An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila
Transposons are silenced in the germline and somatic support cells by piRNA pathways, which remain poorly understood at the molecular level. Here, a novel in vivo assay identifies the function of several factors involved in the biogenesis of somatic piRNAs.
Keywords: Drosophila, piRNA pathway, Piwi, transposon silencing
Abstract
In Drosophila, PIWI proteins and bound PIWI-interacting RNAs (piRNAs) form the core of a small RNA-mediated defense system against selfish genetic elements. Within germline cells, piRNAs are processed from piRNA clusters and transposons to be loaded into Piwi/Aubergine/AGO3 and a subset of piRNAs undergoes target-dependent amplification. In contrast, gonadal somatic support cells express only Piwi, lack signs of piRNA amplification and exhibit primary piRNA biogenesis from piRNA clusters. Neither piRNA processing/loading nor Piwi-mediated target silencing is understood at the genetic, cellular or molecular level. We developed an in vivo RNAi assay for the somatic piRNA pathway and identified the RNA helicase Armitage, the Tudor domain containing RNA helicase Yb and the putative nuclease Zucchini as essential factors for primary piRNA biogenesis. Lack of any of these proteins leads to transposon de-silencing, to a collapse in piRNA levels and to a failure in Piwi-nuclear accumulation. We show that Armitage and Yb interact physically and co-localize in cytoplasmic Yb bodies, which flank P bodies. Loss of Zucchini leads to an accumulation of Piwi and Armitage in Yb bodies, indicating that Yb bodies are sites of primary piRNA biogenesis.
Introduction
Selfish genetic elements such as transposons populate all eukaryotic genomes. Though in a few cases the host benefits from transposons, they overall have a negative impact on the host's reproductive fitness because of their mutagenic character (Slotkin and Martienssen, 2007).
Genetic studies on Drosophila melanogaster illustrate the threat emanating from active transposons. Here, uncontrolled activity of a single transposable element (e.g. the I-element, a retro-element or the P-element, a DNA-element) leads to defects in gametogenesis and sterility (Rubin et al, 1982; Bucheton et al, 1984). Given that the D. melanogaster genome harbours over a 100 different transposon families (Bergman et al, 2006), many of which are still active, a strong selective pressure to silence transposons must exist.
After the discovery of small RNA-silencing pathways, it has become clear that this regulatory mechanism is at the root of transposon control in animals and that different lineages deploy the basic principle of small RNA pathways in different ways to guarantee specific and efficient silencing of selfish genetic elements (reviewed in Girard and Hannon, 2008; Malone and Hannon, 2009).
Although transposon control is important for all cells, their silencing is of pivotal importance in the germline, the only cell lineage that passes its genetic information onto the next generation. Indeed, multi-cellular animals possess a unique small RNA pathway targeted towards silencing selfish genetic elements in their gonads. At the centre of this pathway is a subclass of Argonaute proteins, the so-called PIWI proteins, complexed with 23–30 nt long PIWI-interacting RNAs (piRNAs). Mutations in PIWI proteins result in strong de-repression of transposable elements and lead to widespread defects in gametogenesis and sterility (reviewed in Klattenhoff and Theurkauf, 2008; Malone and Hannon, 2009).
The piRNA pathway is best understood in Drosophila, where insight from three research areas has accumulated. First, studies on transposon biology have uncovered important concepts of the host control system and have also identified a number of essential genetic loci involved (Prud'homme et al, 1995; Aravin et al, 2001; Jensen et al, 2002; Desset et al, 2003; Ronsseray et al, 2003; Sarot et al, 2004; Pelisson et al, 2007). Second, sterility and egg patterning screens have uncovered more than a dozen factors that turned out to be piRNA pathway members (Schupbach and Wieschaus, 1991; Clegg et al, 1997; Lin and Spradling, 1997; Cook et al, 2004; Chen et al, 2007; Pane et al, 2007). And finally, research centred on small RNA pathways and bioinformatics analysis of piRNA populations has allowed combining the diverse genetic findings into a coherent model of transposon control in Drosophila (Aravin et al, 2003; Vagin et al, 2006; Brennecke et al, 2007; Gunawardane et al, 2007). According to this, PIWI proteins are loaded with piRNAs in a Dicer and presumably double-stranded RNA (dsRNA)-independent manner (Vagin et al, 2006). Most piRNAs originate from transposon- and other repeat regions in the genome (Aravin et al, 2003; Saito et al, 2006; Vagin et al, 2006; Brennecke et al, 2007; Gunawardane et al, 2007). A hallmark of the piRNA pathway is that discrete genomic loci (piRNA clusters) that harbour a diverse collection of transposon and repeat fragments are major sources of piRNAs (Brennecke et al, 2007). Another unique feature of the piRNA pathway is the involvement of PIWI proteins in a target-dependent amplification loop (Brennecke et al, 2007; Gunawardane et al, 2007). In this so-called ping-pong cycle, the PIWI proteins Aubergine and AGO3 cleave reciprocally sense (from active elements) and antisense (from piRNA clusters) transcripts, respectively. It is postulated that each cleavage event triggers the production of a novel piRNA from the cleaved RNA. Thus, the ping-pong cycle leads to a preferential amplification of silencing competent piRNAs. Gonad-specific activity of the piRNA pathway, repeat enriched piRNA clusters and signatures of the ping-pong cycle are all conserved features in vertebrates (reviewed in Malone and Hannon, 2009).
Major open questions are how the cell distinguishes transcripts from piRNA clusters, transposons and endogenous genes and how piRNA biogenesis and loading into PIWI proteins is controlled. Equally unclear is how PIWI–piRNA complexes silence the array of selfish genetic elements in the genome. Various studies indicate that post-transcriptional control through target slicing and degradation and transcriptional control through guiding DNA and/or chromatin modifications both have an active function (Carmell et al, 2007; Klenov et al, 2007; Kuramochi-Miyagawa et al, 2008; Lim et al, 2009).
In Drosophila, germline cells and somatic support cells of male and female gonads possess a functional piRNA pathway. However, in somatic support cells, only one of the three PIWI family proteins (Piwi) is expressed and a simplified pathway (the somatic piRNA pathway) lacking the ping-pong cycle is active (Sarot et al, 2004; Pelisson et al, 2007; Lau et al, 2009; Li et al, 2009; Malone et al, 2009; Saito et al, 2009). Deep sequencing of small RNA populations indicates that single-stranded RNAs from a subset of piRNA clusters and to a lower extent also from over a 1000 genes are processed into piRNAs (Lau et al, 2009; Malone et al, 2009; Robine et al, 2009). The most prominent piRNA cluster in somatic support cells is the genetically identified flamenco or COM locus (Prud'homme et al, 1995; Desset et al, 2003). It contains an exceptionally high density of transposon fragments, nearly all of which belong to the gypsy family of retro-elements. As nearly all fragments are oriented antisense to the transcription direction, piRNA processing from flamenco yields almost exclusively antisense piRNAs (Pelisson et al, 2007; Brennecke et al, 2007; Malone et al, 2009). Many gypsy family transposons expressed in somatic support cells encode functional gag, pol and env genes. In the absence of efficient silencing, they form viral particles that invade the neighbouring oocyte potentially through cellular transport vesicles (Pelisson et al, 1994; Chalvet et al, 1999; Leblanc et al, 2000; Brasset et al, 2006).
The somatic cells of the Drosophila gonad are the only described cell type with an active piRNA pathway that lacks the ping-pong cycle. These cells are, therefore, ideally suited for genetic and biochemical approaches towards elucidating the core concepts of the piRNA pathway. Here, we describe a genetic assay that allows for testing any gene of interest for its involvement in the somatic piRNA pathway. We use it to identify the three evolutionarily conserved proteins Armitage (Armi), Zucchini (Zuc) and Yb (King et al, 2001; Cook et al, 2004; Pane et al, 2007) as essential components of primary piRNA biogenesis in Drosophila and link the Yb body, a previously identified cellular structure (Szakmary et al, 2009) to piRNA biogenesis.
Results
An in vivo assay for the identification of novel piRNA pathway components
To facilitate the identification of novel somatic piRNA pathway members, we constructed a tester fly line that could be combined with the Vienna Drosophila RNAi collection (VDRC) (Dietzl et al, 2007). The VDRC library consists of Gal4-inducible UAST lines allowing expression of dsRNA hairpins against >90% of the Drosophila genes. The tester line combines a GAL4 driver specific for somatic support cells with a sensor capable of monitoring the integrity of the somatic piRNA pathway.
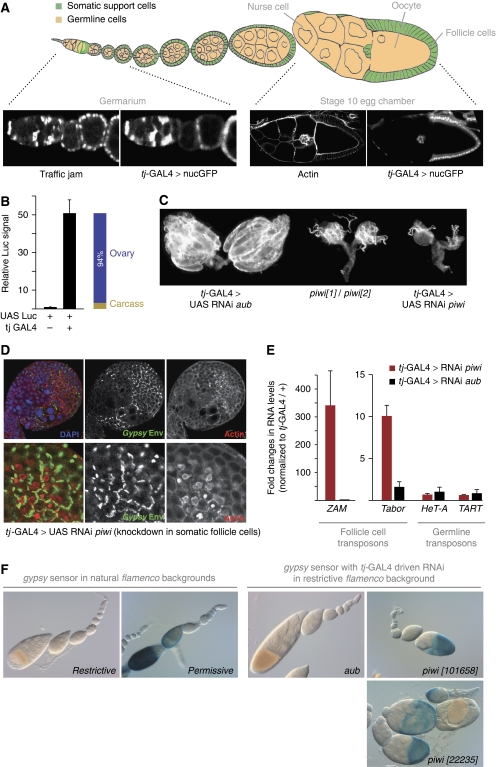
We identified the previously reported traffic jam GAL4 line (tj-GAL4) as an optimal driver for RNAi in somatic support cells (Tanentzapf et al, 2007). GAL4 expression mirrored the restricted expression of the host gene tj to gonadal somatic support cells when crossed to UAST-GFP (Figure 1A) (Li et al, 2003). GFP signal was only detected in somatic support cells of the ovary including somatic stem cells in the germarium. To determine the tissue specificity of tj-GAL4, we crossed the line to UAST-Luciferase and measured reporter activity in whole females, dissected ovaries and carcasses (flies with manually removed ovaries). The tj-GAL4 driver induced Luciferase signal ∼50-fold and on average, 94% of this signal originated from ovaries (Figure 1B).
Figure 1.
An in vivo RNAi-based assay for the somatic piRNA pathway. (A) Cartoon depicting the two major cell types in the ovary with somatic cells in green and germline cells in beige. The tj-GAL4 driver is expressed at all developmental stages in somatic support cells (labelled with Tj) as judged by a cross to UAST-GFP. (B) Relative Luciferase reporter activity is shown from flies of the indicated genotypes (left) and from ovaries and carcasses of tj-GAL4>UAST-Luc flies. (C) Morphology of ovaries from flies expressing dsRNA against piwi and aub through tj-GAL4 compared with ovaries from piwi mutants. (D) Immuno-staining of the retroviral gypsy Env protein (green) in ovaries with tj-GAL4 induced Piwi knockdown. The lower panels show a zoom onto the follicular epithelium. Actin (red) marks the cell cortex. (E) Shown are changes in steady-state RNA levels (n=3) of the endogenous retro-elements ZAM and Tabor upon Piwi (red) or Aubergine (black) knockdown in comparison with tj-GAL4/+ flies. The germline-controlled HeT-A and TART elements serve as controls. (F) Shown is the gypsy-lacZ reporter in restrictive and permissive flamenco backgrounds (left) and in a restrictive background upon RNAi against aub and piwi (two independent hairpins). β-Gal staining (blue) is strongest in the columnar follicle epithelium of stages 9–10 egg chambers.
Crosses to a broad spectrum of RNAi lines targeting essential genes resulted in missing or rudimentary ovaries with no impact on viability (Supplementary Table S1). This is significant, as it has been reported that a number of strong follicle cell GAL4 drivers were incapable of inducing efficient RNAi (Zhu and Stein, 2004). Importantly, tj-GAL4-driven RNAi against piwi, the only undisputed member of the somatic piRNA pathway, resulted in ovaries that phenocopied piwi mutant ovaries (Figure 1C) (Cox et al, 2000). A known target of the somatic piRNA pathway, the retroviral gypsy transposon (Sarot et al, 2004), was de-repressed in these ovaries with a strong accumulation of the gypsy Env protein at the follicle cell cortex (Figure 1D). No staining by the gypsy Env antibody was detectable in wild-type ovaries and staining was restricted to patches of cells in which piwi RNAi was induced clonally (Supplementary Figure S1). Quantitative RT–PCR analysis further indicated that besides gypsy, the retro-elements ZAM and Tabor, two additional targets of the somatic piRNA pathway (Desset et al, 2003; Li et al, 2009; Malone et al, 2009), are strongly de-silenced in piwi RNAi crosses compared with aubergine RNAi crosses (aub shows no detectable expression in follicle cells) (Figure 1E). Notably, RNA levels of the telomeric retro-elements HeT-A and TART, which are sensitive to defects in the germline piRNA pathway (Vagin et al, 2006; Klattenhoff et al, 2009), were unaffected. These results are in line with the abundance of piRNAs against gypsy, ZAM and Tabor and the low level of piRNAs against HeT-A and TART in cultured ovarian somatic cells (OSCs), which are derived from somatic support cells (Supplementary Figure S2) (Niki et al, 2006; Lau et al, 2009).
We combined the tj-GAL4 driver with a gypsy-lacZ reporter that in flies carrying a restrictive allele of the flamenco cluster is fully silenced by the somatic piRNA pathway (Sarot et al, 2004). Lack of the somatic piRNA pathway or homozygosis for a permissive flamenco allele strongly de-silences β-Gal expression (Figure 1F) (Sarot et al, 2004). When the tj-GAL4; gypsy-lacZ line was crossed to an aub-RNAi line, no β-Gal signal was detectable. In contrast, crosses to two independent piwi RNAi lines resulted in strong β-Gal activity (Figure 1F). Thus, given the near genome-wide collection of RNAi lines at the VDRC, almost any gene can be tested for its involvement in the somatic piRNA pathway with a single genetic cross.
Armitage and Zucchini are essential components of the somatic piRNA pathway
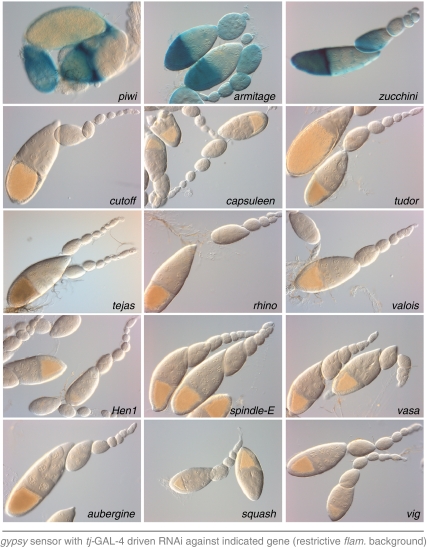
Over a dozen genes have been implicated in the Drosophila piRNA pathway (Boswell and Mahowald, 1985; Gillespie and Berg, 1995; Cox et al, 1998; Harris and Macdonald, 2001; Cook et al, 2004; Vagin et al, 2004; Anne et al, 2007; Chen et al, 2007; Horwich et al, 2007; Lim and Kai, 2007; Pane et al, 2007; Saito et al, 2007; Klattenhoff et al, 2009; Li et al, 2009; Malone et al, 2009; Patil and Kai, 2010). To test their involvement in the somatic piRNA pathway, we crossed the tj-GAL4; gypsy-lacZ tester strain to RNAi lines for all candidates if a VDRC line was available. Most candidates are seemingly not essential components of the somatic piRNA pathway (Figure 2). Consistent with this, loss of spindle-E, squash, aubergine, rhino or vasa does not affect flamenco-derived piRNA levels, indicating their specific involvement in the germline-specific ping-pong amplification cycle (Klattenhoff et al, 2009; Malone et al, 2009). In addition, loss of the piRNA methyl-transferase Hen1 has been reported to impact transposon silencing only very mildly (Horwich et al, 2007). It is of interest to note that lack of symmetrically methylated Arginines (mediated by knocking down the arginine methyl-transferase capsuleen or its cofactor valois) did not lead to detectable sensor activation. Thus, this post-translational modification of PIWI proteins (Kirino et al, 2009; Nishida et al, 2009; Vagin et al, 2009) has no or only a modulatory function in the somatic piRNA pathway, although we cannot exclude the involvement of other arginine methyl transferases.
Figure 2.
Comprehensive analysis of the involvement of piRNA pathway members in the somatic pathway. Shown are β-Gal stainings as readout for gypsy silencing for genetically identified piRNA pathway genes in which a VDRC line was available using gypsy-lacZ and tj-GAL4 in a restrictive flamenco background.
In contrast, the gypsy sensor was strongly de-silenced in the cross to the zucchini (zuc) RNAi line (Figure 2). This is in agreement with published findings that loss of Zuc affects flamenco-derived piRNA levels as well as those derived from the tj-3′UTR, a gene expressed in somatic support cells only (Malone et al, 2009; Robine et al, 2009; Saito et al, 2009).
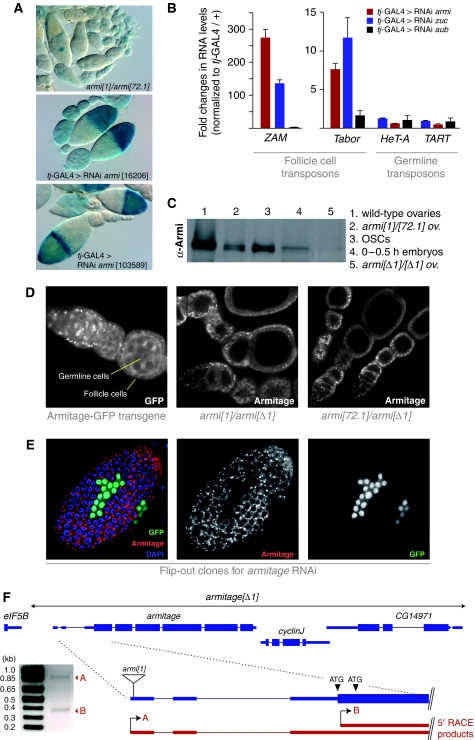
To our surprise, knockdown of armitage (armi) led to a robust de-silencing of the sensor (Figure 2). This contradicts earlier findings that armi mutations specifically affect the germline piRNA pathway (Malone et al, 2009): in armi[1]/armi[72.1] mutants, Piwi-bound piRNAs and nuclear localization of Piwi were only lost in germline cells. Furthermore, flamenco-derived piRNA levels were unchanged in those mutants. In support of the previous genetic results, the gypsy-lacZ sensor was not de-silenced in follicle cells of armi[1]/armi[72.1] ovaries (Figure 3A). In these ovaries, we observed weak β-Gal activity in nurse cells, indicating a slight activity of the gypsy promoter and an Armi-dependent silencing mechanism in ovarian germline cells.
Figure 3.
Armitage is an essential player in the somatic piRNA pathway. (A) gypsy-lacZ reporter activity in armi[1]/armi[72.1] ovaries compared with ovaries in which armi was knocked down using tj-GAL4-mediated RNAi. (B) Shown are fold changes in steady-state RNA levels (n=3) for the ZAM, Tabor, HeT-A and TART retro-elements in ovaries with tj-GAL4-driven RNAi against armi, zuc and aub (control). (C) Western blot analysis showing two Armi isoforms expressed in ovaries and OSCs, one isoform in early embryos, persistent expression of Armi in published mutant alleles and the absence of Armi in the armi[Δ1] allele. (D) Shown are Armi stainings of an ovariole expressing Armi-GFP under its endogenous promoter (left) and of ovarioles trans-heterozygous for the published alleles armi[1] and armi[72.1] over the armi[Δ1] allele. (E) Armi staining (red) of the follicular epithelium in which armi-RNAi has been clonally activated. GFP signal (green) labels cells expressing the armi dsRNA hairpin. (F) Cartoon of the armi genomic locus showing the extent of the armi[Δ1] deletion, the two identified 5′RACE products (red), their respective transcription start sites and the insertion site of the P-element in the armi[1] allele. To the left, an Agarose gel with the two sequenced RACE products is shown.
The armi[1] and armi[72.1] alleles are not null alleles as noted previously (Cook et al, 2004). Nevertheless, they cause a severe reduction of Armi protein in germline cells. No difference in immuno-staining, however, has been detected in somatic follicle cells of these mutants, which has been interpreted as antibody cross-reactivity (Cook et al, 2004). The following observations, however, indicate that Armi is expressed in follicle cells and that it is an essential component of the somatic piRNA pathway. We first verified our knockdown result with an independent RNAi line (Figure 3A). Second, knockdown of Armi led to a strong de-silencing of the ZAM and Tabor retro-elements with no impact on HeT-A and TART (Figure 3B). Similar results were obtained from flies expressing the zuc RNAi construct. We next verified robust Armi expression in somatic support cells by three independent means: first, a transgenic fly line expressing Armi as a C-terminal GFP fusion under its own promoter showed robust GFP signal in germline and somatic cells of the ovary, similar to the published immuno-stainings (Figure 3D) (Cook et al, 2004). Second, the follicle cell staining of Armi was specifically lost in clones of cells expressing the armi RNAi construct (Figure 3E). Third, western analysis showed that ovary lysate contains two Armi isoforms that appear as a duplet and that both isoforms are also present in a lysate from OSCs (cultured somatic cells) (Figure 3C). To test for the specificity of the western signal, we generated a genomic deletion that removes the entire armi locus including two downstream genes (armi[Δ1]). Homozygous armi[Δ1] flies are viable, contain malformed ovaries and do not lay eggs. Extracts from these ovaries lack both Armi isoforms (Figure 3C).
This indicates that the two published alleles armi[1] and armi[72.1] are not null alleles at the protein level. Indeed, both Armi isoforms were detected by western analysis in armi[1]/armi[72.1] flies albeit at ratios that resemble that found in OSCs (Figure 3C). Furthermore, Armi protein was detectable in follicle cells only of armi[1]/armi[Δ1] and armi[72.1]/armi[Δ1] mutant ovaries (Figure 3D). We, therefore, suspected that the armi locus contains two promoters and, therefore, gives rise to two distinct mRNAs encoding the two observed protein isoforms. Indeed, two mRNA isoforms have been observed by northern analysis with only the larger one being severely affected by the published alleles (Cook et al, 2004). We performed 5′RACE analysis on OSC RNA and identified two distinct mRNA 5′ ends with one corresponding to the Flybase annotated transcription start site (TSS) (Figure 3F). The second TSS is a few nucleotides downstream of the annotated ATG start codon and, therefore, is predicted to generate an ∼4 kDa smaller isoform. In this model, the insertion site of the P-element in armi[1] and the small deletion in the armi[72.1] allele, which was derived from armi[1] by imprecise excision are expected to affect only the longer isoform. Although the 5′RACE analysis indicates that both promoters are active in follicle cells, a western analysis from lysate obtained from 0 to 0.5 h old embryos identified only the larger isoform, indicating that the downstream promoter is not active in germline cells (Figure 3C). Our results, therefore, offer a consistent explanation for the discrepancy of the published literature with our result that Armi is an essential component of the somatic piRNA pathway.
Yb co-localizes with Armitage in cytoplasmic foci and is essential for the somatic piRNA pathway
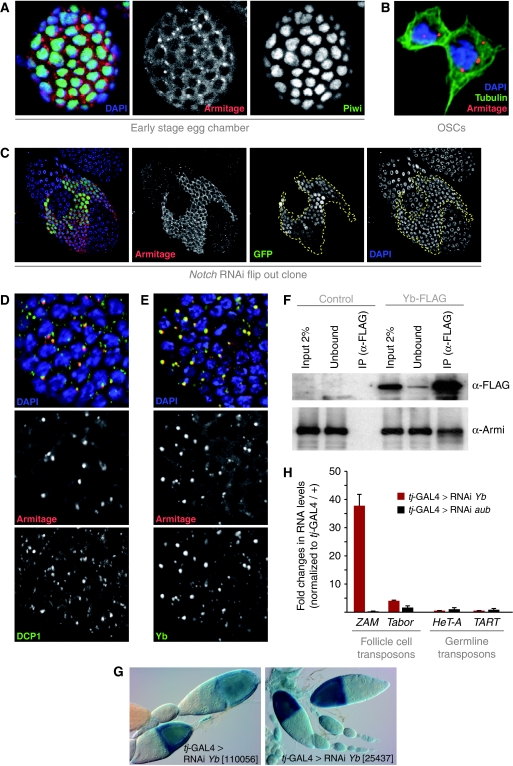
Both GFP tagged and endogenous Armi localized to the cytoplasm of follicle cells and OSCs with a strong enrichment in 1–3 peri-nuclear foci (Figure 4A and B). Within the ovary, Armi foci were much more prominent in the germarium and younger egg chambers and became progressively smaller from stage 5–6 onwards (not shown). At this stage, follicle cells switch from a mitotic cycle to an endo-replication cycle in a Notch-signalling-dependent manner (Deng et al, 2001). Interestingly, disruption of the Notch-signalling pathway by RNAi against Notch led to a strong increase in Armi levels in older (>stage 6), but not in younger egg chambers (Figure 4C; not shown), suggesting that Notch signalling dampens Armi expression in endo-replicating cells.
Figure 4.
Armi localizes to Yb bodies and Yb is an essential pathway component. (A) Shown are immuno-fluorescence stainings for Armi (red) and Piwi (green) in somatic follicle cells. DNA was labelled with DAPI (blue). (B) Immuno-fluorescence staining for Armi (red), Tubulin (green) and DNA (blue) in OSCs. (C) Immuno-fluorescence analysis of Armi (red) in the follicular epithelium of an egg chamber containing a Notch dsRNA-expressing clone (clone marked with GFP (green) and clone borders indicated by the dashed line). DAPI staining (blue) confirms that Notch knockdown impairs endo-replication. (D) Co-labelling of Armi (red) and DCP1-GFP (green) in somatic follicle cells. (E) Co-labelling of Armi (red) and Yb (green) in somatic follicle cells. (F) Co-immuno-precipitation of endogenous Armi with FLAG-tagged Yb. Shown are western blots against FLAG to detect Yb (top) and against endogenous Armi (bottom). Control cells did not express FLAG-Yb. (G) Shown are β-Gal stainings of ovarioles expressing the gypsy reporter and two different dsRNA hairpins against fs(1)Yb with tj-GAL4. (H) Shown are fold changes in steady-state RNA levels (n=3) of ZAM, Tabor, HeT-A and TART retro-elements in ovaries in which fs(1)yb and aub (control) were knocked down through tj-GAL4-mediated RNAi. Values are normalized to a tj-GAL4/+ control.
The distinct localization of Armi in follicle cells was reminiscent of processing bodies (P bodies), cytoplasmic foci involved in RNA degradation and storage (Eulalio et al, 2007a). Co-labelling of Armi and the P-body marker DCP1 indicated that Armi foci are in most cases directly adjacent to P bodies (Figure 4D). There appear to be many more P bodies than Armi foci, but typically the most prominent P bodies flank Armi foci. In germline cells, we observed that areas with Armi staining typically exhibit also strong DCP1 labelling (Supplementary Figure S3A). The significance of this correlation remains to be determined, but we note that knockdown of two critical P-body components (Lsm1 and Me31b) with tj-GAL4 did not lead to defects in gypsy sensor silencing. Moreover, RNAi flip-out clones for Lsm1 did not affect number or integrity of the Armi foci (not shown).
The subcellular localization of Armi resembles that of the Yb protein. Yb is encoded by the fs(1)Yb gene and is a putative RNA helicase that contains a predicted Tudor domain (King and Lin, 1999; Szakmary et al, 2009). Yb is expressed in somatic support cells of the male and female gonad and accumulates in cytoplasmic foci that have been termed Yb bodies (Szakmary et al, 2009). Yb mutants are female sterile and exhibit defects in the maintenance of germline and somatic stem cells (King and Lin, 1999; King et al, 2001). Immuno-fluorescence experiments indicated that Armi and Yb proteins co-localize and accumulate in Yb bodies of follicle cells (Figure 4E) and OSCs (Supplementary Figure S3B). In addition, a specific interaction between the two proteins was observed in co-IP experiments from OSC lysate using FLAG-tagged Yb (Figure 4F). To test for a functional involvement of Yb in the somatic piRNA pathway, we crossed two independent fs(1)Yb RNAi lines to our tester strain and observed strong de-repression of the gypsy reporter (Figure 4G). Depletion of Yb also led to a de-silencing of the ZAM and Tabor retro-transposons, but not the germline-controlled HeT-A and TART elements (Figure 4H). These results identified Yb as an essential component of the somatic piRNA pathway and link the previously described Yb bodies to piRNA biology.
Loss of Armitage, Zucchini and Yb impairs nuclear localization of Piwi
To gain insight into the defects exerted onto the somatic piRNA pathway upon loss of Armi, Zuc and Yb, we performed a systematic analysis of Armi, Piwi and Yb localization in cells mutant for each factor (a lack of reagents prevented localization studies for Zuc).
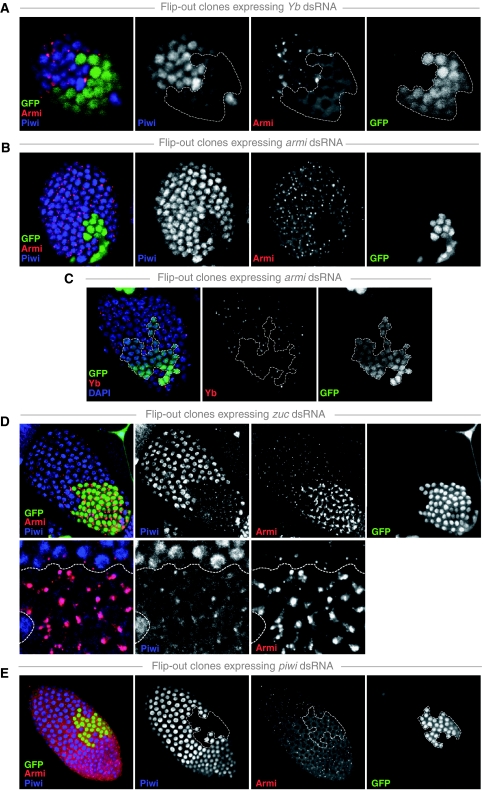
RNAi-mediated knockdown of Yb in follicle cell clones led to a loss of nuclear Piwi accumulation (Figure 5A). Furthermore, no Armi foci were detectable when Yb was depleted by RNAi. Instead, Armi was uniformly distributed throughout the cytoplasm (Figure 5A). Identical results were obtained in FRT-mediated mitotic clones using the Yb[72] loss of function allele (Supplementary Figure S4).
Figure 5.
Comprehensive analysis of Armi, Piwi and Yb localization upon their reciprocal knockdown. (A–E) Shown are surface views of the follicular epithelium of egg chambers containing clones expressing dsRNA hairpins against the indicated genes stained for Piwi (blue), Armi (red) and Yb (red). Clones are marked by the presence of GFP (green) and clone borders are indicated (dashed line). In all rows, the merged RGB image is shown alongside with the individual channels in black and white. In (D), the lower row shows higher magnification views.
RNAi-mediated knockdown of Armi also caused a loss of Piwi from the nucleus (Figure 5B). In addition, Yb bodies were reduced in size, but did not disappear entirely (Figure 5C). We, therefore, analysed Yb localization in armi null mutants and observed a loss of detectable Yb bodies in nearly all cells (Supplementary Figure S5). Thus, Armitage and Yb are each required for their reciprocal localization to Yb bodies and potentially for Yb-body formation per se.
The most interesting effects were seen upon Zuc knockdown. Here, Piwi was not only lost from the nucleus, but instead accumulated in peri-nuclear foci (Figure 5D). Co-labelling experiments showed that these correspond to Armi bodies (Figure 5D, bottom). In addition, the size of Armi foci increased several fold in Zuc mutant cells (Figure 5D). We also observed elevated intensity of Yb foci in these cells, but the effects were much weaker than those observed for Armi (not shown). Identical results were obtained in FRT-mediated mitotic clones using the zuc[HM27] loss of function allele (Supplementary Figure S6). The co-localization of Piwi and Armi in zuc mutant cells suggests a rapid transit of Piwi through Armi/Yb bodies in wild-type cells, as we never observed accumulation of endogenous Piwi protein in cytoplasmic foci. However, OSCs chemically transfected with GFP-tagged Piwi often exhibit co-localization of Piwi with Armi foci, potentially as a consequence of Piwi over-expression (Supplementary Figure S7).
To test whether Armi is required for Piwi's accumulation in Yb bodies, we simultaneously knocked down Armi and Zuc in follicle cell clones. In these cells, Piwi did not accumulate in cytoplasmic foci (Supplementary Figure S8). We note, however, that Yb-body size is severely reduced upon Armi knockdown, thus complicating this interpretation. No defects on Armi localization were seen in cells expressing an RNAi construct against Piwi or in cells homozygous for the piwi[1] loss of function allele (Figures 5E and 8D). This indicates that Armi does not depend on Piwi for its localization to Yb bodies and that Yb-body formation occurs in the absence of Piwi.
Armitage and Piwi interact physically in OSCs and ovaries
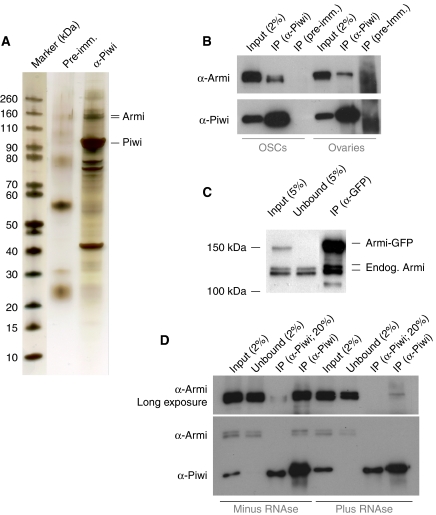
In an attempt to identify Piwi-interacting proteins, we immuno-precipitated endogenous Piwi from OSC lysate using polyclonal antibodies directed against the Piwi N-terminus. Silver staining identified two protein bands at ∼140 kDa that were specific for the Piwi-IP compared with a control IP (Figure 6A). Mass spectrometric analysis identified Armi as the major protein in both bands (32 and 27% sequence coverage). This result is at odds with Armi being a cytoplasmic protein, whereas Piwi is enriched in the nucleus under wild-type conditions. However, our observation that Piwi localizes transiently with Armi to Yb bodies offers an explanation for the observed interaction. Western blot analysis indicated that this interaction is also robust in IPs from ovary lysate (Figure 6B). The major interacting Armi isoform in ovaries is the germline-specific larger isoform, whereas the smaller isoform predominates in OSCs. To test the specificity of the Armi Piwi interaction, we immuno-precipitated transfected FLAG-tagged Piwi from OSC lysate. Again, endogenous Armi protein was only detected in the FLAG-Piwi-IP, but not in the FLAG-IP from non-transfected cells (not shown). Finally, endogenous Piwi was co-immuno-precipitated with Armi using flies expressing C-terminally GFP-tagged Armi under its own promoter (not shown). In this experiment, we also noted that Armi forms oligomeric complexes in vivo, as endogenous Armi protein was efficiently co-immuno-precipitated with GFP-tagged Armi (Figure 6C).
Figure 6.
Piwi and Armitage interact physically. (A) SDS–PAGE of a Piwi immuno-precipitation (IP) alongside a control IP from OSC lysate stained with silver. A molecular weight marker is shown to the left and Piwi and Armi bands are indicated. (B) Western blot showing co-IP of endogenous Armi with Piwi from OSCs and ovaries. Pre-immune serum was used for the control IP. (C) Western blot showing co-IP of endogenous Armi with GFP-tagged Armi from ovary lysate. (D) Western blot showing co-IP of endogenous Armi with Piwi in the presence or absence of RNAse.
An immuno-precipitation of Piwi in the presence of RNAse A and T1 indicates that the interaction between Armi and Piwi depends at least in part on RNA (Figure 6D). Although we cannot exclude incomplete RNA digestion, we suggest that the interaction between Armi and Piwi is direct, but enhanced by RNA. Such a direct interaction is supported by a co-immuno-precipitation of mouse PIWI proteins and the mouse Armi orthologue MOV10L1 in 293 cells that contain no active piRNA pathway (Zheng et al, 2010).
Armitage, Zucchini and Yb are required for primary piRNA biogenesis
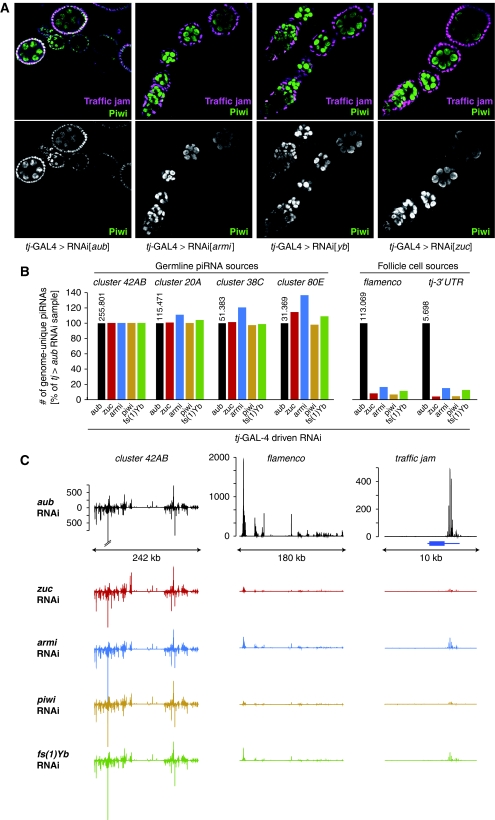
Loss of Armi in germline cells leads to a failure of nuclear Piwi accumulation accompanied by a loss of germline-specific Piwi-bound piRNAs (Malone et al, 2009). A correlation between nuclear localization and intact small RNA biogenesis has also been observed for mouse MIWI2 and Tetrahymena Twi1 (Aravin et al, 2008; Noto et al, 2010; Zheng et al, 2010). We sequenced Piwi-bound piRNAs from ovaries in which we depleted Piwi, Armi, Zuc or Yb in all ovarian somatic support cells by RNAi using the tj-GAL4 driver. RNAi against aub was used as a control. An immuno-fluorescence analysis showed the efficiency of the tj-GAL4-driven knockdown in follicle cells as they lacked Piwi-nuclear accumulation (Figure 7A). We prepared ovary lysates and immuno-precipitated Piwi–piRNA complexes. From these, small RNAs were isolated and used for the construction of cDNA libraries, which were sequenced on the Illumina G2 platform. After removal of small RNAs mapping to abundant cellular non-coding transcripts (rRNAs, tRNAs, snoRNAs), the composition of all libraries was highly similar with ∼84% mapping to annotated repeats (Supplementary Table S2). As Piwi function within germline cells was unaffected, we used the germline-specific piRNA cluster at position 42AB to normalize the individual libraries. Three other peri-centromeric piRNA clusters (cluster 20A, cluster 38C, cluster 80E) with predominant or exclusive piRNA processing in the germline (Lau et al, 2009; Malone et al, 2009) showed similar piRNA levels across all libraries (Figure 7B). In contrast, piRNAs uniquely mapping to the flamenco locus, a piRNA cluster that is exclusively or predominantly processed into piRNAs in somatic support cells, were 6- to 10-fold reduced in piwi, armi, zuc and fs(1)Yb knockdown ovaries. The tj-3′UTR is another source for abundant piRNAs in somatic support cells (Robine et al, 2009; Saito et al, 2009). Levels of tj-derived piRNAs also decreased 6- to 10-fold in all libraries in comparison with the control (Figure 7B). Figure 7C depicts the normalized piRNA profiles (sense and antisense) originating from cluster 42AB, the flamenco cluster and the tj locus.
Figure 7.
piRNA biogenesis defects in somatic cells lacking Armi, Piwi, Zuc and Yb. (A) Shown are ovarioles stained for Piwi (green) and Traffic jam (magenta) from flies expressing the indicated dsRNAs with tj-GAL4. Note the lack of nuclear Piwi in follicle cells that are marked by Tj (aub knockdown serves as negative control). (B) Shown are relative levels of genome-unique Piwi–piRNAs mapping to the indicated piRNA clusters in ovaries of the indicated genotypes. Libraries were normalized through the genome-unique mappers to 42AB, a germline-specific piRNA cluster. The tj-GAL4>aub-RNAi sample serves as control. The respective raw read numbers from the tj>aub_RNAi library are indicated above the black bars. (C) Detailed piRNA profiles (only genome-unique piRNAs are shown) across the piRNA clusters 42AB and flamenco as well as the major genic piRNA precursor tj. The colour code is as in (B) and the annotated tj gene is indicated in blue. The y axis indicates the number of sequences per 200 nt window normalized to 1 million sequenced reads and graphs within a vertical row are shown at the same scale.
We further analysed piRNA levels mapping to transposons, which have been classified as germline dominant (HeT-A, F-element, TART, GATE) or soma dominant (gypsy5, ZAM, Tabor, 412, Idefix) (Li et al, 2009; Malone et al, 2009). Using the same normalization from before (cluster 42AB), piRNA levels for the germline-dominant elements were essentially unchanged in all libraries (Supplementary Figure S9A). In contrast, piRNAs mapping to the soma-dominant elements were strongly reduced in number and those remaining had a higher proportion of sense-derived piRNAs, which in wild-type ovaries make up <5% of all ZAM- and Tabor-specific piRNAs (Supplementary Figure S9A,B). We speculate that the de-silencing of the ZAM and Tabor elements leads to disproportionate piRNA processing directly from their active sense transcripts.
Our combined results show that primary piRNA biogenesis or loading into Piwi is dependent on Armi, Yb and Zuc in ovarian somatic support cells.
Similarities between somatic and germline Piwi pathways
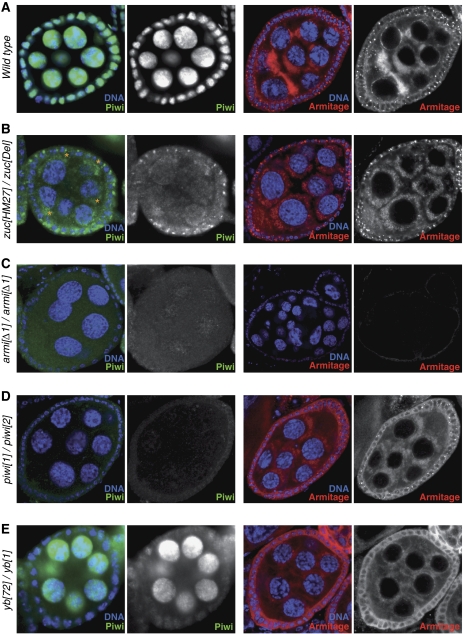
It has been suggested that Piwi biology in soma and germline of the Drosophila ovary differs (Malone et al, 2009). This was based, however, on armi alleles, which based on our data are germline specific. It has also been reported that zuc null mutants exhibit no defects in Piwi-nuclear accumulation (Malone et al, 2009). This prompted us to re-investigate the similarities of the somatic and the germline Piwi pathways using null mutants for armi, piwi, fs(1)Yb and zuc.
Other than previously reported, zuc mutant ovaries exhibited a severe loss of Piwi from the nucleus in somatic and germline cells of the ovary (Figure 8A and B). While Piwi protein accumulated in Yb bodies in follicle cells, it appeared to concentrate in clouds at the nuclear periphery of germline cells (asterisks in Figure 8B). These clouds were also prominently stained for Armi (Figure 8B, right). To test whether this could represent disintegrating nuage, we stained zuc mutant ovarioles for Aub and AGO3, two proteins localizing to nuage. No obvious defects in nuage pattern were detectable based on Aub localization, but we observed frequent accumulation of AGO3 in cytoplasmic foci (Supplementary Figure S10). Loss of Zuc thus leads to a significant co-localization of Piwi and Armi in soma (Yb bodies) and germline (peri-nuclear clouds, distinct from nuage). As expected, loss of Armi de-localized Piwi from the nucleus in germline and soma (Figure 8C). No changes in Armi localization either to Yb bodies in somatic cells or peri-nuclear clouds in germline cells were seen in ovaries lacking Piwi (Figure 8D). These results suggest that Piwi biology is similar in somatic and germline cells of the ovary.
Figure 8.
Similarities of Piwi biology in soma and germline of the ovary. (A–E) Shown are optical sections through mid stage egg chambers of indicated genotypes (left) stained for DNA (blue), Piwi (green) and Armi (red). Peri-nuclear accumulation of Piwi in germline cells of zuc mutant ovaries is marked by asterisks in (B).
The only protein that acts differentially in the two cell types is Yb. Ovaries mutant for Yb lost nuclear Piwi accumulation in somatic but not in germline cells (Figure 8E). Similarly, although somatic Armi localization to Yb bodies was lost, the pattern of Armi in germline cells was wild type (Figure 8E). This is consistent with Yb being specifically expressed in somatic cells of the ovary only (Szakmary et al, 2009), and raises a question to as whether a redundant protein might serve Yb's function in germline cells (see Discussion).
Our immuno-fluorescence analysis indicated that Piwi levels were reduced in cells mutant for armi, zuc or fs(1)Yb. To test this, we performed western blot analysis using ovary extracts obtained from null mutants for each respective gene (Supplementary Figure S11). This indicated indeed strongly reduced Piwi levels in armi and zuc mutants, whereas fs(1)Yb mutants were less affected, consistent with it being only important for Piwi biology in the somatic support cells. No changes in Armi levels were observed in piwi, zuc or fs(1)Yb mutants (not shown). We conclude that failure to load Piwi with piRNAs leads to Piwi destabilization and failure in nuclear accumulation.
Discussion
In this study, we present genetic, cell biological and molecular data that shed light on primary piRNA biogenesis in Drosophila.
We describe a robust in vivo assay that allows the identification of genes with a critical involvement in the somatic piRNA pathway. It takes advantage of the recently constructed genome-wide RNAi library at the VDRC and, therefore, will allow the conduction of a genome-wide RNAi screen. The somatic support cells of the Drosophila ovary are the only described cells containing a piRNA pathway without ping-pong cycle. Elucidation of this pathway will, therefore, not only uncover the basic concepts behind the piRNA biogenesis and silencing machineries. It will also simplify the genetic and functional characterization of identified candidates, aided by the availability of the OSC culture system (Niki et al, 2006). Similar to any screen system, our assay has drawbacks such as availability of an RNAi line, potential off-target effects or inefficient knockdown. However, an RNAi screen has the major advantage that knockdown of genes essential for other vital functions in the cell might still allow the development of ovaries suitable for analysis, whereas EMS generated null alleles would often prevent this.
We used this assay to assign essential functions to the piRNA pathway proteins Armi and Zuc within the somatic pathway and identified Yb as a novel piRNA pathway gene. Detailed cell biological analyses in conjunction with protein interaction studies and piRNA sequencing efforts place all three factors upstream of the active Piwi–piRNA complex, either in piRNA biogenesis or in piRNA loading into Piwi. All three proteins are required for piRNA accumulation and for the nuclear accumulation of Piwi. The cytoplasmic localization of Armi, Yb and Zuc suggests that piRNA biogenesis/loading occurs in the cytoplasm. This is supported by the observation that N-terminally truncated Piwi that cannot translocate into the nucleus is still loaded with piRNAs (Saito et al, 2009). In addition, piRNAs derived from cellular mRNAs typically map to the 3′UTR (Robine et al, 2009), indicating that ribosomal association with those mRNAs did precede their funnelling into piRNA biogenesis.
Our data indicate that unloaded Piwi does not translocate into the nucleus. This is highly reminiscent to the situation in mouse and Tetrahymena. The mouse PIWI family protein MIWI2 as well as the Tetrahymena Twi1 protein accumulate in the cytoplasm under conditions that do not allow piRNA biogenesis or loading into these factors (Aravin et al, 2008; Noto et al, 2010; Zheng et al, 2010). Furthermore, unloaded Piwi appears to be unstable in vivo. This effect is most apparent in ovaries lacking Armi and Zuc, two factors that are essential for Piwi–piRNA biogenesis/loading in soma and germline.
Our results further indicate that aspects of piRNA biogenesis and/or piRNA loading take place in discrete peri-nuclear foci that have been previously termed Yb bodies (Szakmary et al, 2009). The two RNA helicases Armi and Yb localize both to Yb bodies and interact physically. Both proteins are required for their reciprocal localization to cytoplasmic foci and potentially for Yb-body formation per se. A strong argument for the importance of Yb bodies in the piRNA pathway stems from the observation that fs(1)Yb[1] mutant ovaries also show Piwi loss from the nucleus (not shown). In this allele, a conserved Arginine residue in the Tudor domain is mutated. This still allows Yb protein expression, but blocks Yb-body formation (Szakmary et al, 2009). Ultra-structurally, Yb bodies were described as electron dense cytoplasmic spheres that are directly adjacent to an RNA containing electron dense structure (Szakmary et al, 2009). Our localization studies with DCP1 indicate that these previously identified neighbouring structures are likely a subset of cellular P bodies. The functional significance of this is unclear, as we do not detect any defects in gypsy silencing upon knockdown of two central P-body components LSM1 and Me31b, which have been reported to lead to a dispersal of P bodies in Drosophila S2 cells (Eulalio et al, 2007b). Nevertheless, the physical neighbourhood of these two bodies is intriguing and is reminiscent of the neighbouring and/or coinciding localization of a set of piRNA pathway proteins and P-body components in mouse germ cells and Drosophila nurse cells (Aravin et al, 2009; Lim et al, 2009).
Of the three identified proteins, Yb is the only factor that seems specific for the Piwi pathway in somatic support cells. This is consistent with genetic analyses showing that germline-specific mutants for fs(1)Yb are fertile (King et al, 2001). The function of Yb in the somatic support cells is required for maintenance of somatic and indirectly also for germline stem cells (King et al, 2001). In this, fs(1)Yb mutants strongly resemble piwi mutants and indicate once again that the Piwi-dependent piRNA pathway in the somatic support cells is essential for germline stem cell niche maintenance. The specificity of Yb for the somatic cells raises the question, whether there is a related protein with equivalent functions in germline cells. Protein BLAST analysis indeed identified two proteins (CG31755 and CG11133) with similarity to Yb over most of their sequence (Szakmary et al, 2009). Similar to Yb, CG31755 and CG11133 also contain a recognizable Tudor domain downstream of their helicase domain. Both proteins are selectively expressed in ovaries and testes (FlyAtlas). These observations indicate that CG31755 and CG11133 could serve Yb's function within the germline. We note that knockdown of these proteins individually does not impact the somatic piRNA pathway (not shown). CG31755 and CG11133 share considerable similarity to mouse TDRD12, a gene with testis and oocyte-specific expression.
The function of Armi as an essential factor in primary piRNA biogenesis is conserved in mouse (Frost et al, 2010; Zheng et al, 2010). In the absence of the germline-specific orthologue MOV10L1, spermatogenesis is blocked at a similar stage as in MILI mutants and MILI and MIWI2, which is thought to function downstream of MILI in the mouse ping-pong cycle are lacking bound piRNAs. In Drosophila, Armi is per se not required for the ping-pong cycle (Malone et al, 2009), indicating that the function of this RNA helicase is restricted to primary piRNA processing or Piwi loading. Two Armi isoforms are expressed in ovaries with germline cells expressing only the larger one. Although this might suggest functional specialization, we note that the two isoforms show an identical distribution in a glycerol gradient, interact both with Piwi and show identical subcellular localization in follicle cells based on GFP fusions (data not shown).
Similar to Armi, Zuc is well conserved in vertebrates. The mouse orthologue PLD6 is expressed specifically in testes (http://www.biogps.gnf.org) consistent with a function in the piRNA pathway. Zuc contains a single phospho-lipase D domain, which is predicted to confer endo-nucleolytic activity (Zhao et al, 1997). The three important residues for catalysis are conserved in Zuc and a point mutation in the active site (zuc[SG]; Pane et al, 2007) causes a similar de-localization of Piwi from the nucleus as the null mutant (not shown). Our data indicate that Zuc function (potentially endo-nucleolytic generation of piRNA 5′ or 3′ ends) is required to release Piwi and Armi from Yb bodies. We further show that Zuc is essential for Piwi–piRNA biogenesis. This casts doubts on the rather weak effects of zuc mutations on flamenco piRNA biogenesis reported previously (Malone et al, 2009). Genotyping of trans-heterozygous zuc mutants (zuc[HM27]/zuc[Def]) from stocks balanced over CyO is complicated by a dominant wing phenotype of the zuc[Def] allele interfering with confident selection against the CyO balancer. We suspect that this led to a mix of homozygous and heterozygous ovaries analysed in that study. It is unclear, however, how the defects in Piwi localization went unnoticed in such a situation.
The phenotype of armi and zuc mutant ovaries indicates that Piwi biology is similar in germline and somatic support cells. We note that the analysis of piRNAs bound by Piwi and AGO3 also showed a participation of Piwi in the germline-specific ping-pong cycle (Brennecke et al, 2007; Li et al, 2009). However, in armi germline mutants, the ping-pong cycle is still active, yet Piwi is not loaded effectively (Malone et al, 2009). This suggests that primary piRNA biogenesis through Zuc/Armi is the major pathway feeding into Piwi in germline cells.
In summary, we identified the RNA helicases Armi and Yb and the predicted nuclease Zuc as essential components of primary piRNA biogenesis. We further link peri-nuclear Yb bodies to piRNA biogenesis and show that Piwi biology in germline and soma follows a common logic. The described genetic assay system and the availability of the cultured OSC line will be important resources to work towards a mechanistic understanding of piRNA biogenesis and the silencing mechanism, two of the most mysterious open questions in the small RNA field.
Materials and methods
Drosophila stocks
All experiments were performed at 25°C. The used fly strains were
tj-GAL4 (DGRC stock 104055);
flam[restrictive]; gyspy-lacZ (Sarot et al, 2004);
attP2-UAST-Luciferase (Markstein et al, 2008);
RNAi lines were from the Vienna Drosophila RNAi Centre: armi (16205GD; 103589KK), zuc (48764GD; 110430KK), piwi (22235GD; 101658KK, fs(1)Yb (25435GD; 110056KK), tejas (103264KK); Notch (100002KK); no RNAi lines are available at the VDRC for krimp and AGO3.
YFP-Dcp1 (Lin et al, 2006);
armi[1]/TM3 and armi[72.1]/TM3 (Cook et al, 2004);
zuc[HM27]/CyO and Df(2l)PRL/CyO (Pane et al, 2007);
piwi[1]/CyO and piwi[2]/CyO (Lin and Spradling, 1997);
fs(1)Yb[72]/FM6 and fs(1)Yb[1]/FM6 (Swan et al, 2001).
armi[Δ1] is an FRT-mediated deletion based on DGRC stocks e001160 and d07385; it deletes the entire armi, CycJ and CG14971 genes;
Armi-GFP flies carry a genomic rescue construct with an EGFP cassette inserted at the C-terminus by bacterial recombineering;
act-GAL4 flip-out clones were generated using y,w,hsflp122;; act<CD2>GAL4,UAS-GFP; newly enclosed females were heat shocked 1 h at 37°C and dissected 4–5 days later;
FRT-based mitotic clones were based on hsflp122,FRT19A,ubi-GFP and hsflp122;FRT40A armadillo-lacZ; clones were induced by heat-shocking freshly enclosed females on two consecutive days for 1 h at 37°C; flies were dissected 5 days later.
Luciferase assay
Three female flies, carcasses or pairs of ovaries of the genotype [tj-GAL4/+; attP2-UAST-Luciferase/+] and three females of the genotype [attP2-UAST-Luciferase] were homogenized in 100 μl of Glo Lysis Buffer (Promega). A total of 20 μl lysate were mixed 1:1 with Steady-Glo Assay Reagent (Promega). Luminescence was measured with a Synergy Plate Reader after 10 min with a sensitivity of 160 and an integration time of 1 s (results were the average of three experiments).
β-Gal stainings
Ovaries from 2- to 3-day-old flies were dissected in PBS, kept on ice (maximum 30 min), fixed in 0.5% glutaraldehyde/PBS at room temperature (RT; 20 min) and rinsed with PBS. The chromogen reaction was run in staining solution (10 mM PBS, 1 mM MgCl2, 150 mM NaCl, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, 0.1% Triton, 0.1% X-Gal) for 2 h at RT.
Immuno-cytochemistry
Ovaries were dissected in PBS on ice, fixed with 4% paraformaldehyde/PBS with 0.2% Triton X-100 at RT for 20 min and rinsed three times with PBT (PBS with 0.2% Triton X-100). After blocking in BBT (PBT with 0.1% BSA) for 2 h at RT, the ovaries were incubated with primary antibodies in BBT overnight at 4°C. After three PBT washes, secondary antibodies (1:500; Molecular Probes) were incubated 3 h at RT.
OSCs were plated on Concanavalin-A-coated coverslips, fixed with 4% paraformaldehyde in PBS (RT, 20 min), permeabilized with 0.1% Triton X-100 in PBS (10 min) and blocked with 1% BSA in PBS.
Primary antibodies used were α-Tj (1:500, rat; Li et al, 2003), α-Piwi (1:1000, rabbit), α-Piwi (1:1000, mouse; Saito et al, 2006), α-Armi (1:1000, rabbit; Cook et al, 2004), α-β-Gal (1:50, mouse 40-1a from Developmental Studies Hybridoma Bank), α-Yb (1:1000, rabbit; Szakmary et al, 2009), α-gyspy-Env (1:1000, rabbit).
5′ RACE
Total RNA from OSCs was used for the 5′ RACE determination of armi mRNAs (Invitrogen). Primers used were TCCCAGGAATGCTCAAGG (reverse transcription) and CACATCCGACGTGTAGATCCAAG (PCR).
Cell culture
OSCs were cultured as described (Niki et al, 2006). OSCs were transfected with the Cell Line Nucleofector kit V (Amaxa Biosystems), selecting the programme T-029 (K Saito, personal communication).
Piwi-IP MS analysis
OSCs were harvested by trypsinization and lysed in two volumes of lysis buffer (10 mM Hepes pH7, 150 mM NaCl, 5 mM MgCl2 10% glycerol, 1% Triton X-100, complete protease inhibitors (Roche), 1 mM DTT, 1 mM EDTA, 0.1 mM PMSF), followed by manual disruption using a Wheaton dounce homogenizer and a 10 min centrifugation step at 4 degrees. The pellet was re-extracted twice and supernatants were pooled. A total of 3 mg of total protein were incubated (2 h, 4°C) with peptide purified Piwi antibodies (cross-linked to Dynabeads ProteinG (Invitrogen) using a standard dimethyl pimelimidate cross-linking protocol). Pre-immune serum from the same rabbit was used as control. Beads were washed five times with low detergent wash buffer (10 mM Hepes pH7, 150 mM NaCl, 5 mM MgCl2 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 1 mM EDTA, 0.1 mM PMSF), five times with wash buffer (Hepes pH7, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA) and once with 150 mM NaCl. The beads were boiled with SDS sample buffer and analysed by one-dimensional gel electrophoresis and silver staining.
Indicated bands were excised from the gel, processed by a standard in-gel tryptic digest protocol and further analysed by Tandem Mass Spectrometry. Data analysis was with Mascot 2.2.04.
Co-immuno-precipitation
act>3xFLAG-Yb and act>3xFLAG-Piwi constructs were based on the Drosophila Gateway collection (http://www.ciwemb.edu/labs/murphy/Gateway%20vectors.html) and were transfected in OSCs. Non-transfected OSCs were used as negative control. Per sample, a 10 cm plate of OSCs was harvested 4 days after transfection and lysed in 200 μl lysis buffer (10 mM Tris–HCl pH7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Nonidet P40). Lysates were cleared for 10 min at 16 000 g at 4°C and the supernatant incubated for 4 h at 4°C with 7 μg of α-FLAG antibody (SIGMA M2) cross-linked to 30 μl of Invitrogen ProteinG Dynabeads. Beads were washed six times for 10 min with washing buffer (10 mM Tris–HCl pH7.5, 150 mM NaCl, 0.5 mM EDTA) and eluted by boiling in 2 × western blot sample buffer.
Small RNA cloning and analysis
Ovaries were dissected in PBS (on ice). Lysate from ∼100 μl ovaries was obtained by extracting them three times in lysis buffer (10 mM Hepes pH7, 150 mM NaCl, 5 mM MgCl2 10% glycerol, 1% Triton X-100, complete protease inhibitors (Roche), 1 mM DTT, 1 mM EDTA, 0.1 mM PMSF). Piwi was immuno-precipitated using polyclonal rabbit antiserum and small RNAs were extracted with phenol chloroform. Small RNA libraries were generated as previously described (Brennecke et al, 2007) and sequenced using the Illumina G2 platform. Only sequences matching the Drosophila genome (release 5; excluding Uextra) 100% were retained. Libraries were normalized through their unique mappers to the 42AB piRNA cluster to allow for cross-comparisons.
Transposon QPCR analysis
First strand cDNA was obtained by using random primers on Trizol extracted total ovarian RNA from 2- to 3-day-old flies. Quantitative PCR was performed using BioRad IQ SYBR Green Super Mix. Steady-state RNA levels were calculated from the threshold cycle for amplification using the 2−Δ Δ CT method (Livak and Schmittgen, 2001). Fold enrichments are in comparison with a tj-GAL4/+ control sample. Throughout rp49 was used for the normalization and comparable results were obtained by using actin5C for normalization. Average levels and standard deviations were calculated from three biological replicates according to Livak and Schmittgen (2001). Control experiments with template dilution indicated that the efficiencies of target amplification and control genes (rp49 and actin5C) were comparable.
Primers for QPCR analysis were
rp49_for CCGCTTCAAGGGACAGTATCTG
rp49_rev: ATCTCGCCGCAGTAAACGC
actin5C_for: AAGTTGCTGCTCTGGTTGTCG
actin5C_rev: GCCACACGCAGCTCATTGTAG
ZAM_for: CTACGAAATGGCAAGATTAATTCCACTTCC
ZAM_rev: CCCGTTTCCTTTATGTCGCAGTAGCT
Tabor_for: ACGTTGTTCACGACATTAGCCG
Tabor_rev: GGGTTGGTTCGGATCTGACG
HeT-A_for: CGCGCGGAACCCATCTTCAGA
HeT-A_rev: CGCCGCAGTCGTTTGGTGAGT
TART_for: TTTTCCGGATCCAAGTGAAC
TART_rev: TCTGGTCGTCGGAAGTTGTT
Supplementary Material
Acknowledgments
We thank members of the Brennecke laboratory for helpful discussions and A Pelisson for valuable comments on the paper. We are grateful to Andreas Sommer for help with deep sequencing. We thank M Siomi, W Theurkauf, H Lin, D Godt, A Pelisson, M Lin, M Milan, T Schüpbach, M Markstein, the Developmental Studies Hybridoma Bank and the VDRC for flies and antibodies. We thank A Pelisson and A Bucheton for generously sharing their unpublished gypsy Env antibody and M Siomi for OSCs. Small RNA libraries are deposited at GEO (accession no. GSE23560, data sets GSM577958 to GSM577962).
Footnotes
The authors declare that they have no conflict of interest.
References
- Anne J, Ollo R, Ephrussi A, Mechler BM (2007) Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T (2003) The small RNA profile during Drosophila melanogaster development. Dev Cell 5: 337–350 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A (2009) Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabehere D, Ashburner M (2006) Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol 7: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP (1985) tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43: 97–104 [DOI] [PubMed] [Google Scholar]
- Brasset E, Taddei AR, Arnaud F, Faye B, Fausto AM, Mazzini M, Giorgi F, Vaury C (2006) Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology 3: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ (1984) The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell 38: 153–163 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant H, de Rooij D, Bestor T, Hannon GJ (2007) Miwi2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chalvet F, Teysset L, Terzian C, Prud'homme N, Santamaria P, Bucheton A, Pelisson A (1999) Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J 18: 2659–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T (2007) Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol 17: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H (1997) maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development 124: 4661–4671 [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE (2004) The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H (2000) piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Deng WM, Althauser C, Ruohola-Baker H (2001) Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128: 4737–4746 [DOI] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C (2003) COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007a) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E (2007b) P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN (2010) MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA 107: 11847–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA (1995) Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev 9: 2495–2508 [DOI] [PubMed] [Google Scholar]
- Girard A, Hannon GJ (2008) Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM (2001) Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Dramard X, Heidmann T (2002) Regulation of I-transposon activity in Drosophila: evidence for cosuppression of nonhomologous transgenes and possible role of ancestral I-related pericentromeric elements. Genetics 162: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King FJ, Lin H (1999) Somatic signaling mediated by fs(1)Yb is essential for germline stem cell maintenance during Drosophila oogenesis. Development 126: 1833–1844 [DOI] [PubMed] [Google Scholar]
- King FJ, Szakmary A, Cox DN, Lin H (2001) Yb modulates the divisions of both germline and somatic stem cells through piwi- and hh-mediated mechanisms in the Drosophila ovary. Mol Cell 7: 497–508 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z (2009) Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W (2008) Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE (2009) The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA (2007) Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC (2009) Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc P, Desset S, Giorgi F, Taddei AR, Fausto AM, Mazzini M, Dastugue B, Vaury C (2000) Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol 74: 10658–10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD (2009) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D (2003) The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol 5: 994–1000 [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T (2009) piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 186: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Lin MD, Fan SJ, Hsu WS, Chou TB (2006) Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell 10: 601–613 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ (2009) Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP (2006) Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci USA 103: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi MC (2009) Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K (2010) The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell 140: 692–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T (2007) zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T (2010) Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor Domain Protein Tejas. Curr Biol 20: 724–730 [DOI] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A (2007) A novel rasiRNA-mediated silencing pathway downregulates sense gypsy transcripts in the somatic cells of the drosophila ovary. J Virol 81: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG (1994) Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13: 4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A (1995) Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC (2009) A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Josse T, Boivin A, Anxolabehere D (2003) Telomeric transgenes and trans-silencing in Drosophila. Genetica 117: 327–335 [DOI] [PubMed] [Google Scholar]
- Rubin GM, Kidwell MG, Bingham PM (1982) The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell 29: 987–994 [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A (2004) Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E (1991) Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Swan A, Hijal S, Hilfiker A, Suter B (2001) Identification of new X-chromosomal genes required for Drosophila oogenesis and novel roles for fs(1)Yb, brainiac and dunce. Genome Res 11: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H (2009) The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol 185: 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanentzapf G, Devenport D, Godt D, Brown NH (2007) Integrin-dependent anchoring of a stem-cell niche. Nat Cell Biol 9: 1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA (2004) The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol 1: 54–58 [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA (2009) Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Stuckey JA, Lohse DL, Dixon JE (1997) Expression, characterization, and crystallization of a member of the novel phospholipase D family of phosphodiesterases. Protein Sci 6: 2655–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ (2010) Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA 107: 11841–11846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Stein D (2004) RNAi-mediated inhibition of gene function in the follicle cell layer of the Drosophila ovary. Genesis 40: 101–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.