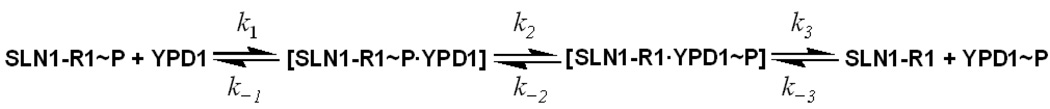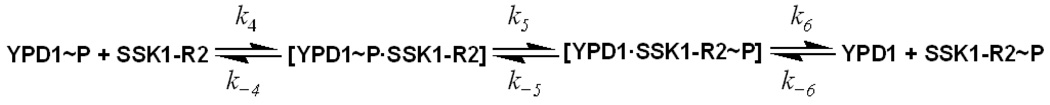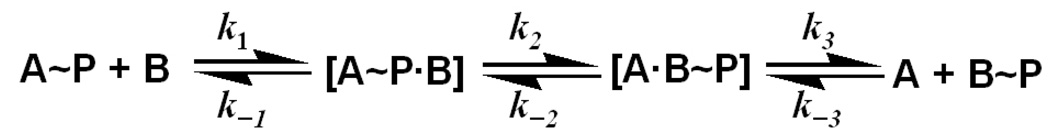Abstract
For both prokaryotic and eukaryotic His-Asp phosphorelay signaling pathways, the rates of protein phosphorylation and dephosphorylation determine the stimulus-to-response time frame. Thus, kinetic studies of phosphoryl group transfer between signaling partners are important for gaining a full understanding of how the system is regulated. In many cases, the phosphotransfer reactions are too fast for rates to be determined by manual experimentation. Rapid quench flow techniques thus provide a powerful method for studying rapid reactions that occur in the millisecond time frame. In this chapter, we describe experimental design and procedures for kinetic characterization of the yeast SLN1-YPD1-SSK1 osmoregulatory phosphorelay system using a rapid quench flow kinetic instrument.
Introduction
Kinetic studies have contributed to our understanding of many two-component signaling systems. For example, the rate of histidine kinase autophosphorylation and subsequent phosphotransfer to the downstream response regulator protein determines how quickly the cell can respond to changes in the environment. Likewise, the rate of hydrolysis of the aspartyl phosphate on the response regulator (due to its intrinsic stability or phosphatase-catalyzed rate) will determine the duration of the cellular response and the return to a pre-stimulus state.
Two-component regulatory systems and the expanded multi-step His-Asp phosphorelay systems are essential for adaptation to a variety of environmental stresses in bacteria and, to a more limited extent, in eukaryotic organisms such as fungi and plants. The number of proteins participating in these His-Asp phosphorelay systems can vary from a minimum of two components, a histidine kinase (HK) and response regulator (RR), to three or more signaling molecules that comprise multi-step phosphorelay systems (West and Stock, 2001, Stock et al., 2000, Parkinson and Kofoid, 1992, Appleby et al., 1996). These systems are regulated by sequential phosphoryl group transfer and hydrolysis reactions as a means of information transfer.
In cases where the phosphotransfer reaction is too rapid to capture manually, a rapid quench flow (RQF11) instrument can be used for monitoring phosphotransfer reactions that occur in the millisecond time frame, measuring the rate constants and other kinetic parameters. This approach has been used to study two-component phosphorelay systems in bacteria (Stewart, 1997, Fisher et al., 1996, Grimshaw et al., 1998) and has been more recently applied to the study of the multi-step phosphorelay system from Saccharomyces cerevisiae (Janiak-Spens et al., 2005, Kaserer et al., 2009). In S. cerevisiae, a branched multi-step phosphorelay system is responsible for adaptation to hyperosmotic, oxidative and other environmental stresses (Hohmann et al., 2007, Posas et al., 1996, Saito and Tatebayashi, 2004). The SLN1-YPD1-SSK1 branch controls the downstream HOG1 MAP kinase cascade that allows cells to adapt to hyperosmotic stress. Under non-osmotic stress conditions, the transmembrane hybrid SLN1 kinase is active and transfers phosphoryl groups from its central kinase domain to its C-terminal receiver domain (referred to as the SLN1-R1 domain). Subsequently, phosphoryl groups are transferred to YPD1, a cytoplasmic histidine-containing phosphotransfer (HPt) protein and then to the response regulator domain on SSK1 (referred to as the SSK1-R2 domain), thereby maintaining SSK1 in a constitutively phosphorylated state. Hyperosmotic stress leads to dephosphorylation of SSK1 and activation of the downstream HOG1 MAP kinase cascade resulting in an increase in intracellular glycerol, a compatible osmolyte that restores homeostasis (Posas and Saito, 1998, Horie et al., 2008). The SLN1-YPD1-SKN7 branch responds primarily to cell wall perturbations (Shankarnarayan et al., 2008, Lu et al., 2003, Li et al., 2002). The SKN7 response regulator is a nuclear localized transcription factor, thus its function is to modulate gene expression in response to environmental conditions (Brown et al., 1993, Brown et al., 1994, Krems et al., 1996).
Previous studies of the multi-step phosphorelay system from S. cerevisiae from our laboratory have centered on structural and functional characterization of the YPD1 HPt protein. Specifically, important information regarding YPD1-RR recognition and binding (Porter and West, 2005, Porter et al., 2003) and X-ray structures of YPD1/SLN1-R1 complexes (Xu et al., 2003, Zhao et al., 2008) have been obtained. In vitro data had shown that YPD1 can form a complex with the phosphorylated SSK1-R2 domain and it was suggested that YPD1 shields the phosphoryl group on SSK1 preventing it from hydrolysis (Janiak-Spens et al., 2000). Further studies focused on the interaction of YPD1 with the response regulator domains associated with SLN1, SSK1, and SKN7 (R1, R2, and R3, respectively) with respect to phosphotransfer, protein binding affinity, specificity of interaction, and characterization of YPD1 mutants (Janiak-Spens et al., 1999, Janiak-Spens et al., 2000, Janiak-Spens and West, 2000, Janiak-Spens et al., 2005, Porter and West, 2005).
The phosphotransfer reactions that comprise the SLN1-YPD1-SSK1 phosphorelay reach steady-state levels within eight seconds (Janiak-Spens and West, 2000), thus necessitating utilization of rapid quench kinetics as a means of studying phosphotransfer rates. The individual phosphoryl transfer reactions between YPD1 and the response regulator domains have been examined kinetically and the individual phosphotransfer rates and dissociation constants were determined and analyzed (Janiak-Spens et al., 2005). The data demonstrated that phosphotransfer from YPD1 to SSK1 was strongly favored over phosphotransfer to SKN7, and phosphotransfer from YPD1 to SSK1 was irreversible. These findings were consistent with the concept that SSK1 is constitutively phosphorylated under normal osmotic conditions.
Moreover, these data led to the hypothesis that upon hyperosmotic stress, when water rapidly effluxes the cell, the increasing ion/solute concentrations inside the cell might disrupt the YPD1·SSK1~P complex. Therefore the effect of osmolyte concentrations on the half-life of phosphorylated SSK1-R2 in the presence and absence of YPD1 and the kinetics of the individual phosphorelay reactions was examined (Kaserer et al., 2009). Our findings suggest that as intracellular osmolyte concentrations increase, the YPD1•SSK1~P complex dissociates thereby facilitating dephosphorylation of SSK1 and activating the HOG1 MAP kinase cascade. Later, when glycerol and other ions reach their highest concentration in the cell, attenuation of the pathway is achieved, in part, because the kinetics of the phosphorelay favor production of SSK1~P and inhibition of the HOG1 pathway.
In this article, we provide a description of the application of rapid quench flow analysis in order to measure kinetic parameters of the SLN1-YPD1-SSK1 osmoregulatory phosphorelay system from S. cerevisiae.
Materials and Methods
A. Protein purification and phosphorylation
Expression and purification of the S. cerevisiae SLN1-HK, SLN1-R1, YPD1, SSK1-R2, and SKN7-R3 proteins has been described in detail in another chapter in this volume (Fassler and West). Here, we will summarize preparation of the radiolabeled phosphorylated protein donors for the purpose of rapid quench kinetic experiments. The phosphorylated SLN1-R1 domain acts as the phosphoryl donor in the first half reaction between SLN1-R1~P and YPD1. GST-linked SLN1-HK (7 µM) bound to glutathione-Sepharose 4B resin is incubated with 7 µM [γ-32P]-ATP for 30 min. Unincorporated [γ-32P]-ATP is then washed from SLN1-HK~P with 50 mM Tris-HCl, pH 8.0, 100 mM KCl, 15 mM MgCl2, 2 mM DTT, and 20% glycerol using 3 consecutive centrifugations (1 min at 1000 × g). The SLN1-R1 protein (18.6 µM) is then added in the same buffer and incubated for 10 min at room temperature in a total volume of 300 µL. Phospho-SLN1-R1 is recovered in the supernatant after gently pelleting the GST-SLN1-HK bound to the resin. EDTA is added to the supernatant to a final concentration of 30 mM to prevent dephosphorylation. Phosphorylated SLN1-R1 is diluted to 0.45 µM in S2 buffer containing 50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mM DTT.
For the second half-reaction, phosphotransfer from YPD1~P to SSK1-R2, a similar protocol can be followed with the following modifications. Incubation of GST-tagged SLN1-HK-R1 (7 µM) and [γ-32P] ATP (7 µM) is for 60 min. The YPD1 protein (18.6 µM) is then added to the reaction mixture.
B. The RQF instrument
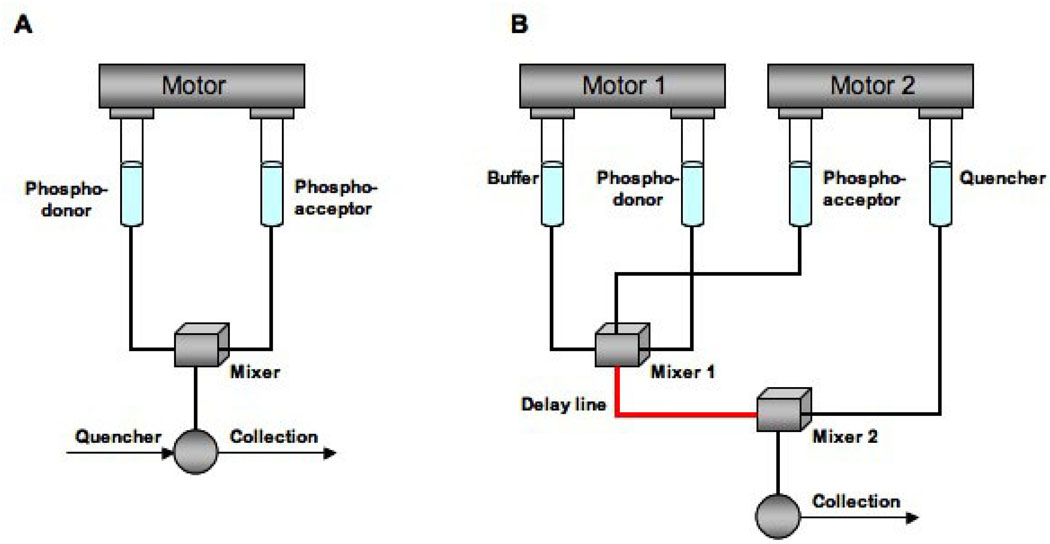
There are several commercially available rapid quench instruments2 and they all consist of a control unit, a drive mechanism, and a mixing chamber. In order to cover a broader time scale of enzymatic reactions, different quench flow devices can be used; however, the principles remain the same. For example, a rapid quench flow device (Fig. 1A) is applicable for times in the millisecond range from 5 msec to 300 msec and a time-delay quench flow (Fig. 1B) for times greater than 300 ms (Barman et al., 2006).
Figure 1.
A) Continuous or rapid quench flow apparatus with a single drive mode. By activation of the drive, phospho-donor and phospho-acceptor are mixed in the mixing chamber. The reaction mixture is aged. The age of the reaction mixture corresponds to t=V/S where V is the volume of the capillary and S is the rate of the flow of the reaction mixture down the capillary tube. The value of t can be controlled by varying V and S which is in the range of 30–300 ms. The reaction mixture is quenched upon sample collection. B) Time-delay mode for the quench flow device. In this case, there are two drives. One is for the buffer and phospho-donor and the second one is for the phospho-acceptor and quencher. By activation of the first drive, phospho-donor and buffer are mixed in the first mixing chamber. Activation of the second drive fills the mixing chamber with phospho-acceptor. The reaction mixture is driven in the delay line and stays there for a certain amount of time. After a specific time-delay, the reaction mixture is expelled from the delay line into a second mixing chamber where it is quenched by the injected quencher and collected for analysis. For this method, the reaction time is in the range of 150 ms to minutes and the reaction mixture is less than 50 µL (Figure adapted from Barman et al, 2006).
A rapid quench flow apparatus makes use of continuous liquid flow, as shown in Fig. 1A, a drive system pushes the plungers at constant speed, enzyme and substrate are mixed in a mixing chamber, and the reaction mixture fills and passes through the capillary tube at a constant speed, S. The reaction time, or age of the mixture, is calculated as t = V/S, where V is the volume of the capillary. At the end of the capillary tube, the reaction is quenched (alternatively, the quenching solution can also be injected into a second mixing chamber with a syringe or by the drive mechanism, Fig. 1B). The quenched sample is then collected and analyzed. Changing the capillary tube (V), and or the drive speed (S), provides for flexibility in the reaction time. Accurate calculation of the rates and other kinetic parameters requires knowledge of reaction time (t) and the dilution factor of the reaction mixture by the quenching solution.
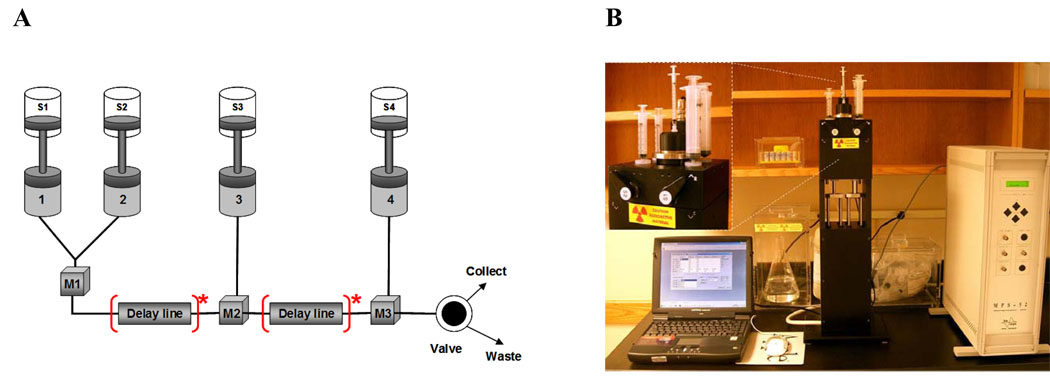
In this article, attention is focused on the SFM-4/Q rapid quench instrument3 from Bio-Logic used in our laboratory (Janiak-Spens et al., 2005, Kaserer et al., 2009). As shown in Fig. 2A, the instrument can be operated with a total of four syringes4, including two reagent syringes (S2 and S3), one syringe for the buffer or optionally the third reagent of the reaction (S1), and one syringe for the quenching solution (S4). The standard syringe volumes for the instrument are 5 mL for S2 and S3, and 20 – 30 mL for S1 and S4. Syringes are interchangeable, which allows custom adjustment of the system. Four independently programmable stepping motors are used to actuate syringes S1, S2, S3, and S4. Motor drive rates are independent, so variable-mixing ratios can be obtained by simply programming the drive sequence.
Figure 2.
A) A schematic representation of the RQF experiment. S1–S4 – syringes; M1–M3 – mixers; 1–4- motors; [Delay line]* - possible location of the delay line. Figure is reproduced from the SFM-4/Q instrument manual with permission from Bio-Logic (www.bio-logic.info). B) The SFM-4/Q rapid quench-flow instrument from Bio-Logic (newer model is the SFM400) showing placement of syringes and valves (inset). Note that users should follow proper safety procedures for working with radioactivity, including shielding while working with the instrument and properly disposing of both liquid and solid radioactive waste.
The reaction mixture can be aged in the delay line allowing various delay times. The volume of the delay line is fixed, however, a variety of delay line volumes are available; the volume can be as low as a few µL. Aging in the delay line can be set by varying the mean flow rate of the syringes (effectively from 1 µL/s to 5 mL/s). The reaction product is mixed with the quenching solution in the mixer and the final mixture (product) is collected via the exit line. The final product is collected through the exit purge port using buffer or the next reaction mixture.
C. Experimental Design
The following aspects of experimental design should be considered in preparation for rapid quench-flow kinetic analysis:
-
What are the major components of the reaction mixture?
For example, the SLN1-YPD1-SSK1 multi-step phosphorelay pathway was subdivided for the rapid quench flow analysis into two half reactions of SLN1-R1~P to YPD1 (scheme 1) and YPD1~P to SSK1-R2 (scheme 2).
-
What protein concentrations are required?
This will depend on the specific activity of the radiolabeled phosphodonor protein preparation. One could determine empirically what the minimal detectable concentration of the protein phosphodonor is based on the amount of radioactivity detectable by phosphorimager analysis (Molecular Dynamics). In the case of SLN1-R1~P, we estimate ~10% of the protein is radiolabeled (Janiak-Spens et al., 2005) and a protein concentration of ~0.45 µM is suitable for the rapid quench experiments.
Scheme 1.
Scheme 2.
-
What quenching solution will be used?
The quenching solution may vary depending on the reaction components used for the experiment. In the case of phosphotransfer analysis of the S. cerevisiae multi-step phosphorelay system the quenching solution contained EDTA to chelate Mg2+ and SDS for protein denaturation for subsequent SDS-PAGE analysis.
-
What enzyme to substrate ratios will be assayed?
For example, in the case of SLN1-YPD1-SSK1 phosphotransfer reactions the following concentration ratios of SSK1-R1 to YPD1 and YPD1 to SSK1-R2 were used: 1:0.5, 1:1, 1:2, 1:5, 1:10 and 1:20.
-
What are the timescales?
Different time scales are expected for different phosphotransfer reactions and this should be determined empirically. The earliest manually detectable time point for the phosphotransfer reaction between SLN1-R1~P and YPD1 was 8 sec (Janiak-Spens and West, 2000). Thus, the following time points were used for both half-reactions: 30, 40, 60, 80, 100, 150 and 300 ms.
-
Will a delay line be used?
The RQF instrument does not require installation of the delay lines if the reaction time scale is within 30–300 ms. However, if the reaction timescale is longer, delay lines should be installed and used for proper aging time.
The RQF final result will always depend on an accurate and precise chemical analysis of the product or intermediate.
A typical experiment
For kinetic analysis of the SLN1-YPD1-SSK1 phosphorelay, the rapid quench flow technique was employed and the two half reactions, SLN1-R1 to YPD1 and YPD1 to SSK1-R2, were analyzed in detail. The following operational mode is given here as an example adapted for the S. cerevisiae SLN1-YPD1-SSK1 multi-step phosphorelay.
Only syringes S2, S3 and S4 are used for the typical experiment (phosphotransfer from SLN1-R1~P to YPD1 or YPD1~P to SSK1-R2), while S1 is filled with buffer but is not used for the reaction5.
Reactants are in S2 (5 mL) and S3 (5 mL) and the quenching reagent is in S4 (20 mL).
A delay line is not necessary.
For collection of the final mixture, the SFM-4/Q instrument has a programmable exit electro-valve, which also provides sample economy. After the final solution is accumulated in the mixer and pushed to the exit valve, the sample is collected in a pipette or syringe. Use of a plastic syringe is recommended for collection of radioactive samples6.
D. Rapid Quench Flow Experiments
The SFM-4/Q instrument (Bio-Logic) was used for the rapid quench flow experiments to determine the phosphotransfer rates of the SLN1-YPD1-SSK1 pathway (Figure 2). The instrument was calibrated via monitoring the base-catalyzed hydrolysis of p-nitrophenylacetate (Gutfreund, 1969) as recommended by the instrument manufacturer (Bio-Logic). The reaction mixture contained 500 µL of 0.625 mM 2,4-dinitrophenyl acetate and 0.237 M NaOH, quenched in 500 µL of 4 M HC1. The base-catalyzed hydrolysis of 2,4-dinitrophenylacetate can be examined over a wide range of rates, generated by changing the concentration of the excess reagent.
After calibration, the SFM-4/Q should be flushed thoroughly with reaction buffers (S1-S3, see below); the syringe drive motors should be run in forward and reverse to release any air bubbles trapped in the system. Blank pre-runs can be conducted to ensure proper volume dispensing during sample collection. The minimal reaction mixture volume is approximately 180 µL for the SLN1-YPD1-SSK1 phosphotransfer reactions. To estimate minimal protein concentration, non-radioactive pre-runs were conducted with 60 µL of SLN1-R1 (1, 0.75 and 0.45 µM) rapidly mixed with 60 µL of YPD1 (0.9 µM) and quenched with 60 µL of S4 buffer (see below) for the first half-reaction; optimal concentrations were 0.45 µM SLN1-R1 or YPD1.
Using the same procedure, the second half-reaction YPD1 to SSK1-R2 was analyzed in similar manner. Blank non-radioactive pre-runs were conducted (60 µL of SSK1-R2 (1, 0.6 and 0.45 µM), 60 µL of YPD1 (0.9 µM) and 60 µL of quenching S4 buffer.
The data sets for the blank experiments were collected in a time-dependent manner (30, 40, 60, 80, 100, 150 and 300 ms) and each experiment included three phosphodonor to phosphoacceptor concentration ratios (SLN1-R1 to YPD1 or YPD1 to SSK1-R2): 1:2, 1:5, 1:10. The concentration of the phosphodonor (SLN1-R1 or YPD1) was kept constant throughout the experimental procedure. Once the blank experiments were completed, the collected samples were inspected by SDS-PAGE to verify even dispensing by the instrument.
When the preliminary analysis was completed, the instrument was filled with the following buffers as designed for the His-Asp phosphorelay system from S. cerevisiae (Figure 2):
S1 – 20 mL of 50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mM DTT;
S3 – 3 mL of the phospho-accepting protein at concentrations of 0.45 to 20 µM were diluted into 50 mM Tris-HCl, pH 8.0, 20 mM MgCl2, 1 mM DTT;
S4 – 20 mL of stop buffer (8% SDS, 80 mM EDTA).
The radiolabeled phosphodonor protein (SLN1-R1 or YPD1; diluted to 0.45 µM) is immediately transferred into the S2 syringe (Figure 2). The diluted phosphorylated SLN1-R1 (60 µL) is then mixed with 60 µL of phospho-accepting protein (YPD1 or SSK1-R2 at 0.45 – 20 µM). Reactions are quenched with 60 µL of the stop buffer after a specified time. To further prevent phosphate hydrolysis, the reaction samples can be frozen or placed on ice prior to gel electrophoresis.
Data sets are collected in the time-dependent manner (30, 40, 60, 80, 100, 150 and 300 ms) and each new experiment contained a different phosphodonor to phospho-accepting concentration ratio as specified above. However, the concentration of the phosphodonor (SLN1-R1 or YPD1) was kept constant for all experiments.
To analyze the results, 30 µL of the quenched reaction was mixed with 10 µL of 4X SDS-PAGE loading buffer (200 mM Tris pH 6.8, 400 mM DTT or β-mercaptoethanol, 8% SDS, 0.4 % bromophenol blue, and 40% glycerol), and then 30 µL samples were loaded onto 15% SDS-PAGE gels7. After gel electrophoresis, wet gels are wrapped in plastic wrap and analyzed using a Phosphorimager (Molecular Dynamics, Storm 840). The phosphotransfer reaction kinetic parameters are quantified on the basis of disappearance of the 32P-label from the phosphodonor protein and the appearance of 32P-label in the phospho-accepting protein as described below.
E. Data Analysis and Interpretation
A phosphotransfer reaction profile or time course is used to extract the kinetic parameters. A representative radiograph image of the raw data is shown in Figure 3. The parameters are quantified on the basis of the disappearance of the band corresponding to the 32P-label from the phospho-donor protein or the appearance of the band corresponding to the 32P-label in the phospho-accepting protein. The phosphotransfer profile is usually time dependent and the time of incubation depends on how fast the reaction occurs, which is usually captured in the range of milliseconds to seconds. After scanning the storage phosphor screen of the gel or membrane containing the proteins of interests, using a pixel processing software (ImageQuant version 5.2 in our case), the total intensities of the pixels corresponding to a specific band is calculated. For volume calculation, ImageQuant subtracts the background value from the intensity of each pixel in the object, and then adds all the values as follows:
| (1) |
where the value of the background is calculated as a local average of all the pixel values in the object outline (ImageQuant reference, version 5.0, Molecular Dynamics, Inc., USA).
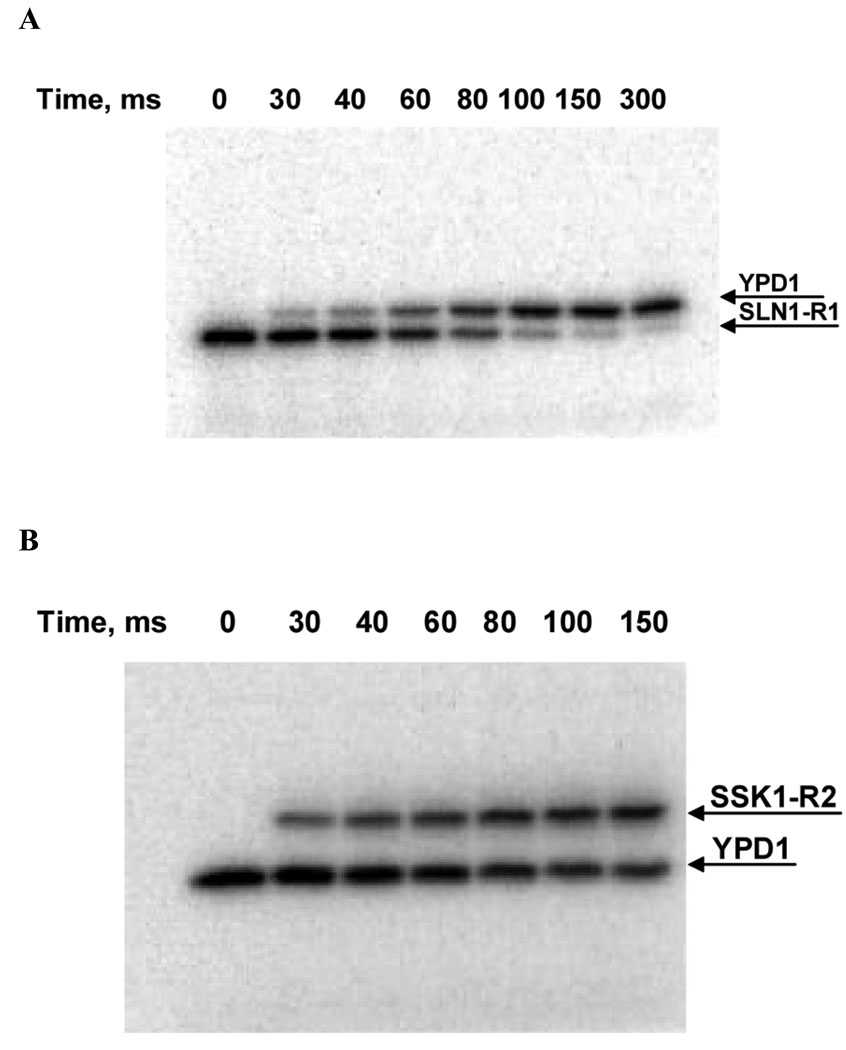
Figure 3.
Phosphotransfer analysis of the multi-step phosphorelay pathway from S. cerevisiae using RQF kinetics. A) The resulting gel radiograph image of the phosphotransfer reaction between phosphorylated receiver domain of SLN1-R1~P (phospho-donor) and histidine phosphotransfer protein YPD1 (phospho-acceptor) as a function of time. Phosphorylated SLN1-R1~P (0.45 µM) was incubated with YPD1 (0.9 µM) for 30, 40, 60, 80, 100, 150 and 300 ms. Each reaction mixture was quenched with quench buffer (8% SDS, 80 mM EDTA) at the specified time point. Each quenched reaction mixture (30 µL) was mixed with 4X SDS-PAGE loading buffer (10 µL) and separated on a 15% SDS-PAGE gel. The radiolabeled bands were quantified using phosphorimager analysis. B) The radiograph image of the phosphotransfer reaction between phosphorylated YPD1 (phospho-donor) and the response regulator domain of SSK1-R2 (phospho-acceptor) as a function of time. Phosphorylated YPD1 (0.45 µM) was incubated with SSK1-R2 (0.9 µM) for 30, 40, 60, 80, 100, 150 ms. Each reaction mixture was quenched with quench buffer and analyzed as described in A.
To determine the percent of the remaining phosphodonor protein, the volume of the corresponding band is divided by the sum of the volumes of the bands for phosphodonor and phosphoacceptor proteins and the fraction value is multiplied by 100. The percent of the remaining phosphodonor protein is then plotted as the natural logarithm vs reaction time (Fig. 4A) (Janiak-Spens et al., 2005). The observed first-order rate constants (kobs) is obtained by fitting each time course (obtained at a particular fixed [S]) to the linear relationship as shown in eq 2,
| (2) |
where A is the amount of the phosphorylated phosphodonor protein at times t and 0, and kobs is the observed first order rate constant obtained using least squares fitting in Excel (Microsoft Office). The purpose of the data plotting as the natural logarithm is to assess data quality and obtain a linear relationship between the dependent (At) and independent variables (t). However, time course data can also be fitted to the exponential form of the eq 3 as shown below
| (3) |
However, the fit does not change the value of kobs in comparison to eq 2. The advantage of fitting to eq 2 is the feasibility of the linear regression as well as better data assessment.
Figure 4.
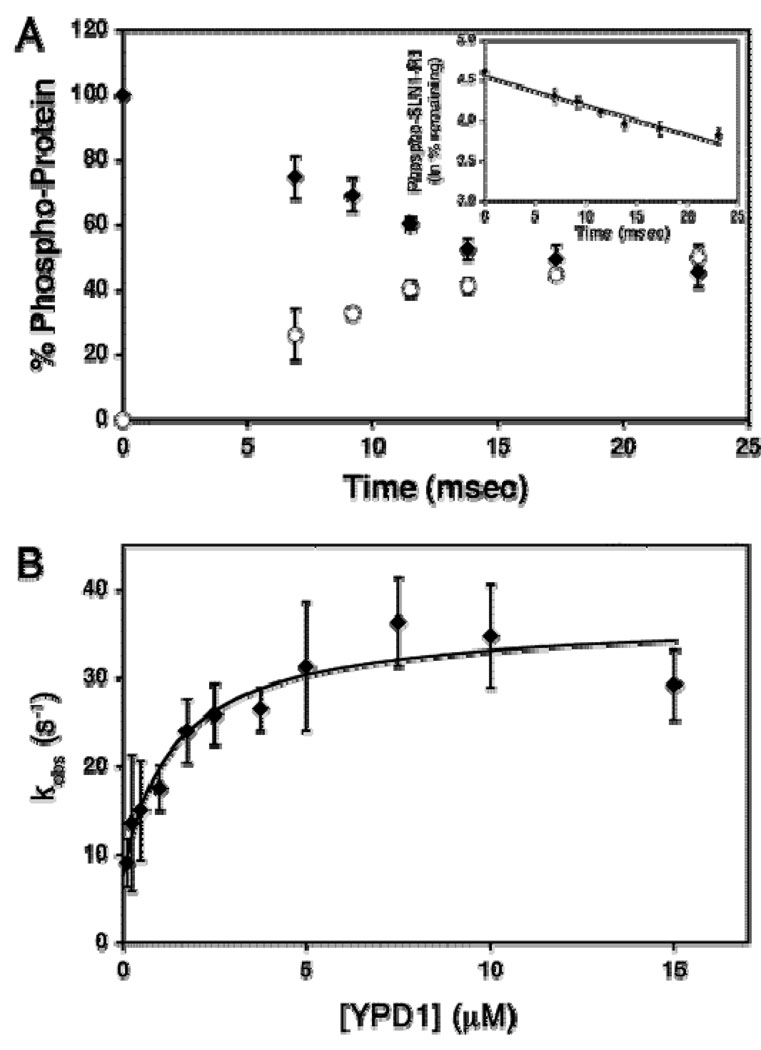
A) A time course of phosphoryl transfer from SLN1-R1~P to YPD1. The percent of the 32P-radiolabeled proteins are shown as follows: (○) YPD1~P, (♦) SLN1-R1~P. Data obtained was from the average of three experiments. The inset shows the percent of the remaining SLN1-R1~P plotted as natural logarithm vs reaction time. The slope of the linear fit (eq 2) is kobs. B) YPD1-dependent rate of the phosphoryl transfer reaction. The plot shows saturation at high YPD1 concentrations and a finite non-zero ordinate intercept (krev). Kd can be defined as the concentration of YPD1 where kobs = ½ kmax, which is 1.4 µM. Data were fitted to eq 2 (Janiak-Spens et al., 2005). Figure was reproduced with permission from the American Chemical Society.
The general procedures for the subsequent data fitting are to take the slope of the line fitted to the raw data using eq 2 (the slope is equal to kobs and is obtained at a particular fixed phospho-acceptor concentration). Then, graph the slopes or kobs (obtained at each fixed substrate concentration) vs. substrate concentration to assess data quality and evaluate the mathematical trend of the data and or the shape of the graph in order to find the proper model and equation for data fitting.
As shown in the Fig. 4B, kobs is a rectangular hyperbolic function of phospho-acceptor concentration with a finite value on the y-axis. Data were fitted to eq 4 (three-parameter rectangular hyperbola), which adheres to the following phosphotransfer reaction model (scheme 3),
| (4) |
where [S] is the concentration of the phospho-accepting protein, kobs is the observed first-order rate constant for the phosphotransfer reaction at a particular [S], kfwd is the maximal forward net rate constant for phosphoryl transfer from the phosphorylated protein to the phospho-acceptor protein, krev is the corresponding maximum reverse net rate constant for the reaction between phospho-donor and phospho-acceptor proteins, and Kd is the dissociation constant of the phospho-donor-acceptor complex (Fig. 4B). The fit can be obtained using any program with data fitting algorithms. In this case, the Enzfitter program was used (version 2.04, Biosoft, Cambridge, U.K.).
Scheme 3.
The limit of eq. 4 when the concentration of the phospho-accepting protein is very high relative to the Kd is as follows:
| (5) |
where kmax is the maximum observed first order rate constant. When the concentration of the phospho-accepting protein is near zero, the limit of eq 4 is equal to krev. The limits of the eq 4 are applicable to Fig. 4B. The values of the kinetic parameters (kfwd, krev, and Kd) can also be obtained under different conditions such as different concentrations of osmolytes (Kaserer et al., 2009) or some other variable. In this case, the values of the kinetic parameters (kfwd, Kd or kfwd/Kd) are plotted against the concentrations of osmolytes and the data interpretation depends on the overall mathematical shape of the plots and their interrelationships.
In general, the kinetics of the phosphotransfer reactions (see examples below) between biological macromolecules follow the enzyme kinetic model (Michaelis-Menten model) and the data analysis is similar. However, different approaches may be taken for the analysis of the raw data. Here, we give three examples of data obtained using the RQF method:
Specificity of the vancomycin resistance kinase VanS for two response regulators, VanR and PhoB (Fisher et al., 1996). In this study, the catalytic efficiency (kcat/KM) was determined for phosphotransfer from VanS to VanR and PhoB with the former reaction showing a 104-fold preference. Both reactions follow Michaelis-Menten kinetics with slight modifications. The raw data for the time course experiments were treated with a nonlinear least-squares regression curve fitting to a rectangular hyberbola.
Phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway (Stewart et al., 1997). Experiments showed a reversible three step mechanism for CheA to CheY phosphotransfer, which includes binding of CheY to CheA~P, rapid phosphotransfer to CheY, and dissociation of CheA·CheY~P complex. The rate of phosphotransfer shows saturation kinetics similar to enzymatic reactions. Raw data were fitted to the equation for a rectangular hyberbola using a nonlinear least-squares method.
Interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis (Grimshaw et al., 1998). Divalent-metal dependent auto-phosphorylation of KinA is the first step in the phosphotransfer reaction. The value of kcat/KM for Spo0F in the formation of Spo0F~P is 57000 fold higher than Spo0A in the formation of Spo0A~P. The authors showed the kinetic mechanism was hybrid ping-pong/sequential with a pronounced (>40 fold) substrate synergism by Spo0F in the auto-phosphorylation of KinA. In this study, reciprocal initial velocities were plotted vs. reciprocal substrate concentrations and fitted by a least square method for some of the data collected.
The three examples above, as well as recent papers from this lab (Janiak-Spens et al., 2005, Kaserer et al., 2009) are typical of the kinetics of the phosphotransfer reactions obtained using the RQF method. They show the versatility of the method in obtaining the data under different conditions.
Conclusion
The highly versatile method of rapid quench flow kinetics was described here with regard to its application to the study of His-Asp phosphotransfer reactions in the yeast osmoregulatory signal transduction pathway. Using this method, data on intermediate species can be obtained in the time range of milliseconds to minutes. Finally, treatment and analysis of the data obtained using the RQF method is facilitated by computer operated instrument modes, software and fitting algorithms.
Acknowledgements
We gratefully acknowledge funding from the NIH (GM59311 to AHW), the Oklahoma Center for the Advancement of Science and Technology (OCAST) (HR 06-123 to AHW) and the Grayce B. Kerr endowment to the University of Oklahoma (to support the research of PFC) for the research described here. We would also like to thank Dr. Fabiola Janiak-Spens for superb technical advice regarding quench flow experiments and experimental procedures and Vidya Kumar for useful feedback on the manuscript.
Footnotes
Abbreviations: ATP, adenosine-5′-triphosphate; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; GST, glutathione-S-transferase; HK, histidine kinase domain; HPt, histidine-containing phosphotransfer; MAP, mitogen-activated protein; PAGE, polyacrylamide gel electrophoresis; RR, response regulator; SLN1-R1, C-terminal response regulator domain of SLN1; SSK1-R2, C-terminal response regulator domain of SSK1; SKN7-R3, C-terminal response regulator domain of SKN7; RQF. rapid quench flow; SDS, sodium dodecyl sulfate.
TgK Scientific (www.tgkscientifc.com), Bio-Logic (www.bio-logic.info), KinTek (www.kintek-corp.com), and AppliedPhotophysics (www.photophysics.com)
The SFM400 is a newer model from Bio-Logic that has replaced the SFM-4/Q instrument, but the basic design and principle of use is the same.
The instrument can also be used with two to three syringes (although the fourth syringe cannot be empty; one can fill it with buffer) and a delay line with either single or double mixing.
The reason for filling syringe S1 is to avoid software interface problems in communicating with the instrument for this specific instrument model.
During a typical experiment, be aware that backpressure from a purge port can push out the collection syringe if it is not held by hand during the collection mode.
Do not boil samples prior to gel loading, as this increases the rate of phosphate hydrolysis.
Reference
- Appleby JL, Parkinson JS, Bourret RB. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- Barman TE, Bellamy SR, Gutfreund H, Halford SE, Lionne C. The identification of chemical intermediates in enzyme catalysis by the rapid quench-flow technique. Cell Mol. Life Sci. 2006;63:2571–2583. doi: 10.1007/s00018-006-6243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Bussey H, Stewart RC. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler JF, West AH. Genetic and biochemical analysis of the SLN1 pathway in Saccharomyces cerevisiae. Meth. Enzymol. doi: 10.1016/S0076-6879(10)71016-8. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SL, Kim S-K, Wanner BL, Walsh CT. Kinetic comparisons of the specificity of the vancomycin resistance kinase VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- Grimshaw CE, Huang S, Hanstein CG, Strauch MA, Burbulys D, Wang L, Hoch JA, Whiteley JM. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- Gutfreund H. Rapid mixing: continuous flow. Meth. Enzymol. 1969;16:229–249. [Google Scholar]
- Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Meth. Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- Horie T, Tatebayashi K, Yamada R, Saito H. Phosphorylated Ssk1 prevents unphosphorylated Ssk1 from activating the Ssk2 MAP kinase kinase kinase in the yeast HOG osmoregulatory pathway. Mol. Cell. Biol. 2008;28:5172–5183. doi: 10.1128/MCB.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak-Spens F, Cook PF, West AH. Kinetic analysis of YPD1-dependent phosphotransfer reactions in the yeast osmoregulatory phosphorelay system. Biochemistry. 2005;44:377–386. doi: 10.1021/bi048433s. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling DP, West AH. Novel role for an HPt domain in stabilizing the phosphorylated state of a response regulator domain. J. Bacteriol. 2000;182:6673–6678. doi: 10.1128/jb.182.23.6673-6678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling JM, Gurfinkel M, West AH. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J. Bacteriol. 1999;181:411–417. doi: 10.1128/jb.181.2.411-417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak-Spens F, West AH. Functional roles of conserved amino acid residues surrounding the phosphorylatable histidine of the yeast phosphorelay protein YPD1. Mol. Microbiol. 2000;37:136–144. doi: 10.1046/j.1365-2958.2000.01973.x. [DOI] [PubMed] [Google Scholar]
- Kaserer AO, Andi B, Cook PF, West AH. Effects of osmolytes on the SLN1-YPD1-SSK1 phosphorelay system from Saccharomyces cerevisiae. Biochemistry. 2009;48:8044–8050. doi: 10.1021/bi900886g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krems B, Charizanis C, Entian K-D. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell. 2002;13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JM-Y, Deschenes RJ, Fassler JS. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Euk. Cell. 2003;2:1304–1314. doi: 10.1128/EC.2.6.1304-1314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Porter SW, West AH. A common docking site for response regulators on the yeast phosphorelay protein YPD1. Biochim. Biophys. Acta. 2005;1748:138–145. doi: 10.1016/j.bbapap.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Porter SW, Xu Q, West AH. Ssk1p response regulator binding surface on histidine-containing phosphotransfer protein Ypd1p. Euk. Cell. 2003;2:27–33. doi: 10.1128/EC.2.1.27-33.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 "two-component" osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Saito H, Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 2004;136:267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- Shankarnarayan S, Malone CL, Deschenes RJ, Fassler JS. Modulation of yeast Sln1 kinase activity by the CCW12 cell wall protein. J. Biol. Chem. 2008;283:1962–1973. doi: 10.1074/jbc.M706877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RC. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry. 1997;36:2030–2040. doi: 10.1021/bi962261k. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Xu Q, Porter SW, West AH. The yeast YPD1/SLN1 complex: Insights into molecular recognition in two-component systems. Structure. 2003;11:1569–1581. doi: 10.1016/j.str.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Zhao X, Copeland DM, Soares AS, West AH. Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog. J. Mol. Biol. 2008;375:1141–1151. doi: 10.1016/j.jmb.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]