Here, the AAV Reference Standard Working Group presents the results of their international collaboration to produce and characterize a recombinant AAV serotype 2 Reference Standard Material. The purpose of this material is to provide a universal standard for particle titer, vector genome titer, and infectious titer for AAV2 gene transfer vectors.
Abstract
A recombinant adeno-associated virus serotype 2 Reference Standard Material (rAAV2 RSM) has been produced and characterized with the purpose of providing a reference standard for particle titer, vector genome titer, and infectious titer for AAV2 gene transfer vectors. Production and purification of the reference material were carried out by helper virus–free transient transfection and chromatographic purification. The purified bulk material was vialed, confirmed negative for microbial contamination, and then distributed for characterization along with standard assay protocols and assay reagents to 16 laboratories worldwide. Using statistical transformation and modeling of the raw data, mean titers and confidence intervals were determined for capsid particles ({X}, 9.18 × 1011 particles/ml; 95% confidence interval [CI], 7.89 × 1011 to 1.05 × 1012 particles/ml), vector genomes ({X}, 3.28 × 1010 vector genomes/ml; 95% CI, 2.70 × 1010 to 4.75 × 1010 vector genomes/ml), transducing units ({X}, 5.09 × 108 transducing units/ml; 95% CI, 2.00 × 108 to 9.60 × 108 transducing units/ml), and infectious units ({X}, 4.37 × 109 TCID50 IU/ml; 95% CI, 2.06 × 109 to 9.26 × 109 TCID50 IU/ml). Further analysis confirmed the identity of the reference material as AAV2 and the purity relative to nonvector proteins as greater than 94%. One obvious trend in the quantitative data was the degree of variation between institutions for each assay despite the relatively tight correlation of assay results within an institution. This relatively poor degree of interlaboratory precision and accuracy was apparent even though attempts were made to standardize the assays by providing detailed protocols and common reagents. This is the first time that such variation between laboratories has been thoroughly documented and the findings emphasize the need in the field for universal reference standards. The rAAV2 RSM has been deposited with the American Type Culture Collection and is available to the scientific community to calibrate laboratory-specific internal titer standards. Anticipated uses of the rAAV2 RSM are discussed.
Introduction
Recombinant adeno-associated viral (rAAV) vectors are rapidly becoming the gene delivery vehicle of choice for gene transfer, with numerous publications describing their use in animal models and more recently in clinical trials (Warrington and Herzog, 2006; Mueller and Flotte, 2008). The movement of the gene therapy field toward the use of rAAV vectors in clinical trials is largely due to the demonstration of long-term transgene expression in animal models with little associated toxicity and good overall safety profiles in humans (Snyder and Flotte, 2002; Moss et al., 2004; Warrington and Herzog, 2006; Maguire et al., 2008; Mueller and Flotte, 2008; Brantly et al., 2009). Most of the historic data involve rAAV serotype 2 vectors, but vector systems based on other AAV serotypes with more efficient gene delivery profiles in specific tissues are currently in human trials (Brantly et al., 2009; Nienhuis, 2009) and their use will likely increase.
A major problem associated with the body of data to date has been the inability to normalize vector doses administered by different investigators to animals and humans. Thus, there is a need for a reference standard that is recognized by the rAAV research community and that is used to normalize laboratory-specific internal reference standards and test vector titers related to common reference standard units. This need is not new to the field of gene therapy and has previously been addressed for adenoviral vectors. The Adenovirus Reference Material Working Group (ARMWG) developed and characterized the adenovirus reference material (ARM) for the purpose of normalizing titers and doses of gene therapy vectors based on adenovirus type 5 (Hutchins, 2002). Following this example, the U.S. Food and Drug Administration (FDA) and the National Institutes of Health (NIH) Recombinant DNA Advisory Committee (RAC), together with the support of the National Gene Vector Laboratory (NGVL), a program of the NIH National Center for Research Resources (NCRR), encouraged academic and industry scientists within the AAV community to form an AAV Reference Standard Working Group (AAVRSWG) charged with the development a high-quality rAAV reference standard material (Snyder and Flotte, 2002). The AAVRSWG is a volunteer organization and comprises members from both industry and universities in nine different countries, and the International Society for BioProcess Technology (www.ISBioTech.org), under the guidance of the FDA and NIH (Moullier and Snyder, 2008; Potter et al., 2008). Although new serotypes of AAV are currently emerging as efficient gene delivery vectors, the AAVRSWG decided that the first AAV reference standard material should be based on the prototypical AAV serotype 2 because this is by far the best characterized serotype. The approach used in the development of this reference standard lays the groundwork for the development of reference standard materials based on other serotypes. Indeed, a second AAVRSWG has been formed for the development of an AAV8 reference material (Moullier and Snyder, 2008).
The AAV2RSWG recognized that the rAAV2 reference standard material (rAAV2 RSM) must be supplied in sufficient quantity to each requestor for use in all necessary tests at each location, be of high quality, and remain stable for an extended period of time. The group drew up guidelines for the production and purification of the rAAV2 RSM, which were carried out at the Vector Core of the University of Florida's Powell Gene Therapy Center (Gainesville, FL) (Potter et al., 2008). The production process involved cotransfection of batches of ten 10-layer Cell Factories containing HEK293 cells with an AAV2 genome/eGFP transgene plasmid and a second plasmid encoding the AAV2 capsid proteins and necessary helper functions. The transfected cells were harvested and the vector was purified by sequential rounds of column chromatography. A Quality Control subcommittee of the AAV2RSWG was formed for the purpose of characterizing the rAAV2 RSM. In consultation with members of the AAV2RSWG, the committee selected the following characterization assays: (1) capsid titer by A20 enzyme-linked immunosorbent assay (ELISA; Progen Biotechnik, Heidelberg, Germany); (2) vector genome titer by quantitative polymerase chain reaction (qPCR); (3) infectious titer by median tissue culture infective dose (TCID50) with qPCR readout and by transduction (green fluorescent protein [GFP] readout); and (4) purity and capsid identification by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
The AAV2 RSM was distributed worldwide to 16 laboratories that volunteered to conduct one or more of the characterization assays. Testing proceeded from July 2008 to March 2009, at which point the quantitative data were collated and statistically analyzed to determine mean titer and confidence intervals. Preliminary analysis showed significant variance and nonnormal data distribution with all of the assays except for particle titer determination. The variance was observed despite providing a standardized protocol and reagents to the testing group and highlights the need within the AAV community for a reference standard with which assay titers can be normalized. Using statistical transformation to better approximate normal distributions and modeling to offset the lack of independence of duplicate assays within an institution, appropriate estimations of the mean titers and confidence intervals have been determined.
Materials and Methods
Reference standard material production
Production and purification of the rAAV2 RSM were carried out at the Vector Core of the University of Florida's Powell Gene Therapy Center between February 2006 and January 2007. The production process has been described in detail elsewhere (Potter et al., 2008) and is briefly summarized here. Production was initiated by cotransfection of HEK293 cells in ten 10-layer Nunc Cell Factories (Thermo Fisher Scientific, Waltham, MA) with plasmid pTR-UF-11, containing the vector genome and eGFP expression cassette (Burger et al., 2004), and the pDG-KanR helper plasmid, a kanamycin-resistant version of pDG (Grimm et al., 1998) at a 1:1 molar ratio, using a calcium phosphate precipitation method. After a 60-hr incubation at 37°C, 5% CO2, transfected cells were washed with phosphate-buffered saline (PBS) and harvested in PBS containing 5 mM EDTA. Samples of cells were combined with spent tissue culture medium and tested for mycoplasma and in vitro adventitious agents. Cells were collected by centrifugation and stored at −20°C until purified. For vector purification, cells were thawed, lysed with 0.5% sodium deoxycholate, treated with Benzonase (Merck, Darmstadt, Germany), and then disrupted by microfluidization. Virions were then purified by STREAMLINE (GE Healthcare Life Sciences, Piscataway, NJ) heparin affinity chromatography. Peak fractions were pooled and applied to a Phenyl Sepharose (GE Healthcare Life Sciences) chromatography column. The flow-through collected from this second purification step was purified and concentrated by sulfopropyl cation-exchange chromatography. Vector was eluted with 5–10 ml of 135 mM NaCl in PBS (equivalent to 285 mM ionic strength) and stored at −80°C. Eighteen batches were prepared and pooled. The purified bulk was diluted to ∼2 × 1011 vector genomes (VG)/ml with 135 mM NaCl in PBS, and sterile filtered into two 1.3-liter portions. This filtered formulated bulk was stored frozen (–80°C) until vialed. One of the 1.3-liter portions of the bulk was thawed and refiltered, and 0.5 ml was dispensed into 2087 vials at the American Type Culture Collection (ATCC, Manassas, VA) to produce VR-1616, the rAAV2 RSM.
Predistribution testing
Mycoplasma
Mycoplasma testing of the production culture cell harvest and of the filtered formulated bulk purified material was performed at a contract testing laboratory (WuXi AppTec, Shanghai, China). For the cell harvest material, medium and supernatant from each production batch were sampled and pooled for testing. A total of 1 × 107 cells in 15 ml of production culture supernatant was tested according to Good Laboratory Practice (GLP), using the “Points to Consider” assay described by the FDA Center for Biologics Evaluation and Research (CBER/FDA, 1993). This assay detects the presence of mycoplasma by both indirect (cell culture) and direct (broth and agar) assays. The test article was incubated with monkey kidney cells, stained with a DNA-binding fluorochrome (Hoechst stain), and evaluated microscopically by epifluorescence. Agar and broth flasks were inoculated with test article and incubated anaerobically and aerobically, respectively. Broths were subcultured onto agar plates on days 3, 7, and 14 days postinoculation. All plates were examined no sooner than 14 days postinoculation. Purified bulk rAAV2 RSM was tested by a modification of the “Points to Consider” assay, in which three cycles of inoculation and incubation on Vero cells precede the assay to allow amplification of mycoplasma. Both harvested material and purified bulk material were also tested, using a PCR assay directed against the 16S rRNA gene of various mycoplasma species (WuXi AppTec).
Bioburden
The presence of aerobes, fungi, spores, and anaerobes in the purified bulk material was quantified by plating on various media under specific incubation conditions (WuXi AppTec). The sensitivity of this assay is <5 colony-forming units (CFU)/ml.
Sterility
The vialed rAAV2 RSM (cat. no. VR-1616; ATCC) was tested for sterility at the Indiana University Vector Production Facility (Indianapolis, IN) according to GLP guidelines. The presence of aerobes, anaerobes, and fungi was tested by direct inoculation of thioglycolate broth, Trypticase soy broth, and Sabouraud dextrose agar and incubation for 14 days at the appropriate temperature. Negative and positive (Bacillus subtilis, Candida albicans, and Bacteroides vulgatus) controls were included.
Endotoxin
The vialed rAAV2 RSM was also tested for endotoxin at the Indiana University Vector Production Facility according to GLP guidelines and using the Limulus amebocyte lysate gel-clotting assay. Test samples were assayed in duplicate and diluted 2-fold with water. The test reagent (100 μl) was added to the rAAV2 RSM dilution and incubated at 37°C for 60 min, and the tube was examined for the presence of a gel clot. Negative, positive, and spiked controls were included. The sensitivity of the assay was 0.06 endotoxin unit (EU)/ml.
Previaling stability study
An rAAV2-GFP vector preparation at 2 × 1011 VG/ml, in the same formulation as the rAAV2 RSM (PBS + 135 mM NaCl), was placed in polypropylene and glass vials (not siliconized) at 0.5 ml per vial. Vials of each type were stored at both temperatures. Vials held at room temperature were assayed for infectious titer after 1 hr, 1 day, 3 days, and 7 days; vials stored at −80°C were assayed for infectious titer at 1 hr, 1 day, 14 days, 35 days, and 124 days (Potter et al., 2008).
RSM handling stability study
The rAAV2 RSM was thawed on ice and aliquoted into siliconized plastic vials. Aliquots were either tested immediately for transducing titer, infectious titer, and vector genome titer or stored at 4°C or −80°C for 3 days and then tested.
rAAV2 RSM handling
For AAV2 RSM characterization, each testing laboratory received two vials from the ATCC on dry ice. On receipt both vials were stored frozen at −70°C to −90°C. One vial was thawed at room temperature while mixing gently and then kept on wet ice. Within 1 hr of thawing, the infectious titer and transducing titer assays were conducted. The remainder of the thawed vial was stored at 4°C and mixed gently on use. Within 5 days of vial thaw, the particle titer, vector genome titer, and purity/identity assays were performed. These steps were repeated for the second vial, starting on a different calendar day.
rAAV2 RSM characterization assays
Brief descriptions of each characterization assay follow. For those wishing to reproduce these assays, detailed protocols can be found at the links specified below or may be requested directly from M. Lock or R. Snyder.
Particle titer
Particle concentration was determined by each laboratory, using four separate dilution series from a single vial in the Progen AAV2 titration ELISA (cat. no. PRATV; Progen Biotechnik), by comparison with a standard curve prepared from a previously titered rAAV2 preparation. See the protocol posted at http://www.isbiotech.org/ReferenceMaterials/pdfs/AAV2_capsid_titer_assay_V2.pdf
Vector genome titer
Vector genome concentration was determined in duplicate, testing one replicate from each of two vials, by quantitative PCR of serial dilutions of rAAV2 RSM against a standard curve of plasmid pTR-UF-11 (MBA-331; ATCC) (Burger et al., 2004) See the protocol posted at http://www.isbiotech.org/ReferenceMaterials/pdfs/AAV2_RSS_genome_copy_titration_QPCR.pdf
Transducing titer
Serial 10-fold dilutions of rAAV2 RSM were made on HeLaRC32 cells (CRL-2972; ATCC) (Chadeuf et al., 2000) and coinfected with adenovirus type 5 (VR-1516; ATCC). Fluorescence microscopy was used to count GFP-expressing cells at 72 hr postinfection. See the protocol posted at http://www.isbiotech.org/ReferenceMaterials/pdfs/AAV2_RSS_Infectious_titer_assays_V2.pdf
Infectious titer
Serial 10-fold dilutions of an rAAV2 reference standard stock (RSS) were made on HeLaRC32 cells (CRL-2972; ATCC) and coinfected with adenovirus type 5 (VR-1516; ATCC). Seventy-two hours postinfection total cell DNA was extracted and analyzed for vector genome copies by qPCR. Input vector genomes were subtracted and TCID50 titers were calculated according to the method of Kärber (Kärber, 1931). See the protocol posted at http://www.isbiotech.org/ReferenceMaterials/pdfs/AAV2_RSS_Infectious_titer_assays_V2.pdf
Purity and identity
The purity and identity of the rAAV2 RSM were evaluated by SDS–PAGE, using SYPRO ruby (Invitrogen, Carlsbad, CA) or silver staining (SilverXpress; Invitrogen). The AAV2 VP1, VP2, and VP3 capsid protein bands were evaluated for their stoichiometry and size. Purity relative to nonvector impurities visible on stained gels was determined. Vector identity was verified by observation of the electrophoretic banding pattern expected for AAV2 and by comparison with positive controls. See the protocol posted at http://www.isbiotech.org/ReferenceMaterials/pdfs/AAV2_RSS_Identity-Purity_assay.pdf
Results
rAAV2 RSM production and predistribution testing
The goal of the AAV2RSWG manufacturing subcommittee was to make a single lot of an rAAV-eGFP vector with a yield of 1 × 1015 vector genomes. Eighteen batches of ten 10-layer cell factories containing 293 cells were transfected and the resulting AAV vector was purified from the transfected cells by sequential heparin affinity, hydrophobic interaction, and cation-exchange chromatography. The final column eluates from the 18 batches prepared were pooled for a total of 150 ml. The genome titer of this purified bulk was assayed by dot–blot assay and, using this method, it was determined that the material contained 5.69 × 1014 VG (Potter et al., 2008). The purified bulks were combined, diluted to ∼2 × 1011 VG/ml, and sterile filtered into two 1.3-liter portions. This filtered formulated bulk was stored frozen (–80°C) in anticipation of vialing. In March 2008, one of the 1.3-liter portions of the bulk stock was thawed, refiltered, and dispensed into 2087 vials at the ATCC to produce the rAAV2 RSM (cat. no. VR-1616). The vials, frozen in the repository at the ATCC, are available for distribution. The other 1.3-liter portion of the bulk material remains frozen at the ATCC, to be dispensed at a later date if demand warrants (Potter et al., 2008).
Before freezing, the filtered formulated bulk was sampled (5 ml) and tested for bioburden by a contract testing laboratory (WuXi AppTec). Aerobes, fungi, spores, and obligate anaerobes all tested negative with an assay sensitivity of <5 CFU/sample. Before distribution, the vialed rAAV2 RSM was tested under GLP guidelines for sterility (aerobes, anaerobes, fungi) and endotoxin at the Indiana University Vector Production Facility. No bacterial or fungal contamination was detected and endotoxin levels were less than 0.06 EU/ml. The production culture cell harvest and the filtered purified bulk material were tested at WuXi AppTec for mycoplasma contamination as detailed in Materials and Methods (GLP “Points to Consider” assay). The harvest material tested positive for Mycoplasma arginini (bovine origin) and the tests were valid (i.e., all controls performed). The filtered purified bulk material was tested for mycoplasma, using a modified assay with increased sensitivity. In this test, a sample of the bulk material was passaged three times on Vero cells before performing the GLP “Points to Consider” assay. The bulk material tested negative for mycoplasma whereas the spike-in controls performed as expected, indicating that the assay was valid. Last, the harvest and bulk materials were tested by PCR (WuXi AppTec) for a 16S rRNA gene region specific to various mycoplasma species. Using this assay, the harvest was confirmed positive and the filtered formulated bulk again tested negative, with all controls performing as expected. Thus whereas the harvested cells were positive for mycoplasma, the purified bulk was negative for viable mycoplasma and no mycoplasma DNA was detected, so it was concluded that the purification process likely separated and/or inactivated the contaminating mycoplasma present in the harvest material (Potter et al., 2008).
Beta testing
The AAV2RSWG Quality Control subcommittee was formed for the purpose of characterizing the rAAV2 RSM. Decisions regarding the characterization assays required were made in consultation with the AAV2RSWG, and members were invited to submit their assay protocols. These protocols were reviewed and a lead protocol was chosen for each assay. The assays chosen included (1) confirmation of the serotype and capsid particle titer by A20 ELISA (Progen Biotechnik); (2) determination of vector genome titer by qPCR; (3) determination of infectious titer by median tissue culture infective dose (TCID50) with qPCR readout and by transduction (GFP readout); (4) evaluation of the purity, capsid subunit stoichiometry, and chemical integrity of the capsid by SDS–PAGE.
During the protocol selection process it was realized that the highest level of assay reproducibility in the various testing laboratories could be ensured only if certain reagents were provided along with the rAAV2 RSM. The reagents, which are now available from the ATCC, include a cell line expressing the AAV2 rep and cap genes (HeLa32; kindly provided by P. Moullier, INSERM UMR649 Nantes, France), a concentrated adenovirus helper virus (the adenovirus type 5 reference standard material, ARM) (VR-1516; ATCC) for the transduction and infectivity assays, and the pTR-UF-11 vector plasmid (kindly provided by S. Zolotukhin, Powell Gene Therapy Center and Division of Cellular and Molecular Therapy, Department of Pediatrics, University of Florida, Gainesville, FL) used to manufacture the AAV2 RSM viral vector. For both genome titer and infectious titer assays, a qPCR primer–probe set directed to the simian virus 40 (SV40) poly(A) sequence and a dilution series specific to the RSM were selected. For the purity and identity assay, commercially available assay reagents with the highest sensitivity were suggested. Using these reagents, the modified assay protocols were beta tested at the University of Pennsylvania Gene Therapy Program against both in-house standards (AAV2.CMV.eGFP and AAV2.CMV.lacZ) and the rAAV2 RSM itself. The genome, infectious, and transduction titers of the in-house standards correlated well with the titers previously established by in-house assays (see Table 2; and data not shown) and vector genome-to-infectious or transduction unit ratios were similar to those published elsewhere (Salvetti et al., 1998; Zolotukhin et al., 1999; Zen et al., 2004). Repeating the beta testing with the rAAV2 RSM allowed appropriate dilution ranges to be established for several of the characterization assays. The finalized protocols were posted at the International Society for BioProcess Technology website (www.ISBioTech.org) and the HeLaRC32 cells (CRL-2972; ATCC), pTR-UF-11 plasmid (MBA-331; ATCC), and ARM (VR-1516; ATCC) assay reagents were made available through the ATCC. The rAAV2 RSM (VR-1616; ATCC) was distributed along with the required reagents and handling instructions to 16 laboratories worldwide (Table 1) that volunteered to conduct one or more of the characterization assays.
Table 2.
Qualification of Reference Standard Material (RSM) Testing Methods Using In-House Standards and Beta Testing of rAAV2 RSM Under Various Storage Conditions
| |
In-house standards |
rAAV2 RSM |
|||
|---|---|---|---|---|---|
| AAV2.CMVe.GFP | AAV2.CB.lacZ | At thaw | 4°Ca | −80°Ca | |
| Transducing titer (GFU/ml) | 1.42 × 1010 | NA | 1.89 × 109 | 1.23 × 109 | 8.38 × 108 |
| Infectious titer (TCID50 IU/ml) | 6.96 × 1010 | 2.42 × 1011 | 1.65 × 1010 | 3.56 × 109 | 6.23 × 109 |
| Physical titer (vector genomes/ml) | 4.53 × 1012 | 2.15 × 1012 | 3.39 × 1010 | 3.39 × 1010 | 3.41 × 1010 |
| Vector genomes: infectious units | 65 | 8.87 | 2.05 | 9.53 | 5.39 |
| Vector genomes: transducing units | 319 | NA | 17.90 | 27.50 | 40.70 |
Abbreviations: GFU, GFP (green fluorescent protein) forming units; NA, not available; TCID50, median tissue culture infective dose.
Assayed 3 days postthawing, after storage at the indicated temperature.
Table 1.
rAAV2 Reference Standard Material Testing Laboratories
| University of Naples Federico II, Italy |
| University of North Carolina Vector Laboratories, USA |
| Universitat Autònoma de Barcelona, Spain |
| Research Institute at Nationwide Children's Hospital, USA |
| University of Florida, USA |
| Laboratoire de Thérapie Génique, France |
| Généthon, France |
| Applied Genetic Technologies, USA |
| International Center for Genetic Engineering and Biotechnology (ICGEB), Italy |
| German Cancer Research Center (DKFZ), Germany |
| University of Pennsylvania, USA |
| Jichi Medical University, Japan |
| Sangamo BioSciences, USA |
| Université Libre de Bruxelles, Belgium |
| Amsterdam Molecular Therapeutics, The Netherlands |
| Children's Hospital of Philadelphia, USA |
Before filling the rAAV2 RSM, a study was conducted with a different lot of rAAV2-GFP vector in the same formulation to evaluate the short-term stability of the vector at room temperature (the filling condition) and at −80°C (the storage condition). Vials were filled with the beta test vector and held at the test temperatures for the time periods indicated (see Materials and Methods) and were then assayed for infectious titer. In all scenarios, a 30–40% drop was observed between the initial titer and the average of all samples taken during the time course and it was assumed that this loss likely indicated absorption to the container surfaces at the low vector concentration (Potter et al., 2008), because these containers were not siliconized. After filling the rAAV2 RSM, a limited study was also conducted to evaluate the stability of the rAAV2 RSM after post-thaw storage at 4°C, and after refreezing at −80°C. The purpose of the study was to determine the appropriate conditions for handling of the RSM once received by the testing laboratory. Transduction titers fell 35 and 56% after storage or refreezing at 4°C and −80°C, respectively, and infectious titer fell 78 and 63%, respectively (Table 2). Conversely, no change was observed in the vector genome titers under the various storage conditions. These results indicated that the postthaw storage conditions were adversely affecting the potency of the rAAV2 RSM and that this decrease was not due to absorption to the siliconized aliquot vials used in this study, because the vector genome titer was unchanged. On the basis of these data the decision was made to test two separate aliquots of the RSM and to determine transducing and infectious titers within 1 hr of thawing a vial.
rAAV2 RSM characterization
The characterization phase of the rAAV2 RSM proceeded from July 2008 to March 2009. On receipt, the RSM was evaluated by the 16 testing laboratories according to the posted protocols, and data were recorded on the assay worksheets provided, which contained the necessary calculations for titer determination. Fifteen laboratories performed the particle titer assay (31 replicates), 16 laboratories performed the genome titer assay (36 replicates), 10 laboratories performed the infectious titer assay (23 replicates), and 12 laboratories performed the transducing titer assay (19 replicates). In some cases (four of the tests), substantial experimental deviation from the posted protocols was noted and these data have been omitted from the statistical analyses.
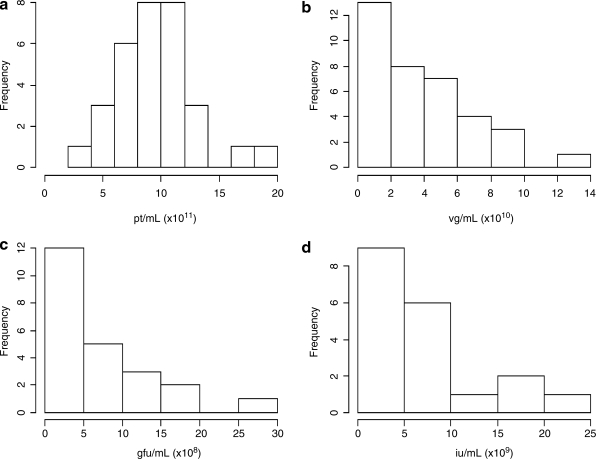
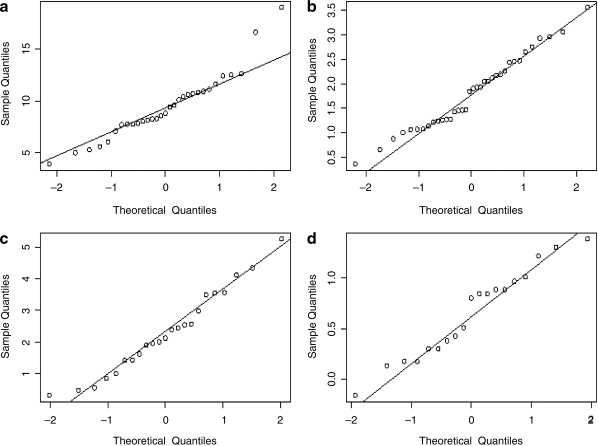
The raw data that emerged from the testing laboratories for the four quantitative titer assays (Table 3) was statistically analyzed to determine true mean titer values and confidence intervals. The distribution was first visualized as histograms (Fig. 1), and it was noted that with the possible exception of the particle titer results, the data do not appear to be normally distributed. Because valid estimation of the mean and confidence interval relies on an underlying normal distribution, it was clear that some form of transformation was warranted. Common statistical transformation methods employed are square root, natural log, and log base 10. Means and intervals are calculated on the transformed data and the results are then back-transformed to the original measurement scale; the data for each assay were processed in this way, using all three transformations. Analysis of the mean and median (for normally distributed data these two values are the same) and skewness and kurtosis estimates confirmed that the transformations were performing as expected (data not shown). Figure 2 shows the results of the most successful transformations for each assay as quantile–quantile plots. In this analysis the quantiles (i.e., 5%, 10%, etc.) obtained from the transformed data are compared with the quantiles that would be expected for a normal distribution. A line that extends through the 25th and 75th quantile is shown on the plots and the nearer the points are to this line, the more normal the data distribution. With the exception of two extreme points, the capsid particle titer assay data appeared normally distributed without the need for transformation. For the vector genome titer and the transduction titer, square root transformation appeared to approximate normal distribution and seemed warranted. For the infectious titer, a log base 10 transformation appeared the most appropriate. The transformed data are summarized in Table 4. For each assay, 2 and 3 standard deviation limits were calculated corresponding to nominal 95 and 99.7% confidence bounds on individual values. Any test result lying outside of the 3 standard deviations was considered to be an outlier. Using this criterion only a single test result (from the particle titer assay) was determined to be an outlier and was removed from the analysis; the values reported in Table 4 exclude this data point.
Table 3.
rAAV2 Reference Standard Material Raw Characterization Data
| Laboratory | Replicate | Particle titer (ELISA) (pt/ml) | Genome titer (qPCR) (VG/ml) | Transducing titer (green cells) (GFU/ml) | Infectious titer (TCID50) (IU/ml)a |
|---|---|---|---|---|---|
| A | 1 | 1.08 × 1012 | 4.68 × 1010 | 1.27 × 109 | |
| 2 | 1.26 × 1012 | 8.77 × 1010 | 1.26 × 109 | ||
| B | 1 | 8.25 × 1011 | 7.03 × 1010 | 3.63 × 108 | 6.32 × 109 |
| 2 | 8.28 × 1011 | 7.58 × 1010 | 1.00 × 108 | 1.36 × 109 | |
| 3 | 9.39 × 1010 | 7.15 × 107 | 2.00 × 109 | ||
| C | 1 | 7.70 × 1011 | 8.60 × 1010 | 9.80 × 106 | |
| 2 | 6.05 × 1011 | 4.19 × 1010 | |||
| D | 1 | 8.80 × 1011 | 1.01 × 1010 | 1.70 × 109 | 1.02 × 1010 |
| 2 | 7.83 × 1011 | 3.71 × 1010 | 2.77 × 109 | 6.96 × 109 | |
| E | 1 | 1.16 × 1012 | 1.58 × 1010 | ||
| 2 | 1.04 × 1012 | 1.14 × 1010 | |||
| 3 | 1.61 × 1010 | ||||
| F | 1 | 1.66 × 1012 | 2.17 × 1010 | 6.60 × 108 | |
| 2 | 1.01 × 1012 | 2.12 × 1010 | 5.70 × 108 | ||
| G | 1 | 1.90 × 1012 | 1.13 × 1010 | 4.02 × 108 | 3.23 × 109 |
| 2 | 9.39 × 1011 | 1.30 × 1010 | 2.67 × 109 | ||
| H | 1 | 2.04 × 1010 | |||
| 2 | 2.13 × 1010 | ||||
| I | 1 | 8.56 × 1011 | 5.93 × 1010 | 2.20 × 107 | |
| 2 | 7.08 × 1011 | 6.10 × 1010 | 2.95 × 107 | ||
| J | 1 | 1.24 × 1012 | 3.39 × 1010 | 1.89 × 109 | 1.65 × 1010 |
| 2 | 1.07 × 1012 | 4.49 × 1010 | 1.22 × 109 | 2.42 × 1010 | |
| K | 1 | 5.29 × 1011 | 3.72 × 1010 | 2.01 × 108 | |
| 2 | 7.75 × 1011 | 3.62 × 1010 | 2.04 × 108 | ||
| L | 1 | 8.13 × 1011 | 1.31 × 109 | 5.96 × 108 | 1.50 × 109 |
| 2 | 4.99 × 1011 | 1.63 × 1010 | 6.48 × 108 | 6.96 × 108 | |
| M | 1 | 1.11 × 1011 | 1.16 × 1010 | 8.88 × 108 | 2.00 × 109 |
| 2 | 1.25 × 1011 | 1.53 × 1010 | 4.52 × 108 | 1.50 × 109 | |
| N | 1 | 3.93 × 1011 | 1.47 × 1010 | 3.82 × 108 | 2.00 × 1010 |
| 2 | 5.59 × 1011 | 7.63 × 109 | 2.62 × 108 | 7.66 × 109 | |
| 3 | 9.58 × 1011 | 4.24 × 109 | 2.40 × 109 | ||
| O | 1 | 1.06 × 1012 | 6.04 × 1010 | ||
| 2 | 1.09 × 1012 | 1.27 × 1011 | |||
| P | 1 | 8.02 × 1011 | 4.20 × 1010 | 7.66 × 109 | |
| 2 | 7.74 × 1011 | 5.10 × 1010 | 6.96 × 109 | ||
| 3 | 4.82 × 1010 | 9.28 × 109 | |||
| Mean | 9.43 × 1011 | 3.82 × 1010 | 6.94 × 108 | 7.00 × 109 | |
| Standard deviation | 3.19 × 1011 | 2.97 × 1010 | 7.03 × 108 | 6.70 × 109 |
Four replicate test results were omitted because of documented experimental deviation from the posted protocols.
FIG. 1.
Histograms displaying the distributions of (a) particle titer (pt/ml), (b) vector genome titer (VG/ml), (c) transducing titer (GFU/ml), and (d) infectious titer (IU/ml).
FIG. 2.
Quantile–quantile plots displaying sample versus normal quantiles of (a) particles per milliliter, (b) square root of vector genomes per milliliter, (c) square root of transducing units per milliliter, and (d) log10 infectious units per milliliter. Lines pass through the 25th and 75th quantiles.
Table 4.
rAAV2 Reference Standard Material Titer Estimates after Transformation
| Titer units (method) | Transformationa | Mean | Lower 95% confidence limit for the mean | Upper 95% confidence limit for the mean | ± 2 SD | ± 3 SD |
|---|---|---|---|---|---|---|
| Particles/ml (ELISA) | Untransformed | 9.11 × 1011 | 8.10 × 1011 | 1.01 × 1012 | 3.73 × 1011–1.45 × 1012 | 1.04 × 1011–1.78 × 1012 |
| Vector genomes/ml (qPCR) | Square root | 3.26 × 1010 | 2.41 × 1010 | 4.25 × 1010 | 8.82 × 108–1.10 × 1011 | 0–1.66 × 1011 |
| Transducing units/ml (green cells) | Square root | 5.29 × 108 | 2.99 × 108 | 8.23 × 108 | 0–2.43 × 109 | 0–3.90 × 109 |
| Infectious units/ml (TCID50) | Log10 | 4.49 × 109 | 2.75 × 109 | 7.29 × 109 | 5.94 × 108–3.39 × 1010 | 2.16 × 108–9.31 × 1010 |
Used to better qualify the assumption of normal distribution for the purpose of determining distributional values.
An assumption we have made when calculating the mean values and confidence intervals is that each test result is independent of another; however, this assumption does not take into account that all institutions submitted duplicate (and in some cases triplicate) test results for the assays. To assess the degree of correlation between the two duplicate samples, Pearson coefficients were determined. These estimates ranged from 0.57 to 0.84 for the four assays (where a value of 1 indicates a perfect correlation). The results indicate that there is a significant correlation within institution and that the assumption that each result is independent is violated. To account for this correlation, the transformed data were modeled, using a linear random effect modeling approach (Littell et al., 2006). This allows for a unique component associated with each institution to be included in the model, under the assumption that these institutional random effects have a mean of zero. When the correlation within an institution is accounted for, the precision of the mean estimate as illustrated by the width of the 95% confidence interval is decreased (Table 5). Taking the transformed, modeled data as the true estimate of the mean, we have arrived at the following determinations for the rAAV2 RSM: the mean particle titer is 9.18 × 1011 particles/ml with 95% confidence that the true value lies in the range of 7.89 × 1011 to 1.05 × 1012 particles/ml; the mean vector genome titer is 3.28 × 1010 vector genomes/ml with 95% confidence that the true value lies in the range of 2.70 × 1010 to 4.75 × 1010 vector genomes/ml; the mean transducing titer is 5.09 × 108 transducing units/ml with 95% confidence that the true value lies in the range of 2.00 × 108 to 9.60 × 108 transducing units/ml; and the mean infectious titer is 4.37 × 109 TCID50 IU/ml with 95% confidence that the true value lies in the range of 2.06 × 109 to 9.26 × 109 TCID50 IU/ml. The mean vector genome titer of 3.28 × 1010 VG/ml is almost 1 log lower than the titer of 2 × 1011 VG/ml assessed for the diluted purified bulk harvest before vialing. The discrepancy between the bulk material and final fill may be due to loss of vector after filtering of the bulk product, to the different assay methods used for the titering (dot–blot vs. qPCR), or a combination of both. The bulk vector was titered at the University of Florida, using the method of dot–blot hybridization to determine the appropriate formulation volume for the final fill. It is possible that the loss, if any, occurred during the final filtration and filling of the dilute reference standard material at the ATCC (diluted nearly 1000 times relative to preparations that are used preclinically or clinically). The product that was vialed and frozen constitutes the reference standard material that was characterized, and that is available to the community.
Table 5.
Final rAAV2 Reference Standard Material Titer Estimates after Transformation and Modeling
| Titer units (method) | Transformationa | Mean | Lower 95% confidence limit for the mean | Upper 95% confidence limit for the mean | ± 2 SD | ± 3 SD |
|---|---|---|---|---|---|---|
| Particles/ml (ELISA) | Untransformed | 9.18 × 1011 | 7.89 × 1011 | 1.05 × 1012 | 3.73 × 1011–1.45 × 1012 | 1.04 × 1011–1.78 × 1012 |
| Vector genomes/ml (qPCR) | Square root | 3.28 × 1010 | 2.70 × 1010 | 4.75 × 1010 | 9.00 × 108–1.04 × 1011 | 0–1.66 × 1011 |
| Transducing units/ml (green cells) | Square root | 5.09 × 108 | 2.00 × 108 | 9.60 × 108 | 0–2.47 × 109 | 0–4.00 × 109 |
| Infectious units/ml (TCID50) | Log10 | 4.37 × 109 | 2.06 × 109 | 9.26 × 109 | 5.15 × 108–3.71 × 1010 | 1.77 × 108–1.08 × 1011 |
Used to better qualify the assumption of normal distribution for the purpose of determining distributional values.
Some important properties of the rAAV2 RSM are indicated by the ratios of the titers (Table 6). The vector genome-to-infectious titer (VG:IU) ratio is often used as a measure of the relative infectivity of the vector, with lower ratios reflecting more infectious preparations. The rAAV2 RSM VG:IU ratio is 7.5, which indicates that the RSM has retained infectivity. The vector genome-to-transduction titer (VG:TU) ratio is 8.6-fold higher than the VG:IU ratio, and this result reflects the different sensitivities of the infectivity and transduction (measuring infectivity and gene expression) assays. Another ratio that is often used is the particle-to-vector genome titer ratio (P:VG). This ratio indicates the ratio of total particles, including both empty and full, to those particles containing the vector genome. The P:VG ratio obtained for the rAAV2 RSM is 28 and indicates a large excess of empty particles. This finding is consistent with the fact that the chromatographic purification process used in the production of the rAAV2 RSM was not designed to separate empty and full particles. One concern is that empty particles may have adversely affected the performance of the rAAV2 RSM in transduction and infectivity assays. However, during beta testing, two triple-transfected CsCl-purified lots (one each of AAV2.CMV.eGFP and AAV2.CMV.lacZ) were tested, using the RSS characterization methods: vector genome (qPCR), TCID50, and where applicable eGFP transduction titering. Because these were CsCl-purified preparations the empty capsid content is lower than in preparations purified by chromatography. The VG:TCID50 IU ratios and VG:TU ratios were similar or greater than those obtained for the reference standard (Tables 2 and 6). Similarly, the VG:TCID50 IU ratios and VG:TU ratios of the reference standard (Table 6) are similar to those reported in the literature for other AAV2 vectors (Salvetti et al., 1998; Zolotukhin et al., 1999; Zen et al., 2004).
Table 6.
rAAV2 Reference Standard Material Titer Ratios
| Ratio | |
|---|---|
| Particles: vector genomesa | 27.99 |
| Vector genomes: infectious units | 7.51 |
| Vector genomes: transducing units | 64.44 |
| Particles: infectious units | 210.07 |
A measure of the ratio of total particles to full particles.
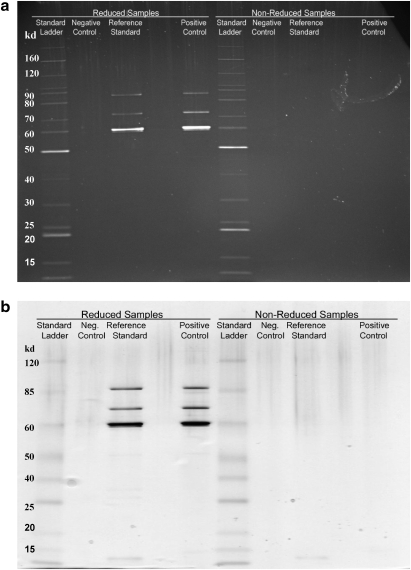
The purity of the rAAV2 RSM was assessed and the capsid identity confirmed by SDS–PAGE analysis. The RSM was examined under both reducing and nonreducing conditions, using SYPRO ruby and silver stains (Fig. 3). Under reducing conditions all proteins including the denatured AAV2 capsids are expected to enter the gel and impurities would be detected as protein bands other than the capsid proteins VP1, VP2, and VP3. Under nonreducing conditions the capsid would remain intact and would not be expected to enter the resolving gel, whereas impurities would enter the gel; proteins that previously comigrated with the capsid proteins on reducing gels would thus be detected. Silver nitrate staining was included because it is capable of detecting DNA, lipid, and carbohydrate impurities as well as nanogram levels of protein (Weiss et al., 2009). SYPRO ruby is a protein-specific fluorescent dye that has a sensitivity close to that of silver stain (Rabilloud et al., 2001; Weiss et al., 2009). In each case the rAAV2 RSM was analyzed alongside an internal laboratory standard AAV2 vector. The consensus data from the 11 testing laboratories that carried out the purity/identity test estimated that the rAAV2 RSM was greater than 94% pure and confirmed that VP1, VP2, and VP3 comigrated with the AAV2 capsid proteins of the internal vector standards (Fig. 3; and data not shown).
FIG. 3.
The rAAV2 RSM was run on SDS–polyacrylamide gels under both reducing and native conditions and then stained with (a) SYPRO ruby or (b) silver stain. An in-house rAAV2 standard was run as a positive control and buffer as a negative control. The lanes for each gel are as follows: (1) benchmark ladder (unstained or prestained)—reduced; (2) negative control—reduced; (3) AAV reference material—reduced; (5) positive control—reduced; (6) benchmark ladder (unstained or prestained)—native; (7) negative control—native; (8) AAV reference material—native; (10) positive control—native.
Discussion
As rAAV vectors more frequently head toward the clinic for gene therapy trials, there is an increasing need to share pharmacokinetic, toxicologic, and efficacy data. This need is currently confounded by the lack of standardization of critical vector parameters such as vector strength and potency. The standardization issues arise because different assays or different protocols for the same assay are often used by individual investigators to measure an identical vector property. The introduction of a widely accepted rAAV reference standard would allow laboratories to characterize AAV vectors in terms of common units, therefore facilitating comparison of doses determined by disparate assays and permitting safe and effective dosage at equivalent levels. Furthermore, efficacy and toxicology data reported in the literature could be used as a guide for initial dosing in animals and humans.
Here we have described the characterization of the first rAAV reference standard, an AAV serotype 2 vector. The goal of the AAV2RSWG was to provide a stable, high-quality, highly characterized RSM that would be both accepted and easily accessed by the AAV research community. As pointed out by FDA officials at the beginning of the effort, a reference standard material does not need to be pure or the “best,” it just needs to be well characterized. Furthermore, there are many examples of viral reference standard materials from the World Health Organization (WHO, Geneva, Switzerland) and the National Institute for Biological Standards and Control (NIBSC, Potters Bar, UK) that are not pure (e.g., poliovirus and hepatitis B virus references). Although the rAAV2 RSM was made in a research vector core and not at a current Good Manufacturing Procedure (cGMP) facility, it was extensively tested for adventitious agents and contaminants. The final rAAV2 RSM product was negative for adventitious agents in all tests to which it was subjected, although the harvest material was exposed to mycoplasma that was cleared and/or inactivated in the purification process, because the purified bulk tested negative for viable mycoplasma and mycoplasma DNA (Potter et al., 2008). Because the rAAV2 RSM is a reference standard to be used in research and quality control (QC) laboratories and is not intended for use in humans, the AAV2RSWG recommended that filling, banking, and characterization proceed. A summary of the mycoplasma testing will be included on the product information sheet supplied with each shipment of the rAAV2 RSM, stating that the reference standard has been exposed to mycoplasma, but is mycoplasma-free. Thus, institutions and companies requesting the rAAV2 RSM will be fully informed and can decide if they want to bring it into their QC laboratories. The reference material is intended to be restricted to QC laboratories, isolated from production suites. In addition, it is envisioned that internal reference standards will be calibrated against the AAV2 RSM one time and then used on a routine basis for product-specific testing.
The short-term stability testing performed on the surrogate AAV2-GFP vector as well as on the final vialed rAAV2 RSM material suggested that some loss of vector potency was occurring on storage. Initially this loss was assumed to be due to absorption to the surfaces of the vial as was seen in the previous study using vials that were not siliconized (Potter et al., 2008); however, when vector genomes were assayed no corresponding loss was seen when using siliconized vials (Table 2). One explanation for the loss of potency observed may be the omission of a stabilizing excipient in the final formulation (Croyle et al., 2001; Wright et al., 2003). The beta test stability results influenced the way the rAAV2 RSM was handled during the testing phase; aliquots were thawed only once and transduction and infectivity assays were performed within 1 hr of this thaw. Regarding future use of the reference material for potency assays, it would seem essential that a similar protocol be followed when normalizing internal reference standards against the rAAV2 RSM. For physical titer assays such as particle and vector genome assays, storage and refreezing are permissible. Plans for assessing the long-term stability of the rAAV2 RSM by yearly testing for capsid protein integrity, infectious titer, transducing titer, and vector genome titer are in place. Data will be reported by the AAV2RSWG through the Reference Standards section of the International Society for BioProcess Technology website (www.ISBioTech.org).
The characterization phase of the rAAV2 RSM project successfully fulfilled the goals of the AAV2RSWG by obtaining mean titers and 95% confidence intervals from a large number of representative assays performed by numerous test centers. The tightest confidence intervals were obtained for the nonbiological assays (particle titer and vector genome titer) whereas the biological assays (infectious titer and transduction titer) gave wider intervals (Table 5). This pattern might be expected because the biological assays are inherently more variable. The tight confidence interval observed for the vector genome titer is relevant because this titer has been used exclusively in dosing regimens and a high degree of precision is important for the use of the rAAV2 RSM in dose standardization.
One obvious trend in the quantitative assay data was the degree of variation between institutions for each assay (Fig. 1 and Table 3) despite the relatively tight correlation of assay results within an institution (Table 3). This poor degree of interlaboratory precision and accuracy was apparent even though attempts were made to standardize the assays by providing detailed protocols and common reagents. The variation may be explained by the use of different reagents (i.e., other than those provided, such as tissue culture media, PCR primers, and PCR mixes), equipment, and/or operator technique. This is the first time that such variation between laboratories has been thoroughly documented and the findings emphasize the need in the field for universal reference standards. This need is especially apparent when it is considered that fundamentally dissimilar tests are often used to measure the same parameter (e.g., qPCR and dot–blot for vector genome titer) and that even when different laboratories use the same assay, different protocols are usually followed. For some assays the variation is not large, with the most important measure, vector genome titer (qPCR), which is almost exclusively used for dosing in preclinical and clinical studies, having low variation (confidence interval of less than 0.5 log). Despite the spread of infectious titers, the mean value represents the best titer based on multiple replicates conducted at the different sites on different test dates.
Because the rAAV2 RSM supply is limited, it is not intended that it be used routinely, but rather for the calibration of laboratory-specific internal reference standards, which can then be run concurrently with test samples in subsequent assays that have been validated. The initial calibration would involve titering the RSM alongside the internal standard in the same assay; the difference between the titer determined in this assay and the accepted titer of the RSM would act as a conversion factor for calculating the titer of the internal standard in reference standard units (RSU). Once the internal standard titer is known in reference standard units per milliliter, the titer of test samples can be calculated similarly in the same units, during subsequent assays. It is envisaged that the RSM will be used in this way for standardizing the genome titer, particle titer, and infectious titer of AAV2 vectors. A prerequisite for qPCR or hybridization-based vector genome/infectivity titering methods would be that the internal AAV standard share enough genome sequence with the rAAV2 RSM for oligonucleotide or labeled probe annealing. Several common transcriptional elements are included in the rAAV2 RSM genome for this purpose and many existing internal reference standards will therefore be candidates for calibration. If this is not the case, new internal standards will need to be produced that harbor DNA elements in common with the AAV2 RSM. For transducing titers, the encoded transgene provides the basis of detection and, therefore, with the exception of GFP-expressing vectors for preclinical studies, these titers will generally not be amenable to standardization using the rAAV2 RSM.
Although the primary intent of the rAAV2 RSM was to provide a reference point for AAV2 serotype vectors it is possible that for nonbiological assays such as vector genome titration, the rAAV2 RSM could be used for the calibration of other AAV serotypes. Because the vector capsid is not directly involved in these types of assays, it might be argued that there is no capsid specificity and that the capsid serotype would not have an impact. As an example, in the vector genome titer assay it might be assumed that different capsids are equally susceptible to PCR heat treatment for liberation of the vector genome. However, conditions would need to be optimized because equal susceptibility of AAV serotypes to heat has not been definitively demonstrated. In addition, proteolysis is often used to liberate the vector genome and it is known that different capsid serotypes have different susceptibilities to protease treatments (Van Vliet et al., 2006). Similarly, serotype-independent methods of determining particle titer (e.g., high-performance liquid chromatography, spectrophotometry) could be calibrated, using the rAAV2 RSM, but the same assumption of capsid independence would apply, and for spectrophotometric measurements the proper extinction coefficient would need to be incorporated (Sommer et al., 2003). If data are available to demonstrate that an assay is indeed capsid independent, then the use of the rAAV2 RSM for other serotypes may well be acceptable, but thorough review with the appropriate regulatory agency is recommended. For biological assays such as infectious titer, the paramount roles of the capsid, the requisite target cell line, and the helper virus preclude the use of the rAAV2 RSM to calibrate other serotypes. For these assays, investigators must await the development of further reference standard materials such as the AAV8 material currently under production (Moullier and Snyder, 2008).
The rAAV2 RSM carries a single-stranded DNA vector genome. Self-complementary AAV vector genomes, generated with a mutation within the terminal repeat (McCarty et al., 2003), have become popular for gene transfer because they bypass the rate-limiting genome conversion of single-stranded to double-stranded DNA during transduction of target cells. The rAAV2 RSM can be used to normalize “in-house” reference standards for both the classic single-stranded vectors and self-complementary vectors. Because self-complementary vectors carry double the genome complement of single-stranded vectors, a simple conversion is necessary when calculating vector genome titers for these two vector types.
In the United States, the FDA Center for Biologics Evaluation and Research (CBER), Office of Cellular, Tissue, and Gene Therapies (OCTGT), Division of Cellular and Gene Therapies (DCGT) recommends reference materials as benchmarking tools for qualifying and validating “in-house” reference standards and assays by comparison with the collective data. It should be noted that it is not the intent of the FDA to standardize assay methods across the field or to require that the values assigned to the rAAV2RSM be duplicated during validation studies. Furthermore, there is no requirement in the United States to follow rAAV2 RSM procedures when assaying particle concentration, genome copy number, or infectious titer. Sponsors of adeno-associated virus-related investigational new drugs (INDs) should consult with the FDA/CBER or appropriate national agency for further guidance. The rAAV2 RSM fulfills many of the requirements of a reference standard material in that it (1) is sufficiently homogeneous and stable with respect to specified properties, (2) is established to be fit for its intended use in measurement, (3) is accompanied by documentation, (4) provides relevant property values that are based on multiple measurements conducted at different locations, and (5) is accompanied with associated measurement uncertainty.
From the outset, the vision of the AAV2RSWG for the rAAV2 RSM was that it would represent the first step toward standardization of AAV-based gene therapy dosing and provide a blueprint for the development of reference standards for other AAV serotypes. This vision is becoming reality through the successful production and characterization reported here, and with the effort to develop the AAV8 reference standard material underway. The requirement that the reference materials be universally accepted by the AAV community has dictated the need for a voluntary communal effort in the production and characterization phases. Despite the numerous drawbacks, difficulties, and delays inherent in this type of approach, the AAV gene therapy community has responded selflessly and with enthusiasm. It is hoped that the ultimate success of this collaboration will inspire future reference standard efforts and contribute to the development and commercialization of AAV-based gene therapeutics.
Acknowledgments
Current and former members of the Executive Committee: Denise Gavin–FDA (nonvoting member); Daniel Rosenblum–NIH NCRR (nonvoting member); Keith Carson–International Society for BioProcess Technology; Parris Burd–Bayer Corporation; Olivier Danos–Généthon; Maritza McIntyre–FDA (nonvoting member); Richard Knazek–NIH NCRR (nonvoting member). Manufacturing committee: Guang Ping Gao–University of Pennsylvania; Philip Cross–Harvard Medical School; Anna Salvetti–CHU Hôtel Dieu, Nantes, France; Sue Washer–Applied Genetic Technologies; Guang Qu–Avigen. Quality committee: Marie Printz–Ceregene; Paul Husak–Cell Genesys; Scott McPhee–Thomas Jefferson University; Jurg Sommer–Avigen; Jim Marich–Cell Genesys. We acknowledge the technical help of the following during the testing phase: Mark Potter–PGTC Vector Core; Gitte Kitlen–PGTC Quality Control; Lynn Combee–PGTC Toxicology Core; Cheryl Roberts–PGTC Toxicology Core; Tanja Finnäs–AMT; Martha Hoekstra–AMT. We are grateful to James Wilson–University of Pennsylvania for supporting the beta testing through NIH grant P30-DK-047757 and an SRA from GlaxoSmithKline. We are also indebted to Peggy Fahnestock and Liz Kerrigan at the ATCC for coordinating the distribution of materials to testing laboratories. This work was supported by NIH grant U42RR11148. We acknowledge the generosity of the ATCC, Nunc, Aldevron, Corning, Fisher Thermo Scientific, the Indiana University Vector Production Facility, HyClone, Mediatech, Progen, and the Williamsburg Bioprocessing Foundation. Richard Surosky is employed by a company that may have interest in these vectors for therapeutic purposes. Richard Snyder owns equity in a gene therapy company that is commercializing AAV for gene therapy applications.
References
- Brantly M.L. Chulay J.D. Wang L. Mueller C. Humphries M. Spencer L.T. Rouhani F. Conlon T.J. Calcedo R. Betts M.R. Spencer C. Byrne B.J. Wilson J.M. Flotte T.R. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C. Gorbatyuk O.S. Velardo M.J. Peden C.S. Williams P. Zolotukhin S. Reier P.J. Mandel R.J. Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- CBER/FDA (Center for Biologics Evaluation and Research, U.S. Food and Drug Administration) Points to consider in the characterization of cell lines used to produce biologicals. 1993. http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/OtherRecommendationsforManufacturers/UCM062745.pdf. [Jul;2010 ]. http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/OtherRecommendationsforManufacturers/UCM062745.pdf
- Chadeuf G. Favre D. Tessier J. Provost N. Nony P. Kleinschmidt J. Moullier P. Salvetti A. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J. Gene Med. 2000;2:260–268. doi: 10.1002/1521-2254(200007/08)2:4<260::AID-JGM111>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Croyle M.A. Cheng X. Wilson J.M. Development of formulations that enhance physical stability of viral vectors for gene therapy. Gene Ther. 2001;8:1281–1290. doi: 10.1038/sj.gt.3301527. [DOI] [PubMed] [Google Scholar]
- Grimm D. Kern A. Rittner K. Kleinschmidt J.A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- Hutchins B. Development of a reference material for characterizing adenovirus vectors. BioProcess. J. 2002;1:25–28. [Google Scholar]
- Kärber G. 50% end-point calculation. Arch. Exp. Pathol. Pharmak. 1931;162:480–483. [Google Scholar]
- Littell R.C. Milliken G.A. Stroup W.W. Wolfinger R.E. SAS for Mixed Models. 2nd. SAS Institute; Cary, NC: 2006. [Google Scholar]
- Maguire A.M. Simonelli F. Pierce E.A. Pugh E.N., Jr. Mingozzi F. Bennicelli J. Banfi S. Marshall K.A. Testa F. Surace E.M. Rossi S. Lyubarsky A. Arruda V.R. Konkle B. Stone E. Sun J. Jacobs J. Dell'Osso L. Hertle R. Ma J.X. Redmond T.M. Zhu X. Hauck B. Zelenaia O. Shindler K.S. Maguire M.G. Wright J.F. Volpe N.J. McDonnell J.W. Auricchio A. High K.A. Bennett J. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.M. Fu H. Monahan P.E. Toulson C.E. Naik P. Samulski R.J. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Moss R.B. Rodman D. Spencer L.T. Aitken M.L. Zeitlin P.L. Waltz D. Milla C. Brody A.S. Clancy J.P. Ramsey B. Hamblett N. Heald A.E. Repeated adeno-associated virus serotype 2 aerosol-mediated cystic fibrosis transmembrane regulator gene transfer to the lungs of patients with cystic fibrosis: A multicenter, double-blind, placebo-controlled trial. Chest. 2004;125:509–521. doi: 10.1378/chest.125.2.509. [DOI] [PubMed] [Google Scholar]
- Moullier P. Snyder R.O. International efforts for recombinant adeno-associated viral vector reference standards. Mol. Ther. 2008;16:1185–1188. doi: 10.1038/mt.2008.125. [DOI] [PubMed] [Google Scholar]
- Mueller C. Flotte T.R. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- Nienhuis A. Dose-escalation study of a self complementary adeno-associated viral vector for gene transfer in hemophilia B. Clinical trial NCT00979238. 2009. http://clinicaltrials.gov/ct2/show/NCT00979238. [Jul;2010 ]. http://clinicaltrials.gov/ct2/show/NCT00979238
- Potter M. Phillipsberg G. Phillipsberg T. Pettersen M. Sanders D. Korytov I. Fife J. Zolotukhin S. Byrne B.J. Muzyczka N. Manufacture and stability study of the recombinant adeno-associated virus serotype 2 vector reference standard. Bioprocess. J. 2008;7:8–14. [Google Scholar]
- Rabilloud T. Strub J.M. Luche S. van Dorsselaer A. Lunardi J. A comparison between SYPRO Ruby and ruthenium II tris(bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics. 2001;1:699–704. doi: 10.1002/1615-9861(200104)1:5<699::AID-PROT699>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Salvetti A. Oreve S. Chadeuf G. Favre D. Cherel Y. Champion-Arnaud P. David-Ameline J. Moullier P. Factors influencing recombinant adeno-associated virus production. Hum. Gene Ther. 1998;9:695–706. doi: 10.1089/hum.1998.9.5-695. [DOI] [PubMed] [Google Scholar]
- Snyder R.O. Flotte T.R. Production of clinical-grade recombinant adeno-associated virus vectors. Curr. Opin. Biotechnol. 2002;13:418–423. doi: 10.1016/s0958-1669(02)00369-5. [DOI] [PubMed] [Google Scholar]
- Sommer J.M. Smith P.H. Parthasarathy S. Isaacs J. Vijay S. Kieran J. Powell S.K. McClelland A. Wright J.F. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Van Vliet K. Blouin V. Agbandje-McKenna M. Snyder R.O. Proteolytic mapping of the adeno-associated virus capsid. Mol. Ther. 2006;14:809–821. doi: 10.1016/j.ymthe.2006.08.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington K.H., Jr. Herzog R.W. Treatment of human disease by adeno-associated viral gene transfer. Hum. Genet. 2006;119:571–603. doi: 10.1007/s00439-006-0165-6. [DOI] [PubMed] [Google Scholar]
- Weiss W. Weiland F. Gorg A. Protein detection and quantitation technologies for gel-based proteome analysis. Methods Mol. Biol. 2009;564:59–82. doi: 10.1007/978-1-60761-157-8_4. [DOI] [PubMed] [Google Scholar]
- Wright J.F. Qu G. Tang C. Sommer J.M. Recombinant adeno-associated virus: Formulation challenges and strategies for a gene therapy vector. Curr. Opin. Drug Discov. Dev. 2003;6:174–178. [PubMed] [Google Scholar]
- Zen Z. Espinoza Y. Bleu T. Sommer J.M. Wright J.F. Infectious titer assay for adeno-associated virus vectors with sensitivity sufficient to detect single infectious events. Hum. Gene Ther. 2004;15:709–715. doi: 10.1089/1043034041361262. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S. Byrne B.J. Mason E. Zolotukhin I. Potter M. Chesnut K. Summerford C. Samulski R.J. Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]