Abstract
Recent studies have suggested a link between inhaled particulate matter (PM) exposure and increased mortality and morbidity associated with pulmonary and cardiovascular diseases. However, a precise understanding of the biological mechanism underlying PM-associated toxicity and pathogenesis remains elusive. Here, we investigated the impact of PM exposure in intracellular stress signaling pathways with animal models and cultured cells. Inhalation exposure of the mice to environmentally relevant fine particulate matter (aerodynamic diameter < 2.5 μm, PM2.5) induces endoplasmic reticulum (ER) stress and activation of unfolded protein response (UPR) in the lung and liver tissues as well as in the mouse macrophage cell line RAW264.7. Ambient PM2.5 exposure activates double-strand RNA-activated protein kinase-like ER kinase (PERK), leading to phosphorylation of translation initiation factor eIF2α and induction of C/EBP homologous transcription factor CHOP/GADD153. Activation of PERK-mediated UPR pathway relies on the production of reactive oxygen species (ROS) and is critical for PM2.5-induced apoptosis. Furthermore, PM2.5 exposure can activate ER stress sensor IRE1α, but it decreases the activity of IRE1α in splicing the mRNA encoding the UPR trans-activator X-box binding protein 1 (XBP1). Together, our study suggests that PM2.5 exposure differentially activates the UPR branches, leading to ER stress-induced apoptosis through the PERK-eIF2α-CHOP UPR branch. This work provides novel insights into the cellular and molecular basis by which ambient PM2.5 exposure elicits its cytotoxic effects that may be related to air pollution-associated pathogenesis.
Keywords: air pollution, unfolded protein response
air pollution is a continuing challenge to public health for the general population since epidemiological studies have linked fine particulate matter (aerodynamic diameter < 2.5 μm, PM2.5) pollution to the increase of cardiovascular mortality and morbidity (8). It is known that, depending on their natural and/or anthropogenic emission sources, air PM pollution is a complex mixture of chemical and/or biological elements, such as metals, salts, carbonaceous material, volatile organic compounds, polycyclic aromatic hydrocarbons, and endotoxins (2, 42). Airborne PM demonstrates an incremental capacity to penetrate into the distal airway units and potentially enter the systemic circulation with diminishing sizes (6, 33). PM2.5, primarily derived from stationary and traffic-related combustion sources, has been linked strongly with cardiovascular disease and metabolic disease (6, 10, 12, 35, 47). Recent studies have addressed the impacts of airborne PM2.5 on the functioning of signaling pathways with significant roles in redox homeostasis and inflammation (14, 48, 49, 54). However, the understanding of molecular and cellular mechanisms underlying PM2.5-associated systemic diseases remains incomplete. Our study here provides novel biological insights into the molecular basis of PM2.5-induced intracellular events, highlighting the activation of a pathophysiological endoplasmic reticulum (ER) stress response upon PM2.5 exposure.
The ER stress response, also called unfolded protein response (UPR), is an intracellular stress signaling from the ER to protect cells from stress caused by the accumulation of unfolded or misfolded proteins (37, 56). In mammalian cells, the ER is the site of folding of membrane and secreted proteins, synthesis of lipids and sterols, and storage of free calcium (56). As a unique protein-folding compartment and dynamic calcium store, the ER is exquisitely sensitive to changes of intracellular homeostasis. Physiological states that increase protein-folding demand, or stimuli that disrupt protein folding reactions, create an imbalance between the protein-folding load and capacity of the ER. This can cause the accumulation of unfolded or misfolded proteins in the ER lumen (the condition referred to “ER stress”). To ensure the fidelity of protein folding and to handle the accumulation of unfolded or misfolded proteins, the ER has evolved a group of signal transduction pathways: the UPR to alter transcriptional and translational programs. The basic UPR pathways in mammalian cells consist of three main signaling cascades initiated by three primary ER stress sensors: IRE1α (inositol-requiring 1α), PERK (double-strand RNA-activated protein kinase-like ER kinase), and ATF6 (activating transcription factor 6) (38). Under ER stress, PERK kinase phosphorylates translation initiation factor 2α (eIF2α) to shut down protein synthesis, whereas IRE1α functions as an RNase to splice the mRNA encoding X-box binding protein 1 (XBP1), leading to transcriptional reprogramming of the stressed cells. Under ER stress, ATF6 is also activated to induce gene expression to help the ER recover from the stress. The combined effect of the UPR pathways is to remodel the cells to adapt to the stress. However, if the ER stress cannot be resolved, the UPR will induce apoptosis to eliminate the stressed cells. In addition to those that are strictly involved in protein-folding process, the UPR can also be triggered by pathogenic infection, chemical insult, inflammatory stimuli, and even differentiation of specialized cell types (56). Recently, accumulating evidence suggested that the UPR signaling is critical for health and disease. Under physiological or pathophysiological conditions, the UPR can modulate cell metabolism, inflammation, and programmed cell death, leading to the pathogenesis of a variety of diseases, such as cardiovascular disease, diabetes, neurodegenerative disease, and cancer (56).
In this study, we used “real-world” PM2.5 exposure system, the mobile “Ohio's Air Pollution Exposure System for the Interrogation of Systemic Effects (OASIS-1)” (46, 55), to perform subchronic whole body exposure of the mice to environmentally relevant PM2.5 or filtered air (FA). To elucidate the molecular basis of ambient PM2.5-associated cytotoxicity, we investigated the intracellular stress-signaling pathways in the animal model or cultured cells that were exposed to PM2.5. We demonstrated that PM2.5 exposure differentially activates the UPR branches that lead to ER stress-induced apoptosis in the mice exposed to PM2.5. Further studies revealed that PM2.5-induced stress response relies on the production of reactive oxygen species (ROS). The findings on PM2.5-induced ER stress signaling pathways will not only contribute to our better understanding of molecular and cellular mechanisms by which PM2.5 elicits its cytotoxic effects but also will be informative to the prevention and treatment of air pollution-induced systemic diseases.
EXPERIMENTAL PROCEDURES
Materials.
Chemicals were purchased from Sigma unless indicated otherwise. Synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Antibodies against phosphorylated or total eIF2α, phosphorylated PERK, phosphorylated and total c-Jun NH2-terminal kinase (JNK), and total IRE1α proteins were purchased from Cell Signaling Technologies (Danvers, MA). Antibodies against total PERK, ATF4, C/EBP homologous protein (CHOP), ATF3, GADD34, and HA were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Antibodies against GRP78/binding immunoglobulin protein (BiP) and GRP94 were from Stressgen (Ann Arbor, MI). The antibody against caspase-3 antibody was purchased from Invitrogen. The antibody against phosphorylated double-stranded RNA-dependent protein kinase (PKR) was from Signalway Antibody (Pearland, TX). Antibody against α-tubulin was from Sigma. Plasmid PERK K618A pcDNA was from Addgene (ID2185) (16). Plasmids PERK pcDNA and pCGN-ATF6 were kindly provided by Dr. David Ron and Dr. Ron Prywes, respectively (16, 52). The PM2.5 particles for the in vitro experiments were extracted from the filters that were collected from the exposure system during the time period when the mice were exposed, as previously described (48, 49, 55). The stock PM2.5 solution (5 mg/ml in PBS) was stored in −80°C.
Animal model.
Six-week-old C57BL/6 male mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and were equilibrated for 2 wk before experimental enrollment. CHOP knockout mice (59) were initially backcrossed onto the Taconic Farms (Hudson, NY) B6C3H F1 strain for 11 generations (43) before being mated within the littermates. The mice were housed in cages with regular chow in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal housing facility. The Committees on Use and Care of Animals at the Ohio State University approved all experimental procedures.
“Real-world” exposure of animals to ambient PM2.5.
Mice were exposed to concentrated ambient PM2.5 or FA from June to August 2008 in a mobile trailer “Ohio's Air Pollution Exposure System for the Interrogation of Systemic Effects (OASIS)-1” in Columbus, OH, where most of the PM2.5 component is attributed to long-range transport (55). The concentrated PM2.5 was generated using a versatile aerosol concentration enrichment system (VACES) developed by Sioutas et al. (41) and modified by Chen and Nadziejko (11). The mice were exposed to concentrated PM2.5 at nominal 10× ambient concentrations for 6 h per day, 5 days per week for a total of 10 wk, as detailed previously (46, 49). The control (FA) mice in the experiment were exposed to an identical protocol with the exception of a high-efficiency particulate-air filter positioned in the inlet valve position to remove all of the PM2.5 in the filtered air stream. On the final day of the exposure, the mice were euthanized and tissue samples were collected for further studies.
Analysis of PM2.5 concentration and composition.
The concentrations of PM2.5 in ambient air and in the chambers were monitored in filters that retained PM2.5 using an oscillating microbalance (model 1400, Tapered-Element Oscillating Microbalance, Rupprecht and Patashnick, East Greenbush, NY), as described previously (55). Briefly, PM2.5 were collected from the exposure chambers and weighed in a temperature- and humidity-controlled weighing room using an oscillating microbalance (model 1400, Tapered-Element Oscillating Microbalance). The weight was used to calculate exposure concentrations. The analysis of PM2.5 composition was performed by RTI following compendium Environmental Protection Agency method IO-3.3 using X-ray fluorescence spectroscopy [www.epa.gov/ttnamti1/files/ambient/inorganic] (55).
Cell culture, in vitro exposure to PM2.5, and adenoviral transduction of cells.
Murine monocytic-macrophage cell line RAW264.7 (American Type Culture Collection) were cultured in FG12 media supplemented with l-glutamine and 10% fetal bovine serum in a 5% CO2 atmosphere at 37°C. For PM2.5 in vitro exposure experiments, the Teflon filters used for gravimetric and elemental analyses were placed downstream of the cyclone inlet of the “OASIS-1” exposure system to collect ambient particulates as previously described (28, 48, 49). PM2.5 was collected at the time when the mice were exposed. The stock PM2.5 solution (5 mg/ml in PBS) was stored in −80°C. The frozen PM2.5 aliquots were thawed and briefly sonicated before being added into the cell culture media for the in vitro experiments. Cells were treated with PM2.5 at the concentrations ranging from 50 to 400 μg/ml, and the same amount of PBS was added in the control group. For the adenoviral transduction experiments, preparation of replication-incompetent adenovirus expressing manganese superoxide dismutase (Mn-SOD) or dominant-negative N17Rac1 was as previously described (9, 25, 39). RAW264.7 cells cultured in six-well plates were infected with adenovirus expressing Mn-SOD, N17Rac1, or control β-galactosidase at a multiplicity of infection of 150 when the cells were about 60% confluent. The in vitro PM2.5 exposure with the transduced cells was performed at 36 h after the infection.
Animal exposure to ambient PM2.5 by intranasal inhalation.
Ambient PM2.5 in 25 μl PBS (1.6 μg/g body wt) or 25 μl sterile PBS (control) were intranasally delivered to 3-mo-old male mice over a 5-min period twice for 5 days (3, 47). PM2.5 particles were collected from the filters installed in the “OASIS-1” system during the “real-world” PM2.5 exposure period. For the intranasal delivery, the mice were anesthetized by intraperitoneal injection of pentobarbital sodium (50 mg/kg body wt). On the fifth day after the first administration of PM2.5, the mice were euthanized and tissue samples were collected for further studies.
Transmission electron microscopy.
The lung and liver tissues from PM2.5-exposed or FA-exposed mice were fixed with 2% formaldehyde and 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer. The fixed lung and liver tissue for electron microscopy were prepared, imaged, and scanned as described previously (36, 40).
Immunohistochemistry staining and DNA fragmentation analysis for apoptosis.
Paraffin-embedded tissue sections (4 μm) were blocked with 0.5% H2O2 in methanol to reduce endogenous peroxidase activity. The sections were incubated with anti-BiP/GRP78 antibody (1:100) overnight at 4°C. Sections were developed in Zytomed peroxidase substrate (Zytomed, Berlin, Germany) and counterstained with hematoxylin. To visualize the apoptotic events in the lung and liver tissue sections of mice exposed to PM2.5 or FA, we used a fluorescent ApoAlert DNA fragmentation assay kit (Clontech) to stain DNA fragmentation with the paraffin-embedded tissue section slides according to the manufacture's instruction (57). The apoptotic cells exhibited green fluorescence using a standard fluorescein filter set (520 ± 20 nm). All cells were stained with propidium iodide (PI) and displayed strong red cytoplasmic fluorescence.
Western blot analyses.
To determine expression levels of GRP78/BiP, GRP94, CHOP, PERK, inositol-requiring 1-α (IRE1α), α-tubulin, and phosphorylated and total eIF2α proteins, total cell lysates were prepared from either lung or liver tissue or cultured macrophages using Nonidet-40 (NP-40) lysis buffer (1% NP-40, 50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 0.05% SDS, 0.5 mM Na vanadate, 100 mM NaF, 50 mM β-glycerophosphate, and 1 mM phenylmethylsulfonyl fluoride) supplemented with protease inhibitors (EDTA-free Complete Mini, Roche). Denatured proteins were separated by SDS-PAGE on 10% Tris-glycine polyacrylamide gels and transferred to a 0.45-mm polyvinyl difluoride (PVDF) membrane (GE Healthcare). Membrane-bound antibodies were detected by an enhanced chemiluminescence detection reagent (GE Healthcare). Because PERK and IRE1α are very low-abundance proteins in most cell types, we detected phosphorylated and unphosphorylated PERK or IRE1α by IP-Western blot analysis using an anti-PERK or IRE1α antibody (15, 57). The total protein lysates from the liver tissue (250 mg tissue/ml lysate buffer) were immunoprecipitated with the anti-PERK or IRE1α antibody, followed by the Western blot analysis with the same antibody.
Semiquantitative reverse transcription (RT)-PCR analysis of Xbp1 mRNA splicing and quantitative real-time RT-PCR analysis.
The RT-PCR analysis of Xbp1 mRNA splicing and quantitative real-time RT-PCR analysis were as previously described (57). Briefly, total cellular RNA was prepared using TRIzol reagent as instructed by the manufacturer (Invitrogen). Total RNA was reverse transcribed to cDNA using a random primer (Applied Biosystems). For semiquantitative RT-PCR analysis of Xbp1 mRNA splicing, 25 ng cDNA was used for each reaction. The forward primer for PCR amplification of spliced and total mouse Xbp1 mRNA is 5-ACACGCTTGGGAATGGACAC-3, and the reverse primer is 5-CCATGGGAAGATGTTCTGGG-3. PCR products were separated by electrophoresis on a 2.5% agarose gel and visualized by ethidium bromide staining. The size of amplified unspliced Xbp1 mRNA is 170 bp, and the size of amplified spliced Xbp1 mRNA is 144 bp. For real-time PCR analysis, the reaction mixture containing cDNA template, primers, and SYBR Green PCR Master Mix (Invitrogen) was run in a Stratagene MX3000P Real-Time PCR System (Stratagene). The sequences of primers for examining the regulated IRE1-dependent decay (RIDD) were previously as described (18) The other real-time PCR primer sequence information is shown in Table 1. Fold changes of mRNA levels were determined after normalization to internal control β-actin RNA levels.
Table 1.
Sequences of real-time PCR primers
| Gene | Forward | Reverse |
|---|---|---|
| HO-1 | CACGCATATACCCGCTACCT | CCAGAGTGTTCATTCGAGA |
| Xbp1(s) | GAGTCCGCAGCAGGTG | GTGTCAGAGTCCATGGGA |
| GPX1 | CTCACCCGCTCTTTACCTTCCT | ACACCGGAGACCAAATGATGTACT |
| SOD1 | GCGGTGAACCAGTTGTGTTGTC | CAGTCACATTGCCCAGGTCTCC |
| SOD2 | GCGGTCGTGTAAACCTCAT | CCAGAGCCTCGTGGTACTTC |
| Pdi-P5 | ACAGAAGCTGGAGCCCAGTA | CCGAAAGACAGTTCCCTGAG |
| ATF4 | ATGGCCGGCTATGGATGAT | CGAAGTCAAACTCTTTCAGATCCATT |
| CHOP | CTGCCTTTCACCTTGGAGAC | CGTTTCCTGGGGATGAGATA |
| BiP | CATGGTTCTCACTAAAATGAAAGG | GCTGGTACAGTAACAACTG |
| GRP94 | AATAGAAAGAATGCTTCGCC | TCTTCAGGCTCTTCTTCTGG |
| P58(ipk) | TCCTGGTGGACCTGCAGTACG | CTGCGAGTAATTTCTTCCCC |
| Edem1 | TGGAATTTGGGATTCTGAGC | CTGCAGTCCAGGGAAGAAAG |
| β-Actin | GATCTGGCACCACACCTTCT | GGGGTGTTGAAGGTCTCAAA |
HO-1, heme oxygenase-1; Xbp1, X box binding protein 1; GPX1, glutathionine peroxidase-1; SOD1, -SOD2, superoxide dismutase-1 and -2, respectively; Pdi-P5, protein disulfide isomerase-P5; ATF4, activating transcription factor 4; BiP, binding immunoglobulin protein; GRP94, glucose-regulated protein 94; P58, protein kinase inhibitor of 58 kDa Edem1, enhancing α-mannosidase-like protein 1.
Dihydroethidium fluorescence of liver tissue.
Dihydroethidium (DHE), an oxidative fluorescent dye, was used to detect superoxide in segments of frozen liver tissue as described previously (31). Briefly, fresh unfixed segments of liver tissue were frozen in OCT compound, and transverse sections (10 μm) were generated with a cryostat and placed on glass slides. Sections were then incubated in a light-protected chamber at room temperature for 30 min with 10 μmol/l DHE (Molecular Probes). Images were obtained with the use of a Zeiss laser scanning confocal microscope equipped with a krypton-argon laser. The excitation wavelength was 488 nm, and emission fluorescence was detected with the use of a 585-nm long-pass filter.
Statistics.
Experimental results are shown as means ± SE (for variation between animals or experiments). The mean values for biochemical data from the experimental groups (PM2.5 exposure verse filtered air) were compared by a paired or unpaired, two-tailed Student's t-test. Statistical tests with P < 0.05 were considered significant.
RESULTS
PM2.5 exposure induces both oxidative stress and ER stress in mouse lung and liver tissues.
To elucidate in vivo effect of subchronic PM2.5 exposure, male C57BL/6J mice were exposed to concentrated ambient PM2.5 for 10 wk in the mobile trailer “OASIS-1” exposure system composed of the midwestern regional background in Columbus, OH, where most of the PM2.5 is attributed to long-range transport (46, 55). During the exposure time period, the mean daily ambient PM2.5 concentration at the study site was 6.5 ± 4.8 μg/m3, whereas the mean concentration inside the PM2.5 exposure chamber was 74.6 μg/m3. Because the mice were exposed 6 h a day, 5 days a week, the equivalent PM2.5 concentration to which the mice were exposed in the chamber “normalized” over the 10-wk period was 11.6 μg/m3 after taking into account nonexposed time and weekends [the annual average PM2.5 National Ambient Air Quality Standard (NAAQS) of 15.0 μg/m3 (13)]. The control mice in the experiment were exposed to an identical protocol with the exception of a high-efficiency particulate-air filter positioned in the inlet valve position to remove all of the PM2.5 in the filtered air stream. The X-ray fluorescence spectroscopic analysis of PM2.5 composition in the exposure chamber revealed higher concentration of a range of metals (55). The major composition of PM2.5 included alkali metals (K and Na), alkaline earth metals (Mg and Ca), transition metals (Fe and Zn), and nonmetals (S) (Table 2).
Table 2.
Major composition of PM2.5 in the exposure chamber
| Category | Chemical | Concentration, ng/m3 |
|---|---|---|
| Alkali metals | K | 308.3 ± 75.1 |
| Na | 375.0 ± 91.7 | |
| Alkaline earth metals | Mg | 50.5 ± 16 |
| Ca | 220.1 ± 54.1 | |
| Sr | 20.3 ± 3.5 | |
| Transition metals | Fe | 385.0 ± 99.1 |
| Zn | 115.9 ± 29.5 | |
| Poor metals | Al | 53.0 ± 27.5 |
| Sn | 55.0 ± 18.9 | |
| Pb | 19.9 ± 4.1 | |
| Lanthanoids | Sm | 3.3 ± 1.8 |
| Metals | Eu | 1.5 ± 0.4 |
| Nonmetals | S | 9167.5 ± 913.1 |
| Si | 833.4 ± 167.3 |
Twelve particulate matter (aerodynamic diameter < 2.5 μm) (PM2.5) samples were collected from June to August, 2008 when the animals were exposed to PM2.5 through the Ohio's Air Pollution Exposure System for the Interrogation of Systemic Effects (OASIS-1) system. The samples were subjected to X-ray fluorescence spectroscopy for composition analysis. The values on the left column represent the concentrations of major PM2.5 chemicals (means ± SD, n = 12).
To evaluate the intracellular impact of PM2.5 exposure, we performed transmission electron microscopy analysis of the cell ultrastructure with the lung and liver tissue samples from the mice exposed to PM2.5 or FA. The lung bronchoalveolar lavage macrophages and liver macrophages (Kupffer cells) of the mice exposed to PM2.5 displayed an increased number of mitochondria and expansion of ER compartment when compared with those from the mice exposed to FA (Fig. 1, A and B). This suggested increased ER and mitochondria activities in the macrophages stimulated by PM2.5 exposure. It has been proposed that PM2.5 may cause oxidative stress due to their features of small diameters and high surface area that have potential ability in ROS generation (46, 49). To test whether inhaled PM2.5 causes oxidative stress in vivo, we examined the redox states in the lung and liver tissues of mice exposed to PM2.5 or FA. DHE, an oxidative fluorescent dye, was used to detect superoxide in segments of frozen liver tissue. The result of DHE staining indicated that superoxide production was markedly increased in the liver of PM2.5-exposed mice compared with that in FA-exposed mice (Fig. 1, C and D). Because lung tissue has to be perfused by fixatives to keep the alveolar space patent for tissue sectioning, we cannot obtain appropriate frozen lung tissue sections for DHE staining. However, we utilized quantitative real-time RT-PCR analysis to determine the redox state in the lung tissue of mice exposed to PM2.5 or FA. Exposure to PM2.5 stimulates oxidative stress in the lung tissue, as evidenced by upregulation of the mRNAs encoding antioxidant enzymes, including glutathione peroxidase (Gpx)-1, Heme oxygenase-1 (HO-1), superoxide dismutase (SOD)-1, and SOD2 (Fig. 1E).
Fig. 1.
Particulate matter (aerodynamic diameter <2.5 μm, PM2.5) particles are retained intracellularly and induces oxidative stress in vivo. A and B: transmission electron micrographs of lung and liver macrophages from the mice exposed to PM2.5 or filtered air (FA) for 10 wk. E, ER; M, mitochondria. C: dihydroethidium (DHE) staining of liver tissue sections from the mice exposed to FA or PM2.5. Frozen liver tissue sections were stained with DHE (10 μmol/l). The oxidative red fluorescence (Texas red) was detected by a Zeiss fluorescence microscope. D: DHE signals were quantified by counting the number of positive-stained nuclei in 8 random fields. Microscopic interference contrast was used to exclude positive signals from noncell origin. The percentages of DHE-positive nuclei (compared with total nuclei) were shown. Data are shown as means ± SE for 6 animals per group. **P < 0.01. P values are shown for statistically significant differences. E: quantitative real-time RT-PCR analysis of the expression of mRNAs encoding glutathione peroxidase-1 (Gpx-1), heme oxygenase-1 (HO-1), superoxide dismutase-1 (SOD1), and SOD2 in the lung tissue of the mice exposed to FA or PM2.5. Fold changes of mRNA levels were determined after normalization to internal control β-actin RNA levels. For each comparison group, the mRNA level of one FA-exposed mouse was defined as 1 and was used to calculate the fold changes of mRNA levels for the other mice. Each bar denotes the means ± SE (n = 6 mice/group). **P < 0.01.
Recent evidence suggested that ER stress is linked to the alteration of redox states (17, 30, 50, 56). To determine whether PM2.5 can induce ER stress in vivo, we examined the induction of ER stress markers in various organs and tissues of the animals exposed to PM2.5. We first visualized the induction of ER stress marker GRP78/BiP in the tissues of the mice exposed to PM2.5 or FA by immunohistochemistry staining. BiP is an ER chaperone that is induced by ER stress and has been proposed to function as a master regulator for the activation of ER stress sensors (37). Immunohistochemistry staining showed that levels of BiP were significantly increased in the lung and liver tissues of the mice exposed to PM2.5 when compared with those in the mice exposed to FA (Fig. 2, A–D), suggesting the induction of ER stress in these tissues. In comparison, ER stress marker BiP was only marginally induced in other vulnerable tissues including aorta and spleen of mice exposed to PM2.5 (data not shown). To confirm the induction of ER stress in the lung and liver tissues by PM2.5 exposure, we examined the expression of ER stress markers in these tissues by Western blot analysis and quantitative real-time RT-PCR assay. Western blot analyses revealed that the levels of ER chaperones BiP and GRP94 were significantly increased in the lung and liver tissues of mice exposed to PM2.5 compared with those of FA controls, thus confirming the effect of PM2.5 on inducing ER stress in these tissues (Fig. 2, E and F).
Fig. 2.
PM2.5 exposure induces ER stress in the lung and liver tissues. A–D: immunohistochemistry staining of lung and liver tissue sections for binding immunoglobulin protein (BiP) expression. The BiP signals were developed with peroxidase substrate reaction (brown signal). The slides were counterstained with hematoxylin. Magnification: ×400. B and D are the percentages of BiP-staining-positive cells in the lung or liver tissue sections of the mice exposed to PM2.5 or FA. The numbers of positive- and negative-stained cells were counted in 8 random fields per sample. The percentages were calculated by normalizing BiP-staining-positive cells to the total cells. Data are shown as means ± SE for 6 animals per group. *P < 0.05. P values are shown for statistically significant differences. E: Western blot analyses for the expression levels of glucose-regulated protein 94 (GRP94) and BiP proteins in the lung tissue of the mice exposed to FA or PM2.5. Denatured lung protein lysates (150 μg per sample) are separated on a 10% Tris-glycine polyacrylamide gel. Levels of α-tubulin protein were determined as internal controls. The values below the gels represent the normalized protein signal intensities. The protein band signals were quantified by using NIH Image J software. F: Western blot analyses for the expression levels of GRP94 and BiP proteins in the liver tissue of the mice exposed to FA or PM2.5. Denatured liver protein lysates (80 μg per sample) are separated on a 10% Tris-glycine polyacrylamide gel. Levels of α-tubulin protein were determined as internal controls. The values below the gels represent the normalized protein signal intensities. For A–D, the experiments were repeated at least three times, and the representative data are shown.
PM2.5 activates the UPR pathway mediated through PERK/eIF2α.
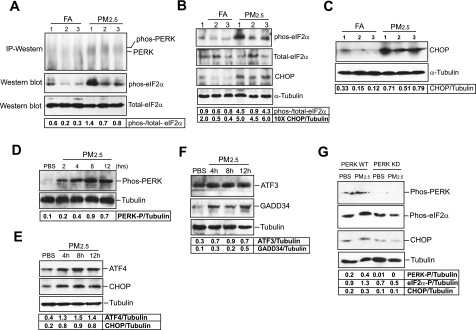
Next, we examined the activation of UPR pathways induced by PM2.5. Under ER stress, ER stress sensor PERK is activated to phosphorylate translation initiation factor eIF2α, a key step in attenuation of protein translation (38). However, ER stress-induced phosphorylation of eIF2α can lead to selective translation of specific mRNAs encoding functions in modulating amino acid metabolism, antioxidative stress response, and apoptosis (37, 56). To evaluate activation of the UPR pathways under PM2.5 exposure, we first determined the levels of phosphorylated PERK and its substrate, phosphorylated eIF2α, in the liver or lung tissue of the mice exposed to PM2.5. Because PERK is a very low-abundance protein in most cell types and tissues, it is very difficult to detect phosphorylated PERK by regular Western blot analysis (15). To detect activated PERK in mouse tissue samples, we first concentrated the PERK protein by IP using an anti-total PERK antibody and then performed Western blot analysis using the same antibody. The result indicated that the levels of phosphorylated PERK were increased in the liver tissue of the mice exposed to PM2.5 compared with that in the mice exposed to FA (Fig. 3A). Consistently, the ratio of phosphorylated to total eIF2α levels was increased in the liver tissue of the mice exposed to PM2.5 (Fig. 3A). Moreover, we can detect an increase in ratio of phosphorylation to total eIF2α levels in the lung tissue of the mice exposed to PM2.5 (Fig. 3B). Furthermore, C/EBP homologous transcription factor CHOP/GADD153 is one of the downstream targets in PERK/eIF2α-mediated UPR pathway (59). Under prolonged or severe ER stress, phosphorylated eIF2α can selectively induce expression of CHOP, which subsequently activates ER stress-associated apoptosis. In correlation with eIF2α phosphorylation, the expression levels of CHOP were increased in the liver and lung tissues of the mice under PM2.5 exposure (Fig. 3, B and C), suggesting that PM2.5 exposure activated the UPR pathway through PERK/eIF2α, leading to the expression of ER stress-induced pro-apoptotic factor CHOP.
Fig. 3.
PM2.5 exposure activates the unfolded protein response (UPR) pathway through protein kinase-like ER kinase (PERK)/eIF2α and C/EBP homologous protein (CHOP) in vivo and in vitro. A: immunoprecipitation (IP)-Western blot and Western blot analyses for PERK and phosphorylated and total eIF2α proteins in the livers of the mice exposed to FA or PM2.5. Denatured liver protein lysates (80 μg per sample) are separated on a 10% Tris-glycine polyacrylamide gel. For PERK IP-Western blot analysis, an anti-total PERK antibody was used. The values below the gels represent the ratios of phosphorylated eIF2α to total eIF2α protein signals. B: Western blot analyses for phosphorylated and total eIF2α and CHOP proteins in the lung tissue of the mice exposed to FA or PM2.5. Denatured lung protein lysates (150 μg per sample) are separated on a 10% Tris-glycine polyacrylamide gel. Levels of α-tubulin protein were determined as internal controls. The values below the gels represent the ratios of phosphorylated to total eIF2α and normalized CHOP protein signal intensities. C: Western blot analysis for expression of CHOP protein in the livers of the mice exposed to FA or PM2.5. Denatured liver protein lysates (80 μg per sample) are separated on a 10% Tris-glycine polyacrylamide gel. Levels of α-tubulin protein were determined as internal controls. D: Western blot analysis for phosphorylated PERK in RAW264.7 cells transfected with the vector expressing wild-type PERK. The cells were incubated with PM2.5 (50 μg/ml) for various time intervals. An antiphosphorylated PERK antibody was used for the Western blot analysis. E and F: Western blot analyses for levels of ATF4, CHOP, ATF3, and GADD34 in RAW264.7 cells incubated with PM2.5 (50 μg/ml) for various time intervals. G: Western blot analysis for levels of phosphorylated PERK, phosphorylated eIF2α, and CHOP in the RAW264.7 cells expressing wild-type PERK or PERK kinase dominant negative. RAW264.7 cells were transduced with the vector expressing wild-type PERK (PERK-WT) or PERK kinase dominant negative K618A (PERK KD), and then challenged with PM2.5 (50 μg/ml) or vehicle PBS buffer for 6 h. Levels of phosphorylated PERK were determined by using an anti-phosphorylated PERK antibody. For A–G, the experiments were repeated at least three times, and the representative data are shown. The values below the gels represent the normalized protein signal intensities.
It has been known that three other mammalian kinases, including the double-stranded RNA (dsRNA)-dependent protein kinase (PKR), the heme-regulated eIF2α kinase (HRI), and eukaryotic translation initiation factor 2α kinase 4 (GCN2), can phosphorylate eIF2α, leading to translational attenuation (22). To clarify possible activation of the other eIF2α kinases by PM2.5, we first examined whether PM2.5 can induce phosphorylation of PKR in a mouse macrophage cell line RAW264.7. PKR is known to be induced in macrophages and required for pathogen-induced macrophage apoptosis (20). As a positive control, we treated RAW264.7 cells with lipopolysaccharide (LPS) to induce phosphorylation of PKR (see online Supplemental Fig. 1 at the AJP-Cell website). In contrast to LPS treatment, PM2.5 failed to induce phosphorylation of PKR in RAW264.7 cells (Supplemental Fig. 1). Additionally, we could not detect phosphorylated PKR in the liver tissue of the mice exposed to PM2.5 for 10 wk. Similarly, we could not detect PM2.5-induced phosphorylation of HRI or GCN2 in the mice or in RAW264.7 cells (data not shown).
Induction of CHOP through PERK-mediated UPR contributes to PM2.5-induced apoptosis.
To further elucidate the effect of PM2.5 on the activation of PERK-mediated UPR pathway, we expressed wild-type PERK in RAW264.7 cells and then treated the cells with PM2.5 particles for various time intervals. After the treatments, we examined phosphorylation of PERK in transfected cells by using an anti-phosphorylated PERK antibody. Along with PM2.5 treatments, levels of phosphorylated PERK were increased in a time-dependent manner (Fig. 3D). In nontransfected RAW264.7 cells, in vitro PM2.5 exposure increased expression of the downstream targets of PERK-mediated UPR pathway, including activating transcription factor (ATF) 4, CHOP, ATF3, and growth arrest and DNA damage-inducible protein 34 (GADD34) (Fig. 3, E and F). These results further confirmed that PM2.5 can induce PERK-mediated UPR pathway. Furthermore, to evaluate the requirement of PERK for PM2.5-induced phosphorylation of eIF2α and induction of CHOP, we suppressed the eIF2α kinase activity of PERK in RAW264.7 cells by expressing a dominant negative form of PERK, PERK K618A (16). The RAW264.7 cells expressing wild-type PERK or PERK kinase dominant negative were treated with PM2.5 for 6 h. Western blot analysis showed that levels of phosphorylated PERK, phosphorylated eIF2α, and CHOP in the PERK kinase activity-suppressed cells were significantly lower than that in the control cells expressing wild-type PERK after PM2.5 treatments (Fig. 3G). This result confirmed the critical role of PERK in PM2.5-induced eIF2α phosphorylation and CHOP induction.
Next, to determine PM2.5-associated cytotoxicity, we stained DNA fragmentation, a hall marker of apoptosis, to reveal apoptotic cells in the lung tissues of mice exposed to FA or PM2.5. DNA fragmentation activities were significantly increased in the lung tissues of mice exposed to PM2.5 (Fig. 4A). The percentage of apoptotic cells in the lung tissues of the mice exposed to PM2.5 was approximately sevenfold higher than that in the mice exposed to FA (Fig. 4B). Furthermore, to determine whether ER stress-induced pro-apoptotic factor CHOP is critical for PM2.5-induced apoptosis, we exposed CHOP knockout and wild-type control mice to ambient PM2.5 through intranasal inhalation of PM2.5 particles (1.6 μg/g body wt) collected during the time of animal exposure. After the PM2.5 challenge, levels of cleaved caspase-3, the executioner of apoptosis, were significantly increased in the wild-type control mice (Fig. 4C). In contrast, levels of cleaved caspase-3 were only marginally changed in the CHOP knockout mice after PM2.5 challenge, suggesting that CHOP, the target of PERK-mediated UPR pathway, plays a key role in PM2.5-induced apoptosis.
Fig. 4.
PM2.5 exposure causes ER stress-induced apoptosis in the lung and liver tissues of mice exposed to PM2.5 or FA. A: DNA fragmentation assay with the lung tissue sections from the mice exposed to PM2.5 or FA. The mice exposed to PM2.5 or FA were euthanized on the final day of exposure, and the lung tissues were fixed for preparing paraffin-embedded sections. The tissue sections were stained for DNA fragmentation using a fluorescent TUNEL staining kit. Fragmented DNAs were stained for green fluorescence, and the cell cytoplasm was stained for red fluorescence. Magnification: ×200. B: quantification of DNA fragmentation in the lung tissues of mice exposed to PM2.5 or FA. The percentages of apoptotic cells were quantified by counting the cells exhibiting positive DNA fragmentation staining in 8 random fields per sample. Data are shown as means ± SE for 6 animals per group. **P < 0.01. P values are shown for statistically significant differences. C: Western blot analysis for caspase-3 in the lung tissues of CHOP knockout or control mice after intranasal inhalation of PM2.5 or vehicle PBS. CHOP knockout (KO) or wild-type (WT) control mice took up ambient PM2.5 in 25 μl PBS (1.6 μg/g body wt) or 25 μl sterile PBS through intranasal inhalation twice for 5 days. Western blot analysis was performed with the mouse lung tissues to detect both the precursor and cleaved caspase-3 proteins. The values below the gels represent the normalized cleaved caspase-3 protein signal intensities.
PM2.5-induced eIF2α phosphorylation and CHOP induction relies on the production of ROS.
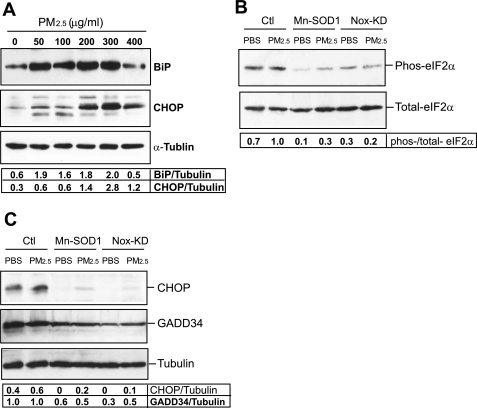
To further elucidate the role and mechanism of PM2.5 in the activation of ER stress response, we incubated RAW264.7 cells with culture media containing PM2.5 particles at different concentrations ranging from 50 to 400 μg/ml. It should be clarified that the effect of in vitro PM2.5 exposure, even at “low” levels, is not comparable to the in vivo effect of PM2.5 exposure through “real-world” PM delivery system. Here, we used the in vitro PM2.5 exposure system to elucidate the cytotoxic effect and biochemical mechanism of PM2.5 in ER stress response. Our result indicated that incubation with 50 μg/ml PM2.5 for 6 h was sufficient to induce the initial phase of ER stress response, as evidenced by the increased expression of the ER chaperone BiP (Fig. 5A). The late phase of ER stress response, represented by the expression of ER stress-induced apoptotic factor CHOP, was gradually induced along with the increase of PM2.5 concentrations and reached a peak level when the cells were treated with 300 μg/ml of PM2.5 (Fig. 5A). Recent evidence suggested that ER stress and oxidative stress are ultimately linked and that ROS may function as a messenger for oxidative stress-induced ER stress (17, 30, 50, 56). To explore the mechanism by which PM2.5 exposure induces ER stress response, we tested the hypothesis that ROS serves as the messengers to mediate PM2.5-induced ER stress response. To test whether ROS is required for PM2.5-induced UPR signaling, we utilized the adenoviral system to express Mn-SOD or dominant negative NADPH oxidase to suppress ROS production in RAW264.6 cells that were exposed to PM2.5. Mitochondria-localized Mn-SOD plays an important role in protecting against oxidative stress (9, 27). It has been demonstrated that disruption of cytosolic and mitochondrial ROS production can be achieved by overexpression of Mn-SOD (25–27). Alternatively, NADPH oxidase is the main enzymatic source for ROS production. Overexpression of dominant negative N17Rac1, the small GTPase component of NADPH oxidase, has been shown to inhibit NADPH oxidase activity, resulting in a lack of ROS generation (27, 39). RAW264.7 cells overexpressing Mn-SOD or dominant negative N17Rac1 were exposed to PM2.5 to test the requirement of ROS in PM2.5-induced ER stress response. PM2.5 exposure increased levels of phosphorylated eIF2α, CHOP, and GADD34 in the control RAW264.7 cells transduced with adenovirus expressing β-galactosidase (Fig. 5, B and C). In comparison, the levels of phosphorylated eIF2α, CHOP, and GADD34 in the RAW264.7 cells expressing Mn-SOD or dominant negative N17Rac1 were much less than that in the control cells after PM2.5 treatment (Fig. 5, B and C). These results indicated that ROS, produced through mitochondrial electron transport and/or NADPH oxidase pathways, is critical for the activation of PM2.5-induced UPR pathway through phosphorylated eIF2α and CHOP.
Fig. 5.
PM2.5-induced eIF2α phosphorylation and CHOP induction depends on the production of reactive oxygen species (ROS). A: Western blot analysis of BiP and CHOP expression levels in RAW264.7 cells in response to in vitro exposure to PM2.5 particles at different concentrations ranging from 50 to 400 μg/ml for 6 h. Levels of α-tubulin protein were determined as internal controls. B and C: Western blot analyses for phosphorylated and total eIF2α, CHOP, and GADD34 in RAW264.7 cells under in vitro exposure to PM2.5 or PBS. RAW264.7 cells were infected by adenovirus expressing Mn-SOD1, dominant-negative N17Rac1, or the control vector for 36 h, followed by the incubation with PM2.5 (300 μg/ml) or PBS for 6 h. For A–C, the experiments were repeated at least three times, and the representative data are shown. The values below the gels represent the normalized protein signal intensities.
PM2.5 exposure activates ER stress sensor IRE1α but suppresses its activity in splicing the Xbp1 mRNA.
In addition to the PERK/eIF2α-mediated UPR pathway, we also examined whether PM2.5 exposure can induce the other UPR pathways mediated through ER stress sensors IRE1α and ATF6, respectively. IRE1α is the most conserved ER stress sensor that transduces the UPR signaling through activating the transcription factor XBP1 (37, 56). In response to ER stress, IRE1α endoribonuclease can specifically splice a 26-bp small intron from the Xbp1 mRNA, resulting in the synthesis of a potent UPR trans-activator (37, 38). IP-Western blot analysis showed increased levels of phosphorylated IRE1α in the liver tissues of mice exposed to PM2.5 when compared with that exposed to FA, suggesting that PM2.5 exposure can induce activation of IRE1α (Fig. 6A). Furthermore, we examined Xbp1 mRNA splicing, a reliable marker for detecting IRE1α RNase activity, in the liver tissues. Because the liver is an organ that plays major roles in metabolism and detoxification, “constitutive” or physiological activation of IRE1α/XBP1-mediated UPR in the normal liver has been proposed (1, 24, 56). Indeed, through quantitative real-time RT-PCR analysis, we were able to detect basal induction of spliced Xbp1 mRNA in mouse livers under FA (Fig. 6B). Surprisingly, despite the activation of IRE1α, the levels of spliced Xbp1 mRNA were modestly reduced in the liver tissue of the mice exposed to PM2.5 (Fig. 6B). We also examined expression of spliced XBP1 protein in the liver tissue of mice exposed to PM2.5 or FA. Western blot analysis showed that spliced XBP1 protein levels in the liver of mice exposed to PM2.5 were slightly decreased compared with that in the mice exposed to FA (Fig. 6C). Consistent with these observations, expression of the XBP1-dependent UPR target genes, including ERdj4, enhancing α-mannosidase-like protein 1 (Edem1), and protein kinase inhibitor of 58 kDa (P58 ipk), was not increased upon PM2.5 exposure (Fig. 6B). In comparison, expression of the UPR target genes under the PERK/eIF2α pathway, including Atf4 and CHOP, was increased in the livers of the mice exposed to PM2.5 compared with that in the mice exposed to FA (Fig. 6B).
Fig. 6.
PM2.5 exposure can activate ER stress sensors IRE1α and ATF6. A: IP-Western blot analysis of IRE1α in the livers of mice exposed to FA or PM2.5. The values below the gels represent the ratios of phosphorylated IRE1α to total IRE1α. B: quantitative real-time RT-PCR analysis for expression levels of the spliced Xbp1 (xbp1s), ERdj4, Edem1, p58(ipk), Bip, Grp94, Pdi-P5, CHOP, and Atf4 mRNAs in the livers of mice exposed to FA or PM2.5. Fold changes of mRNA levels were determined after normalization to internal control β-actin mRNA levels. For each comparison group, the mRNA level of one FA-exposed mouse was defined as 1 and was used to calculate the fold changes of mRNA levels for the other mice. Each bar denotes the mean ± SE (n = 6 mice/group). *P < 0.05. P values are shown for statistically significant differences. C: Western blot analysis of spliced XBP1 protein in the livers of mice exposed to FA or PM2.5. D: quantitative real-time RT-PCR analysis for expression levels of the Pmp22, Col6, HgNat, Blos1, Scara3, and PdgfR mRNAs in the livers of mice exposed to FA or PM2.5. Fold changes of mRNA levels were determined after normalization to internal control β-actin mRNA levels. For each comparison group, the mRNA level of one FA-exposed mouse was defined as 1 and was used to calculate the fold changes of mRNA levels for the other mice. Each bar denotes the mean ± SE (n = 6 mice/group). *P < 0.05. E: Western blot analysis for ATF6 cleavage in RAW264.7 cells expressing full-length ATF6 after PM2.5 challenge. RAW264.7 cells were transfected with the vector expressing HA-tagged full-length ATF6. The transfected cells were treated with PM2.5 (50 μg/ml) for various time intervals. ATF6 P90 represents the full-length ATF6, and ATF6 P50 represents the cleaved form of ATF6. The experiment was repeated three times, and the representative data are shown.
In addition to splicing the Xbp1 mRNA, IRE1α is known to activate JNK-mediated pathway (51) and the RIDD (18, 19) under ER stress conditions. In the liver tissue of mice exposed to PM2.5, levels of phosphorylated JNK were significantly increased compared with that in the mice exposed to FA (Supplemental Fig. 2). Furthermore, quantitative real-time RT-PCR analysis demonstrated that levels of the RIDD targets, including the Pmp22, Col6, HgNat, Blos1, Scara3, and PdgfR mRNAs (18), were all decreased in the livers of mice exposed to PM2.5 compared with that in the mice exposed to FA (Fig. 6D), indicating the activation of RIDD pathway upon PM2.5 exposure. Together, our results suggested that PM2.5 exposure can activate IRE1α-mediated pathways, including the JNK pathway and RIDD, although it reduces the activity of IRE1α in splicing the Xbp1 mRNA. To determine whether PM2.5 exposure can activate ATF6-mediated UPR pathway, we examined expression of ATF6-specific UPR target genes, including Bip, Grp94, and Pdi-P5 (23, 53), in the liver tissues of mice exposed to PM2.5 or FA. The levels of the Bip, GRP94, and Pdi-P5 mRNAs were increased in the livers of the mice exposed to PM2.5 compared with that in the mice exposed to FA (Fig. 6B), implicating the activation of ATF6-mediated UPR pathway upon PM2.5 exposure. Because of the difficulty in detecting activation/cleavage of endogenous ATF6 by using currently available antibodies, we transduced RAW264.7 cells to express a HA-tagged full-length ATF6 protein for detecting PM2.5-induced ATF6 activation/cleavage. In response to PM2.5, levels of cleaved ATF6 in the RAW264.7 cells expressing full-length ATF6 were increased (Fig. 6E), thus confirming the activation of ATF6 upon PM2.5 exposure.
To further assess the effect of PM2.5 exposure on Xbp1 mRNA splicing, RAW264.7 cells were exposed to PM2.5 particles for up to 6 h. The levels of spliced and nonspliced forms of Xbp1 mRNAs were determined by semiquantitative RT-PCR. Incubation of the cells with PM2.5 failed to induce splicing of the Xbp1 mRNA (Fig. 7A). Moreover, we exposed the cells to PM2.5 particles for 30 min before the cells were treated with tunicamycin (TM), a reagent that can strongly induce ER stress by disrupting N-linked protein glycosylation. Under the PM2.5 exposure, the cells expressed reduced levels of spliced Xbp1 mRNA in response to TM treatment (Fig. 7A). To verify the specificity of PM2.5 exposure in inhibiting Xbp1 mRNA splicing, we examined the phosphorylation of eIF2α in response to PM2.5 exposure and/or TM treatment. The levels of phosphorylated eIF2α were increased in RAW264.7 cells upon the exposure to either PM2.5 alone or PM2.5 plus TM (Supplemental Fig. 3), suggesting that PM2.5 suppressed Xbp1 mRNA splicing while it induced eIF2α phosphorylation. In addition to macrophages, we also tested the effect of PM2.5 exposure on the suppression of Xbp1 mRNA splicing in cultured mouse hepatocyte cell line H2.35. Under PM2.5 exposure, the splicing of the Xbp1 mRNA was not detectable (Fig. 7B). In contrast, the spliced Xbp1 mRNA was readily detected in response to TM treatment. Furthermore, we examined TM-induced Xbp1 mRNA splicing in the cells that were pretreated with PM2.5 particles. Supporting the suppressive effect of PM2.5 on the Xbp1 mRNA splicing, mouse hepatocytes that were exposed to PM2.5 expressed diminished levels of spliced Xbp1 mRNA under the treatment of TM (Fig. 7B). Taken together, PM2.5 exposure appears to suppress the activity of IRE1α in splicing the Xbp1 mRNA. Since functional XBP1 plays essential roles in normal differentiation and function of specialized cell types and in remodeling cells to adapt to cellular stress (37, 56), PM2.5 may be involved in disease pathogenesis through inhibiting Xbp1 mRNA splicing. The mechanism by which PM2.5 suppresses splicing of Xbp1 mRNA is an intriguing phenomenon to be elucidated.
Fig. 7.
PM2.5 exposure suppresses the activity of IRE1α in splicing the Xbp1 mRNA. A: detection of unspliced and spliced forms of Xbp1 mRNAs in macrophage cell line RAW264.7 exposed to PM2.5 and/or TM by semiquantitative RT-PCR analysis. RAW264.7 cells were exposed to PM2.5 (300 μg/ml) for 30 min and then treated with tunicamycin (TM, 2 μg/ml) for the time course as indicated. RAW264.7 cells treated with vehicle PBS buffer or with TM for 6 h were included as controls. The size of amplified unspliced Xbp1 mRNA is 170 bp, and the size of amplified spliced Xbp1 mRNA is 144 bp. Levels of β-actin mRNA were determined as internal controls. B: detection of unspliced and spliced forms of Xbp1 mRNAs in mouse hepatocyte cell line H2.35 exposed to PM2.5, TM, or PM2.5 plus TM by semiquantitative RT-PCR analysis. H2.35 cells were exposed to PM2.5 (300 μg/ml) or TM (2 μg/ml) for 2 to 6 h. Additionally, H2.35 cells were exposed to PM2.5 (300 μg/ml) for 30 min and then treated with TM (2 μg/ml) for 2 to 6 h. The cells treated with vehicle PBS buffer were included as the control. Levels of β-actin mRNA were determined as internal controls. For A and B, the experiments were performed at least in triplicate, and the representative data are shown. C: a schematic diagram depicting ER stress response induced by PM2.5 in mouse lung and liver tissues.
DISCUSSION
In this study, we characterized intracellular stress signaling induced by ambient PM2.5 exposure in vivo and in vitro (Fig. 7C). It has been demonstrated that inhaled air pollution particles in the fine or ultrafine range, such as PM2.5, can transgress into the systemic circulation and are linked strongly with the pathogenesis of metabolic and cardiovascular disease (7, 32, 34, 46, 47). Additionally, liver is the major organ responsible for the detoxification of chemical compounds and lipid metabolism. Accordingly, we propose that the lung and liver are two primary organs that airborne PM2.5 targets and where PM2.5 elicits its cytotoxic effects. Our study here implicated that subchronic exposure to ambient PM2.5 induces ER stress and selective activation of the UPR signaling pathways, triggering ER stress-induced apoptosis through the PERK-eIF2α-CHOP UPR branch in the lung and liver tissues. The UPR signaling induced by PM2.5 exposure relies on reactive oxygen intermediates. These results provide novel insights into the molecular and cellular basis by which PM2.5 exposure alters cell physiology and elicits its cytotoxic effects.
Our study demonstrated that the physiological UPR induced by PM2.5 exposure is distinct from those that are induced by pharmacological induction. Under the treatments of ER stress-eliciting chemicals, such as TM, thapsigargin, and dithiothreitol, all the UPR pathways are activated (38). However, PM2.5 exposure differentially activates the UPR pathways. While activation of PERK-mediated phosphorylation of eIF2α and induction of CHOP is a predominant feature of PM2.5-triggered UPR signaling (Fig. 3), PM2.5 exposure negatively affects the activity of IRE1α in splicing the Xbp1 mRNA, resulting in a slight decrease in the level of spliced XBP1 protein in the liver tissue of the mice exposed to PM2.5 (Fig. 6, B and C). The differential regulation of the UPR pathways by PM2.5 was reflected by expression profiles of the UPR target genes. Expression of PERK-mediated UPR targets, including phosphorylated eIF2α, ATF4, ATF3, CHOP, and GADD34, was increased in response to PM2.5 exposure. In comparison, expression of IRE1α/XBP1-mediated UPR target genes, including ERdj4, Edem1, and P58 (ipk), was not upregulated by PM2.5 exposure (Fig. 6B). The suppression effect of PM2.5 on IRE1α-mediated splicing of the Xbp1 mRNA was further confirmed by the observation that PM2.5 treatment significantly reduced TM-induced Xbp1 mRNA splicing (Fig. 7, A and B). However, IRE1α was activated upon PM2.5 exposure, as evidenced by increased levels of phosphorylated IRE1α protein in the liver tissue of PM2.5-exposed mice (Fig. 6A). The other IRE1α-mediated pathways, including JNK pathway and RIDD, were also activated in the liver of mice exposed to PM2.5 (Fig. 6D and Supplemental Fig. 2). It is possible that IRE1α is activated, but its RNase activity in splicing the Xbp1 mRNA was specifically suppressed by one or multiple PM2.5 components. Interestingly, a recent paper has reported that the components from cigarette smoke can cause similar effect on inhibiting Xbp1 mRNA splicing (21). Because spliced Xbp1 mRNA encodes a functional UPR trans-activator that plays important protective roles in response to ER stress, the impairment of the UPR pathway through the Xbp1 mRNA splicing may have considerable detrimental effects on the pathogenesis of the diseases induced by PM2.5 exposure. Indeed, our study implicated PM2.5 caused cytotoxicity, as evidenced by increased caspase-3 cleavage and DNA fragmentation in the mice exposed to PM2.5 (Fig. 4). It is known that the UPR can provide the stressed cells with survival or death signals depending on the type and duration of ER stress (58). It is likely that PM2.5-induced UPR leads to ER stress-associated apoptosis by promoting the PERK-eIF2α-CHOP UPR branch while suppressing the IRE1α-XBP1 UPR branch.
PM2.5 exposure can also activate ER stress sensor ATF6, as evidenced by the upregulation of ATF6-dependent UPR target genes in the mouse liver exposed to PM2.5 (Fig. 6B) and increased ATF6 cleavage in PM2.5-treated RAW264.7 cells (Fig. 6E). This may explain why expansion of ER compartment can be observed in Kupffer cells of the mice exposed to PM2.5 where splicing of the Xbp1 mRNA was suppressed (Fig. 1B). It has been documented that enforced expression of spliced XBP1 can lead to a proliferation of ER membrane by increasing the synthesis of phosphatidylcholine (44). However, recent evidence suggested that XBP1 and ATF6 play redundant roles in stress-induced ER expansion. ATF6 can induce XBP1-independent ER expansion by increasing phosphatidylcholine synthesis (5). Therefore, PM2.5-triggered activation of ATF6, but not XBP1, may contribute to ER expansion observed in the mouse liver tissue exposed to PM2.5.
Our study also suggested that the PM2.5-triggered stress signaling pathway, represented by eIF2α phosphorylation and CHOP induction, depends on the production of ROS (Fig. 5). It has been hypothesized that increased production of intracellular ROS can target on ER calcium channels and chaperones, leading to Ca2+ release to the cytosol. Ca2+ release from the ER causes ER stress and UPR activation. On the other hand, increased cytosolic Ca2+ stimulates mitochondrial metabolism and subsequent production of more ROS, which can further amplify the UPR (56). Based on our study, the activation of the UPR signaling by PM2.5 exposure represents a pathophysiological system supporting this hypothesis. Given detrimental effects of the prolonged UPR, our results suggest that there might be some potential in using nontoxic antioxidants to alleviate the toxic effects from air particulate pollution.
The PM exposure system “OASIS-1” used in this study is a versatile aerosol concentration enrichment system that can concentrate ambient fine and ultrafine PM particles at nominal 10-folds (11, 41). After taking into account the nonexposed time, the equivalent PM2.5 concentration to which the mice were exposed over the 10-wk exposure period was 11.6 μg/m3. The average PM2.5 concentration over the subchronic exposure period is within the annual average PM2.5 National Ambient Air Quality Standard (NAAQS) of 15.0 μg/m3 (13). It has been demonstrated that the PM2.5 size and composition distribution does not change before (ambient) and after the concentration (11, 29, 45). Therefore, the composition and size of PM2.5 particles collected in the exposure chamber air may reflect that of nonconcentrated PM2.5 present in the ambient air. Indeed, the “OASIS-1” system is one of several limited prototype systems in the United States that allow us to perform the studies on animal models that recapitulate true personal, long-term exposure to environmentally relevant PM2.5 (11, 29, 47). The State of Ohio, where the animals were exposed, is one of the “perfect” states to study the effects of PM2.5 on human health, since “Ohio has serious and widespread air pollution problem, and had failed to meet the national ozone and particulate matter annual standards” (4). Therefore, our approach represents a direction to elucidating physiological cell stress signaling that is relevant to a real public health problem. Although much remains to be done, the information provided by our study is not only important to understand the molecular basis of air pollution-associated pathogenesis but will also be informative to the prevention and potentially treatment of air pollution-associated diseases.
GRANTS
This work was partly supported by new faculty research funding from the Wayne State University and the American Heart Association Grants 0635423Z and 09GRNT2280479 (to K. Zhang) and K01ES016588, R21ES017412, and R56ES018900 from NIH (to Q. Sun), and R01NS4378 from NIH (to A. Gow). The whole body exposure was performed in facilities at the Ohio State University that was supported by NIH Grants R01ES013406 and R01ES015146 (to S. Rajagopalan) and R01NS43783 (to A. Gow). K. Zhang is a recipient of American Heart Association Scientist Development Award.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Morton Lippmann for his critical reading and edits of this manuscript. We also acknowledge the Campus Microscopy and Imaging Facility at The Ohio State University for their support of Transmission electron microscopy.
REFERENCES
- 1.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27: 53–66, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Alfaro-Moreno E, Martinez L, Garcia-Cuellar C, Bonner JC, Murray JC, Rosas I, Rosales SP, Osornio-Vargas AR. Biologic effects induced in vitro by PM10 from three different zones of Mexico City. Environ Health Perspect 110: 715–720, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arantes-Costa FM, Lopes FD, Toledo AC, Magliarelli-Filho PA, Moriya HT, Carvalho-Oliveira R, Mauad T, Saldiva PH, Martins MA. Effects of residual oil fly ash (ROFA) in mice with chronic allergic pulmonary inflammation. Toxicol Pathol 36: 680–686, 2008. [DOI] [PubMed] [Google Scholar]
- 4.American Lung Association State of the Air 2009 http://www.lungusa2.org/sota/2009/SOTA-2009-Full-Print.pdf); 2008Highlights of Recent Research on Particulate Air Pollution: Effects of Long-Term Exposure (http://www.lungusa.org/site/c.dvLUK9O0E/b.36864/k.9F49/State_of_the_Air.htm).
- 5.Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, Sriburi R, Frank M, Jackowski S, Kaufman RJ, Brewer JW. ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci 122: 1626–1636, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brook RD, Franklin B, Cascio W, Hong Y, Howard G, Lipsett M, Luepker R, Mittleman M, Samet J, Smith SC, Jr, Tager I. Air pollution and cardiovascular disease: a statement for healthcare professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation 109: 2655–2671, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Brook RD, Jerrett M, Brook JR, Bard RL, Finkelstein MM. The relationship between diabetes mellitus and traffic-related air pollution. J Occup Environ Med 50: 32–38, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Brunekreef B, Holgate ST. Air pollution and health. Lancet 360: 1233–1242, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Chen AF, O'Brien T, Tsutsui M, Kinoshita H, Pompili VJ, Crotty TB, Spector DJ, Katusic ZS. Expression and function of recombinant endothelial nitric oxide synthase gene in canine basilar artery. Circ Res 80: 327–335, 1997 [DOI] [PubMed] [Google Scholar]
- 10.Chen JC, Schwartz J. Metabolic syndrome and inflammatory responses to long-term particulate air pollutants. Environ Health Perspect 116: 612–617, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen LC, Nadziejko C. Effects of subchronic exposures to concentrated ambient particles (CAPs) in mice. V CAPs exacerbate aortic plaque development in hyperlipidemic mice. Inhal Toxicol 17: 217–224, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Dubowsky SD, Suh H, Schwartz J, Coull BA, Gold DR. Diabetes, obesity, and hypertension may enhance associations between air pollution and markers of systemic inflammation. Environ Health Perspect 114: 992–998, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.American Lung Association Air Quality. http://www.lungusa.org/site/c.dvLUK9O0E/b.33691/k.FE38/Air_Quality.htm.).
- 14.Gomez-Mejiba SE, Zhai Z, Akram H, Pye QN, Hensley K, Kurien BT, Scofield RH, Ramirez DC. Inhalation of environmental stressors & chronic inflammation: autoimmunity and neurodegeneration. Mutat Res 674: 62–72, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5: 897–904, 2000 [DOI] [PubMed] [Google Scholar]
- 16.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Hsu LC, Park JM, Zhang K, Luo JL, Maeda S, Kaufman RJ, Eckmann L, Guiney DG, Karin M. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature 428: 341–345, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen E, Stinson A, Shan L, Yang J, Gietl D, Albino AP. Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC Cancer 8: 229, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman RJ. Regulation of mRNA translation by protein folding in the endoplasmic reticulum. Trends Biochem Sci 29: 152–158, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320: 1492–1496, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Crockett E, Wang DH, Galligan JJ, Fink GD, Chen AF. Gene transfer of endothelial NO synthase and manganese superoxide dismutase on arterial vascular cell adhesion molecule-1 expression and superoxide production in deoxycorticosterone acetate-salt hypertension. Arterioscler Thromb Vasc Biol 22: 249–255, 2002 [DOI] [PubMed] [Google Scholar]
- 26.Li L, Watts SW, Banes AK, Galligan JJ, Fink GD, Chen AF. NADPH oxidase-derived superoxide augments endothelin-1-induced venoconstriction in mineralocorticoid hypertension. Hypertension 42: 316–321, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Luo JD, Wang YY, Fu WL, Wu J, Chen AF. Gene therapy of endothelial nitric oxide synthase and manganese superoxide dismutase restores delayed wound healing in type 1 diabetic mice. Circulation 110: 2484–2493, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Maciejczyk P, Chen LC. Effects of subchronic exposures to concentrated ambient particles (CAPs) in mice. VIII Source-related daily variations in in vitro responses to CAPs. Inhal Toxicol 17: 243–253, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Maciejczyk P, Zhong M, Li Q, Xiong J, Nadziejko C, Chen LC. Effects of subchronic exposures to concentrated ambient particles (CAPs) in mice. II The design of a CAPs exposure system for biometric telemetry monitoring. Inhal Toxicol 17: 189–197, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci USA 105: 18525–18530, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller FJ, Jr, Gutterman DD, Rios CD, Heistad DD, Davidson BL. Superoxide production in vascular smooth muscle contributes to oxidative stress and impaired relaxation in atherosclerosis. Circ Res 82: 1298–1305, 1998 [DOI] [PubMed] [Google Scholar]
- 32.Nemmar A, Hoet PH, Vanquickenborne B, Dinsdale D, Thomeer M, Hoylaerts MF, Vanbilloen H, Mortelmans L, Nemery B. Passage of inhaled particles into the blood circulation in humans. Circulation 105: 411–414, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Nemmar A, Hoylaerts MF, Hoet PH, Dinsdale D, Smith T, Xu H, Vermylen J, Nemery B. Ultrafine particles affect experimental thrombosis in an in vivo hamster model. Am J Respir Crit Care Med 166: 998–1004, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Nurkiewicz TR, Porter DW, Barger M, Millecchia L, Rao KM, Marvar PJ, Hubbs AF, Castranova V, Boegehold MA. Systemic microvascular dysfunction and inflammation after pulmonary particulate matter exposure. Environ Health Perspect 114: 412–419, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neill MS, Veves A, Sarnat JA, Zanobetti A, Gold DR, Economides PA, Horton ES, Schwartz J. Air pollution and inflammation in type 2 diabetes: a mechanism for susceptibility. Occup Environ Med 64: 373–379, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phelps RG, Bernstein LE, Harpaz N, Gordon RE, Cruickshank FA, Schwartz E. Characterization of a dermal derived malignant mesenchymal tumor arising in ultraviolet irradiated mice. Am J Pathol 135: 149–159, 1989 [PMC free article] [PubMed] [Google Scholar]
- 37.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Seshiah PN, Weber DS, Rocic P, Valppu L, Taniyama Y, Griendling KK. Angiotensin II stimulation of NAD(P)H oxidase activity: upstream mediators. Circ Res 91: 406–413, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Sicari MC, Lebwohl M, Baral J, Wexler P, Gordon RE, Phelps RG. Photoinduced dermal pigmentation in patients taking tricyclic antidepressants: histology, electron microscopy, and energy dispersive spectroscopy. J Am Acad Dermatol 40: 290–293, 1999 [DOI] [PubMed] [Google Scholar]
- 41.Sioutas C, Koutrakis P, Burton RM. A technique to expose animals to concentrated fine ambient aerosols. Environ Health Perspect 103: 172–177, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soukup JM, Becker S. Human alveolar macrophage responses to air pollution particulates are associated with insoluble components of coarse material, including particulate endotoxin. Toxicol Appl Pharmacol 171: 20–26, 2001 [DOI] [PubMed] [Google Scholar]
- 43.Southwood CM, Garbern J, Jiang W, Gow A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron 36: 585–596, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167: 35–41, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y, Sipin MF, Spencer MT, Qin X, Moffet RC, Shields LG, Prather KA, Venkatachari P, Jeong C, Kim E, Hopke PK, Gelein RM, Utell MJ, Oberdorster G, Berntsen J, Devlin RB, Chen LC. Real-time characterization of the composition of individual particles emitted from ultrafine particle concentrators. Aerosol Sci Tech 40: 19, 2006 [Google Scholar]
- 46.Sun Q, Wang A, Jin X, Natanzon A, Duquaine D, Brook RD, Aguinaldo JG, Fayad ZA, Fuster V, Lippmann M, Chen LC, Rajagopalan S. Long-term air pollution exposure and acceleration of atherosclerosis and vascular inflammation in an animal model. JAMA 294: 3003–3010, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Sun Q, Yue P, Deiuliis JA, Lumeng CN, Kampfrath T, Mikolaj MB, Cai Y, Ostrowski MC, Lu B, Parthasarathy S, Brook RD, Moffatt-Bruce SD, Chen LC, Rajagopalan S. Ambient air pollution exaggerates adipose inflammation and insulin resistance in a mouse model of diet-induced obesity. Circulation 119: 538–546, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Q, Yue P, Kirk RI, Wang A, Moatti D, Jin X, Lu B, Schecter AD, Lippmann M, Gordon T, Chen LC, Rajagopalan S. Ambient air particulate matter exposure and tissue factor expression in atherosclerosis. Inhal Toxicol 20: 127–137, 2008 [DOI] [PubMed] [Google Scholar]
- 49.Sun Q, Yue P, Ying Z, Cardounel AJ, Brook RD, Devlin R, Hwang JS, Zweier JL, Chen LC, Rajagopalan S. Air pollution exposure potentiates hypertension through reactive oxygen species-mediated activation of Rho/ROCK. Arterioscler Thromb Vasc Biol 28: 1760–1766, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666, 2000 [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem 275: 27013–27020, 2000 [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 13: 365–376, 2007. [DOI] [PubMed] [Google Scholar]
- 54.Yang W, Omaye ST. Air pollutants, oxidative stress and human health. Mutat Res 674: 45–54, 2009 [DOI] [PubMed] [Google Scholar]
- 55.Ying Z, Yue P, Xu X, Zhong M, Sun Q, Mikolaj M, Wang A, Brook RD, Chen LC, Rajagopalan S. Air pollution and cardiac remodeling: a role for RhoA/Rho-kinase. Am J Physiol Heart Circ Physiol 296: H1540–H1550, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 454: 455–462, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K, Kaufman RJ. Identification and characterization of endoplasmic reticulum stress-induced apoptosis in vivo. Methods Enzymol 442: 395–419, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem 279: 25935–25938, 2004 [DOI] [PubMed] [Google Scholar]
- 59.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev 12: 982–995, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.