Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks
Defined biochemical and genetic assays show that the MRX complex promotes recruitment of resection nuclease in part by counteracting Ku binding at double strand breaks, providing insight into the switch from initial end processing to processive long-range resection during homologous recombination repair.
Keywords: double-strand break, Ku, Mre11, resection, Saccharomyces cerevisiae
Abstract
Single-stranded DNA constitutes an important early intermediate for homologous recombination and damage-induced cell cycle checkpoint activation. In Saccharomyces cerevisiae, efficient double-strand break (DSB) end resection requires several enzymes; Mre11/Rad50/Xrs2 (MRX) and Sae2 are implicated in the onset of 5′-strand resection, whereas Sgs1/Top3/Rmi1 with Dna2 and Exo1 are involved in extensive resection. However, the molecular events leading to a switch from the MRX/Sae2-dependent initiation to the Exo1- and Dna2-dependent resection remain unclear. Here, we show that MRX recruits Dna2 nuclease to DSB ends. MRX also stimulates recruitment of Exo1 and antagonizes excess binding of the Ku complex to DSB ends. Using resection assay with purified enzymes in vitro, we found that Ku and MRX regulate the nuclease activity of Exo1 in an opposite way. Efficient loading of Dna2 and Exo1 requires neither Sae2 nor Mre11 nuclease activities. However, Mre11 nuclease activity is essential for resection in the absence of extensive resection enzymes. The results provide new insights into how MRX catalyses end resection and recombination initiation.
Introduction
DNA double-strand breaks (DSBs) occur spontaneously during normal cell growth or upon exposure to genotoxic chemicals or ionizing radiation. Failure to repair DSBs could cause cell cycle arrest, mutagenesis, gross chromosomal rearrangements, cell death and tumourigenesis. Cells rely on two major pathways to repair DSBs; homologous recombination (HR) and non-homologous end joining (NHEJ) (reviewed in Shrivastav et al, 2008). The first step in HR is the processing of DNA ends by 5′ to 3′ degradation, and the resulting 3′ single-stranded DNA (ssDNA) becomes the pivotal intermediate for strand-exchange protein binding and homology search (Krogh and Symington, 2004; Pardo et al, 2009). This resection process also signals DNA-damage-induced cell cycle checkpoints and commits a DSB to a specific repair path, as more degradation shifts the balance towards HR (Lee et al, 1998; Aylon et al, 2004; Ira et al, 2004; Mimitou and Symington, 2008; Bernstein and Rothstein, 2009). DNA end resection is tightly controlled by cell type and growth conditions and often is a critical regulation point for eliciting proper DNA-damage response and repair (Ira et al, 2004; Lee and Myung, 2009). Notably, end processing is not just limited to broken chromosome ends, but applies to natural chromosome ends known as telomeres, as proper end resection is essential for telomere maintenance and integrity (Hackett and Greider, 2003).
Given the important function of end resection in cellular DNA damage responses, elucidating the molecular mechanisms of end resection has been the subject of an intense research effort for the last decade. The most recent studies discovered that end resection is a multi-step process that can be divided into two distinct stages: the initial resection, followed by an extensive, long-range end resection (Mimitou and Symington, 2008; Zhu et al, 2008). Heterotrimeric Mre11/Rad50/Xrs2 (MRX) and Sae2 proteins are responsible for the onset of end resection (Mimitou and Symington, 2008; Zhu et al, 2008). Inactivation of one or more of these genes results in the accumulation of un-resected ends, but the ends that do initiate resection are resected efficiently, at a rate indistinguishable from that observed in wild-type cells. In contrast, Sgs1/Top3/Rmi1 (STR)/Dna2 and Exo1 comprise two distinct pathways of end resection that follow the initial end resection and are responsible for the majority of end resection (Gravel et al, 2008; Liao et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008; Budd and Campbell, 2009). Accordingly, inactivation of Sgs1 and/or Exo1 results in a distinct end resection pattern that is different from that seen in mre11, rad50 or xrs2 mutants; resection immediately adjacent to a break remains largely intact, but the resection of several kilobase (kb) pairs distal from a break site is dramatically reduced. The reduction in end resection also accompanies sizable declines in the recombination frequency between non-allelic sequences and single-strand annealing repair between direct repeats 25 kb apart (Mimitou and Symington, 2008; Zhu et al, 2008).
Besides the aforementioned nucleases and helicases, evidence also suggests that the Ku heterodimer regulates HR through inhibition of DNA end processing (Lee et al, 1998; Zhang et al, 2007; Clerici et al, 2008). Ku exhibits an exceptionally strong double-stranded DNA end-binding activity by forming a ring-like structure that clamps on a DSB end (Walker et al, 2001). It is attractive to speculate that Ku may sterically interfere with the binding and/or subsequent activity of the resection enzyme(s). Furthermore, evidence indicates that DSB resection by Exo1 could be repressed by Ku (Wasko et al, 2009). Exo1 performs resection at telomeric regions in the absence of Ku (Maringele and Lydall, 2002), and deletion of Ku suppresses the hypersensitivity of mre11 or rad50 mutants to DSBs in an Exo1-dependent manner (Tomita et al, 2003). However, it is unknown whether Ku interferes with Exo1 at the DNA ends and if so, how a cell responds to the inhibition by Ku to perform efficient end resection.
Identification of new factors involved in end resection highlights many unresolved questions pertaining to the basic mechanisms of end resection. Why do cells rely on multiple nucleases to process DNA ends? How do MRX and Sae2 catalyse the onset of end resection? How is the switch from the initiation of end resection to the more extensive resection accomplished? To address some of these fundamental questions, we investigated how MRX/Sae2 modulate the stable association of Exo1 and Dna2 at an HO endonuclease-induced DSB using chromatin immunoprecipitation (ChIP) assays. The results indicate that MRX facilitates the loading of Exo1 onto the DSB substrate partly through suppressing excess Ku accumulation at DNA ends. MRX also controls access of Dna2 to the DNA break in a Ku-independent manner. The results provide clues to the early resection events, and they reveal how these nucleases coordinate their actions to achieve optimal levels of end resection.
Results
MRX complex facilitates association of Exo1 and Dna2 to DNA break
In Saccharomyces cerevisiae, end resection is catalysed by multiple nucleases, wherein the combined function of the MRX complex and Sae2 is primarily limited to the onset of end resection, whereas the extensive resection requires Sgs1, Dna2 and Exo1 proteins. The question is then how a cell achieves the transition from the early to long-range resection. We hypothesize that the MRX complex and the Sae2 protein may have functions in this transition by assisting the assembly of the Sgs1/Dna2 and/or Exo1 resection complex at DNA breaks. To test this premise, we examined whether the association of Exo1-myc, Dna2-myc and Sgs1-myc at DNA ends depends on a functional MRX complex using ChIP assays with anti-myc antibodies in mre11Δ and/or rad50Δ mutants carrying a galactose-inducible HO endonuclease gene. HO endonuclease cleaves once per entire genome at the MAT locus of chromosome III. The DSB is not repaired by HR because the homologous sequences HMR and HML are deleted in these strains (Figure 1A). In addition, rejoining of the DSB by NHEJ restores the HO-recognition sequence, and will lead to repetitive cleavage when HO is persistently induced. Only a small percentage of cells (∼0.2%) are able to repair DSBs by imprecise NHEJ by generating a mutation within the HO-recognition site.
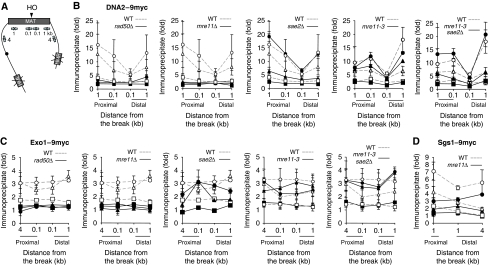
Figure 1.
The MRX facilitates binding of Dna2 and Exo1 at a DSB. The enrichment of Dna2–9myc (B), Exo1–9myc (C) or Sgs1–9myc (D) flanking the HO-induced DSB at the MAT locus (A) in wild-type (dotted lines, open symbols), rad50Δ, sae2Δ, mre11Δ or mre11-3 mutants (solid lines, filled symbols) were shown at indicated times post-HO induction (1 h: square, 2 h: triangle and 3 h: circle). Fold immunoprecipitate represents the ratio of the anti-myc IP PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control and the amount of input DNA. Arrows indicate primers used for ChIP assay. The mean values±s.d. from three independent experiments are shown.
We found that the recruitment of Dna2-myc and Exo1-myc to the HO-induced DNA break is severely repressed in mre11Δ or rad50Δ mutants at 1, 2 or 3 h after galactose induction (Figure 1B and C). Deletion of MRE11 also caused a moderate reduction in the recruitment of Sgs1 to a DSB (Figure 1D), likely reflecting the resection defect in this mutant. The results suggest that the MRX complex mediates recruitment of nucleases Dna2 and Exo1 to DNA ends.
Mre11 nuclease activity and Sae2 are dispensable for Exo1 and Dna2 recruitment at DNA break
Biochemically, the MRX complex exhibits double-strand 3′ to 5′ exonuclease and single-strand endonuclease activities (Paull and Gellert, 1998; Usui et al, 1998; Moreau et al, 1999; Trujillo and Sung, 2001). Sae2 is also an endonuclease that acts on ssDNA and stimulates Mre11 exonuclease activity (Lengsfeld et al, 2007). Therefore, we considered the possibility that the MRX complex and/or Sae2 recruit Dna2 and/or Exo1 by using their nuclease activities to produce limited ssDNA at DNA ends. To test this premise, we examined loading of the myc-tagged Dna2 or Exo1 at the HO-induced DSB in the nuclease-defective mre3-11 mutant or sae2Δ mutant using ChIP assays with anti-myc antibody. We found that Dna2-myc and Exo1-myc binding at the DSB remains largely unchanged in the mre11-3 mutant, suggesting that the nuclease activity of Mre11 is not required for recruitment of Dna2 and Exo1 nucleases to DSB (Figure 1B and C). Similarly, Sae2 is dispensable for the recruitment of either nuclease to the DSB (Figure 1B and C). Finally, in sae2Δ mre11-3 double-mutant, recruitment of Dna2 or Exo1 remains comparable with wild-type cells (Figure 1B and C).
Mre11 and Dna2 nucleases have redundant functions in processing HO-induced DSB in yeast
In budding yeast, the nuclease domain of Mre11 has very limited, if any, function in HO-induced DSB end resection and repair. Indeed, mre11 nuclease mutants show only mild sensitivity to IR when compared with complete MRE11 deletion mutant (Bressan et al, 1998, 1999; Moreau et al, 1999; Tsubouchi and Ogawa, 2000; Lee et al, 2002). However, the Mre11 nuclease domain has a crucial function in DSB resection/repair in fission yeast (Williams et al, 2008) and mammals (Jazayeri et al, 2006; Buis et al, 2008). CtIP, the fission yeast and human orthologue of Sae2 is also an indispensable component of DSB-damage response, along with MRN (Limbo et al, 2007; Sartori et al, 2007). In contrast, the budding yeast Sae2 does not directly interact with MRX, but rather, it acts as a structure-specific nuclease, processing hairpins and terminal DNA adducts (reviewed in Mimitou and Symington, 2009).
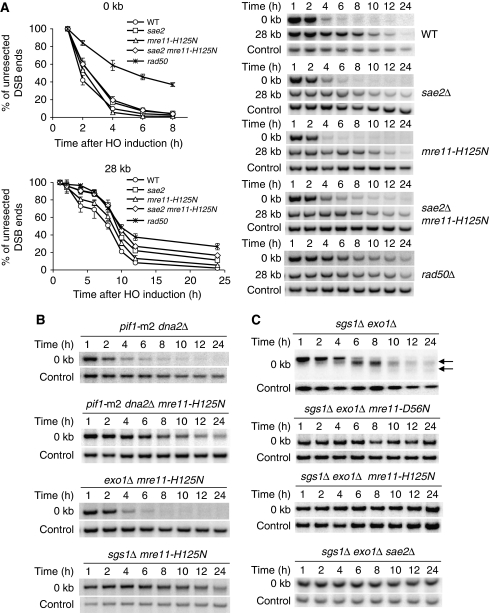
There are several possibilities to explain the different functions of the Mre11 nuclease domain in these organisms. One is that Sae2, which was shown to have nuclease activity itself (Lengsfeld et al, 2007), can substitute for Mre11, whereas human CtIP cannot. This leaves Mre11 as the primary nuclease in the MRN/CtIP complex (Buis et al, 2008). Alternatively, Dna2 can substitute Mre11 nuclease for processing 5′ strands even at the very ends of the DSB in budding yeast, whereas human Dna2 has more limited functions in end resection. Finally, in budding yeast, but not in fission yeast or mammals, Mre11 could initiate the resection process by loading Dna2 and Exo1 nucleases without initial cleavage. To test the first possibility, we compared resection side-by-side in wild-type, rad50Δ, sae2Δ, mre11-H125N and double-mutant sae2Δ mre11-H125N strains. We examined the initiation and long-range resection by a Southern blot hybridization-based assay using two DNA probes, MATa and FEN2, to detect the removal of the 5′ strand immediately proximal to the break and 28 kb distal from the DNA break, respectively (Zhu et al, 2008). As shown in Figure 2A, the most dramatic defect in initial resection is observed in the rad50Δ mutants. In the single-mutant sae2Δ cells, we observed a mild defect in initiation of resection, whereas the resection in the mre11-H125N cells is comparable with that seen in the wild-type cells. Finally, resection in the double-mutant sae2Δ mre11-H125N is indistinguishable from the sae2Δ single mutant. Given that Dna2 and Exo1 are loaded efficiently in the double-mutant sae2Δ mre11-3 (Figure 1B and C), we conclude that Mre11 and Sae2 nucleases do not perform redundant functions in resection of HO breaks.
Figure 2.
Mre11 and Sae2 nucleases have non-redundant functions in resection of HO-induced DSB. (A) The 5′-strand resection was analysed at the break and 28 kb away from the DSB in indicated mutants. Plots showing average percentage of unprocessed 5′ strand at these sites are shown. (B) The 5′-strand resection was analysed in indicated mutants by Southern blot hybridization. Dna2 and Mre11 nucleases have redundant functions in initial resection. (C) The 5′-strand resection at the break was analysed in mutants lacking both extensive resection enzymes and either Mre11 or Sae2 nuclease by Southern blot hybridization. Products of limited resection occurring in the absence of Exo1 and Sgs1 are indicated by arrows.
Next, we tested whether Dna2 can substitute for Mre11 nuclease activity in initial end resection by creating mre11-H125N dna2Δ pif1-m2 mutant and performing a Southern blot hybridization-based assay. The pif1-m2 mutation suppresses the lethality of the dna2Δ, but the pif1-m2 mutation by itself does not affect the rate of resection (Zhu et al, 2008). As shown in Figure 2B, initial resection is more defective in mre11-H125N dna2Δ pif1-m2 than in dna2Δ pif1-m2. Similarly, initial end resection is more defective in sgs1Δ mre11-H125N than in sgs1Δ, whereas exo1Δ mre11-H125N is no more defective than exo1Δ. The observations that mre11-H125N dna2Δ pif1-m2 cells are more sensitive to ionizing radiation, MMS, phleomycin and HU than each single gene mutant further supports the idea that Mre11 and Dna2 have redundant functions in resection (Budd and Campbell, 2009) (Supplementary Figure S1). Together, our results suggest that (i) budding yeast MRX activates extensive resection enzymes independently of its nuclease activity. The nuclease activity of Mre11 cannot be substituted by Sae2; (ii) in budding yeast, Dna2 can substitute Mre11 in processing DSB ends. These results may explain the basis of far less severe phenotypes of yeast mre11 nuclease mutants compared with those in human cells.
Mre11 nuclease with Sae2 resect DNA ends in the absence of extensive resection enzymes
Previously, we and others have shown that the MRX complex together with the Sae2 nuclease produces a few hundred nucleotides of ssDNA at the break when extensive resection enzymes Sgs1 (or Dna2) and Exo1 are absent (Mimitou and Symington, 2008; Zhu et al, 2008). Here, we examined whether, in the absence of extensive resection enzymes, Sae2- and Mre11-dependent end resection is dependent on the nuclease activity of Mre11. To address this question, we examined end resection in strains lacking both Sgs1 and Exo1, but expressing nuclease-deficient alleles of mre11 (mre11-H125N and mre11-D56N) from its own genomic locus. We found that the residual resection in the absence of Sgs1 and Exo1 depends on Mre11 nuclease activity, because expression of the nuclease-deficient allele of Mre11 failed to produce cleavage products in the Southern blot-based resection assay (Figure 2C). These results suggest that in the absence of Sgs1 and Exo1, both Mre11 and Sae2 nucleases are needed to generate short ssDNA. This ssDNA, however, is insufficient for normal levels of DSB repair and checkpoint activation (Gravel et al, 2008; Zhu et al, 2008).
MRX complex suppresses excess Ku protein accumulation at DNA breaks
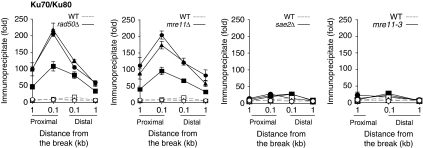
We next sought to determine how the MRX complex facilitates the recruitment of Exo1 and Dna2 at DNA breaks. Previously, we found that the absence of MRE11 induces a significant (∼20-fold) accumulation of Ku and Lif1 proteins at DNA breaks in budding yeast cells (Zhang et al, 2007; Wu et al, 2008; Figure 3). In contrast, the deletion of SAE2 or the expression of the nuclease-deficient mre11 caused only a modest (<2.5-fold) accumulation of Ku proteins at the DSB (Figure 3; Supplementary Figure S2). These results suggest that the level of Ku proteins at a DSB inversely correlates with the level of Exo1 and/or Dna2 proteins. Being an avid end-binding protein, Ku may interfere with the binding of Exo1 or Dna2 to DNA ends, and the MRX complex may neutralize the inhibitory effect of Ku by limiting its accumulation at DNA breaks. According to this model, we anticipated that deletion of both MRE11/RAD50 and YKU70 would restore the binding of Exo1-myc and/or Dna2-myc at DNA break.
Figure 3.
MRX suppresses excess Ku protein recruitment at a DSB. Kinetics of Ku recruitment to the HO-induced DSB in wild-type (dotted lines, open symbols), rad50Δ, mre11Δ, sae2Δ and mre11-3 mutants (solid lines, filled symbols) were determined by ChIP assays. The values at 1 h (square), 2 h (triangle) and 3 h (circle) are shown. Fold immunoprecipitate represents the ratio of the Ku IP PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control and the amount of input DNA. Data represent the mean±s.d. of three or more independent experiments.
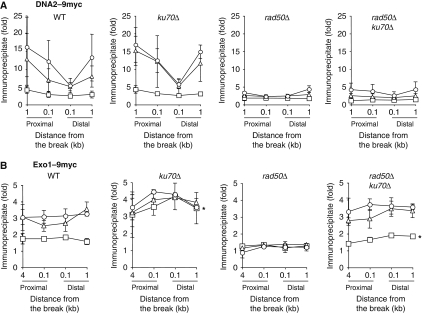
Consistent with this prediction, we found that deletion of YKU70 dramatically elevates the binding of Exo1-myc, but only modestly increases the levels of Dna2-myc at the DNA break, suggesting that Ku mostly inhibits the association of Exo1 to DNA breaks (Figure 4A and B). Furthermore, deletion of both YKU70 and RAD50 allows Exo1 to bind to the DNA break at a level comparable with that in wild-type cells (see Figure 4B). On the contrary, deletion of YKU70 allows only a modest increase in the binding of Dna2 at the DNA break in rad50Δ (see Figure 4A; Supplementary Figure S3). These results suggest that excess Ku proteins at DNA ends impede the binding of Exo1 at DNA ends. In addition, MRX likely has an important function in neutralizing the inhibitory effect of Ku on Exo1.
Figure 4.
Ku inhibits the recruitment of Exo1 at a DSB. The enrichment of Dna2-9myc (A) and Exo1-9myc (B) flanking the HO-induced DSB at the MAT locus in the wild-type, yku70Δ, rad50Δ and yku70Δ rad50Δ mutants were shown at indicated times post-HO induction (1 h: square, 2 h: triangle and 3 h: circle). Fold immunoprecipitate was calculated as described in Figure 1. The mean values±s.d. from three independent experiments were shown. IP values at 1 h post-HO expression was marked with ‘*' to highlight the level of Exo1-9myc recruitment in yku70Δ and yku70Δ rad50Δ mutants.
Deletion of Ku improves resection and HR in the absence of the MRX complex
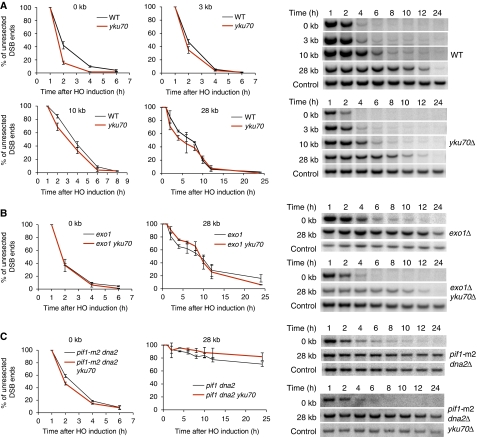
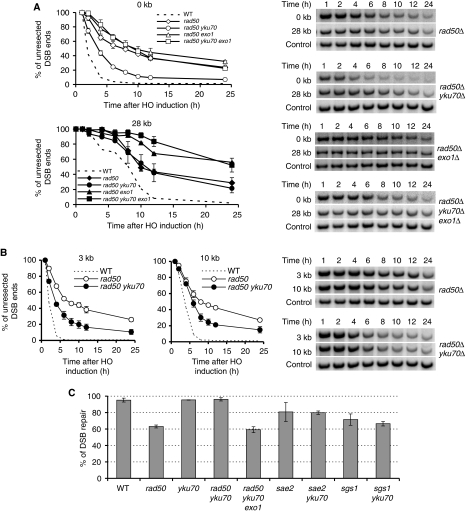
The results presented above suggest that increased level of Ku observed in rad50 mutant cells may explain the basis of their DSB resection defect. To test this premise, we examined whether the deletion of YKU70 suppresses the resection defect in mrx mutants. We expected that Exo1-dependent resection would be restored in the rad50Δ yku70Δ double mutant. Consistent with the previous reports (Clerici et al, 2008; Lee et al, 2008), initiation of end resection is increased in the yku70Δ mutant (Figure 5A). Two hours after HO expression, about 25% more cells resected 5′ strands next to the break compared with wild-type cells. Resection is also faster at distances 3, 10 and 28 kb from the break, although the difference is not as dramatic as those at the very ends of the DSB. These results suggest that Ku proteins primarily impede the initiation of resection (Figure 5A). To test whether faster resection close to the break in yku70Δ strains is dependent on Exo1 or Dna2, we examined resection in exo1Δ yku70Δ and in dna2Δ pif1-m2 yku70Δ mutant using Southern blot-based resection assay (Figure 5B and C). As shown in Figure 5B, Exo1 is required for the elevated resection observed in yku70Δ. Deletion of DNA2 alone slightly decreases the initiation of resection, but very severely decreases extensive resection (Zhu et al, 2008). Resection is also modestly improved in dna2Δ pif1-m2 yku70Δ (Figure 5C), suggesting that both Exo1 and Dna2 contribute to the fast resection in yku70Δ. Importantly, we found that deletion of YKU70 in rad50Δ dramatically (but not fully) improved end resection close to the break and partially improved resection 3 and 10 kb away from the break. At 28 kb away from the break, resection was comparable in rad50Δ and rad50Δ yku70Δ mutants (Figure 6A and B). The increased resection close to the break in rad50Δ yku70Δ mutants was dependent on Exo1 and Dna2 (Figure 6A; Supplementary Figure S4).
Figure 5.
Ku complex inhibits Exo1-dependent resection. (A–C) The 5′-strand resection was analysed at the break and 28 kb away from the DSB in indicated mutants. Plots are showing the average percentage of unprocessed 5′ strands at these sites. Resection in wild-type and yku70Δ mutants were compared at 3 and 10 kb away from the break.
Figure 6.
Deletion of YKU70 suppresses resection and repair defects of rad50Δ. (A) Initial resection at the break and long-range resection measured 28 kb from DSB were analysed in indicated mutants. Southern blots are shown. (B) Resection in rad50Δ and rad50Δ yku70Δ were compared at 3 and 10 kb away from the break. (C) Analysis of DSB repair through gene conversion in an ectopic recombination assay using indicated mutants.
Previous studies showed that the repair of HO-induced breaks by HR is reduced (∼2-fold) in mrx mutants (Ivanov et al, 1994). To test whether elimination of Ku also suppresses DSB repair defect in mrx mutants, we constructed a yku70Δ rad50Δ mutant carrying the MATa and the MATa-inc sequences at chromosome V and III, respectively. The ectopic recombination frequency between MAT sequences was measured upon induction of HO endonuclease (Ira et al, 2003; Zhu et al, 2008) (Figure 6C). We found that the improved end resection in yku70Δ rad50Δ results in the complete suppression of a DSB repair defect (Supplementary Figure S1). Deletion of YKU70 did not improve mild DSB repair defects in sgs1Δ or sae2Δ. As expected, the improved repair in rad50Δ yku70Δ was almost entirely dependent on Exo1 (Figure 6C). Collectively, the results suggest that one of the primary functions of the MRX complex in end resection is to antagonize excessive binding of Ku proteins at DNA ends and to recruit Dna2 and Exo1 nucleases.
Ku inhibits Exo1-dependent resection in the absence of the MRX complex in vitro
To elucidate the effect of Ku on Exo1- and MRX-dependent resection, we expressed and purified recombinant yeast Exo1 protein and subjected to the nuclease activity tests using a linear duplex DNA as a substrate in the presence or absence of the recombinant yeast Ku heterodimer and/or the MRX complex. The 5′-nucleolytic degradation was quantitatively monitored by separating the reaction mixtures in native agarose gel electrophoresis and analysed by staining of the DNA with SYBR green and by Southern blot hybridization with a strand-specific RNA probe for the 3′ strand at the break site (Figure 7A). We discovered that Ku proteins strongly inhibit 5′-strand degradation by Exo1 (compare lanes 2 and 4). Furthermore, addition of MRX to the reaction can partially neutralize the inhibitory effect of Ku on Exo1-mediated 5′ degradation of duplex DNA ends, as seen by the increase in ssDNA in the presence of MRX and Sae2 (Figure 7A, compare lanes 4 and 8). The results are congruent with the model that MRX catalyses an initial stage of end resection by suppressing the excess Ku protein binding at DNA ends and stimulating the exonuclease activity of Exo1.
Figure 7.
Ku and MRX complexes oppositely regulate the nuclease activity of Exo1. (A) DNA resection assays were performed with linearized DNA as a substrate (cut with SphI) and analysed by native agarose gel electrophoresis and SYBR green staining for the double-stranded DNA (bottom panel) followed by non-denaturing Southern hybridization with a strand-specific RNA probe for the 3′ strand, as previously described (Hopkins and Paull, 2008) (top panel). Reactions contained 10 ng (0.35 nM) pNO1 DNA, 2 nM yeast wt Exo1, MRX (8.3 or 25 nM), Sae2 (8.6 or 26 nM) and 10 nM Ku heterodimer as indicated. The position of single-stranded DNA in the gel was marked as ‘ss', and that of the un-resected plasmid is marked as double stranded, ‘ds'. Migration of molecular weight markers (kb) are shown in the lane marked ‘M'. (B) A model describing how MRX and Sae2 proteins facilitate end resection at HO breaks.
Discussion
Four distinct sets of nucleases and their associated protein components (MRX, Sae2, STR/Dna2 and Exo1) are required for initiation and long-range end resection in budding yeast (Mimitou and Symington, 2008; Zhu et al, 2008; Budd and Campbell, 2009). The important question is then what entails the initial stage of end resection, and how do MRX and Sae2 contribute to this process? More broadly, why do cells evolve to retain multi-step end resection? The goal of this study is to address some of these questions by elucidating the function of MRX and Sae2 protein in the assembly of Sgs1/Dna2 and Exo1 at the HO-induced DSB. Our results suggest that MRX and Sae2 proteins facilitate end resection at HO breaks in budding yeast by three distinct manners (Figure 7B): neutralize the inhibitory effect of Ku, promote stable association of Exo1 and Dna2 to DNA ends, and catalyse limited end resection itself when extensive resection enzymes are absent.
Mre11 as anti-Ku factor
Previously, we and other groups reported that the absence of MRX causes a dramatic increase in the Ku protein level at DNA ends using ChIP assays (Zhang et al, 2007; Wu et al, 2008). Amassing Ku proteins at DNA ends in the absence of Mre11 or Rad50 does not reflect solely on the resection defect in these mutants, because inhibition of end resection by other means, such as arresting cells at G1 or inhibition of Cdk1 by excess Sic1 production, did not result in the same level of increase in the Ku binding at DNA ends (Zhang et al, 2007) (Supplementary Figure S5). Rather, we suggest that the MRX complex has a more direct function in repressing excess Ku (and likely repressing Dnl4/Lif1 as well) build-up at DNA ends.
We found that excess Ku proteins at DNA ends severely impede the binding of Exo1 at DNA ends. Thus, one function of MRX is to repress Ku accumulation at DNA ends and thereby stimulates Exo1 recruitment and resection activity. Indeed, deletion of MRE11 causes severe end resection and DSB repair defects, which are largely restored by the deletion of Ku, thereby improving Exo1-dependent resection and repair (Bressan et al, 1999). Deletion of YKU70 also offsets severe hypersensitivity to MMS, phleomycin and HU in mre11Δ (Supplementary Figure S1). Supporting our observation in live cells, we further showed that in in vitro end resection reconstituted by purified Exo1 and the model DNA substrate, the nuclease activity of Exo1 is strongly inhibited by Ku and that the inclusion of MRX can partially overcome this inhibition.
Exo1 is a 5′ exonuclease that can target both linear duplex and ssDNA for degradation (Tran et al, 2004). Therefore, Exo1 is theoretically capable of initiating end resection from duplex DNA ends without additional assistance from other nucleases or helicases. However, the inability of Exo1 to gain access to Ku-occupied DNA ends renders MRX an essential factor for early resection because MRX suppresses Ku accumulation at DNA ends. Correspondingly, Ku has been implicated in inhibiting Exo1 in telomeric DNA processing (Maringele and Lydall, 2002; Bertuch and Lundblad, 2004; Wasko et al, 2009), which likely shares some of the features of DSB end processing. The effect of excess Ku on Exo1 binding may also account for why over-expression of Exo1 can only render partial improvement of the end resection defect in mre11Δ (Tsubouchi and Ogawa, 2000; Lee et al, 2002); excess Ku proteins in mre11Δ may act as a barrier for Exo1 activity even when over-produced. Exo1 can still contribute to resection in a limited way even in the absence of MRX complex as double-mutant exo1Δ mre11Δ show a much more dramatic defect in resection than each single mutant (Tsubouchi and Ogawa, 2000).
How does the MRX complex suppress Ku protein accumulation at DNA ends? We found that the nuclease activity of Mre11 or Sae2 is dispensable for Ku repression. In addition, a brief survey of a few available rad50 or mre11 mutants for their ability to suppress excess Ku binding at DNA ends did not reveal any apparent biochemical activities of MRX complex responsible for this function (data not shown). Only the null mutants deficient in DNA binding or MRX complex formation showed this defect, raising the possibility that structural integrity of MRX complex at DNA ends may be important for suppressing Ku accumulation. Previous structural studies suggest that Ku proteins form a ring-shaped molecule and binds to DNA ends by threading through its hole in the middle (Walker et al, 2001). Ku then slides further inside to leave the ends open for further Ku loading (de Vries et al, 1989). MRX also binds to DNA ends, bridging the broken ends together by the long coiled-coil structure of Rad50 (Chen et al, 2001; Hopfner et al, 2001; Trujillo and Sung, 2001). Therefore, MRX may repress excess Ku binding at DNA ends by competing for the DNA end and thereby setting the upper limit for Ku binding. We further speculate that MRX uniquely positions itself at DNA ends to limit the number of Ku molecules bound for the end-joining reaction.
Lastly, if MRX regulates Exo1 recruitment solely by neutralizing inhibitory action of Ku, we expect that the level of Exo1 in yku70Δ mre11Δ should be identical to that in yku70Δ because MRX should become dispensable for Exo1 recruitment in the absence of Ku proteins. However, the level of Exo1 at DSB in yku70Δ mre11Δ is substantially less than that in yku70Δ (see the IP value marked by ‘*' in Figure 4B). This suggests that Mre11 has yet another function in the recruitment of Exo1 at DNA break besides neutralizing Ku-mediated inhibition on the binding of Exo1 to DNA ends. The biochemical nature of this additional Exo1 recruitment function of MRX is presently unknown.
Mre11 promotes binding of Dna2 at DNA ends
In addition to its function as an anti-Ku factor, MRX possesses yet another activity in assisting the loading of Dna2 at DNA ends. Such an activity is clearly distinct from its anti-Ku function, because deletion of YKU70 in the mre11Δ or rad50Δ strains did not substantially improve the binding of Dna2 at HO-cleaved ends in the ChIP assays. In addition, reduced loading of Dna2 in mrx mutants is not due to the low amount of ssDNA or the RPA proteins in yku70Δ rad50Δ, in which resection is largely restored to that of wild-type cells (Figure 6A and data not shown).
Dna2 participates in lagging-strand DNA synthesis, processing Okazaki fragments by cleaving displaced 5′ flaps (Bae et al, 2001; Ayyagari et al, 2003; Kao et al, 2004). Therefore, we surmise that Dna2 weakly associates with unprocessed duplex DNA; MRX may catalyse limited strand unwinding at DNA ends to make ends more amenable for the stable association of Dna2. Previous biochemical studies showed ATP-dependent partial-strand unwinding activity in the recombinant human MRN complex, and such an activity should be particularly suited for this function (Paull and Gellert, 1999). However, no evidence is yet available that MRX facilitates Dna2 binding at DNA ends by strand unwinding.
Function of Mre11 nuclease activity in DNA end resection
We found that efficient association of Dna2 or Exo1 to DNA ends does not rely on the nuclease activities of Mre11 or Sae2. Instead, the nuclease activities encoded by Mre11 or Sae2 are required for limited end resection when Sgs1 and Exo1 are inactivated. This is in striking contrast to fission yeast and mammalian cells, in which both Mre11 nuclease domain and Sae2 orthologue CtIP are essential for DSB ends resection (Buis et al, 2008; Williams et al, 2008). This difference between budding yeast and other organisms cannot be explained by the fact that Sae2 nuclease activity is absent in human beings, as we show that Sae2 nuclease does not have redundant function with Mre11 nuclease. Surprisingly, we instead found that Dna2 substitutes Mre11 in initial resection in S. cerevisiae. Therefore, phenotypic differences of mre11 nuclease mutants in budding, fission yeast, and mammalian cells may be explained by different functions of human and yeast Dna2 in the end resection process. The function of Dna2 in resection in human cells was not extensively studied, but it is likely that human Dna2 may have more limited functions in end resection. The nuclease activities of Mre11 and Sae2 become important when broken DNA ends require additional processing, such as those bound by Spo11 or other covalent adducts (Furuse et al, 1998; Usui et al, 1998; Bressan et al, 1999; Moreau et al, 1999; Neale et al, 2005). Dna2 cannot fully compensate for Mre11 nuclease processing such ‘dirty' DNA ends.
In summary, our results suggest that each nuclease complex has evolved to deal with specific DNA or DNA–protein structures during DNA end resection. MRX and Sae2 are evolved to have highly specialized functions for initial end resection by neutralizing high-affinity end-binding Ku proteins and facilitating stable association of Exo1 and Dna2 at DNA ends. Sgs1/Dna2 and Exo1 may be optimized for long-range end resection at the expense of initiating end resection on their own. This may explain, in part, why multiple sets of nucleases are required for end resection throughout evolution.
Materials and methods
Yeast strains and plasmids
Most yeast strains used in this study are derivatives of JKM139 (hoΔ MATa hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG'ura3-52 ADE3::GAL::HO) and the genotypes of the strains are listed in the Supplementary Table 1. Plasmids expressing wild-type Mre11 and mre11-H125N were generated by swapping the SacI/KpnI digestion fragment of pRS414-Mre11 and pRS414-mre11-H125N onto pRS416 (Moreau et al, 1999). The mre11-3 was generated by replacing mre11Δ::KANr with the PCR product amplifying mre11-3 allele and the URA3 marker gene placed at the 3′ of MRE11 open reading frame from the plasmid, pTAP8, generously provided by Petrini group. The mre11-3 mutation was validated by digesting PCR products carrying MRE11 gene with BspHI followed by sequencing of the entire MRE11 open reading frame. The wild-type MRE11, but not mre11-3 should be cleaved by BspHI.
Chromatin immunoprecipitation
ChIP assays were performed as described previously (Zhang et al, 2007), using an antibody specific to the myc epitope tag, Ku heterodimer, Rpa1, Mre11 or Rad50. The immunoprecipitated DNA was analysed by real time quantitative PCR using multiple sets of primers that anneal 0.1, 1.0 and/or 4.0 kb proximal and distal from the DSB, as well as primers specific for the PRE1 gene situated on chromosome V as a control. The antibodies for Ku were a generous gift from Dr Alan Tomkinson and the antibodies for Mre11 and Rad50 were from Dr Patrick Sung. The antibodies for Rpa1 were a gift from Dr Steven Brill.
DSB end resection and DSB repair assays
Resection was analysed at HO endonuclease-induced non-repairable breaks at MAT locus at chromosome III in haploid strains as previously described (Zhu et al, 2008). Analysis was performed at least three times in each mutant strain. To determine the viability in response to a repairable DSB, cells carrying ectopic recombination donor sequences described in detail previously (Ira et al, 2003) were grown overnight in 2% raffinose medium and appropriate culture dilutions were plated onto YEPD and YEP-galactose plates. The proportion of viable cells was estimated by dividing the number of colony forming units on YEP-galactose plates by that on YEPD plates.
Protein expression and purification
Recombinant yeast Mre11, Rad50 and Xrs2 complexes were co-expressed in Sf21 insect cells, and purified as previously described (Bhaskara et al, 2007). Exo1 was expressed using the Bac-to-Bac system (Invitrogen) in Sf21 cells and purified as described (MN and TP, manuscript submitted). Sae2 was expressed in E. coli and purified as described (Lengsfeld et al, 2007). Yeast Ku heterodimer was a gift from Dr Alan Tomkinson.
Nuclease assays
Nuclease assays with 4.4 kb unlabelled plasmid DNA substrate (Figure 7A) contained 10 ng plasmid DNA (pNO1, Topogen) linearized at a single site with SphI (NEB), and 25 mM MOPS pH 7.0, 50 mM NaCl, 1 mM DTT, 5 mM MgCl2 and 0.5 mM ATP in 10 μl reactions and were incubated at 30°C for 1 h. Reactions were stopped by the addition of 0.2% SDS and 10 mM EDTA, incubated with 1 μg proteinase K at 37°C for 15 min, analysed on 0.8% native agarose gels run in 1X Tris–acetate–EDTA (40 mM Tris–acetate, 1 mM EDTA) buffer. Agarose gels were stained with SYBR green (Invitrogen) and imaged with a Typhoon system (GE), then washed into 20 × SSC (3 M NaCl, 0.3 M sodium citrate) and DNA was transferred by capillary action onto nylon membranes (NEN) overnight in 20 × SSC. Membranes were probed with RNA complementary to the 3′ strand of the DNA substrate in a 500 bp region on one end, adjacent to the SphI site. The probes were internally labelled with [α-32P]CTP (NEN) and were made using Riboprobe System T7 (Promega) and purified with RNAeasy extraction (Qiagen) according to the manufacturer's instructions. After extensive washing at 42 and 65°C, the membranes were analysed by phosphorimager (GE).
Supplementary Material
Acknowledgments
We thank S Brill, J Haber, J Campbell and A Tomkinson for gifts of strains, plasmids, antibodies and purified proteins, and D Villarreal for an editorial help. We also thank the members of the SEL, TTP and GI laboratory for helpful discussion. This work is funded by grants from NIH to SEL (GM071011, 3R01 GM071011), TTP (CA094008) and GI (GM080600 and 3R01GM080600). SEL is a scholar of the Leukemia and Lymphoma Society. Studies performed in the Paull laboratory were also supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Gomes XV, Gordenin DA, Burgers PM (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J Biol Chem 278: 1618–1625 [DOI] [PubMed] [Google Scholar]
- Bae SH, Bae KH, Kim JA, Seo YS (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461 [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Rothstein R (2009) At loose ends: resecting a double-strand break. Cell 137: 807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V (2004) EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics 166: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT (2007) Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell 25: 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Baxter BK, Petrini JH (1999) The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol 19: 7681–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL (2009) Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One 4: e4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP (2008) The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries E, van Driel W, Bergsma WG, Arnberg AC, van der Vliet PC (1989) HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J Mol Biol 208: 65–78 [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K (1998) Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J 17: 6412–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Greider CW (2003) End resection initiates genomic instability in the absence of telomerase. Mol Cell Biol 23: 8450–8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473–485 [DOI] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT (2008) The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE (1994) Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol 14: 3414–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA (2004) On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J Biol Chem 279: 15014–15024 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Lee SE (2008) Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454: 543–546 [DOI] [PubMed] [Google Scholar]
- Lee SE, Bressan DA, Petrini JH, Haber JE (2002) Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 1: 27–40 [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lee SE, Myung K (2009) Faithful after break-up: suppression of chromosomal translocations. Cell Mol Life Sci 66: 3149–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, Yan H (2008) Identification of the Xenopus DNA2 protein as a major nuclease for the 5′ → 3′ strand-specific processing of DNA ends. Nucleic Acids Res 36: 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S (2005) Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A (2009) DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Erdeniz N, Symington LS, Liskay RM (2004) EXO1-a multi-tasking eukaryotic nuclease. DNA Repair (Amst) 3: 1549–1559 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Sung P (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H (2000) Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716 [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614 [DOI] [PubMed] [Google Scholar]
- Wasko BM, Holland CL, Resnick MA, Lewis LK (2009) Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair (Amst) 8: 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE (2008) Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE (2007) Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 14: 639–646 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.