Abstract
Apical reabsorption from the urine has been shown to be important for such processes as the maintenance of critical metabolites in the blood and the excretion of nephrotoxic compounds. The solute carrier (SLC) transporter OAT4 (SLC22A11) is expressed on the apical membrane of renal proximal tubule cells and is known to mediate the transport of a variety of xenobiotic and endogenous organic anions. Functional characterization of genetic variants of apical transporters thought to mediate reabsorption, such as OAT4, may provide insight into the genetic factors influencing the complex pathways involved in drug elimination and metabolite reclamation occurring in the kidney. Naturally occurring genetic variants of OAT4 were identified in public databases and by resequencing DNA samples from 272 individuals comprising 4 distinct ethnic groups. Nine total nonsynonymous variants were identified and functionally assessed using uptake of three radiolabeled substrates. A nonsense variant, R48Stop, and three other variants (R121C, V155G, and V155M) were found at frequencies of at least 2% in an ethnic group specific fashion. The L29P, R48Stop, and H469R variants displayed a complete loss of function, and kinetic analysis identified a reduced Vmax in the common nonsynonymous variants. Plasma membrane levels of OAT4 protein were absent or reduced in the nonfunctional variants, providing a mechanistic reason for the observed loss of function. Characterization of the genetic variants of reabsorptive transporters such as OAT4 is an important step in understanding variability in tubular reabsorption with important implications in innate homeostatic processes and drug disposition.
Keywords: SLC22A11, SNP, kidney, pharmacogenetics
understanding the role of organic anion transporters (OATs) in the kidney has become increasingly important given their demonstrated roles in drug disposition, adverse drug interactions, and xenobiotic-induced toxicities (7, 11, 23). Currently, the majority of this research has focused on the basolateral membrane transporters OAT1 (SLC22A6) and OAT3 (SLC22A8), which mediate the initial uptake of xenobiotics from the blood into the proximal tubule cells in the process of renal secretion. A number of studies have shown that these transporters can facilitate the uptake of numerous drugs and toxins (1), likely providing a significant mechanism for renal toxicity, and that these transporters in particular respond to renal injury (34). While renal secretion of endogenous small molecules and xenobiotics is an important facet of the physiological and pharmacological functions of the kidney, the reabsorption of organic anions from the urine is also a substantial process that occurs in the kidney. Passive and facilitated reabsorption can influence both the pharmacokinetic profile of a drug and the circulating levels of important metabolic intermediates and precursors in the blood (10, 12). To date, comparatively little has been done to evaluate and understand the potential impact of apical organic anion transporters that play a role in reabsorptive mechanisms of drugs and other xenobiotics in the renal proximal tubule.
OAT4 (SLC22A11) is a solute carrier (SLC) family transporter which functions primarily as an organic anion/dicarboxylate exchanger (6). SLC22A11 transcripts are expressed at appreciable levels in the kidney, similar to OCT2 (SLC22A2) and OCTN2 (SLC22A5) but at a substantially lower level compared with OAT1 and OAT3 (14, 21). OAT4 has been localized to the apical membrane of renal proximal tubules, accordant with its function as a reabsorptive transporter in the kidney (6). Previous studies have demonstrated that OAT4 is capable of transporting or interacting with both endogenous compounds and xenobiotics including steroid sulfates (4), nonsteroidal anti-inflammatory drugs (25), angiotensin II and leukotriene receptor antagonists (32), prostaglandins (15), and uric acid (12) and thus could affect homeostatic processes occurring in the kidney and the body as a whole as well as impact the efficacy or safety of commonly used drugs. Similarly, OAT4 is expressed on the basolateral membrane of the syncytiotrophoblast of the placenta and is postulated to be important in the removal of steroid sulfates from the fetal compartment and thereby promoting appropriate steroid signaling and minimizing toxic effects for the developing fetus (4, 26, 35). Prior studies have also identified associations between genetic variants of OAT4 and the clearance of the loop diuretic torsemide (30) as well as plasma uric acid levels (16, 29), suggesting that function-altering variants of OAT4 may be responsible for differences observed in the clearance of xenobiotics and the reabsorption of important endogenous metabolites.
Previous studies have identified a number of genetic variants in OAT4, including multiple variants that result in a change in the amino acid sequence. Variants have been described in studies that examined a small number of individuals from diverse ethnic groups (31) and also in specific populations such as osteoporotic Korean women (19), the latter of which identified a nonfunctional variant only in those suffering from osteoporosis. A recent article by Zhou, et al. has functionally characterized a number of previously described naturally occurring genetic variants of OAT4 (31), however, these variants were discovered by analyzing a limited number of DNA samples. The current study analyzed a large number of DNA samples (n = 272) from 4 distinct ethnic groups to identify coding variants in OAT4 in healthy individuals. A number of variants were identified and underwent subsequent functional assessments using model substrates of OAT4. Many of these variants demonstrated altered functionality and may significantly affect the renal excretion of a number of xenobiotics and anionic metabolites.
MATERIALS AND METHODS
Reagents.
Radiolabeled [3H]estrone sulfate was purchased from PerkinElmer Life Sciences (Waltham, MA). Radiolabeled [3H]ochratoxin A and [14C]uric acid were purchased from Moravek (Brea, CA). All other chemicals were purchased from Sigma (St. Louis, MO). All cell culture media and reagents were purchased from the University of California Cell Culture Facility (San Francisco, CA).
Identification of OAT4 variants.
Nonsynonymous variants of OAT4 were identified from submissions in the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/snp) or through resequencing of genomic DNA as part of the Studies of Pharmacogenetics in Ethnically Diverse Populations (SOPHIE) project. DNA samples from the SOPHIE project were collected and analyzed in 68 healthy individuals from 4 ethnic groups: Caucasian, African-American, Asian-American, and Mexican-American. Variants in the coding and flanking intronic regions in OAT4 were identified following methodologies described previously (18). A complete list of OAT4 variants found in the SOPHIE project can be accessed at http://pharmacogenetics.ucsf.edu/. Use of genomic DNA from healthy volunteers was performed according to procedures reviewed and approved by the UCSF Committee on Human Research, and informed consent was obtained from all subjects enrolled in the study.
Computational analysis.
All complete mammalian OAT4 sequences were downloaded from the ENSEMBL database (http://www.ensembl.org). Alignments of OAT4 protein sequences were created using MUSCLE 3.6 (5), and the resulting alignment was visualized using the BioEdit software suite. Prediction of OAT4 variant effects was done using preexisting tools including SIFT (20), PolyPhen (22), PhD-SNP (3), and SNAP (2), which use a variety of sequence and evolutionary analysis methods to predict whether a variant would be potentially deleterious to the function of the protein.
Cell line construction.
A full-length human OAT4 transcript (GenBank Accession No. NM_018484) was PCR amplified from cDNA synthesized from a commercially available adult kidney RNA sample purchased from Clontech (Mountain View, CA). The reference OAT4 coding sequence was subcloned into the mammalian expression vector pcDNA5/FRT (Invitrogen, Carlsbad, CA), and variants of OAT4 were created using site-directed mutagenesis utilizing Pfu Ultra DNA polymerase (Stratagene, La Jolla, CA) and the pcDNA5/FRT-OAT4 vector as a template. All vector sequences were verified via direct sequencing of the complete open reading frame, including the variant locations. Cell lines stably transfected with pcDNA5/FRT (empty vector control), reference OAT4, or its variants were created using FLP-In-293 cells (Invitrogen) as previously described (28). All stably transfected cell lines were grown in a humidified 37°C incubator with 5% CO2 and cultured in DMEM H-21 supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml hygromycin B.
RNA isolation and quantitative RT-PCR.
Stable cell lines expressing reference OAT4 or its variants were grown in six-well poly-d-lysine-coated plates (BD Biosciences, San Jose, CA) until reaching 90–95% confluency. Upon reaching confluency, cell culture media was aspirated and the cells were washed once with cold PBS. After removal of the PBS wash, TRIzol reagent (Invitrogen) was added to the cells and RNA was isolated following the manufacturer's standard protocol. RNA purity and quality was assessed spectrophotometrically and by examination of 28S and 18S bands via agarose gel electrophoresis. cDNA was synthesized using a Superscript III Reverse Transcription kit (Invitrogen) from 1 μg of purified total RNA samples following the manufacturer's standard protocol. The resulting cDNA was then diluted and used for quantitative determination of SLC22A11 mRNA levels.
Quantitative RT-PCR (qRT-PCR) was performed using Taqman reagents and specific primer and probe sets for human SLC22A11 and GAPDH (Applied Biosystems, Foster City, CA). Reactions were performed in a 384-well plate with a 10-μl reaction volume using an ABI 7900HT Fast Real-time PCR system (Applied Biosystems) using the default instrument settings. Expression levels were determined from three independent biological samples using the ΔΔCt method after normalization to endogenous levels of GAPDH. The results were expressed as a percentage of the mRNA level determined for the OAT4 reference cell line.
Protein isolation and Western blotting.
Total protein was isolated from OAT4-expressing cells by culturing them to 90–95% confluency in T75 flasks and lysing them using radioimmunoprecipitation assay buffer (Sigma) following the manufacturer's standard protocol. Plasma membrane protein was isolated using a cell surface biotinylation kit (Pierce Biotechnology, Rockford, IL) following the manufacturer's standard protocol with slight modifications. Briefly, stably transfected OAT4 reference and variant cells lines were grown in T75 cell culture flasks, washed with ice-cold PBS, and incubated with Sulfo-NHS-SS-Biotin (Pierce Biotechnology) for 60 min at 4°C with gentle shaking. Concentrated Tris buffer was added to each flask to a final concentration of ∼50 mM to quench any remaining unreacted Sulfo-NHS-SS-Biotin. The cells were subsequently scraped off the flask, washed with ice-cold PBS supplemented with protease inhibitor cocktail (Pierce Biotechnology), and lysed. Biotinylated protein was isolated by incubating the lysate with streptavidin-linked agarose for 60 min at room temperature and eluted using 50 mM dithiothreitol in standard SDS-PAGE buffer. Protein concentration was determined using a bicinchoninic acid (BCA) assay (Pierce Biotechnology).
Comparable amounts of protein from each cell line were run on a 4–20% gradient Tris·HCl polyacrylamide gel and transferred to an Immobilon polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA). The membranes were incubated in 5% nonfat milk in Tris-buffered saline buffer with 0.05% Tween (TBST) for 1 h followed by incubation with 5 μg/ml of rabbit anti-human polyclonal OAT4 antibody (US Biological, Swampscott, MA) overnight at 4°C. Membranes were subsequently washed with TBST buffer and incubated with a 1:2,000 dilution of a goat anti-rabbit IgG horseradish peroxidase-coupled secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted in 5% nonfat milk in TBST buffer for 1 h at room temperature. Following secondary antibody incubation, the membrane was washed with TBST buffer and protein detection was performed using ECL Plus reagents (GE Healthcare, Piscataway, NJ). The membranes were stripped for 45 min using a β-mercaptoethanol-based buffer, and the procedure was repeated using an antibody specific to the α-subunit of the Na+-K+-ATPase (US Biological) at a 1:1,000 dilution to assess gel loading.
Uptake studies.
Cellular uptake of canonical substrates by OAT4 and its variants was determined using [3H]estrone sulfate, [3H]ochratoxin A, and [14C]uric acid. Cells were seeded on 24-well poly-d-lysine-coated cell culture plates (BD Biosciences) in standard cell culture media as described previously, and experiments commenced 24 h after cell seeding. Uptakes were performed at 37°C for 2 min (estrone sulfate and ochratoxin A) or 4 min (uric acid) and began after the media was removed and cells were washed with warm Cl-free uptake buffer (125 mM sodium gluconate, 4.8 mM potassium gluconate, 1.2 mM K2HPO4, 1.2 mM MgSO4, 1.3 mM calcium gluconate, and 25 mM HEPES/Tris; pH ≈ 7.4) for 3 min. Experiments were initiated by incubating the cells with warm Cl-free uptake buffer containing radiolabeled compounds as indicated in the figure legends. At the completion of the incubation time period, cells were quickly washed three times with ice-cold choline buffer (128 mM choline chloride, 4.73 mM KCl, 1.25 mM MgSO4, 1.25 mM CaCl2, and 5 mM HEPES/Tris; pH ≈ 7.4). Any remaining buffer was aspirated, and the cells were lysed with 1 ml of warm 0.1% SDS/0.1 N NaOH solution for a minimum of 45 min. Intracellular levels of radioactive compound were determined using liquid scintillation counting. All resulting levels of substrate uptake were normalized to the total protein in each well using a BCA protein assay. All uptake experiments were performed in triplicate and using empty vector-transfected cell lines to establish background levels of uptake in HEK-293 cells. Statistically significant differences in uptake were evaluated using Graphpad Prism 5.1 (Graphpad, La Jolla, CA) using a one-way ANOVA followed by a Dunnett's post hoc test with a false discovery rate of 0.05.
Kinetic studies were performed in a similar fashion with supplemental amounts of unlabeled substrates included in the uptake buffer to produce the final concentration of substrate. At least three independent experiments were performed using empty vector-transfected cells and OAT4 reference and variant cell lines. Kinetic values (Km and Vmax) were determined using nonlinear regression analysis in Graphpad Prism 5.1 (Graphpad) after subtracting the uptake observed in the empty vector cell line from the uptake in stably transfected OAT4 cell lines at each concentration. Curves were fit to the Michaelis-Menten equation V = Vmax [S]/(Km + [S]) where V is the rate of uptake, Vmax is the maximal uptake rate of the substrate, Km is the substrate concentration at half the Vmax, and [S] is the substrate concentration.
RESULTS
Variant identification.
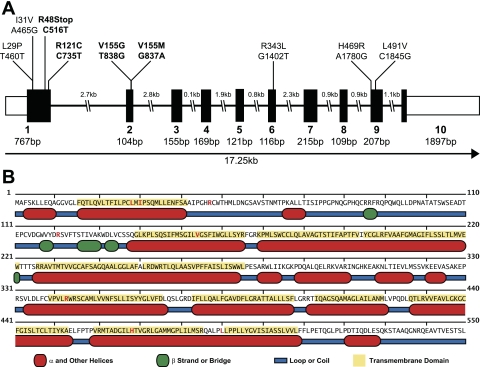
Variants of OAT4 were identified for further study by both querying the dbSNP database and by resequencing 272 DNA samples from 4 distinct ethnic groups within the SOPHIE project. Resequencing of the 10 exons and adjacent flanking regions of SLC22A11 within SOPHIE identified 27 variable positions, which included 7 nonsynonymous and 6 synonymous variants with the remaining 14 variants occurring in intronic regions. Two additional nonsynonymous variants were identified in the dbSNP database. The majority of the nonsynonymous variants occurred in the first two exons of SLC22A11, as shown in Fig. 1A. Additionally, five of the nine amino acid-altering variants were found in predicted transmembrane helix regions, while two were found in the large extracellular loop occurring between transmembrane domain 1 and 2. The L491V variant occurred in the small loop region between the predicted transmembrane helix 11 and 12. The location of these variants is further detailed in Fig. 1B and Table 1. Four of the nonsynonymous variants (R121C, R343L, H469R, and L491V) were previously unidentified, while the remaining three (R48Stop, V155M, and V155G) have been identified in either independent resequencing efforts (31) or as part of the HapMap and 1000 Genomes projects. The novel variant R121C occurred at a frequency of 2.3% in African-Americans within the SOPHIE project. Similarly, the R48Stop and V155M variants were present at higher frequencies (2.3 and 1.7%, respectively) in the SOPHIE cohort than previously reported (31). However, the V155G variant, while a singleton in the Mexican-American arm of SOPHIE, was found at 2.3% in the Han Chinese group within the HapMap genotyping project. All other nonsynonymous variants were found as singletons. All of the nonsynonymous variants identified in Mexican-Americans were found as singletons, while no nonsynonymous variants were found in the Asian-American samples within SOPHIE; the majority of OAT4 nonsynonymous variants were found in a single ethnic group. A complete listing of the allele frequencies of these variants is shown in Table 2. Despite the apparent discrepancy in allele frequency between the SOPHIE results and other genotyping projects, these results suggest that nonsynonymous variants of OAT4 are not uncommon and may occur at higher frequencies than previously reported. Two nonsynonymous variants were previously listed in dbSNP (L29P and I31V), and while these variants lacked allele frequency data they were reported by multiple sequencing groups, suggesting that they are not sequencing artifacts. However, they were not found in any of the ethnic groups comprising SOPHIE or in the HapMap or 1000 Genomes projects. Of note, OAT4 possesses variants that result in two possible alternate amino acids at the same position and a nonsense change that produces a stop codon early in the protein sequence, both of which are considered rare events when variation within the SLC22 transporter family is examined (27).
Fig. 1.
Location of nonsynonymous variants in human organic anion transporter OAT4. A: sites of nonsynonymous variants in the exonic structure of OAT4. Bold indicates variants that occur at a frequency of at least 1% in at least 1 ethnic group. B: location of nonsynonymous variants in OAT4 secondary structural elements based on predictions using MINNOU (http://minnou.cchmc.org/). Red amino acids indicate the positions of OAT4 nonsynonymous variants.
Table 1.
Nonsynonymous variants of human SLC22A11
| ID | Exon | Genomic Position | mRNA Position | Base Change | Protein Position | Amino Acid Change |
|---|---|---|---|---|---|---|
| rs11231819 | 1 | 64080133 | 460 | T→C | 29 | Leu→Pro |
| rs11231820 | 1 | 64080138 | 465 | A→G | 31 | Ile→Val |
| rs35008345 | 1 | 64080189 | 516 | C→T | 48 | Arg→Stop |
| PMT-2945 | 1 | 64080408 | 735 | C→T | 121 | Arg→Cys |
| rs61744144 | 2 | 64083252 | 837 | G→A | 155 | Val→Met |
| rs12785832 | 2 | 64083253 | 838 | T→G | 155 | Val→Gly |
| PMT-2917 | 6 | 64089355 | 1402 | G→T | 343 | Arg→Leu |
| PMT-2910 | 9 | 64093723 | 1780 | A→G | 469 | His→Arg |
| PMT-2911 | 9 | 64093788 | 1845 | C→G | 491 | Leu→Val |
ID indicates either dbSNP or Pharmacogenetics of Membrane Transporters (PMT) IDs.
Table 2.
Allele frequency of nonsynonymous SLC22A11 variants
| 1K Genomes MAF |
PMT MAF |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Variant | dbSNP MAF/Ethnicity | CEU | CHB-JPT | YRI | AA | CA | AS | ME |
| rs11231819 | Leu29Pro | NA* | NA | NA | NA | NA | NA | NA | NA |
| rs11231820 | Ile31Val | NA* | NA | NA | NA | NA | NA | NA | NA |
| rs35008345 | Arg49Stop | 0.029/CA+AA | NA | NA | NA | 0.000 | 0.023 | 0.000 | 0.008 |
| PMT-2945 | Arg121Cys | NA | NA | NA | NA | 0.023 | 0.000 | 0.000 | 0.000 |
| rs61744144 | Val155Met | NA* | 0.000 | 0.000 | 0.027 | 0.015 | 0.000 | 0.000 | 0.000 |
| rs12785832 | Val155Gly | 0.023/CHB | NA | NA | NA | 0.000 | 0.000 | 0.000 | 0.007 |
| PMT-2917 | Arg343Leu | NA | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.007 |
| PMT-2910 | His469Arg | NA | NA | NA | NA | 0.000 | 0.008 | 0.000 | 0.000 |
| PMT-2911 | Leu491Val | NA | NA | NA | NA | 0.000 | 0.000 | 0.000 | 0.007 |
ID indicates either dbSNP or PMT IDs. MAF, minor allele frequency; CA, Caucasian; AA, African-American; AS, Asian-American; ME, Mexican-American; CHB, Han Chinese from Beijing, China; CEU, Northern and Western European ancestry from Utah; CHB-JPT, Han Chinese from Beijing, China and Japanese from Tokyo, Japan; YRI, Yoruban from Ibadan, Nigeria; NA, not identified in the project or database.
Presence of the variant in the database but lacking any frequency data.
Conservation of OAT4 and in silico functional assessment.
A large number of variants found to possess altered function have been shown to occur in evolutionarily conserved positions in membrane transporters (17, 27). When the OAT4 nonsynonymous variants were mapped to multiple alignments of available complete OAT4 sequences, including human, chimpanzee, macaque, bovine, and canine orthologs, only two variants occurred in positions of conservation (R343L and L491V). However, and not without expectation, all of the variable positions are conserved when primate sequences only are examined. Alignment of primate OATs expressed in the kidney (OAT4, OAT3, and OAT1) identified an additional position of conservation in OAT4 (L29P).
Further analysis using various computational algorithms designed to predict the functional consequences of amino acid substitutions was undertaken as well; four different algorithms were used to predict potential functional effects of variants of OAT4. These algorithms, which base their evaluations on calculations of protein stability, amino acid substitution matrices, and evolutionary constraints, by in large predicted that most of the variants identified in OAT4 would result in a change in protein function. The results of these predictions are shown in Table 3. In fact, the only variants consistently predicted to have no functional consequences were I31V and L491V, at the extreme termini of OAT4. There was also a consensus among the four methods that the L29P, R121C, and R343L variants would cause a detrimental change in function. The R48Stop variant was not included in these analyses, as the accepted assumption is that a premature stop codon will undoubtedly cause a change in protein function due to the truncation of the protein. There was also no obvious correlation between the conservation status of the residue and the predicted effect of these variants; variants at both conserved and unconserved sites were predicted to be detrimental to the function of OAT4.
Table 3.
Predicted effect of nonsynonymous variants on OAT4 function
| Prediction of Variant Effect |
|||||||
|---|---|---|---|---|---|---|---|
| ID | Variant | Mammalian Conservation* | OAT Family Conservation† | SIFT | PolyPhen | PhD-SNP | SNAP |
| rs11231819 | Leu29Pro | Unconserved | Conserved | Damaging | Damaging | Disease | Nonneutral |
| rs11231820 | Ile31Val | Unconserved | Unconserved | Neutral | Benign | Neutral | Neutral |
| rs35008345 | Arg49Stop | Unconserved | Unconserved | ND | ND | ND | ND |
| PMT-2945 | Arg121Cys | Unconserved | Unconserved | Damaging | Damaging | Disease | Nonneutral |
| rs61744144 | Val155Met | Unconserved | Unconserved | Damaging | Damaging | Neutral | Neutral |
| rs12785832 | Val155Gly | Unconserved | Unconserved | Damaging | Benign | Disease | Nonneutral |
| PMT-2917 | Arg343Leu | Conserved | Conserved | Damaging | Damaging | Disease | Nonneutral |
| PMT-2910 | His469Arg | Unconserved | Unconserved | Neutral | Damaging | Disease | Nonneutral |
| PMT-2911 | Leu491Val | Conserved | Conserved | Neutral | Benign | Neutral | Neutral |
ID indicates either dbSNP or PMT IDs. OAT, organic anion transporter; ND, not determined.
Consists of human, chimpanzee, gorilla, macaque, dog, and cow OAT4 sequences.
Consists of human, chimpanzee, gorilla, and macaque OAT1, OAT3, and OAT4 sequences.
Functional effects of OAT4 nonsynonymous variants.
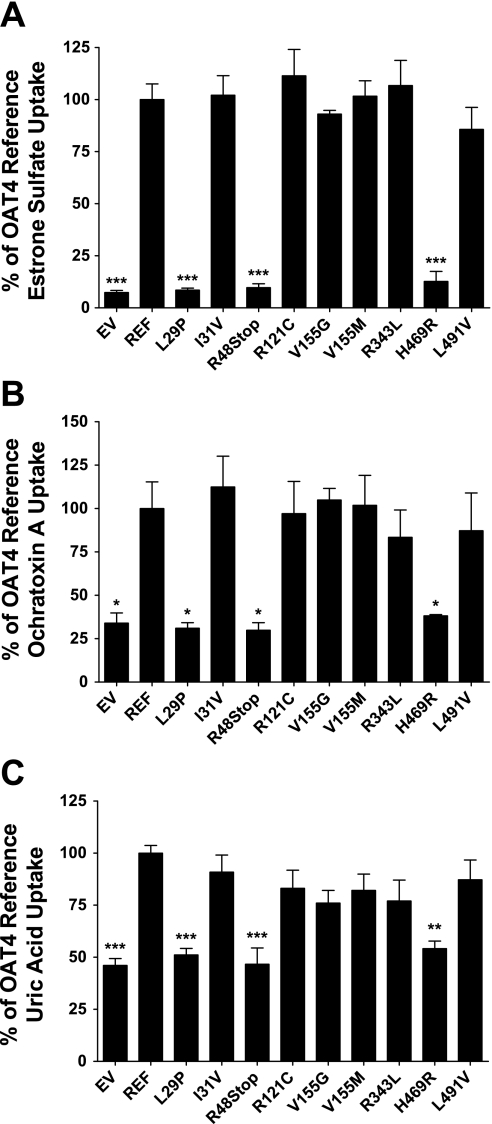
The functional consequences of nonsynonymous OAT4 variants were evaluated using stably transfected HEK-293 cells overexpressing either reference OAT4 or one of the coding variants. The effect of all nine nonsynonymous variants was investigated using uptake assays with the canonical OAT4 substrates estrone sulfate, ochratoxin A, and uric acid. Preliminary experiments demonstrated that the uptake of all three substrates was in the linear range up to 5 min (data not shown); all subsequent experiments were performed within this time frame of linear uptake. Incubation with estrone sulfate for 2 min produced the highest level of intracellular substrate accumulation with 13.8 ± 1.59-fold greater levels in the OAT4 reference cell line compared with empty vector-transfected cells (P ≤ 0.001). Compared with the estrone sulfate uptake in the OAT4 reference cell line, the L29P, R48Stop, and H469R variant cell lines all demonstrated drastically reduced levels of uptake (P ≤ 0.001). These levels were comparable to that in empty vector-transfected cells, strongly suggesting that these variants result in the complete loss of function of OAT4 (Fig. 2A). Similar patterns of uptake were also observed in transiently transfected HEK-293 cells incubated with estrone sulfate (data not shown). The remaining variants did not show a noticeable decrease in estrone sulfate uptake and were all comparable to the reference sequence level of uptake. Uptake of ochratoxin A and uric acid, while not as extensive as that of estrone sulfate, consistently produced two- to threefold greater intracellular accumulation in reference cells than in empty vector cells after 2 min of substrate incubation. The effect of the variants was similar to that observed in the estrone sulfate uptake experiments (Fig. 2, B and C). Specifically, the L29P, R48Stop, and H469R variants all showed a complete loss of function and their levels of uptake were comparable to that observed in the empty vector cell line (P ≤ 0.05 for ochratoxin A; P ≤ 0.001 for uric acid). None of variants displayed differential uptake due to the substrate used, which is suggestive that none of these variants cause a substrate selectivity change in OAT4.
Fig. 2.
Functional characterization of genetic variants of OAT4 using uptake of canonical substrates. OAT4 mediated uptake of estrone sulfate (A), ochratoxin (B), and uric acid (C). Experiments were performed for 2 (estrone sulfate and ochratoxin A) or 4 min (uric acid) at 37°C using 17 nM [3H]estrone sulfate, 200 μM [3H]ochratoxin A, or 400 μM uric acid (100 μM [14C]uric acid+300 μM unlabeled uric acid). Uptake values are expressed as a percentage of OAT4 reference sequence uptake and were normalized to total protein in each well. Values are means ± SE of 3 independent experiments. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05 compared with reference uptake levels using 1-way ANOVA and Dunnett's post hoc test.
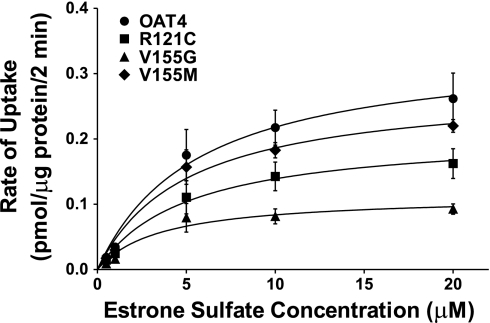
Four nonsynonymous variants in OAT4 were found with frequencies >1% in at least one ethnic group. Of these variants, only the R48Stop variant was found to impair OAT4 uptake capacity. Kinetic analysis of the remaining variants (R121C, V155G, and V155M) using estrone sulfate was performed to ascertain whether there were subtle differences in function in these higher frequency variants that were not observable in uptake assays. As shown in Fig. 3 and Table 4, all of these high-frequency variants showed altered kinetics compared with the reference cell line. While all of the Km values were close to the reference cell line value of 5.65 μM, the R121C and V155G variants both had statistically significant decreases in their Vmax values, with the V155G variant having a Vmax of 0.113 ± 0.0138 pmol·μg protein−1·min−1 and the R121C variant having a Vmax of 0.209 ± 0.0264 pmol·μg protein−1·min−1 compared with the reference value of 0.341 ± 0.0442 pmol·μg protein−1·min−1 (P ≤ 0.001 and P ≤ 0.05, respectively). The ratios of Vmax to Km for all of the variants were not substantially different from the reference cell line, however. Kinetic values were not determined for the complete loss-of-function variants due to the inability to measure any increase in uptake beyond that observed in the empty vector cell line.
Fig. 3.
Saturable uptake of estrone sulfate in HEK-293-Flp cells overexpressing OAT4 and 3 high-frequency nonsynonymous variants. Experiments were performed for 2 min at 37°C using 20 nM [3H]estrone sulfate and unlabeled estrone sulfate for the remainder; uptake values were normalized to total protein and adjusted for the uptake of estrone sulfate in empty vector-transfected cells. Curves were generated using nonlinear regression with automatic removal of outliers in Graphpad Prism 5.1.
Table 4.
Kinetic parameters of estrone sulfate uptake by common OAT4 nonsynonymous variants
| Variant | Km, μM | Vmax, pmol·μg protein−1·min−1 | Vmax:Km Ratio, pmol·mg protein−1 ·min−1·μM−1 |
|---|---|---|---|
| REF | 5.65 ± 2.02 | 0.341 ± 0.0442 | 60.4 ± 12.2 |
| R121C | 5.07 ± 1.84 | 0.209 ± 0.0265* | 41.2 ± 8.98 |
| V155G | 3.52 ± 1.41 | 0.113 ± 0.0138† | 31.6 ± 9.27 |
| V155M | 5.01 ± 1.16 | 0.280 ± 0.0224 | 55.7 ± 10.7 |
Values are means ± SE.
P < 0.05.
P < 0.001.
Effect of variants on OAT4 mRNA and protein expression.
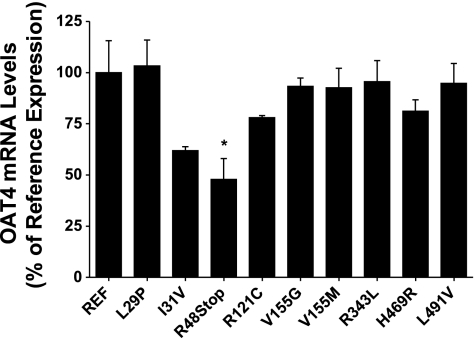
The underlying mechanism occurring in the loss-of-function variants and the kinetic variability observed in the most common OAT4 nonsynonymous variants was investigated initially using qRT-PCR. The level of expression of SLC22A11 in each of the cell lines was determined in relation to the expression of the control gene GAPDH. The expression of SLC22A11 mRNA was routinely 30,000- to 40,000-fold higher in the stably transfected cell lines than in the empty vector-transfected cell lines. While the I31V, R48Stop, and R121C variants showed slight decreases in mRNA levels compared with the reference cell line, only the R48Stop variant mRNA levels were significantly lower (P ≤ 0.05), as shown in Fig. 4. The use of stable cell lines created using the Flp-In system ensured that any differences in mRNA levels would most likely not be due to multiple integration sites of the expression vector, positional effects from random integration in the genome, or variability in the amount of vector successfully transfected transiently.
Fig. 4.
mRNA expression levels of OAT4 and its variants in stably transfected HEK-293-Flp cells. Total RNA was extracted from each cell line, reverse transcribed to cDNA, and subsequently used to determine mRNA levels of each variant of OAT4 using quantitative real-time PCR using a Taqman assay specific to human OAT4. Expression levels were normalized to endogenous expression of GAPDH and are shown as a percentage of reference OAT4 expression. Values are means ± SE from 2 independent experiments.
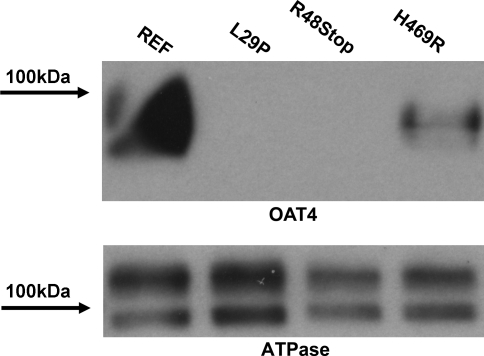
Plasma membrane protein levels resulting from each of the nonfunctional OAT4 variants were assessed using biotinylation of surface proteins in HEK-293 stably transfected cells expressing the reference and variant forms of OAT4, followed by Western blotting. Figure 5 shows a representative blot of the nonfunctional variants of OAT4 with an expected band detected at ∼90 kDa. The reference OAT4 band was the most intense, whereas minimal to no OAT4 protein was detected in either the L29P or the R48Stop cell lines (lanes 3 and 4). It should be noted that the OAT4 antibody is based on an epitope of the protein that occurs at the C terminus of OAT4, thus making the R48Stop variant undetectable. Repeat experiments using alternative antibodies derived from epitopes near the N terminus were unsuccessful. OAT4 was detected in the H469R variant cell line, albeit at an apparently lower level than the reference cell line. Similar results were also obtained in blots of total protein, wherein no protein was detected in either the L29P or R48Stop variants and the H469R variant showed decreased levels compared with the reference cell line. Reprobing the blot with an antibody specific for the Na+-K+-ATPase α-subunit revealed the abundance of membrane protein in the sample and approximately even loading of protein (Fig. 5). Additionally, reprobing of the blot with a β-actin antibody showed very faint to no bands in the plasma membrane fraction whereas abundant staining was present in the total protein blot, indicating that the membrane fraction was largely free of cytoplasmic protein (data not shown).
Fig. 5.
Levels of plasma membrane protein in HEK-293-Flp cells expressing reference and nonfunctional forms of OAT4. Top: plasma membrane protein was isolated via biotinylation, separated using SDS-PAGE and probed with an antibody specific to OAT4. Bottom: the same membrane was stripped and reprobed with an antibody specific to the α-subunit of the Na+-K+-ATPase.
DISCUSSION
Human OAT4 is present on the apical membrane of the renal proximal tubule and, as such, is known or suspected to be an important factor in the disposition of a variety of drugs and biologically relevant metabolites. Accordingly, several studies have focused on the identification and characterization of natural variants that occur in OAT4 (19, 31). Unlike these previous studies, however, this study used DNA samples from a large number of unrelated, ethnically diverse subjects to provide greater confidence in allele frequencies and to identify potential population-specific variants in OAT4. Overall, the variation in the amino acid sequence of OAT4 was low, limited to 12 total variable positions and resulting in only 7 nonsynonymous variants, only 4 of which occur on more than one chromosome and none of which occur at frequencies >3%. While this is a low level of variability, these levels are similar to that observed in other renal SLC22 family transporters, including OAT1 (9) and OAT3 (8). The extent of amino acid conservation at variant positions was also relatively low, with only two completely conserved variant locations compared with multiple mammalian species (R343L and L491V) and only three sites compared with other human OATs expressed in the kidney (L29P, R343L, and L491V). Furthermore, the majority of OAT4 nonsynonymous variants were predicted to be deleterious based on four distinct prediction algorithms. However, when examined using uptake of multiple substrates, the uptake results were concordant with only a third of the computational predictions. Examination of the kinetic properties of the higher frequency variants, all predicted to be damaging to some extent, also found slight reductions in Vmax, however, reductions in Vmax values may reflect differences in surface expression of OAT4. Despite indications of intolerance to variation in OAT4 based on low variability and computational methodologies, neither the conservation of specific residues nor the change in physiochemical properties appeared to be strong predictors of functional consequences as evidenced by the R343L and L491V variants, which occurred at conserved sites yet retained complete functionality, and the three most common variants which, despite a slight reduction in Vmax, were largely functional. This suggests that the conservation of residues is important to the preservation of function among orthologous OAT4 proteins but that the precise residues responsible for OAT4's specific functional capacity are not fully defined nor understood. Recent studies examining the role of histidine and glycine residues in OAT4 have made similar assessments and shed light on potentially critical regions for OAT4 function; systematic alteration of these residues produced distinct functional phenotypes related to membrane trafficking and substrate recognition (36, 37).
Of the nonsynonymous variants examined in this study, three resulted in a complete loss of function (L29P, R48Stop, and H469R), whereas the remainder demonstrated either no change in uptake (I31V, R343L, and L491V) of model substrates or only modest changes in the kinetic properties (R121C, V155G, and V155M) of estrone sulfate uptake. This “all-or-nothing” phenotype is unique among the SLC22 transporters examined thus far in that there were no observed variants possessing intermediate function or altered substrate specificity.
It is interesting to note that one of the highest frequency nonsynonymous variants observed in this study is the replacement of an arginine with a stop codon at position 48 (R48Stop) immediately after the first transmembrane domain. While the introduction of stop codons is an exceedingly rare event, and such nonsense variants are selected against for obvious reasons (24), variants producing premature termination of an SLC22 membrane transporter have been observed in OAT3 (8) and OCTN1 (28). In contrast, both of these variants had an exceedingly low frequency, unlike the R48Stop variant in OAT4, which occurs at 2.3% in Caucasians and was also observed in the Mexican-American group as a singleton. Whole genome surveys of stop codons have recently found that while comprising an exceedingly small percentage of known variants, they do occur throughout the genome; these variants are believed to more often than not result in a severe truncation of the protein, and some are readily identified as strongly causal alleles for certain diseases such as Legionnaires' disease-induced pneumonia (13). Additionally, it is well recognized that premature termination variants such as R48Stop are subject to nonsense-mediated mRNA decay (NMD). The fact that the mRNA of the R48Stop variant was reduced by nearly 50% compared with the expression of reference OAT4 suggests that NMD may be a mechanism contributing to the observed decreased level of mRNA expression, although this was not specifically investigated in the current study. The remaining mRNA that is translated would be expected to produce a severely truncated protein incapable of transporting any of the model substrates, consistent with the observed variant effect in Fig. 2, A–C. Of note, a recent paper by Zhou et al. (38) described a R48Y variant occurring in OAT4 that results in a complete loss of function as well as a potentially altered glycosylation state. However, this variant was not identified in the larger SOPHIE, HapMap, or 1000 Genomes projects; rather, a stop codon was identified at position 48 of OAT4 in these and other smaller sequencing projects (31). While the impact in a clinical context of the premature termination codon resulting from the R48Stop variant is unknown, it would be expected that individuals possessing this variant would have altered reabsorption of small molecules such as estrone sulfate, uric acid, and renally eliminated drugs that are substrates of OAT4.
The L29P variant is also of interest due to its complete loss of function. This variant occurs within the first transmembrane domain of OAT4, and while it was not found in the SOPHIE or 1000 Genomes projects, it was identified before these efforts by two separate sequencing groups and was deposited in dbSNP. This position is not conserved in other mammalian OAT4 sequences but is conserved among other primate OATs, including OAT1 and OAT3, suggesting that this position may be important for the translation of the protein, protein stability, or for localization to the plasma membrane in primates. The mRNA expression of the L29P variant was similar to the reference, but no plasma membrane protein was detected in the L29P cell line, which explains its loss of function. The mechanism by which a leucine-to-proline change at this position results in a loss of protein expression on the plasma membrane may be related to general protein instability or a failure of the protein to be inserted in the membrane; extreme angles induced by proline residues in helical structures may result in drastically unstable or significantly impaired proteins (33). Conversely, the presence of a leucine residue at this position may also confer protein stability based on the conserved nature of this N-terminal residue in other primate OATs. Further inquiry would be needed to determine whether this variant is detrimental due to the introduction of a proline residue or the loss of a leucine residue.
Within the set of singleton variants, only the H469R variant elicited a complete loss of function phenotype. Systematic replacement of histidine residues in OAT4 by Zhou et al. (36), however, did find that the conversion of these residues to alanine resulted in the preservation of comparable uptake activity of estrone sulfate in the majority of mutants compared with unmodified OAT4. This suggests that histidine residues are not critical to the optimal function of OAT4. However, the presence of an arginine at this position resulted in a complete loss of function of OAT4 in the present study, which can be partly explained by the loss of a significant fraction of OAT4 on the plasma membrane, as shown in Fig. 5. This suggests that while this position may not be of critical importance to OAT4 function in general, the presence of a residue with the properties of arginine may be detrimental to protein stability or trafficking.
Three common variants of OAT4 also displayed altered kinetic properties when tested with estrone sulfate. While the Km values of each of these variants for estrone sulfate were all comparable to the reference value determined for OAT4, the Vmax for each of these variants was lower than that of the reference with the V155G variant, demonstrating the greatest difference at ∼33% of the reference value. Consistent with the results of this study, Zhou et al. (38) observed a reduction in function of the V155G variant and a concomitant reduction in protein level, although the degree of decreased uptake was greater than in the current study. Each of these variants had frequencies >2% in at least one ethnic group, suggesting that a number of individuals possessing these variants may also be subject to a lower absolute level of reabsorption of OAT4 substrates, although the effect may not be readily observable in human subjects.
The impact of transporters located on the apical membrane of proximal tubule cells has been demonstrated previously but is a growing area of interest with numerous implications in pharmacokinetics and pharmacogenetics. The characterization of naturally occurring genetic variants of these transporters, such as OAT4, is a necessary and informative step to better define the potential genetic causes of drug-induced renal toxicities and perturbations in the urinary excretion of endogenous metabolites. This study provides an extensive examination of variants found to occur naturally in four ethnically diverse populations and the way in which these variants may affect the ability of OAT4 to control the levels of drug and metabolite substrates reabsorbed from the urine. Three nonsynonymous variants of OAT4 demonstrated a complete loss of function in uptake experiments with multiple chemically diverse substrates, while an additional three variants that occurred at a frequency of at least 2% displayed reduced Vmax values in the transport of estrone sulfate. A loss of protein expression and/or localization to the plasma membrane was a hallmark of the loss-of-function variants and can explain, at least in part, the observed lack of function in cellular uptake studies. Future studies examining the impact of nonsynonymous variants on the pharmacokinetics of drug substrates of OAT4 in a clinical setting would be particularly interesting considering the high frequency of the loss-of-function variants and would provide useful information regarding the affect of transporters primarily involved in the reabsorption of small molecules in the body.
GRANTS
This study was supported by National Institutes of Health (NIH) Grants GM61390 and GM36780. This project was also supported by NIH/NCRR UCSF-CTSI Grant UL1 RR024131, and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
REFERENCES
- 1.Bakhiya N, Monien B, Frank H, Seidel A, Glatt H. Renal organic anion transporters OAT1 and OAT3 mediate the cellular accumulation of 5-sulfooxymethylfurfural, a reactive, nephrotoxic metabolite of the Maillard product 5-hydroxymethylfurfural. Biochem Pharmacol 78: 414–419, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res 35: 3823–3835, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22: 2729–2734, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Cha SH, Sekine T, Kusuhara H, Yu E, Kim JY, Kim DK, Sugiyama Y, Kanai Y, Endou H. Molecular cloning and characterization of multispecific organic anion transporter 4 expressed in the placenta. J Biol Chem 275: 4507–4512, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekaratanawong S, Anzai N, Jutabha P, Miyazaki H, Noshiro R, Takeda M, Kanai Y, Sophasan S, Endou H. Human organic anion transporter 4 is a renal apical organic anion/dicarboxylate exchanger in the proximal tubules. J Pharmacol Sci 94: 297–304, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Enomoto A, Niwa T. Roles of organic anion transporters in the progression of chronic renal failure. Ther Apher Dial 11, Suppl 1: S27–S31, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Erdman AR, Mangravite LM, Urban TJ, Lagpacan LL, Castro RA, de la Cruz M, Chan W, Huang CC, Johns SJ, Kawamoto M, Stryke D, Taylor TR, Carlson EJ, Ferrin TE, Brett CM, Burchard EG, Giacomini KM. The human organic anion transporter 3 (OAT3; SLC22A8): genetic variation and functional genomics. Am J Physiol Renal Physiol 290: F905–F912, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Fujita T, Brown C, Carlson EJ, Taylor T, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Fujita K, Castro R, Chen CW, Lin ET, Brett CM, Burchard EG, Ferrin TE, Huang CC, Leabman MK, Giacomini KM. Functional analysis of polymorphisms in the organic anion transporter, SLC22A6 (OAT1). Pharmacogenet Genomics 15: 201–209, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Ganapathy ME, Brandsch M, Prasad PD, Ganapathy V, Leibach FH. Differential recognition of beta -lactam antibiotics by intestinal and renal peptide transporters, PEPT 1 and PEPT 2. J Biol Chem 270: 25672–25677, 1995 [DOI] [PubMed] [Google Scholar]
- 11.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L. Membrane transporters in drug development. Nat Rev Drug Discov 9: 215–236, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagos Y, Stein D, Ugele B, Burckhardt G, Bahn A. Human renal organic anion transporter 4 operates as an asymmetric urate transporter. J Am Soc Nephrol 18: 430–439, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, Skerrett SJ, Beutler B, Schroeder L, Nachman A, Ozinsky A, Smith KD, Aderem A. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med 198: 1563–1572, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilgendorf C, Ahlin G, Seithel A, Artursson P, Ungell AL, Karlsson J. Expression of thirty-six drug transporter genes in human intestine, liver, kidney, and organotypic cell lines. Drug Metab Dispos 35: 1333–1340, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Kimura H, Takeda M, Narikawa S, Enomoto A, Ichida K, Endou H. Human organic anion transporters and human organic cation transporters mediate renal transport of prostaglandins. J Pharmacol Exp Ther 301: 293–298, 2002 [DOI] [PubMed] [Google Scholar]
- 16.Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M, Mangino M, Albrecht E, Wallace C, Farrall M, Johansson A, Nyholt DR, Aulchenko Y, Beckmann JS, Bergmann S, Bochud M, Brown M, Campbell H, Connell J, Dominiczak A, Homuth G, Lamina C, McCarthy MI, Meitinger T, Mooser V, Munroe P, Nauck M, Peden J, Prokisch H, Salo P, Salomaa V, Samani NJ, Schlessinger D, Uda M, Volker U, Waeber G, Waterworth D, Wang-Sattler R, Wright AF, Adamski J, Whitfield JB, Gyllensten U, Wilson JF, Rudan I, Pramstaller P, Watkins H, Doering A, Wichmann HE, Spector TD, Peltonen L, Volzke H, Nagaraja R, Vollenweider P, Caulfield M, Illig T, Gieger C. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet 5: e1000504, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, Kroetz DL, Ferrin TE, Clark AG, Risch N, Herskowitz I, Giacomini KM. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA 100: 5896–5901, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leabman MK, Huang CC, Kawamoto M, Johns SJ, Stryke D, Ferrin TE, DeYoung J, Taylor T, Clark AG, Herskowitz I, Giacomini KM. Polymorphisms in a human kidney xenobiotic transporter, OCT2, exhibit altered function. Pharmacogenetics 12: 395–405, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Lee WK, Kwak JO, Hwang JS, Suh CK, Cha SH. Identification and characterization of single nucleotide polymorphisms of SLC22A11 (hOAT4) in Korean women osteoporosis patients. Mol Cells 25: 265–271, 2008 [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812–3814, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura M, Naito S. Tissue-specific mRNA expression profiles of human ATP-binding cassette and solute carrier transporter superfamilies. Drug Metab Pharmacokinet 20: 452–477, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30: 3894–3900, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson EE, Rankin GO. Human renal organic anion transporters: characteristics and contributions to drug and drug metabolite excretion. Pharmacol Ther 109: 399–412, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Sawyer SL, Berglind LC, Brookes AJ. Negligible validation rate for public domain stop-codon SNPs. Hum Mutat 22: 252–254, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Takeda M, Khamdang S, Narikawa S, Kimura H, Hosoyamada M, Cha SH, Sekine T, Endou H. Characterization of methotrexate transport and its drug interactions with human organic anion transporters. J Pharmacol Exp Ther 302: 666–671, 2002 [DOI] [PubMed] [Google Scholar]
- 26.Ugele B, St-Pierre MV, Pihusch M, Bahn A, Hantschmann P. Characterization and identification of steroid sulfate transporters of human placenta. Am J Physiol Endocrinol Metab 284: E390–E398, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Urban TJ, Sebro R, Hurowitz EH, Leabman MK, Badagnani I, Lagpacan LL, Risch N, Giacomini KM. Functional genomics of membrane transporters in human populations. Genome Res 16: 223–230, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urban TJ, Yang C, Lagpacan LL, Brown C, Castro RA, Taylor TR, Huang CC, Stryke D, Johns SJ, Kawamoto M, Carlson EJ, Ferrin TE, Burchard EG, Giacomini KM. Functional effects of protein sequence polymorphisms in the organic cation/ergothioneine transporter OCTN1 (SLC22A4). Pharmacogenet Genomics 17: 773–782, 2007 [DOI] [PubMed] [Google Scholar]
- 29.van der Harst P, Bakker SJ, de Boer RA, Wolffenbuttel BH, Johnson T, Caulfield MJ, Navis G. Replication of the five novel loci for uric acid concentrations and potential mediating mechanisms. Hum Mol Genet 19: 387–395, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Vormfelde SV, Schirmer M, Hagos Y, Toliat MR, Engelhardt S, Meineke I, Burckhardt G, Nurnberg P, Brockmoller J. Torsemide renal clearance and genetic variation in luminal and basolateral organic anion transporters. Br J Clin Pharmacol 62: 323–335, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G, Bhatnagar V, Wen G, Hamilton BA, Eraly SA, Nigam SK. Analyses of coding region polymorphisms in apical and basolateral human organic anion transporter (OAT) genes [OAT1 (NKT), OAT2, OAT3, OAT4, URAT (RST)]. Kidney Int 68: 1491–1499, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Yamashita F, Ohtani H, Koyabu N, Ushigome F, Satoh H, Murakami H, Uchiumi T, Nakamura T, Kuwano M, Tsujimoto M, Sawada Y. Inhibitory effects of angiotensin II receptor antagonists and leukotriene receptor antagonists on the transport of human organic anion transporter 4. J Pharm Pharmacol 58: 1499–1505, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Yohannan S, Yang D, Faham S, Boulting G, Whitelegge J, Bowie JU. Proline substitutions are not easily accommodated in a membrane protein. J Mol Biol 341: 1–6, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, Yang X, Li J, Wu J, Peng WX, Dong XQ, Zhou SF, Yu XQ. Upregulation of rat renal cortical organic anion transporter (OAT1 and OAT3) expression in response to ischemia/reperfusion injury. Am J Nephrol 28: 772–783, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Zhou F, Illsley NP, You G. Functional characterization of a human organic anion transporter hOAT4 in placental BeWo cells. Eur J Pharm Sci 27: 518–523, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Zhou F, Pan Z, Ma J, You G. Mutational analysis of histidine residues in human organic anion transporter 4 (hOAT4). Biochem J 384: 87–92, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou F, Tanaka K, Pan Z, Ma J, You G. The role of glycine residues in the function of human organic anion transporter 4. Mol Pharmacol 65: 1141–1147, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Zhou F, Zhu L, Cui PH, Church WB, Murray M. Functional characterization of nonsynonymous single nucleotide polymorphisms in the human organic anion transporter 4 (hOAT4). Br J Pharmacol 159: 419–427, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]