Proteomic and functional analysis of the mitotic Drosophila centrosome
Combining mass spectrometric analyses, localization studies and RNAi loss-of-function screening, this study provides a comprehensive characterization of centrosomes from fly embryos, including identification of new proteins essential for centrosome maintenance and validation of their conserved function in human cells.
Keywords: cell cycle, centrosome, Drosophila, mitosis, proteomics
Abstract
Regulation of centrosome structure, duplication and segregation is integrated into cellular pathways that control cell cycle progression and growth. As part of these pathways, numerous proteins with well-established non-centrosomal localization and function associate with the centrosome to fulfill regulatory functions. In turn, classical centrosomal components take up functional and structural roles as part of other cellular organelles and compartments. Thus, although a comprehensive inventory of centrosome components is missing, emerging evidence indicates that its molecular composition reflects the complexity of its functions. We analysed the Drosophila embryonic centrosomal proteome using immunoisolation in combination with mass spectrometry. The 251 identified components were functionally characterized by RNA interference. Among those, a core group of 11 proteins was critical for centrosome structure maintenance. Depletion of any of these proteins in Drosophila SL2 cells resulted in centrosome disintegration, revealing a molecular dependency of centrosome structure on components of the protein translation machinery, actin- and RNA-binding proteins. In total, we assigned novel centrosome-related functions to 24 proteins and confirmed 13 of these in human cells.
Introduction
Detailed biochemical and functional information about the centrosome is critical for a better understanding of basic cellular organization, cell division, developmental processes and diseases resulting from loss or abnormal function of centrosomal proteins (Khodjakov and Rieder, 2001; Badano et al, 2005; Januschke and Gonzalez, 2008). However, an in-depth biochemical characterization of the eukaryotic microtubule-organizing centre has been hampered mainly by its low cellular abundance. Bioinformatic and proteomic studies have identified components of the yeast spindle pole body (Wigge et al, 1998) and of the Chlamydomonas basal body (Li et al, 2004; Keller et al, 2005), which is the structural and functional homologue of the centriole. In Drosophila, genetic approaches and genome-wide RNAi screening have identified a series of centrosomal proteins (Bettencourt-Dias and Glover, 2007; Goshima et al, 2007; Dobbelaere et al, 2008) but remained short of a comprehensive proteomic characterization of the centrosome. In higher eukaryotic cells, centrosome components have been identified and characterized through bulk isolation methods (Komesli et al, 1989; Moritz et al, 1995; Palazzo and Vogel, 1999; Lange et al, 2000) and by combining mass spectrometry (MS) with protein correlation profiling (Andersen et al, 2003).
Three classes of proteins are thought to be required for the maintenance of centrosome structure (Bornens, 2002; Lange, 2002). First, proteins of the centrosomal core structure, the centriole, as shown by depletion or inactivation of Ana1, Ana2, Asl, Bld10, Sas-4, Sas-6 and Spd-2 (Basto et al, 2006; Dix and Raff, 2007; Goshima et al, 2007; Varmark et al, 2007; Rodrigues-Martins et al, 2007b; Blachon et al, 2008; Mottier-Pavie and Megraw, 2009; Stevens et al, 2009, 2010). Second, certain proteins of the pericentriolar material (PCM), such as Cnn (Li and Kaufman, 1996), are essential for centrosome integrity (Megraw et al, 1999), potentially linking the centriole to other PCM components in Drosophila (Lucas and Raff, 2007). Proteins of the small γ-tubulin ring complex (γ-TuSC), namely Grip84, Grip91 and γ-tubulin (Oegema et al, 1999), form an integral part of the PCM structure (Colombie et al, 2006; Verollet et al, 2006) in addition to their role in microtubule nucleation. Third, factors that regulate PCM recruitment at the interphase-mitosis transition, such as the cell cycle kinases Polo, Cdc2/Cdc2c and AuroraA as well as the kinases SAK, Grp, Mei41 and the ubiquitin ligase complex SCF, which control centrosome duplication and segregation (Bettencourt-Dias and Glover, 2007). In contrast to the mechanisms mediating PCM increase, little is known about the regulation of PCM reduction that occurs during the mitosis to interphase transition, in differentiation (Tassin et al, 1985; Manandhar et al, 2000) and upon viral infection (Ploubidou et al, 2000; Jouvenet and Wileman, 2005; Ferralli et al, 2006).
The diverse functions of the centrosome, especially the regulatory ones, are also reflected by centrosomal components shared between the centrosome and other cell organelles/compartments (Kalt and Schliwa, 1993). A number of molecules previously described as components of the nucleus, the focal adhesion complexes or diverse membrane compartments have been subsequently localized at the centrosome and found to exert a centrosome-related function. In turn, several centrosomal proteins have been additionally localized at other cell organelles and have been shown to perform also non-centrosomal functions. Examples for the former are axin that is also found in the nucleus (Fumoto et al, 2009), which is implicated in centrosome segregation, β-catenin (Bahmanyar et al, 2008), an adherens junction/nuclear protein and component of the wnt signalling pathway that is involved in microtubule nucleation, HEF1 (Law et al, 1998; Pugacheva and Golemis, 2005), integrin-linked kinase (Fielding et al, 2008) and focal adhesion kinase (Park et al, 2009). In contrast, the centriolar protein centrin-2, which is required for centriolar duplication (Salisbury et al, 2002), has recently been identified as a component of the nuclear pore, where it is implicated in mRNA and protein export (Resendes et al, 2008). Furthermore, the γ-TuRC is recruited to unattached kinetochores by the nucleoporin Nup107–160 complex regulating microtubule nucleation at the kinetochore (Mishra et al, 2010). Taken together, the identification of novel centrosome-independent functions and non-centrosomal subcellular localizations of known centrosomal proteins point to a tight coordination of centrosome structure and function with basic cellular processes that control for example cell cycle regulation and cell growth (Sibon et al, 2000; Doxsey, 2001; Lange, 2002).
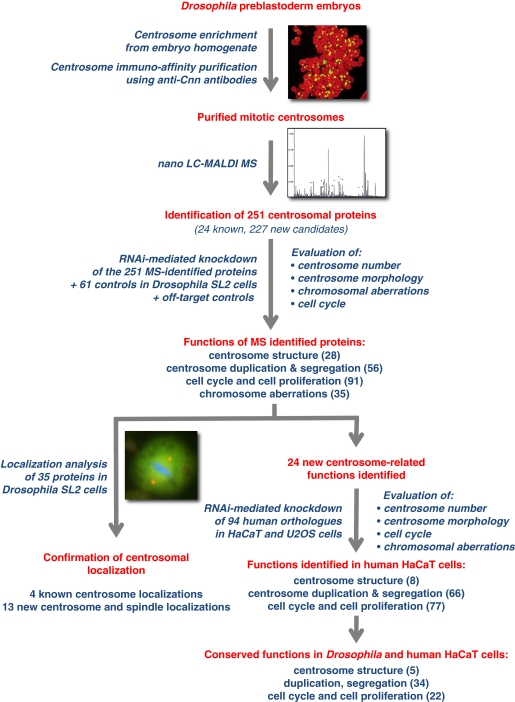
This study describes the identification of the proteome of the Drosophila mitotic centrosome and its functional characterization (Figure 1). Centrosome immunoisolation from Drosophila preblastoderm embryos was followed by MS identification of the organelle's protein components. Subsequently, RNAi in Drosophila SL2 cells was used in order to determine the function of the identified proteins in centrosome duplication/segregation and structure maintenance, chromosome segregation and cell cycle progression, by analysing 15 different phenotypic parameters. Within the group of proteins functioning in centrosome structure maintenance, factors that upon depletion resulted in a striking loss of PCM (‘0' centrosome phenotype) were of particular interest. As centrosome stability is frequently compromised in human cancer cells or upon viral infection, this group was characterized in more detail. In addition, a total of eight new centrosome and five new spindle localizations were independently confirmed, through tagging and antibody approaches. Moreover, functional characterization of the human orthologues in HaCaT cells identified five proteins with conserved function in centrosome structure maintenance. Most significantly, this study identified novel biochemical and functional links connecting proteins previously implicated in RNA binding, translational control and components of the actin cytoskeleton with the centrosome; thus revealing novel and remarkable regulators of centrosome structure maintenance.
Figure 1.
Experimental approach and main findings of the proteomic and functional characterization of the early preblastoderm Drosophila centrosome. Drosophila preblastoderm embryo extract was used as starting material for the immunoisolation of mitotic centrosomes, followed by the identification of the centrosomal proteome components by mass spectrometry. The 251 identified proteins plus 61 controls were characterized by RNAi-mediated knockdown in Drosophila SL2 cells. Fifteen centrosomal, chromosomal and cell cycle features were analysed using immunofluorescence microscopy or FACS. Subsequently, localization analysis was performed (GFP-, TAP-tag expression and immunolocalization in SL2 cells) for the MS-identified proteins whose functional inhibition resulted in a ‘0' centrosome phenotype, for proteins with coiled-coil domains and for control proteins. Functional conservation of the identified proteins was confirmed in human HaCaT and U2OS cells. (Main experimental steps are shown in red colour, experimental procedures and main findings are shown in blue).
Results and discussion
Identification of 251 centrosomal candidate proteins from immunoisolated Drosophila embryo centrosomes
One of the major drawbacks in the identification of the centrosome proteome has been the limited quantity and relatively low purity of centrosome preparations. Here, we used immunoisolation following sucrose gradient centrifugation (Lange et al, 2000; Lehmann et al, 2005) to improve the enrichment of centrosome proteins (Supplementary Figure S1). The resulting preparations were analysed by liquid chromatography–MS. MS analysis of the immunoisolated centrosomes identified 251 proteins, of which 24 have been localized to the Drosophila centrosome in previous studies. All in all we MS-identified 65% (24/37) of all components that were previously localized to the centrosome and 37% (35/96) of proteins that were previously implicated in centrosome-related processes (Supplementary Table S3; http://flybase.bio.indiana.edu/). The MS data including identified peptides are presented in Supplementary Table S1. The fact that we identified low abundance centriolar proteins (Spd-2, Sas-4), centrosomal core components (e.g. Cnn, γ-TuRC proteins) and transiently associated centrosomal mitotic kinases (e.g. Aur, Polo) confirmed the enrichment of our centrosome preparations. However, although our work is likely to cover a major part of the structural centrosome proteins, we cannot exclude that we missed a fraction of low abundant proteins such as the centriolar proteins Ana-1, Ana2 and Asl or proteins only transiently associated with the centrosome such as Cp190 (Oegema et al, 1995), proteins that were not MS identified by our approach.
We identified 17 proteins as contaminants (Supplementary Tables S1 and S3). Major contaminants, as identified in our mock isolation were the highly abundant yolk proteins (Yp1, Yp2 and Yp3), Act5C, betaTub56D and Ef1alpha48D (for a complete list see Supplementary Tables S1 and S3). The identification of the major centrosomal proteins γ-tubulin and Cnn in the negative control sample is likely to be a result of the control beads being exposed to highly concentrated centrosome-enriched sucrose fractions during the immunoisolation (Supplementary data).
Of the 251 candidate centrosome proteins identified here, 222 have known human orthologues according to the Ensembl database (Supplementary Table S3). Of these orthologues, 100 were also identified in the proteomic analysis of the human centrosome (Andersen et al, 2003) (Supplementary Table S2). Thus, the overlap of the proteins identified by MS analysis of centrosomal preparations in the two studies is ∼45%.
Functional characterization and classification of centrosomal candidate proteins
To test the function of the MS-identified proteins in centrosome structure maintenance, duplication and/or segregation and cell cycle control, we carried out an in-depth immunofluorescence microscopy analysis of SL2 cells depleted for all 251 candidates by dsRNA-mediated silencing (Boutros et al, 2004; Bartscherer et al, 2006). In addition, we analysed the phenotypes resulting from depletion of 61 control proteins selected from the UniProt database (http://www.uniprot.org/) through the search terms centrosome and Drosophila (Supplementary Table S3). Of these 61 proteins, 13 were previously localized to the centrosome according to the FlyBase database. The RNAi phenotypes of these controls served as a phenotypical reference list for our subsequent functional characterization (Supplementary Table S3). Off-target effects were evaluated both bioinformatically and through an additional repetition of RNAi experiments for functionally important proteins using alternative dsRNA sequences (Supplementary Table S4; Supplementary data). The high statistical cutoff levels implemented (significance level <0.0001) allowed the robust identification of molecules functioning in centrosome structure maintenance or centrosome duplication/segregation, albeit at the cost of potentially overlooking relatively weak phenotypes. For an overview of the different phenotypic classes and their assignment to the categories centrosome duplication/separation and/or centrosome structure see Supplementary data, section ‘Phenotypic scoring parameters'.
The advantage of our biochemical approach is demonstrated by the relatively high hit rate in comparison to genome-wide RNAi screens. Our hit rate was 9.6% considering the identification of 24 (out of 251 analysed) new proteins that upon RNAi-mediated depletion produced a centrosome structure or duplication/segregation phenotype. Using the flybase database release FB2007_1 (Dmel Release 5.2) for better comparability with previous screens mentioned below, we achieve a hit rate of 10.8%. In comparison, genome-wide screens in Drosophila identified 1.4% hits (205 relevant out of 14 425 analysed; Goshima et al, 2007) and 0.3% hits (32 relevant out of 13 059 analysed; Dobbelaere et al, 2008) important for mitotic spindle assembly and centrosome maturation, respectively. Using also a biochemical approach to identify microtubule-binding proteins, Hughes et al (2008) achieved a hit rate of 16% (13 relevant out of 83 analysed). However, the total number of new protein functions identified with a whole genome approach was significantly higher in the case of the Goshima screen (Goshima et al, 2007). Hence, these different types of approaches are complementary and all contribute significantly to the identification and functional characterization of centrosome and spindle-associated proteins. The overlap of different relevant genomic and biochemical studies with our study is listed in Supplementary Table S3.
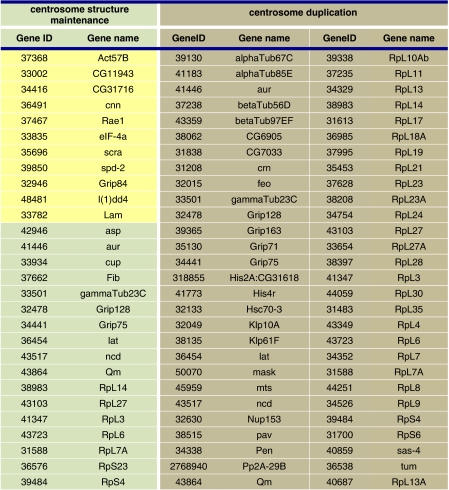
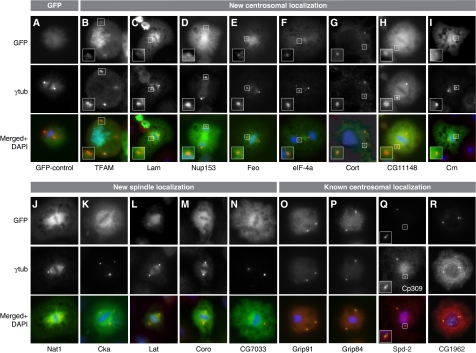
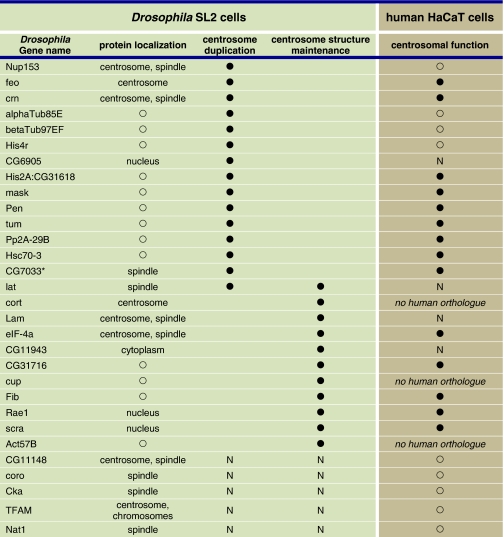
We selected (Table I; Supplementary Table S3) a core group of proteins for localization studies because their depletion resulted in a striking ablation of PCM (‘0' centrosome phenotype) in our RNAi assay and/or because they were annotated (http://www.ebi.ac.uk/interpro/) to possess multiple coiled-coil domains, a common feature of centrosome proteins. On the basis of these criteria, we carried out N- and C-terminal GFP and tandem affinity purification (TAP) tagging for 35 of the MS-identified proteins (Supplementary Tables S5 and S6). In addition, we generated primary antisera specific for four of the selected proteins and obtained additional sera from other groups (Supplementary data). All in all, 34 GFP- or TAP-fusion proteins (Supplementary Table S5) could be expressed in SL2 cells, of which 12 localized to the centrosome and 5 to the mitotic spindle (Figure 2; Supplementary Figures S2 and S3; Supplementary Table S5). Furthermore, we found microtubule, nuclear and cytoplasmic localization (Supplementary Table S5). Primary antisera specific for five of the selected proteins confirmed the localization of the endogenous proteins to the centrosome or the mitotic spindle, respectively. In total, we identified eight new centrosome and five new spindle localizations (Table II) in SL2 cells of Drosophila proteins initially identified from the syncytial blastoderm embryo. The presence of spindle-associated proteins in our preparations is not surprising as tethering of spindle and centrosome proteins by microtubule minus ends and molecular motors brings both spindle proteins to the centrosome and vice versa. For example, D-TACC is targeted to both centrosomes and microtubules through its C-terminal region (Gergely et al, 2000) and is required for centrosomal recruitment of Msps, a microtubule-associated protein that mediates stabilization of centrosomal microtubules (Lee et al, 2001). We may have failed to localize some of the MS-identified proteins to the SL2 centrosome (Supplementary Table S5) because these proteins might possess a lower affinity to the centrosome in cultured cells as compared with the highly mitotic syncytial blastoderm embryo from which we isolated the centrosomes for MS analysis.
Table 1. Functional classification of the RNAi phenotypes in SL2 cells after depletion of MS-identified proteins.
|
Figure 2.
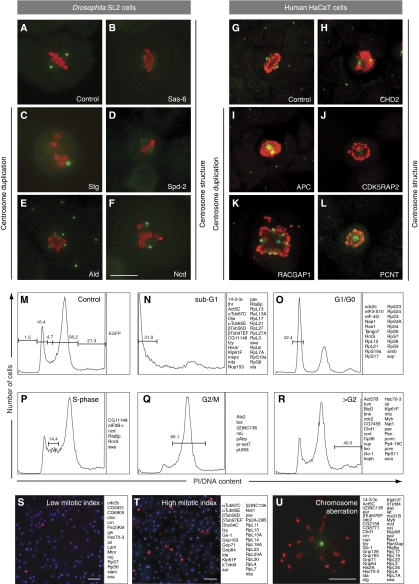
Confirmation of centrosomal or spindle localization of candidate MS-identified proteins and controls. Stable expression of GFP-fusion proteins in SL2 cells identifies new centrosomal and spindle localization of proteins whereas the GFP control shows uniform distribution (A). A centrosome associated localization was identified for TFAM (B), Lam (C), Nup153 (D) (transient expression), Feo (E), eIF-4a (F), Cort (G), CG11148 (H) and Crn (I). Not previously known was the spindle localization of the proteins Nat1 (J), Cka (K), Lat (L), Coro (M) and CG7033 (N). Positive controls confirm known centrosomal localization of Grip91 (O), Grip84 (P), Spd-2 (Q), CG1962 (R). The GFP-tag is shown in green (upper panels, A–R), antibody staining against γ-tubulin (middle panels, A–P, R) and Cp309 (Q) in red and superimposition of both images with DNA labelled by DAPI in blue (lower panels, A–R). The inserts in B–I and Q show a magnification of the area of the respective image marked with a white box.
Table 2. Localization and function of the new centrosomal and spindle proteins, in SL2 and HaCaT cells.
|
Structural aberrations of the centrosome involve three main classes of proteins: DNA/RNA-binding factors, translational control components and actin-interacting molecules
Although the acquisition of PCM (centrosome maturation) has been studied in detail (Palazzo et al, 2000; Dobbelaere et al, 2008), less is known about the reverse process, namely reduction of PCM during the mitosis to interphase transition. This process is likely to be inhibited in cancer cells, which harbour hypertrophic centrosomes (Lingle et al, 1998; Nigg, 2002). The inactivation of regulatory kinases, which generally induce PCM increase when activated, is not the only factor mediating PCM reduction. Additional postulated factors are posttranslational modifications of PCM components (e.g. ubiquitination, dephosphorylation) or the recruitment of interphase-specific centrosomal proteins (Hansen et al, 2002; Graser et al, 2007).
In order to identify centrosomal components functioning in PCM acquisition and structure maintenance (28/251) (Table I), we classified the effect of protein depletion into three categories of abnormal centrosome structure (Figure 3B, D and F): (i) zero centrosomes (11/251), (ii) small centrosomes (5/251) and (iii) fuzzy centrosomes (8/251) (Supplementary Table S3). In addition, for four proteins, we detected a mixed phenotype upon depletion that included at least one centrosome structural phenotype (Supplementary Table S3). The most striking structural phenotype was PCM ablation (‘0' centrosome phenotype) that resulted from depletion of Act57B, eIF-4a, CG11943, CG31716, Lam, Rae1 and Scra, none of which have previously been reported to be centrosome-related proteins. Very similar phenotypes were observed upon depletion of several known centrosomal components: Spd-2, a regulator of PCM recruitment (Dix and Raff, 2007; Giansanti et al, 2008), the major core PCM components Cnn (Li and Kaufman, 1996; Megraw et al, 1999), Grip84 (Oegema et al, 1999; Colombie et al, 2006) and l(1)dd4 (Barbosa et al, 2000) (Table I; Supplementary Table S3).
Figure 3.
Functional characterization of 251 MS-identified Drosophila centrosome candidate proteins plus 61 controls and 94 human orthologues identified centrosomal and cell cycle functions. (A–L) Examples of the two phenotypic classes, aberrant centrosome structure (B, D, F, H, J, L) or centrosome duplication/segregation (C, E, I, K) revealed by RNAi-mediated knockdown in SL2 and HaCaT cells. The RNAi target protein is indicated within each panel. Anti-γ-tubulin (green) and anti-phospho-histone 3 (red) antibodies were used to label centrosomes and mitotic chromosomes, respectively. (M–R) Examples of the cell cycle distribution profiles, determined by FACS analysis of dsRNA-treated SL2 cells. The RNAi target proteins whose depletion is inducing each phenotype are listed on the right of the corresponding cell cycle distribution profile. (M) Control (EGFP dsRNA-treated cells) cell cycle distribution, (N) Sub-G1, (O) G1/G0, (P) S-phase, (Q) G2/M, (R) more than G2 DNA content. (S, T) Representative fields of SL2 cells displaying low (S) or high (T) mitotic index following dsRNA treatment. The RNAi target proteins whose depletion is inducing each phenotype are listed on the right of the corresponding image. DAPI (blue) and anti-phospho-histone 3 antibodies (red) were used to label DNA and mitotic chromosomes, respectively. (U) Example of a cell showing an abnormal chromosome segregation phenotype. The RNAi target proteins, whose depletion results in an aberrant chromosome segregation phenotype are listed on the right of the image. Anti-γ-tubulin (green) and anti-phospho-histone 3 (red) antibodies were used to label centrosome and mitotic chromosomes, respectively. Scale bars represent 10 μm in (F, U), and 20 μm in (S, T). A complete list of all Drosophila and human proteins and the result of their functional analysis can be found in Supplementary Table S3.
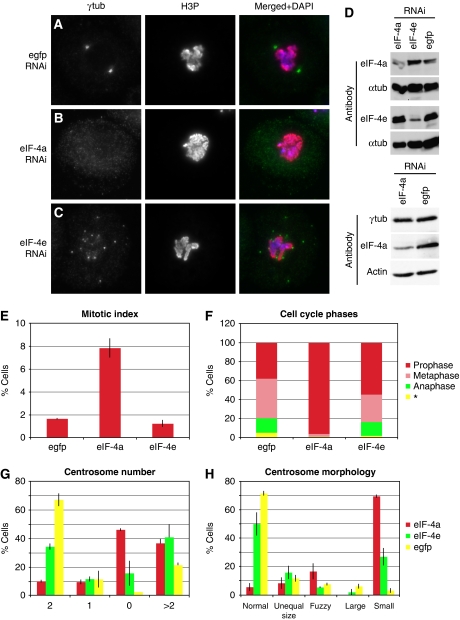
Interestingly, three proteins shown here to function in maintaining centrosome structure have previously been suggested to be implicated in RNA binding and initiation of protein translation: Rae1 (Sitterlin, 2004) CG31716 and eIF-4a (Lasko, 2000; Palacios et al, 2004). We investigated in more detail the consequence of eIF-4a knockdown on PCM and centrioles. Depletion of eIF-4a resulted in reduction of centrosomal γ-tubulin, Cp309 (Kawaguchi and Zheng, 2004; Martinez-Campos et al, 2004) and Spd-2 (Dix and Raff, 2007), but not of Asl (Varmark et al, 2007; Blachon et al, 2008), Bld10 (Blachon et al, 2009), Ana1, Ana2 (Goshima et al, 2007; Stevens et al, 2010), Sas-4 or Sas-6 (Rodrigues-Martins et al, 2007a) in our experiments (Figure 4; Supplementary Figure S4). Therefore, eIF-4a depletion results in removal of PCM components but not of core centriolar proteins, suggesting that eIF-4a is required for PCM cohesion and might be involved in the recruitment of PCM to the centriole. To rule out the possibility that the observed PCM reduction is a secondary effect caused by global inhibition of protein translation, we inhibited translation in two experiments: first, by knockdown of eIF-4e, which is the core component of the translation initiation complex eIF-4F and mediates mRNA cap binding, the first step of translation initiation (Gingras et al, 1999). eIF-4e was not detected in our centrosome preparations. Second, by treatment with sublethal concentrations of cycloheximide, an inhibitor of translation elongation. Although knockdown of eIF-4a produced a ‘0' centrosome phenotype in 46% of cells (Figure 4A–C and G), RNAi-mediated depletion of eIF-4e (confirmed by western blotting; Figure 4D) resulted in elevated centrosome number (>2) in a large proportion of cells (40%) when compared with the control (21%). Cycloheximide-mediated inhibition of protein translation (Supplementary Figure S5) resulted in elevated centrosome numbers, a phenotype similar to eIF-4e depletion but distinctly different to the eIF-4a phenotype.
Figure 4.
The eukaryotic initiation factor 4a has a centrosome and cell cycle related function. As compared with the control (A), depletion of eIF-4a results in SL2 cells with small or no centrosomes as judged by staining with an γ-tubulin antibody (B, G, H), high mitotic index (E) and an accumulation of prophase cells (F), whereas eIF-4e knockdown led to cells with many centrosomes (C, G), normal mitotic index (E) and normal distribution of mitotic phases (F). Western blotting shows protein depletion by RNAi using anti-eIF-4a and eIF-4e antibodies and a stable protein level of γ-tubulin following eIF-4a knockdown. α-Tubulin and actin are used as loading controls (D). The distinct differences between the eIF-4a and eIF-4e RNAi phenotypes strongly suggest that the effect on the centrosome resulting from depletion of eIF-4a is most likely not a consequence of global inhibition of translation. The γ-tubulin reduction at the centrosome concomitant with an unaltered overall protein level in the cell indicates a mislocalization of γ-tubulin rather than disturbed translation (F, *could not be determined).
These data support the hypothesis that the centrosome phenotype caused by eIF-4a depletion is not due to the general inhibition of translation. Consistent with this, western blot analysis of total cell lysates following eIF-4a RNAi, a treatment that abolishes centrosomal localization of γ-tubulin (Figure 4B), detected no change of the total level of γ-tubulin (Figure 4D). In addition, detailed cell cycle analysis of eIF-4a-depleted cells revealed a massive accumulation in prophase (Figure 4F). This phenotype was much less pronounced in eIF-4e-depleted cells. These additional phenotypical differences are consistent with the notion that PCM loss by eIF-4a depletion is mechanistically distinct from the global inhibition of protein translation, indicating a regulatory or structural role for this helicase at the centrosome.
eIF-4e has been previously localized to the centrosome and was identified as microtubule-binding protein (Hughes et al, 2008). The fact that we identified eIF-4a, but not eIF-4e in the embryo centrosome preparations could be due to different affinities to the centrosome. Interestingly, both eIF-4e (Wilhelm et al, 2003) and eIF-4a (Palacios et al, 2004) were previously shown to be part of protein complexes implicated in microtubule-dependent mRNA localization and translocation in the Drosophila oocyte. Finally, components of the eIF-3 translation initiation complex are involved in spindle assembly, as their depletion leads to short and monopolar spindles (Somma et al, 2008). Taken together, these results support a centrosome-related function of RNA-binding proteins that cannot be explained by their role in global mRNA translation alone. It is likely that both regulation of centrosome structure by RNA-binding proteins and local mRNA translation at the centrosome/spindle are required for proper mitotic function (Liska et al, 2004; Blower et al, 2005). However, cap-dependent mRNA translation has been reported to be inhibited during mitosis in higher eukaryotes (Scharff and Robbins, 1966). In contrast, several proteins that are required during mitosis are translated by a mechanism that involves internal ribosome entry sites (IRES) rendering translation independent of 5′cap (Qin and Sarnow, 2004). Hence, a specific molecular impairment of this translational switch from cap-dependent translation to IRES-dependent translation that regulates the expression of selected mRNAs in mitosis (Barna et al, 2008) or the requirement of only parts of the initiation complexes (Pestova et al, 1996) would be alternative explanations for the phenotypes observed. Hence, one possible model that could explain our observations concerning the eIF-4a knockdown phenotype would involve a mechanism allowing regulated translation of specific mRNAs at the centrosome/spindle.
The second group of proteins, depletion of which results in a ‘0' centrosome phenotype, includes Act57B and Scra, which were previously described to be involved in cytokinesis (Thomas and Wieschaus, 2004). This result suggests that actin-related processes are not only required for centrosome separation in interphase (Stevenson et al, 2001) or clustering in mitosis (Kwon et al, 2008) but also regulate centrosome structure in SL2 cells. Other actin-related proteins identified in this work, for example CG1962 (Centrocortin) (Kao and Megraw, 2009) and Coro (Bharathi et al, 2004) had no effect on centrosome structure upon RNAi-mediated depletion, although localization to centrosome and spindle was demonstrated for CG1962 (Figure 2R; Supplementary Figures S2C and S3E) and Coro (Figure 2M; Supplementary Figure S2O).
Reduction of PCM (small centrosome phenotype) was the consequence of RNAi-mediated silencing of Cup, Fib and RpS4 (Supplementary Table S3). In addition, depletion of the ribosomal proteins Qm, RpL27, RpL3, RpL6, RpL7A, RpS23 led to fuzzy centrosome appearance. Some of these ribosomal proteins (Supplementary Table S3) have been described to regulate microtubule dynamics indirectly affecting spindle elongation (Goshima et al, 2007). We propose an additional function in the maintenance of centrosome structure for these proteins but cannot exclude an indirect effect as a centrosome localization of these proteins could not be confirmed.
Centrosome duplication and/or segregation function
Centrosome duplication and segregation depend on a conglomerate of different molecules including regulatory kinases, microtubule minus- and plus end-directed molecular motor proteins, the E3 ubiquitin ligase system, centrosomal linker proteins and a series of PCM components (Hinchcliffe and Sluder, 2001; Lim et al, 2009). This is reflected both in the composition of our centrosome preparation that contained proteins related to each of the classes mentioned above and in results of the subsequent functional analysis (Table I; Supplementary Table S3). RNAi-mediated depletion of 56 MS-identified proteins resulted in single and/or abnormally large and/or >3 centrosomes, indicating malfunction of centrosome duplication and/or segregation (Table I; Supplementary Table S3). The group, depletion of which resulted in a single-large centrosome, was the largest (24), comprising proteins known to affect centrosome duplication and segregation: α-/β-tubulins, γ-TuRC (Colombie et al, 2006; Verollet et al, 2006), Tum (Zavortink et al, 2005), Mts (Snaith et al, 1996) and the motor proteins Klp10A, Klp61F, Ncd (Endow et al, 1994; Barton et al, 1995; Goshima et al, 2007). Unexpectedly, knockdown of Hsc70-3 phenocopies five different phenotypic parameters including centrosome and chromosome segregation plus cytokinesis phenotypes of the two molecular motor proteins Pav (Adams et al, 1998) and Klp61F (Wilson et al, 1997), indicating that these three proteins might participate in the same pathway. Recently, a combined role for Hsc70 and Kinesin-1 in the control of axonal transport was demonstrated in mice (Terada et al, 2010). Together with our results, this suggests a critical function of Hsc70 in the regulation of disease relevant motor-dependent transport processes (Gunawardena et al, 2003) that include centrosome and chromosome segregation.
We identified a function in centrosome duplication and segregation for a group of proteins (CG6905, CG7033, Crn, ribosomal proteins) previously implicated in transcription, translation, RNA processing and chaperoning based on sequence similarities or biochemical studies (Mount and Salz, 2000; Raisin-Tani and Leopold, 2002; Monzo et al, 2010) (Table I; Supplementary Table S3). Of these, we localized CG7033 and Crn to the spindle, midbody and centrosome (Figure 2; Supplementary Figures S2 and S3; Table II). CG7033 has a putative RNA helicase domain and a role in mitotic spindle organization (Goshima et al, 2007; Hughes et al, 2008). In our experiments, CG7033 was MS identified as component of the centrosome preparations. The localization of Crn and CG7033 at centrosome and spindle, respectively, is consistent with either a regulatory or structural role in the process of centrosome duplication and/or segregation. On the basis of sequence homology, CG7033 has been suggested to be part of the TCP (Hughes et al, 2008; Monzo et al, 2010), a chaperonin complex for actin and tubulins (Liang and MacRae, 1997). A centrosome duplication/segregation phenotype (single centrosome) was furthermore observed in cells depleted for proteins of the large ribosome subunit (Supplementary Table S3). The identification of factors implicated in processes related to protein translation, as observed in our experiments, could argue for an indirect effect due to loss of protein expression. In contrast, our control experiments showed that global inhibition of protein translation through cycloheximide (Supplementary Figure S5) results in overreplication of centrosomes. Taken together, these results suggest that the identified proteins implicated in RNA processing and translation are linked to a pathway required for centrosome duplication or segregation (see also section on protein translation above).
Links of the centrosome to cell cycle progression, proliferation and cell viability
We characterized the effect of all MS-identified proteins (251) and all control proteins (61) on cell cycle progression to correlate the detected centrosome phenotypes with cell cycle regulation and to elucidate possible links of centrosomal proteins to cell proliferation pathways (Supplementary Table S3; Figure 3M–T). DNA-content and mitotic index analysis by FACS and phospho-histone H3 labelling, respectively, revealed three major phenotypic groups (Figure 3M–T). These were characterized by enrichment of cells with either (i) sub-G1-phase DNA content, indicating decreased viability (27/251); (ii) higher than G2 DNA content (19/251), suggesting cytokinesis defects and (iii) an increased number of phospho-histone H3-positive cells, indicating mitotic arrest (27/251). Subsequently, we correlated centrosome aberration phenotypes with cell cycle deregulation phenotypes (Supplementary Table S3): of proteins previously described to be centrosome-related control proteins (Supplementary Table S3) functioning in centrosome segregation, we found AlphaTub67C, Fzy, Klp61F, Mts, Pav, Thr to also affect cell viability. In addition, depletion of CG11148 (a protein we localized to the centrosome; Figure 2H; Supplementary Figures S2F and S3B), Nup153 (Figure 2D; Supplementary Figure S2J), His4R and Ote resulted in an increased number of sub-G1 cells. Interestingly, most of the cell viability affecting knockdowns resulted in a ‘1' centrosome phenotype, suggesting that the processes of centrosome duplication/segregation and cell survival are interdependent in the majority of cases examined here.
Expected cytokinesis defects (higher than G2 DNA content) were observed after knockdown of Pav (Adams et al, 1998), Scra and Tum (Somma et al, 2008). A strong cytokinesis defect was induced by depletion of the Heph protein, which contains a RNA recognition motif and is involved in Notch signalling (Dansereau et al, 2002), suggesting an unexpected function of Heph in the cell division pathway.
Mitotic arrest was observed after RNAi of 27 centrosomal candidate proteins (Supplementary Table S3). The majority of cells arrested in mitosis had a single centrosome, suggesting that depletion of these proteins (19) blocked both centrosome duplication/segregation and mitotic progression. We confirmed that depletion of the γ-TuRC, a minor subgroup (4/27) of this phenotypic class, leads to mitotic arrest (Müller et al, 2006; Verollet et al, 2006).
Highest functional conservation between fly and human is observed for proteins functioning in centrosome duplication and segregation
The total number of centrosome proteins remains open. Andersen et al (2003) identified 114 centrosome/centrosome candidate proteins in human interphase centrosome preparations. The centrosomeDB database (Nogales-Cadenas et al, 2009) lists 383 centrosome-related human genes based on a compilation from the literature and homology to centrosome genes identified in various organisms. Indeed, taking different proteomic, bioinformatics and genetic studies into account, an estimate of over 300 centrosome candidate proteins has been proposed (Bettencourt-Dias and Glover, 2007). However, there are uncertainties attached to these numbers due to the fact that many proteins are only transiently associated with the centrosome (Kalt and Schliwa, 1993). In addition, the origin of the centrosome (isolated, e.g., from established cell lines largely in interphase or from highly mitotic embryonic tissue) is likely to contribute to major differences in the types and number of proteins identified.
We tested all human orthologues of MS-identified proteins that yielded a centrosomal and/or chromosome aberration phenotype in the SL2 RNAi assay for functional conservation, by short interfering RNA (siRNA)-mediated silencing in human cells (Supplementary Table S3). We analysed the knockdown effect on centrosome and cell cycle for 71 of these proteins. In addition, we included 23 controls known to localize to the centrosome and/or to fulfill centrosome-related functions, of which 12 resulted in a centrosomal phenotype after knockdown in SL2 cells. We confirmed a conserved centrosome-related function for 42 proteins (Supplementary Table S3).
The largest functional conservation occurs in the class of centrosome duplication and segregation (34), whereas fewer proteins (5) had a conserved function in maintaining centrosome structure (Supplementary Table S3). In the group of proteins, depletion of which affects centrosome duplication/segregation are the two γ-TuRC components 76P (Grip75) and TUBGCP5 (Grip128), molecular motors KIF11 (Klp61F) and KIF2 (Klp10A) together with HSPA5 (Hsc70-3), the regulatory proteins CDC20 (Fzy), CDC25C (Stg), PPP2CA (Mts), PPP2R1A (Pp2A-29B) and several ribosomal proteins. The proteins CDK5RAP2 (Cnn), TUBG1 (γ-tubulin), 76P (Grip75), TUBGCP2 (Grip84) and TUBGCP3 (l(1)dd4) were found to be required for centrosome structure maintenance in both SL2 and HaCaT cells.
Analysis of 10 selected human orthologues in U2OS cells confirmed a function in the maintenance of centrosome structure for NUP205 (CG11943), CEP192 (Spd-2) and CNOT4 (CG31716) (Supplementary Figure S6; Supplementary Table S3). Overall, the conservation in the class of proteins that function in centrosome structure maintenance was about 29% (18% in HaCaT cells). The functional conservation between HaCaT und SL2 was highest (∼72%) in the class of proteins relevant for centrosome duplication and/or segregation. These data are consistent with previously published results from RNAi screens comparing the osteosarcoma cell line U2OS and the cervix cancer-derived HeLa cells with Drosophila cell cultures (Kittler et al, 2007). The previously published overlap between human and Drosophila RNAi screens was 38% (Kittler et al, 2007). The lower level of phenotypic overlap between SL2 and human cells in the category centrosome structure maintenance could alternatively be explained by a high level of redundancy within this functional group of proteins in human cells.
Our MS analysis of immunopurified centrosomes identified 251 proteins of which 222 had human orthologues annotated in the Ensembl database. All in all, 100 of these orthologues (45%) (Supplementary Table S2) were previously identified in preparations of human centrosomes (Andersen et al, 2003). This overlap of the two data sets includes 20 known centrosomal proteins, 3 centrosome candidates and 1 novel centrosomal protein as classified by Andersen et al (2003). Given the overall diversity between the two organisms, this relative large overlap confirms the validity of our MS analysis.
In summary, the functional characterization of the Drosophila embryo centrosome proteome assigned a novel function to 24 proteins, required for maintaining centrosome structure and for centrosome duplication and segregation. We identified 11 proteins that were assigned a previously not described function in maintaining centrosome structure (Table II). Depletion of seven of these proteins resulted in PCM ablation. Interspecies comparison revealed that mainly proteins involved in the processes of centrosome duplication and segregation are functionally conserved. Through the proteomic and functional characterization of the early Drosophila embryo centrosome, this work provides a resource for further molecular characterization of the mechanisms mediating centrosome duplication/segregation and centrosome structure maintenance as well as the implication of the centrosome in signalling pathways, cellular processes and the development of diseases.
Materials and methods
Further details of all experimental procedures can be found in Supplementary data.
Centrosome isolation
Embryo homogenate was prepared from Drosophila preblastoderm stage embryos and centrosomes were enriched through sucrose gradients centrifugation according to Moritz et al (1995). Subsequent immunoisolation of centrosomes was performed as described previously (Lehmann et al, 2005), with modifications detailed in Supplementary data.
Nano LC-MALDI MS
Nano LC-MALDI MS was performed according to Mirgorodskaya et al (2005). In brief, peptides were separated on an 1100 Series Nanoflow LC system (Agilent Technologies). Mass analysis of positively charged peptide ions was performed on an Ultraflex II LIFT MALDI-TOF/TOF mass spectrometer (Bruker Daltonics). Protein identification was performed using the Mascot software (Matrixscience), searching the FlyBase sequence database.
RNA interference and phenotype analysis
RNAi knockdown in Drosophila SL2 and siRNA knockdown in human HaCaT or U2OS cells were each performed in two independent experiments, followed by immunofluorescence labelling of the cells or processing for FACS analysis. For SL2 and U2OS cells, in each experiment, on average, n=100 mitotic cells were analysed for centrosome number and shape, n=2000 SL2 cells were analysed for mitotic index calculation and n=35 000 SL2 cells were subjected to DNA-content analysis by FACS. For HaCaT cells, in each experiment, on average, n=550 mitotic cells were analysed for centrosome number and area plus centrosomal γ-tubulin content, whereas n=29 000 cells were subjected to mitotic index and DNA-content analysis. The values measured were normalized to the corresponding average value of the quadruplicate negative control wells on the plate. Phenotypes were quantified using three different software algorithms.
Data evaluation of RNA interference experiments
Each mitotic cell was assigned to phenotypic categories of centrosome number and morphology as shown in Figure 3 and specified in Supplementary data. The resulting phenotype distributions of the two independent experiments were averaged and compared with the average distribution of the negative controls, by means of a non-parametric two-tailed χ2 test. A significant deviation from the control distribution was assigned for significance levels P<0.0001 (list of P-values in Supplementary Table S4). For the knockdowns thus determined to cause significant effects on centrosome number, the phenotype was identified as the category that showed more than two-fold increase compared with the negative control. If this threshold was exceeded for two or more categories, a mixed phenotype was assigned, unless one of these categories was more than two-fold the abundance of the second highest.
For all other data analysis, the values measured were normalized to the corresponding average value of the quadruplicate negative control wells on the plate. Phenotypes were considered to be statistically significant when a z-score ⩾3 was obtained in both independent experiments and, for mitotic index and DNA-content analysis in HaCaT, when, in addition, the average z-score was ⩾6. The values (z-scores) listed in Supplementary Table S4 represent the average distances between individual knockdowns and control, determined as described above, in fold s.d. of the negative controls.
Supplementary Material
Acknowledgments
We thank CB Chien, M Gatti, D Glover, J Gopalakrishnan, G Gonzalez, G Goshima, T Kaufman, K Kwan, T Megraw, T Orr-Weaver, T Avidor-Reiss, N Sonenberg and Y Zheng for the gift of antibodies and DNA constructs; K Nierhaus for advice on protein translation control experiments; J Hamann and D Schudde for assistance with HT methods and our colleagues for comments on the manuscript. The work was funded by (i) BL laboratory: Berliner Senat für Kultur, Wissenschaft und Forschung, EFRE; NGFN2 SMP Protein; NGFN Plus, IG Mutanom; EU. (ii) MB laboratory: DFG, European Commission; HFSP. (iii) JG laboratory: Technologiestiftung Berlin (TSB); the Structural Fonds of the European Union within the project 2D/3D-ProteinChips, NGFN2 SMP Protein. Bruker Daltonics in Bremen is acknowledged for scientific collaboration. (iv) AP laboratory: Leibniz Association, Joint Initiative for Research and Innovation.
Author contributions: EM, NG and JG carried out the MS work, EM, JG and FD the MS data analysis. HM, SS, VL, KH, DS, TK and BL performed all other experiments. HM, SS, DS, AP and BL analysed the data. HL, RH, AP, MB and BL supervised the experiments. BL designed the experiments and wrote the paper. All authors discussed the data and commented and contributed to the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM (1998) Pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev 12: 1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Badano JL, Teslovich TM, Katsanis N (2005) The centrosome in human genetic disease. Nat Rev 6: 194–205 [DOI] [PubMed] [Google Scholar]
- Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH Jr, O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI (2008) beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev 22: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa V, Yamamoto RR, Henderson DS, Glover DM (2000) Mutation of a Drosophila gamma tubulin ring complex subunit encoded by discs degenerate-4 differentially disrupts centrosomal protein localization. Genes Dev 14: 3126–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D (2008) Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NR, Pereira AJ, Goldstein LS (1995) Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol Biol Cell 6: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M (2006) Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125: 523–533 [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW (2006) Flies without centrioles. Cell 125: 1375–1386 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM (2007) Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev 8: 451–463 [DOI] [PubMed] [Google Scholar]
- Bharathi V, Pallavi SK, Bajpai R, Emerald BS, Shashidhara LS (2004) Genetic characterization of the Drosophila homologue of coronin. J Cell Sci 117: 1911–1922 [DOI] [PubMed] [Google Scholar]
- Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A, Avidor-Reiss T (2009) A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics 182: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T (2008) Drosophila asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics 180: 2081–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K (2005) A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121: 223–234 [DOI] [PubMed] [Google Scholar]
- Bornens M (2002) Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14: 25–34 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Colombie N, Verollet C, Sampaio P, Moisand A, Sunkel C, Bourbon HM, Wright M, Raynaud-Messina B (2006) The Drosophila gamma-tubulin small complex subunit Dgrip84 is required for structural and functional integrity of the spindle apparatus. Mol Biol Cell 17: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansereau DA, Lunke MD, Finkielsztein A, Russell MA, Brook WJ (2002) Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development 129: 5553–5566 [DOI] [PubMed] [Google Scholar]
- Dix CI, Raff JW (2007) Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol 17: 1759–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J (2008) A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S (2001) Re-evaluating centrosome function. Nat Rev 2: 688–698 [DOI] [PubMed] [Google Scholar]
- Endow SA, Chandra R, Komma DJ, Yamamoto AH, Salmon ED (1994) Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J Cell Sci 107(Pt 4): 859–867 [DOI] [PubMed] [Google Scholar]
- Ferralli J, Ashby J, Fasler M, Boyko V, Heinlein M (2006) Disruption of microtubule organization and centrosome function by expression of tobacco mosaic virus movement protein. J Virol 80: 5807–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AB, Dobreva I, McDonald PC, Foster LJ, Dedhar S (2008) Integrin-linked kinase localizes to the centrosome and regulates mitotic spindle organization. J Cell Biol 180: 681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumoto K, Kadono M, Izumi N, Kikuchi A (2009) Axin localizes to the centrosome and is involved in microtubule nucleation. EMBO Rep 10: 606–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW (2000) D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J 19: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M (2008) Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol 18: 303–309 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 13: 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N (2007) Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Nigg EA (2007) Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci 120: 4321–4331 [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS (2003) Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron 40: 25–40 [DOI] [PubMed] [Google Scholar]
- Hansen DV, Hsu JY, Kaiser BK, Jackson PK, Eldridge AG (2002) Control of the centriole and centrosome cycles by ubiquitination enzymes. Oncogene 21: 6209–6221 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G (2001) ‘It takes two to tango': understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev 15: 1167–1181 [DOI] [PubMed] [Google Scholar]
- Hughes JR, Meireles AM, Fisher KH, Garcia A, Antrobus PR, Wainman A, Zitzmann N, Deane C, Ohkura H, Wakefield JG (2008) A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol 6: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gonzalez C (2008) Drosophila asymmetric division, polarity and cancer. Oncogene 27: 6994–7002 [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Wileman T (2005) African swine fever virus infection disrupts centrosome assembly and function. J Gen Virol 86: 589–594 [DOI] [PubMed] [Google Scholar]
- Kalt A, Schliwa M (1993) Molecular components of the centrosome. Trends Cell Biol 3: 118–128 [DOI] [PubMed] [Google Scholar]
- Kao LR, Megraw TL (2009) Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr Biol 19: 937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S, Zheng Y (2004) Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol Biol Cell 15: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR III, Marshall WF (2005) Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol 15: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL (2001) Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol 153: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, Wagner J, Surendranath V, Richter C, Bowen W, Jackson AL, Habermann B, Hyman AA, Buchholz F (2007) Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol 9: 1401–1412 [DOI] [PubMed] [Google Scholar]
- Komesli S, Tournier F, Paintrand M, Margolis RL, Job D, Bornens M (1989) Mass isolation of calf thymus centrosomes: identification of a specific configuration. J Cell Biol 109: 2869–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D (2008) Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22: 2189–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BMH (2002) Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr Opin Cell Biol 14: 35–43 [DOI] [PubMed] [Google Scholar]
- Lange BMH, Bachi A, Wilm M, Gonzalez C (2000) Hsp90 is a core centrosomal component and is required at different stages of the centrosome cycle in Drosophila and vertebrates. EMBO J 19: 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P (2000) The drosophila melanogaster genome: translation factors and RNA binding proteins. J Cell Biol 150: F51–F56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SF, Zhang YZ, Klein-Szanto AJ, Golemis EA (1998) Cell cycle-regulated processing of HEF1 to multiple protein forms differentially targeted to multiple subcellular compartments. Mol Cell Biol 18: 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW (2001) Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol 3: 643–649 [DOI] [PubMed] [Google Scholar]
- Lehmann V, Müller H, Lange BMH (2005) Immunoisolation of centrosomes from Drosophila melanogaster. Curr Protoc Cell Biol 29: 3.17.11–13.17.13 [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N et al. (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552 [DOI] [PubMed] [Google Scholar]
- Li K, Kaufman TC (1996) The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 85: 585–596 [DOI] [PubMed] [Google Scholar]
- Liang P, MacRae TH (1997) Molecular chaperones and the cytoskeleton. J Cell Sci 110: 1431–1440 [DOI] [PubMed] [Google Scholar]
- Lim HH, Zhang T, Surana U (2009) Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol 19: 325–333 [DOI] [PubMed] [Google Scholar]
- Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL (1998) Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci USA 95: 2950–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liska AJ, Popov AV, Sunyaev S, Coughlin P, Habermann B, Shevchenko A, Bork P, Karsenti E (2004) Homology-based functional proteomics by mass spectrometry: application to the Xenopus microtubule-associated proteome. Proteomics 4: 2707–2721 [DOI] [PubMed] [Google Scholar]
- Lucas EP, Raff JW (2007) Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol 178: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar G, Simerly C, Schatten G (2000) Centrosome reduction during mammalian spermiogenesis. Curr Top Dev Biol 49: 343–363 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW (2004) The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC (1999) The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126: 2829–2839 [DOI] [PubMed] [Google Scholar]
- Mirgorodskaya E, Braeuer C, Fucini P, Lehrach H, Gobom J (2005) Nanoflow liquid chromatography coupled to matrix-assisted laser desorption/ionization mass spectrometry: sample preparation, data analysis, and application to the analysis of complex peptide mixtures. Proteomics 5: 399–408 [DOI] [PubMed] [Google Scholar]
- Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M (2010) The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 12: 164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzo K, Dowd SR, Minden JS, Sisson JC (2010) Proteomic analysis reveals CCT is a target of Fragile X mental retardation protein regulation in Drosophila. Dev Biol 340: 408–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA (1995) Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol 130: 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier-Pavie V, Megraw TL (2009) Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell 20: 2605–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Salz HK (2000) Pre-messenger RNA processing factors in the Drosophila genome. J Cell Biol 150: F37–F44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H, Fogeron ML, Lehmann V, Lehrach H, Lange BMH (2006) A centrosome-independent role for gamma-TuRC proteins in the spindle assembly checkpoint. Science 314: 654–657 [DOI] [PubMed] [Google Scholar]
- Nigg EA (2002) Centrosome aberrations: cause or consequence of cancer progression? Nat Rev 2: 815–825 [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Abascal F, Diez-Perez J, Carazo JM, Pascual-Montano A (2009) CentrosomeDB: a human centrosomal proteins database. Nucleic Acids Res 37: D175–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Whitfield WG, Alberts B (1995) The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol 131: 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y (1999) Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol 144: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E (2004) An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757 [DOI] [PubMed] [Google Scholar]
- Palazzo RE, Vogel JM (1999) Isolation of centrosomes from Spisula solidissima oocytes. Methods Cell Biol 61: 35–56 [DOI] [PubMed] [Google Scholar]
- Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X (2000) Centrosome maturation. Curr Top Dev Biol 49: 449–470 [DOI] [PubMed] [Google Scholar]
- Park AY, Shen TL, Chien S, Guan JL (2009) Role of focal adhesion kinase Ser-732 phosphorylation in centrosome function during mitosis. J Biol Chem 284: 9418–9425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CU (1996) Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 16: 6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M (2000) Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J 19: 3932–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Golemis EA (2005) The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol 7: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Sarnow P (2004) Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem 279: 13721–13728 [DOI] [PubMed] [Google Scholar]
- Raisin-Tani S, Leopold P (2002) Drosophila crooked-neck protein co-fractionates in a multiprotein complex with splicing factors. Biochem Biophys Res Commun 296: 288–292 [DOI] [PubMed] [Google Scholar]
- Resendes KK, Rasala BA, Forbes DJ (2008) Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol 28: 1755–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM (2007a) DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol 17: 1465–1472 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M (2007b) Revisiting the role of the mother centriole in centriole biogenesis. Science 316: 1046–1050 [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Suino KM, Busby R, Springett M (2002) Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol 12: 1287–1292 [DOI] [PubMed] [Google Scholar]
- Scharff MD, Robbins E (1966) Polyribosome disaggregation during metaphase. Science 151: 992–995 [DOI] [PubMed] [Google Scholar]
- Sibon OC, Kelkar A, Lemstra W, Theurkauf WE (2000) DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol 2: 90–95 [DOI] [PubMed] [Google Scholar]
- Sitterlin D (2004) Characterization of the Drosophila Rae1 protein as a G1 phase regulator of the cell cycle. Gene 326: 107–116 [DOI] [PubMed] [Google Scholar]
- Snaith HA, Armstrong CG, Guo Y, Kaiser K, Cohen PT (1996) Deficiency of protein phosphatase 2A uncouples the nuclear and centrosome cycles and prevents attachment of microtubules to the kinetochore in Drosophila microtubule star (mts) embryos. J Cell Sci 109: 3001–3012 [DOI] [PubMed] [Google Scholar]
- Somma MP, Ceprani F, Bucciarelli E, Naim V, De Arcangelis V, Piergentili R, Palena A, Ciapponi L, Giansanti MG, Pellacani C, Petrucci R, Cenci G, Verni F, Fasulo B, Goldberg ML, Di Cunto F, Gatti M (2008) Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet 4: e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW (2010) Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol 188: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Wainman A, Gergely F, Raff JW (2009) Ana3 is a conserved protein required for the structural integrity of centrioles and basal bodies. J Cell Biol 187: 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson VA, Kramer J, Kuhn J, Theurkauf WE (2001) Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nat Cell Biol 3: 68–75 [DOI] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M (1985) Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol 100: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada S, Kinjo M, Aihara M, Takei Y, Hirokawa N (2010) Kinesin-1/Hsc70-dependent mechanism of slow axonal transport and its relation to fast axonal transport. EMBO J 29: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Wieschaus E (2004) src64 and tec29 are required for microfilament contraction during Drosophila cellularization. Development 131: 863–871 [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C (2007) Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol 17: 1735–1745 [DOI] [PubMed] [Google Scholar]
- Verollet C, Colombie N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B (2006) Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol 172: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ (2003) Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol 163: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PG, Fuller MT, Borisy GG (1997) Monastral bipolar spindles: implications for dynamic centrosome organization. J Cell Sci 110: 451–464 [DOI] [PubMed] [Google Scholar]
- Zavortink M, Contreras N, Addy T, Bejsovec A, Saint R (2005) Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J Cell Sci 118: 5381–5392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.