jung HI, Prevost AT, Blow JJ, Stoeber K, Williams GH (2004) DNA replication licensing in somatic and germ cells. J Cell Sci 117: 5875–5886 [DOI] [PubMed] [Google Scholar]
- Feng D, Tu Z, Wu W, Liang C (2003) Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res 63: 7356–7364 [PubMed] [Google Scholar]
- Furstenthal L, Swanson C, Kaiser BK, Eldridge AG, Jackson PK (2001) Triggering ubiquitination of a CDK inhibitor at origins of DNA replication. Nat Cell Biol 3: 715–722 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA (1989) Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634 [DOI] [PubMed] [Google Scholar]
- Hsieh SY, Hsieh PS, Chiu CT, Chen WY (2004) Dickkopf-3/REIC functions as a suppressor gene of tumor growth. Oncogene 23: 9183–9189 [DOI] [PubMed] [Google Scholar]
- Huang H, Tindall DJ (2007) Dynamic FoxO transcription factors. J Cell Sci 120: 2479–2487 [DOI] [PubMed] [Google Scholar]
- Jackson PK (2008) Stopping replication, at the beginning. Nat Chem Biol 4: 331–332 [DOI] [PubMed] [Google Scholar]
- Jinno S, Hung SC, Yamamoto H, Lin J, Nagata A, Okayama H (1999) Oncogenic stimulation recruits cyclin-dependent kinase in the cell cycle start in rat fibroblast. Proc Natl Acad Sci USA 96: 13197–13202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S, Yageta M, Nagata A, Okayama H (2002) Cdc6 requires anchorage for its expression. Oncogene 21: 1777–1784 [DOI] [PubMed] [Google Scholar]
- Katayama K, Nakamura A, Sugimoto Y, Tsuruo T, Fujita N (2008) FOXO transcription factor-dependent p15(INK4b) and p19(INK4d) expression. Oncogene 27: 1677–1686 [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL (2009) The nature of cell-cycle checkpoints: facts and fallacies. J Biol 8: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kakusho N, Yamada M, Kanoh Y, Takemoto N, Masai H (2008) Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene 27: 3475–3482 [DOI] [PubMed] [Google Scholar]
- Kingsbury SR, Loddo M, Fanshawe T, Obermann EC, Prevost AT, Stoeber K, Williams GH (2005) Repression of DNA replication licensing in quiescence is independent of geminin and may define the cell cycle state of progenitor cells. Exp Cell Res 309: 56–67 [DOI] [PubMed] [Google Scholar]
- Krude T, Jackman M, Pines J, Laskey RA (1997) Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell 88: 109–119 [DOI] [PubMed] [Google Scholar]
- Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, Chang B, Duong T, Goodearl AD, Gearing DP, Sokol SY, McCarthy SA (1999) Functional and structural diversity of the human Dickkopf gene family. Gene 238: 301–313 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Jo M, Rho SB, Park K, Yoo YN, Park J, Chae M, Zhang W, Lee JH (2009) Dkk3, downregulated in cervical cancer, functions as a negative regulator of beta-catenin. Int J Cancer 124: 287–297 [DOI] [PubMed] [Google Scholar]
- Machida YJ, Teer JK, Dutta A (2005) Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem 280: 27624–27630 [DOI] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W (2008) Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135: 662–678 [DOI] [PubMed] [Google Scholar]
- Matsuzawa SI, Reed JC (2001) Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell 7: 915–926 [DOI] [PubMed] [Google Scholar]
- Mechali M (2001) DNA replication origins: from sequence specificity to epigenetics. Nat Rev Genet 2: 640–645 [DOI] [PubMed] [Google Scholar]
- Miller CT, Gabrielse C, Chen YC, Weinreich M (2009) Cdc7p-Dbf4p regulates mitotic exit by inhibiting Polo kinase. PLoS Genet 5: e1000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M, Sasaki T (1999) Detection of chromatin-bound PCNA in cultured cells following exposure to DNA-damaging agents. Methods Mol Biol 113: 577–582 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Tenca P, Sola F, Carpani D, Brotherton D, Albanese C, Santocanale C (2004) Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res 64: 7110–7116 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C (2006) Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem 281: 10281–10290 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, Patton V, Alzani R, Ciavolella A, Sola F, Molinari A, Volpi D, Avanzi N, Fiorentini F, Cattoni M, Healy S et al. (2008) A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 4: 357–365 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Onishi T (1978) Isolation and purification of nucleoli and nucleolar chromatin from mammalian cells. Methods Cell Biol 17: 141–161 [DOI] [PubMed] [Google Scholar]
- Niehrs C (2006) Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25: 7469–7481 [DOI] [PubMed] [Google Scholar]
- Pabla N, Huang S, Mi QS, Daniel R, Dong Z (2008) ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem 283: 6572–6583 [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA (1998) The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723 [DOI] [PubMed] [Google Scholar]
- Rodier G, Montagnoli A, Di ML, Coulombe P, Draetta GF, Pagano M, Meloche S (2001) p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J 20: 6672–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Acebes S, Proctor I, Loddo M, Wollenschlaeger A, Falzon M, Prevost AT, Sainsbury R, Williams GH, Stoeber K (2010) Targeting DNA replication before it starts: Cdc7 as a therapeutic target in p53-mutant breast cancers. Am J Pathol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ (2002) Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene 21: 6624–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords R, Mahalingam D, O’Dwyer M, Santocanale C, Kelly K, Carew J, Giles F (2010) Cdc7 kinase—a new target for drug development. Eur J Cancer 46: 33–40 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC (2008) Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev 22: 1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H (1999) A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol Cell Biol 19: 5535–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenca P, Brotherton D, Montagnoli A, Rainoldi S, Albanese C, Santocanale C (2007) Cdc7 is an active kinase in human cancer cells undergoing replication stress. J Biol Chem 282: 208–215 [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D (1999) Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1: 20–26 [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Wharton SB, Hibberd S, Eward KL, Crimmins D, Jellinek DA, Levy D, Stoeber K, Williams GH (2004) DNA replication licensing and cell cycle kinetics of oligodendroglial tumours. Br J Cancer 91: 262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Harvey K, Kong L, Newport J (2002) Xic1 degradation in Xenopus egg extracts is coupled to initiation of DNA replication. Genes Dev 16: 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734 [DOI] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 21: 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.