Abstract
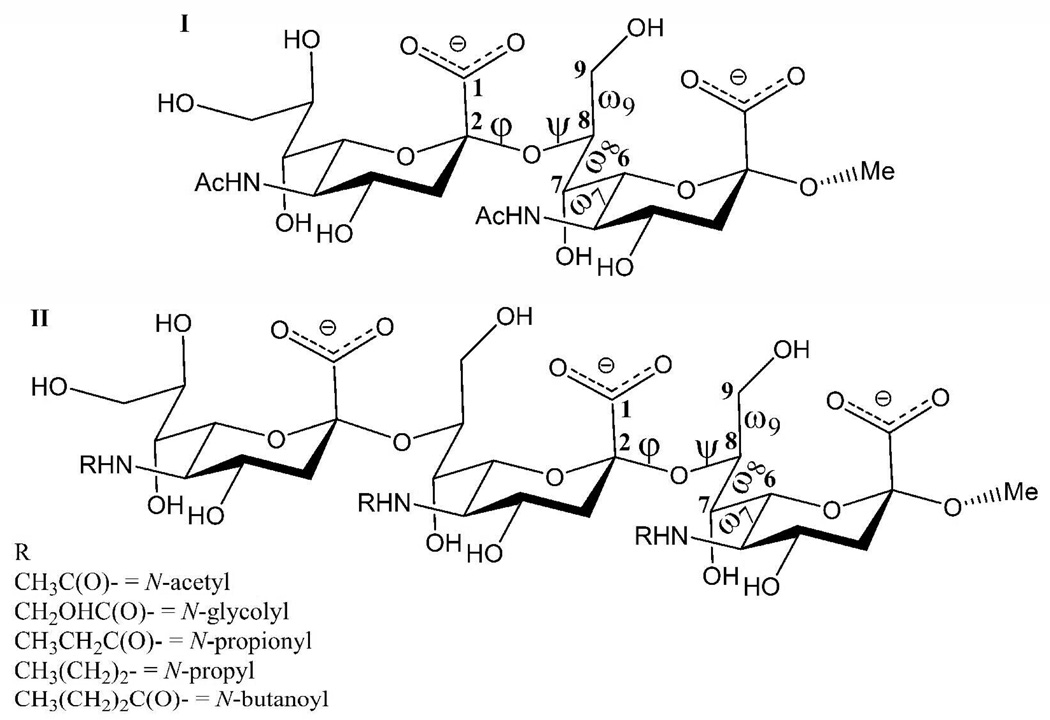
The conformational properties of di- and trisaccharide fragments of the polysialic acid O-antigen capsular polysaccharide (CPS) of Neisseria meningitidis B (NmB), have been investigated by a combination of solution phase NMR spectroscopy and explicit-solvent molecular dynamics (MD) simulations. Simulations employing 100 ns of conventional MD, as well as 160 ns of replica exchange MD (REMD), with the GLYCAM06 force field were shown to be in agreement with experimental NMR scalar J-coupling and nOe values. The presence of conformational families has been determined by monitoring inter-glycosidic torsion angles, by comparing structural superimpositions, as well as via a Bayesian statistical analysis of the torsional data. Attempts to augment the immunogenicity of NmB CPS often involve chemical modifications of the N-acetyl moiety. Here the effects of these chemical group modifications on the conformational properties of the trisialoside have been probed via REMD simulations of the N-glycolyl, N-propionyl, N-propyl and N-butanoyl analogs. Although there were conformational families unique to each non-native analog, the chemical modifications resulted in largely equivalent overall conformational phase-spaces compared to the native trisialoside. On the basis of the conformational distributions, these shared conformational properties suggest that a recurrent global conformational epitope may be present in both the native and chemically modified CPS fragments. Explanations are therefore provided for monoclonal antibody cross-reactivity, in terms of recognition of a shared global CPS conformation, as well as for lack of cross-reactivity, in terms of fine structural differences associated with the N-acyl groups, which may be dominant in highly matured antibody responses.
Neisseria meningitidis (Nm) is a gram negative bacterium that is responsible for septicemia and meningococcal disease in neonates. (1) Annually, about 500,000 cases of meningococcal meningitis are reported worldwide, with 2600 occurrences in the United States. Ten percent of infections result in deaths, while survivors may suffer from seizures, hearing impairments and brain damage. (2) Thirteen strains have been classified serologically, based on the identity of the polymeric repeat units and the types of glycosidic linkages in the O-antigen capsular polysaccharides (CPS) that envelop the surface of the bacteria. (3) Of these thirteen strains, five strains, A, B, C, W-135 and Y, have been identified as the main causative agents of virulent invasive meningococcal disease, Table 1. (4–7)
Table 1.
CPS structures of the five most virulent N. meningitidis serotypes
The lack of cross-reactivity between anti-CPS sera indicates that despite the fact that several Nm capsules bear striking carbohydrate sequence similarities, they must display unique antigenic determinants or epitopes. That is, each CPS must either have a unique overall 3D conformation, or must display its immunodominant regions in unique 3D configurations. For example, serogroups B (NmB) and C (NmC), which are both linear homopolymers of α-N-acetylneuraminic acid (Neu5Ac) containing α-(2,8) and α-(2,9) glycosidic linkages, respectively, elicit antibodies that do not cross-react with each other’s CPS. (3) It should be noted that variations in the level and position of O-acetylation also exist between the serotypes, (Table 1). (5–7) It is remarkable that, despite the apparent sensitivity to carbohydrate sequence, cross-reactivity of a human monoclonal antibody (IgMNOV) with the NmB CPS and poly(Adenine) and other polynucleotides has been observed, (8) and attributed to similar conformational properties and spatial distribution of anionic charges between the carboxyl groups of the B polymer and the phosphate groups of the polynucleotide. (9) This result serves to illustrate a recurrent phenomenon, namely that IgM class antibodies often show broader cross-reactivity than seen with more highly affinity matured IgG antibodies, and it is necessary to consider this factor when discussing CPS reactivities. Each NmB CPS may also display differences in the apparent size of the epitope. For example, in binding inhibition assays a minimum of approximately ten residues of the NmB CPS was required for effective inhibition of antisera binding to the intact polysaccharide antigen, while for strain C, maximal inhibition occurred with a more conventional hapten length of four or five residues. (10) In addition to antigenic variations arising from differences in glycosidic linkage types, epitope size may also be altered by chemical modifications. In the case of NmB, inhibition studies indicated that reduction of the carboxylic acids to primary alcohols (sialitols) led to a decrease in the epitope size to approximately six residues. (11) The requirement in NmB for an oligosaccharide length of at least ten residues, to inhibit antibody binding to the native CPS, has led to the postulation that this CPS adopts an extended conformational epitope. (10) Extended epitopes are generally assumed to reflect the presence of a conformational requirement for binding, giving rise to the concept of a conformational epitope. All extended epitopes should therefore be conformational, but given the fact that oligosaccharides often exist in well defined conformations, even a short sequence may have a conformational requirement for binding. This is in contrast to the case of short peptide epitopes.
Currently, two CPS-based conjugate vaccines against Nm are available: (12) a bivalent form, composed of groups A and C, and a tetravalent form containing A, C, Y and W-135. A conjugate vaccine against NmB is not currently available due to the poor immunogenicity of its CPS, which is structurally equivalent to the polysialic acid (PSA) autoantigen present in the glycoproteins of human neural cell adhesion molecules, and is therefore not immunostimulatory. (13) The corollary to this structural similarity is that antibodies that bind to theNmB CPS can also cross-react with PSA, (14) raising a question as to whether or not a CPS-based vaccine would have undesirable side effects. However, studies of human maternal and placental cord sera have found that "natural" anti-PSA antibodies were present in almost all matched pairs of sera; (15) suggesting that increased levels of anti-NmB CPS antibodies elicited by vaccination would not be pathologic. (15) It has also been hypothesized that the weak immunogenicity of NmB CPS-based vaccines arises from the formation of transglycosidic lactone rings, between the hydroxyl group at C9 and the carboxylic acid in the preceding residue. (16) Lactonization, could alter the antigenicity and immunogenicity of the CPS by changing its conformation and charge distribution, (16) and indeed it has been observed that as little as 9% lactone content was sufficient to attenuate the antigenic properties of the CPS. (16) Recent NMR studies of the NmB CPS displayed on E. coli K12 cell surfaces, under physiological conditions, showed no evidence of lactone formation, confirming that it is not a biologically common form of the CPS. (17)
Several attempts to develop immunogenic NmB vaccines have focused on modifications of the amido group, for example by replacement of the N-acetyl with N-glycolyl (N-Gc), Npropionyl (N-Pr), N-propyl (N-Prop) or N-butanoyl (N-But) moieties. (11, 18–22) Somewhat perplexing cross-reactivities, as well as varying levels of protection and bactericidal activity have emerged from immunizations with these conjugate vaccines. Several of the modified polysaccharides appear to present an extended epitope similar to that found in NmB, as judged by cross-reactivity with monoclonal antibodies known to display specificity for the NmB extended epitope. For example, the cross-reactivity of the N-Pr CPS with the well-characterized bactericidal NmB monoclonal antibody 735 (IgG) indicates that both the N-Pr analog and the native CPS share a similar extended epitope of approximately ten residues. (22) However, some CPS analogs, such as the N-Prop derivative, bind poorly to mAb 735, (18) suggesting that the N-Prop analog differs significantly from the native CPS, either in overall conformation or in local contacts involving the N-acyl groups. Notably, the presence of an extended epitope is not sufficient to ensure cross-reactivity among the modified immunogens and or antisera. For example, although the N-Pr analog and the native CPS both bind to mAb 735, (22) immunization with the N-Pr conjugate, while giving a strong antibody titre against the N-Pr CPS, gave little cross-reaction with NmB CPS. (21) Similarly, a bactericidal monoclonal antibody, IgG 13D9, generated by immunization with the N-Pr conjugate vaccine showed no affinity for the native NmB CPS, (22) although it did cross-react with the N-But CPS. (23) Subsequent analysis confirmed that, mAb 13D9 does bind to the bacterial surface of Nm, leading to its bactericidal activity, but does not bind to NmB CPS. (22) Thus, the bactericidal activity of mAb 13D9 can only be understood by accepting that it recognizes an epitope on the bacterial surface, other than that for which the vaccine was designed. (22)
As an alternative to modification of the NmB CPS, attempts have been made to stimulate antibody production against both the NmB and NmC CPS by immunizing with vaccines made from the CPS of E. coli K92. The K92 CPS consists of alternating α-(2,8) and α-(2,9) linkages, and conjugate vaccines using this CPS could therefore be expected to produce by default anti-NmB (2,8) or NmC (2,9) responses. (15, 24) Although vaccination with K92 CPS conjugates did yield anti-NmC responses, they were poorer than direct immunization with NmC conjugate. (15) Antisera from K92 vaccinations showed only a very modest if any affinity for NmB CPS. (15, 21) In an attempt to enhance the immune response to K92 conjugate, following on the results with chemically modified NmB CPS, the N-Pr analog of K92 was generated. However, immunization with N-Pr K92 conjugate did not lead to any improvement in cross-reactivity, but rather the antisera lost all affinity for both NmB and NmC CPS. (21) In contrast to the case of NmB, the NmC CPS does not display an extended epitope, (21) and the lack of affinity for NmB displayed by the E. coli K92 antisera has been ascribed to the absence of an extended epitope in K92. (21) The origin of the deleterious effect of N-propionylation on the ability of K92 conjugate to elicit anti-NmC antibodies remains unclear, particularly since the N-Pr K92 antisera retained the ability to recognize native K92 CPS.
The potential relevance of an extended conformational epitope to the antigenicity, immunogenicity and bactericidal activity of NmB CPS (10) has stimulated numerous investigations of the conformational properties of this CPS. (9, 11, 18, 25, 26) Understanding the structural features that determine the conformational properties that mediate the specificity and antigenicity of interactions between this CPS and bactericidal antibodies, could provide a basis for the interpretation of the antigenic properties of related structures and possibly for the rational development of more immunogenic and bactericidal vaccines against NmB. Any chemical modification that alters the conformational epitope of a CPS-based immunogen may lead to attenuated antigenicity or unexpected cross-reactivity. By conserving the epitope conformation responsible for generating bactericidal antibodies (the bactericidal epitope) it should be possible to categorize putative immunogens not only in terms of their antigenicity, but also in terms of their potential to elicit bactericidal antibodies. However, the flexibility of NmB in particular, as well as its homopolymeric sequence, currently prevents a direct resolution of the NMR data into definitive conformational families. The linkage regions between the residues of the NmB CPS may be characterized by four conformationally significant rotatable bonds, Figure 1. The presence of these flexible dihedral angles (φ, ψ, ω7, ω8) renders the characterization of the 3-D structure and dynamics the NmB CPS particularly challenging. When conformational variations are significant, such as in carbohydrates, the risk of generating virtual 3D conformations from the use of NMR restraints to guide the structural refinement are significant. (27) MD simulations can be employed to provide a structural basis for interpreting the NMR data or for a priori investigations of the effects of chemical modifications on the size and shape of the CPS epitopes. For example, MD and NMR studies have been used to elucidate the role of sialic acid residues in determining the overall conformation of the CPS from type III group B Streptococcus. (28) For the specific case of the NmB, molecular mechanics potential energy maps and MD simulations have been reported for the α-(2,8)-disialoside (9, 21) and α-(2,8)-tetrasialoside, (26) respectively, in the gas phase. Explicit-solvent MD simulations employing simulation times of 3 ns (21) and 1 ns initiated from nine different low-energy conformations (29) of the α-(2,8)-disialoside, and 530 ps for the α-(2,8)-tetrasialoside (26) have also been reported. However, it is now well established that the life-times of rotamers in glycosidic linkage regions can span more than the 10-ns time frame. (28, 30) In order to identify all of the significant conformational states, as well as to determine their populations, simulations on the order of 100 ns are typically required. (28, 30, 31)
Figure 1.
Schematic representation of the α-2,8-linked N-acetylneuraminic acid dimer (I) and trimer (II). The dihedral angles are defined as follows: φ = C1-C2-O8’-C8’, ψ = C2-O8’-C8’-C7’, ω9 = O9-C9-C8-O8, ω8 = O8-C8-C7-O7 and ω7 = O7-C7-C6-O6.
Here, we characterize the conformational properties of methyl α-(2,8)-di- (I) and trisialoside (II) haptens from the CPS of the NmB, employing data from 100-ns MD simulations performed with a force field (GLYCAM06) (32) that has been specifically developed for carbohydrate simulations. The theoretical conformations were then independently assessed for their experimental accuracy through comparison with new NMR nOe and J-coupling data for I and II. Additionally, replica exchange MD (REMD) simulations were performed for I and II and the N-Gc, N-Pr, N-Prop and N-But analogs of II, Figure 1. The REMD protocol was recently shown to lead to excellent agreement with experimental populations for simulations of glycerol, (33) suggesting that this approach may be appropriate for determining the conformational properties of the glyceryl side chains in the Neu5Ac residues, which determine the conformational properties of NmB. Our goal is to probe the effects of chemical modification on hapten conformation, and to provide a structural interpretation for the observed cross-reactivities or lack thereof between antibodies raised against the modified and native NmB CPS.
Materials and methods
1. Computational methods
1.1 MD simulations
The strain B haptens (methyl α-(2,8)-di- and trisialosides) were built from the GLYCAM (32) library of monosaccharide residues using the XLEaP module of AMBER8. (34) The appropriate number of Na+ counter-ions was added to neutralize the haptens using XLEaP, and the complete systems were immersed in pre-equilibrated boxes of 670 and 743 TIP3P (35) water molecules for the di- and trisialoside, respectively. Nonbonded van der Waals and electrostatic scaling factors for 1–4 interactions were set to unity (SCEE = SCNB = 1). Long range electrostatic interactions were computed with particle-mesh Ewald summation, (36) with a nonbond cut-off distance of 8 Å. The constant pressure-temperature ensemble (nPT) was employed under periodic boundary conditions, with box dimensions of 30 × 33 × 35 Å and 35 × 31 × 36 Å for the di- and trisaccharide systems, respectively. Initially, the solvent configurations were energy minimized through 1000 cycles (50 steepest descent and 950 conjugate gradient minimization), while the CPS fragments were restrained. Subsequently, the entire system was energy minimized with the same protocol. Energy minimization was followed by a short simulated annealing stage, wherein the entire system was heated from 5 K to 300 K over 50 ps, then cooled to 5 K over another 50 ps. Initial atomic velocities were assigned from a Boltzmann distribution at 5 K. Prior to the MD production cycle, the temperature of the system was raised to 300 K over 70 ps and held at that temperature for a further 80 ps. The production cycle ensued for 100 ns. A 2-fs time step was utilized to integrate the equations of motion, with hydrogen-containing bonds constrained to their equilibrium lengths using the SHAKE algorithm. (37)
1.2 Replica exchange MD (REMD) simulations
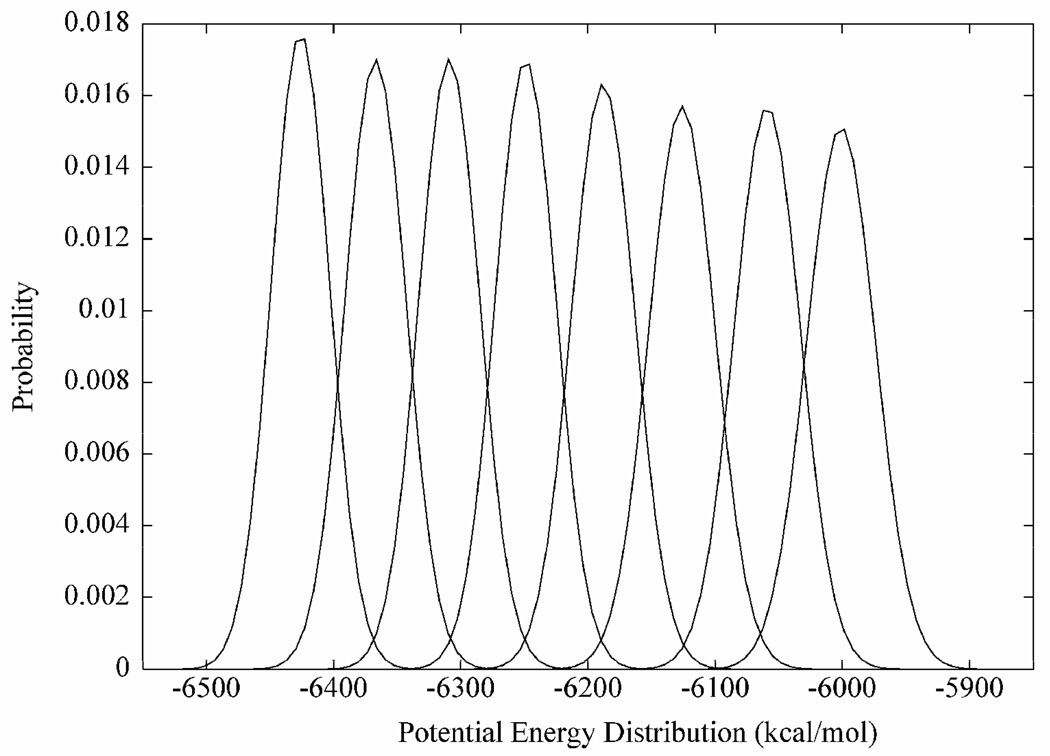
In REMD simulations, multiple replicas of a system are simulated concurrently at progressively higher target temperatures that increase exponentially. (38, 39) At specified intervals, the energies of adjacent replicas are swapped based on an acceptance probability. (39) Prior to performing the REMD simulations, the system was equilibrated following the same procedures outlined in the MD section. To derive the target temperatures employed in the present simulations, an exponential equation was fit to the range of sample temperatures given in the replica exchange section of the AMBER9 manual: 269.5, 300, 334, 371.8, 413.9, 460.7, 512.9 and 570.9 K. The exponential equation obtained from the fit was:
| [1] |
Eight approximate initial target temperatures (299.9, 308.0, 316.4, 324.9, 333.7, 342.8, 352.1 and 361.6 K) were generated by interpolating between the sample temperatures in incremental x-values of 0.25 starting from x = 2.0. From the initial single equilibration stage eight simulations were spawned by equilibrating to the eight approximate target temperatures noted above, followed by 10 ns of production dynamics under the constant volume-temperature (nVT) ensemble. Gaussian distributions (39) of the potential energies from these simulations were plotted, in order to confirm that energy overlaps occurred between successive replicas within the temperature range, Figure 2. Employing the lowest target temperature (300 K) from the simulations, and a swap acceptance probability of 0.2, the final target temperatures (300, 305, 310, 315, 320, 326, 331 and 337 K) were obtained through an iterative process described previously. (39) Prior to committing the replicas to exchange dynamics, each of the eight systems was heated to its target temperature. The hybrid solvent REMD method (38) was employed retaining 100 closest water molecules during each replica exchange process. An exchange was attempted every 500th step for a total of 500 × 40000 steps, resulting in an overall simulation time of 8 × 20 ns for the eight replicas. A 1-fs time step was utilized to integrate the equations of motion, and a 10-kcal/mol restraint force constant was applied to the carbohydrate ring atoms to prevent the rings from distorting at high temperatures.
Figure 2.
Gaussian potential energy distributions indicating the feasibility of the temperatures employed in the replica-exchange simulation
1.3 Bayesian cluster analysis
Cartesian coordinates from the MD simulations were collected every 5 ps for subsequent analysis with either the CARNAL or PTRAJ modules of AMBER8. In addition to typical analysis of torsion angle distributions, (30) global conformational states were also identified by determining transitions between Markov states, via a Bayesian statistical analysis of the glycosidic torsion angles. (40) A 25-component mixture of univariate Gaussians was used as a fuzzy partition of the data space. The centers and the widths of these Gaussians were optimized by a maximum likelihood approach. (41) Subsequently, the transition matrix for the 25 partition volumes was constructed. The two partition volumes connected by the fastest transitions were iteratively united. Thus, 25 Markov models and associated time-scales of the fastest transitions between the states of the respective model were obtained. In this approach, if the time scale (τl) of model l is much larger than the time scale of the subsequent model l+1 (τl >> τl+1), then model l+1 contains rapidly mixing states, while model l contains slowly varying metastable states. In that case, model l represents a plausible conformational state of the system. This enables the definition of several relevant state models for the observed trajectory, (40) and identifies global relationships between glycosidic torsion angles that may not otherwise be readily determined.
1.4 Predicting nOe intensities from the MD trajectories of the di- and trisialosides
2D nOe build-up intensities for the di- and trisialosides were predicted from MD simulations by performing a full relaxation matrix analysis with the CORMA program. (42) A sample of 2500 snapshots, extracted at regular intervals from the MD trajectories, was utilized for the predictions. Correlation times of 0.5 and 0.55 ns were employed for the di- and trisialosides, respectively.
2. NMR spectroscopy
Synthetic samples of the methyl α-2,8-di- and trisialosides were purchased from Dr. Nicolai Bovin (Lectinity Holdings, Lappeenranta, Finland). Samples were lyophilized from D2O then redissolved in 99.96% D2O. Data were collected on Varian Inova 500, 800 and 900 MHz spectrometers at 25°C, except for some nOe data which were also collected at 10°C. All pulse programs were from the standard Varian library. Data were processed with Gaussian window functions, linear prediction, and zero-filling prior to Fourier transformation. For each sample, a COSY dataset was collected with 1582 × 1024 complex points and acquisition times of 0.40 and 0.258 seconds in F2 and F1, respectively. An HSQC dataset was collected with 789 × 400 complex points and acquisition times of 0.2 and 0.03 seconds in F2 and F1, respectively. Assignments are listed in Table 2. A second high-resolution HSQC spectrum was collected with the carbon decoupler turned off to allow a long acquisition time of 0.7 seconds during T2, in order to obtain proton-proton coupling data. 1D transient nOe experiments with mixing times of 100 to 800 ms were performed on the H3 axial signals in the Neu5Ac residues (H3ax) to determine the linear nOe buildup regime. Data was then collected, using a 300-ms mixing time, on those signals that had adequate spectral dispersion. Additional nOe data were obtained by acquiring a 2D NOESY spectrum utilizing a zero-quantum suppression element (43) and a mixing time of 300 ms. Data were collected with 420 × 2048 complex points and acquisition times of 0.459 and 0.09 s in F2 and F1. 1D data were processed with Varian VNMRJ software (Varian, Inc), and 2D data were processed and analyzed using NMRPipe software, (44) while volume integrations were performed with the NMRView software. (45)
Table 2.
Proton chemical shifts of methyl α-(2,8)-di- and trisialoside, acquired at 25°C in D2O, relative to internal DSS.
| Methyl α-(2,8)-disialoside | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H3ax | H3eq | H4 | H5 | H6 | H7 | H8 | H9 | NAc | OMe | |
| b | 1.565 | 2.612 | 3.552 | 3.783 | 3.813 | 3.858 | 4.194 | 4.133, 3.693 | 2.059 | 3.310 |
| a | 1.726 | 2.759 | 3.666 | 3.825 | 3.610 | 3.581 | 3.905 | 3.872, 3.635 | 2.022 | |
| Methyl α-(2,8)-trisialoside | ||||||||||
| c | 1.572 | 2.607 | 3.531 | 3.801 | 3.838 | 3.890 | 4.144 | 4.141, 3.668 | 2.070 | 3.310 |
| b | 1.697 | 2.674 | 3.573 | 3.795 | 3.602 | 3.844 | 4.154 | 4.182, 3.674 | 2.062 | |
| a | 1.739 | 2.753 | 3.656 | 3.833 | 3.601 | 3.573 | 3.928 | 3.890, 3.634 | 2.022 | |
2.1 Chemical shift assignments
Assignments were confirmed with COSY and HSQC data, Table 2–Table 3. Identification of the reducing, middle and terminal Neu5Ac residues was based on the existence of nOes between the H3ax proton and the OMe aglycon, and between the H3ax/eq and H8 protons of the preceding residue, as well as in the relative downfield chemical shift of C8 due to the glycosidic linkage. N-Acetyl methyl groups were assigned based on nOes to protons H5 and H7. Chemical shift data were consistent between I and II as can be seen by comparing the reducing and terminal residues of the di- and trisialosides, Table 2–Table 3.
Table 3.
Carbon chemical shifts of methyl α-(2,8)-di- and trisialoside, acquired at 25°C in D2O. Referenced indirectly to DSS.
| Methyl α-(2,8)-disialoside | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| C3 | C4 | C5 | C6 | C7 | C8 | C9 | NAc | OMe | |
| b | 43.06 | 70.48 | 55.09 | 76.81 | 72.17 | 81.19 | 64.26 | 25.08 | 54.49 |
| a | 43.25 | 71.15 | 54.40 | 75.30 | 70.81 | 73.36 | 65.26 | 24.83 | |
| Methyl α-(2,8)-trisialoside | |||||||||
| C3 | C4 | C5 | C6 | C7 | C8 | C9 | NAc | OMe | |
| c | 42.95 | 70.67 | 55.19 | 76.37 | 71.87 | 80.71 | 64.10 | 25.16 | 54.54 |
| b | 42.88 | 70.88 | 55.09 | 76.47 | 72.08 | 81.10 | 64.18 | 25.09 | |
| a | 43.24 | 71.30 | 54.44 | 75.34 | 70.94 | 74.45 | 65.28 | 24.81 | |
2.2 nOe measurements
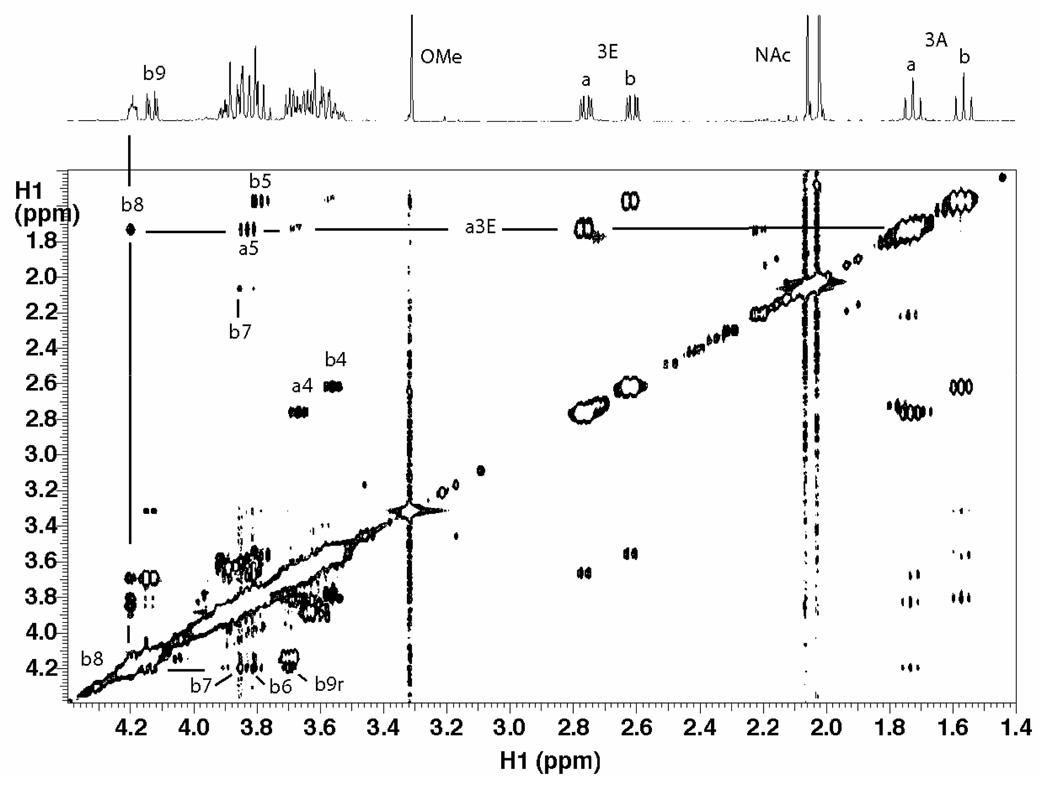
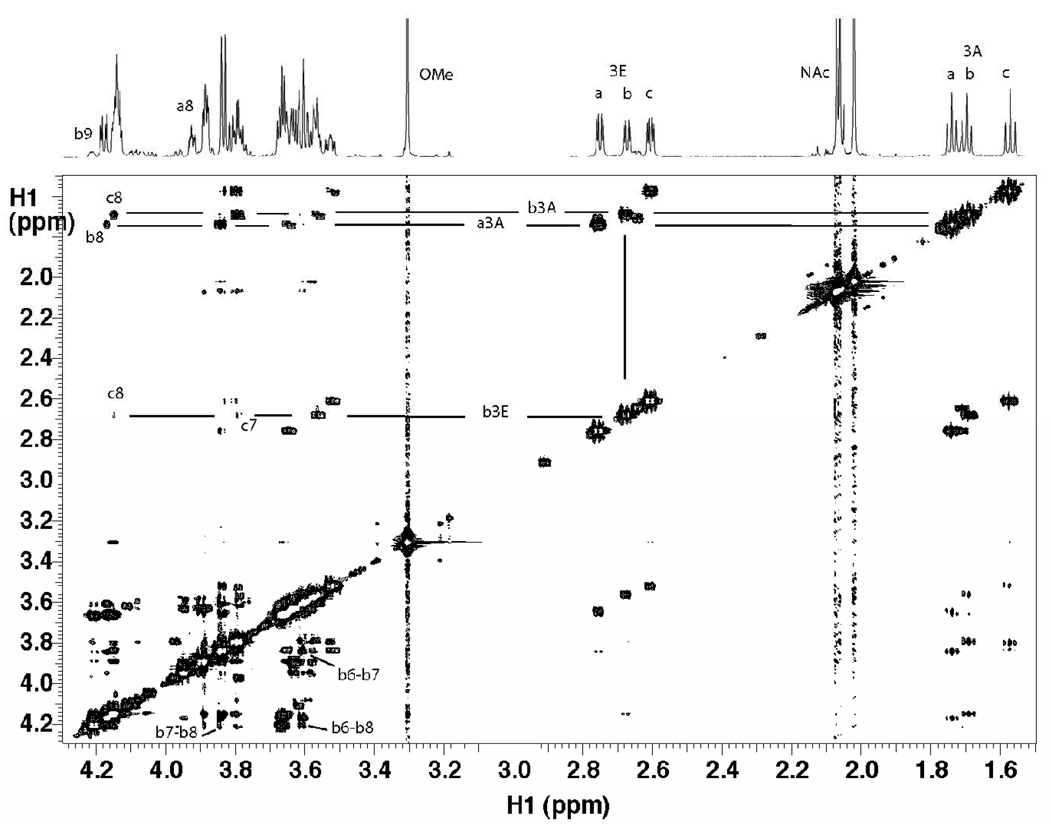
The nOe data were collected at high field (800 and 900 MHz) and 10°C to promote a stronger negative nOe. The 2D NOESY spectra of the disialosyl and trisialosyl haptens are shown in Figure 3 and Figure 4, respectively. Due to the high degree of spectral overlap, only a limited number of nOes could be confidently quantified from the many observed. A 1D selective NOESY spectrum of the trisaccharide, Figure 5, clearly indicated a long-range nOe between protons H9 of the middle residue and H7 of the reducing residue. Peaks in 2D NOESY datasets, processed using NMRView, were integrated by defining boundary boxes around the peaks and summing points within the box according to the NMRView analysis function. 1D nOe data were integrated using standard Varian functions. Integrated values were used for distance calculations based on the isolated spin-pair approximation, wherein the H3eq to H3ax distance was set to 1.79 Å. (9) Cross-peaks originating from the same source proton were then compared for their respective distances.
Figure 3.
NOESY spectrum of disialoside collected at 10°C and 800 MHz with a mixing time of 300 ms. A few cross-peaks used in determining the linkage conformation are indicated.
Figure 4.
NOESY spectrum of trisialoside collected at 10°C and 800 MHz with a mixing time of 300 ms. A few key cross-peaks used in determining the linkage conformation are indicated. Notably, weak cross-peaks between the equatorial H3 proton of the middle residue (bH3eq) and H7 and H8 of the reducing residue (cH7 and cH8) can be observed. Despite substantial overlap of signals, distinct cross-peaks can be measured for some ring protons (e.g. bH6-bH8) of the middle residue.
Figure 5.
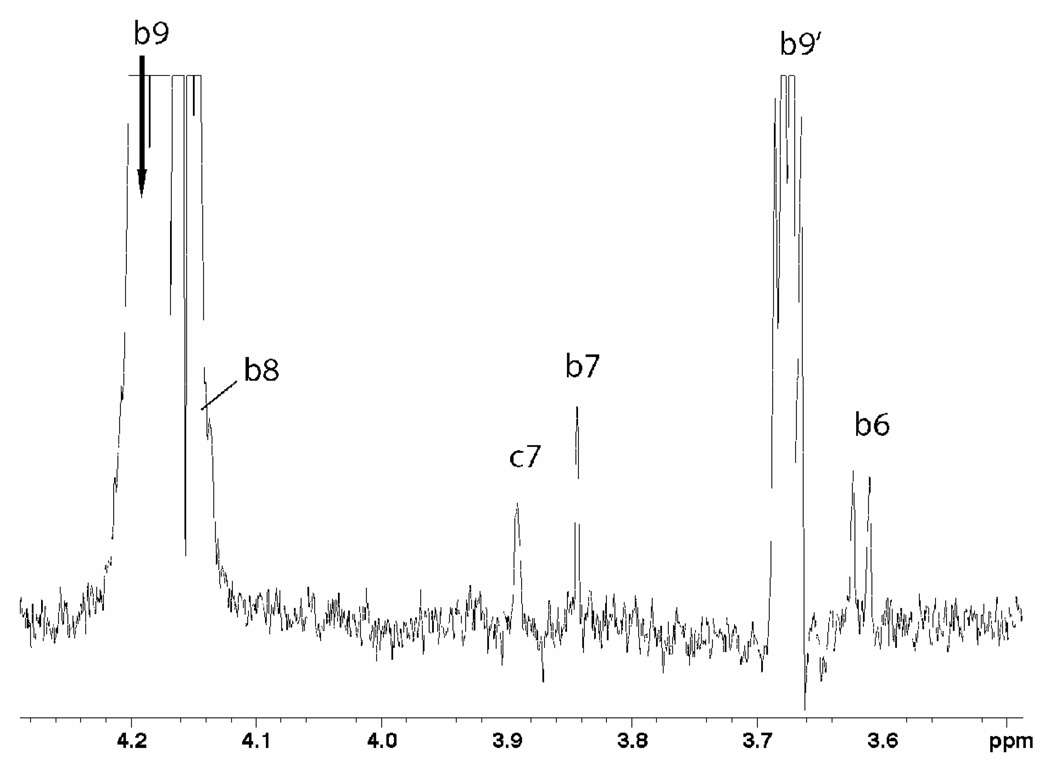
The 1D nOe of trisialoside. Expected intra-residue nOes from proton H9 of the middle residue (bH9) to adjacent protons (bH6, bH7, bH8, bH9’) are shown, in addition to a long-range nOe to the H7 of the reducing residue (cH7).
2.3 J-coupling measurements
Extracting accurate scalar coupling constants from strongly coupled protons frequently found in carbohydrates can be achieved by a combination of selective 1D TOCSY spectra, followed by a simulation of the resulting spin system. In the NmB haptens, selective TOCSY methods were limited by the poor transfer from protons H6 to H7, and from overlapping resonances that prevented access to the important H7, H8 and H9 protons. Thus a 2D HSQC spectrum was acquired with high digital resolution in the proton dimension. The high-resolution HSQC data were Fourier transformed with additional linear prediction and zero-filling in T2 and T1 in order to maximize digital resolution to 1.0 Hz per point. Traces through the carbon-proton peaks provided, in most cases, proton sub-spectra showing first-order coupling. J-values were measured directly from these traces.
3. Results and Discussion
3.1 NMR and conformational properties
The conformational properties of the α-(2,8)-linked Neu5Ac residues are largely determined by the rotatable bonds between the pyranose rings. (9, 46) Previous MD simulations of I indicated some pyranose ring interconversions from the predominant 2C5 chair conformation, (29) which were rationalized only via non-aqueous phase experimental and molecular mechanics studies. However, there were no indications that these ring fluctuations influenced the conformational properties of the dimer. (29) The predominant ring configuration was determined from the scalar 3JHH-values of the ring protons. In the 2C5 conformation the dihedral angles between protons H3ax-H4, H4-H5 and H5-H6 are anti, while that between H3eq-H4 is gauche. These rotamer orientations give rise to high and low magnitude 3J-values for protons in the anti- and gauche-orientations, respectively. The theoretical scalar J-coupling constants were computed employing a Karplus equation that takes into account the electronegativities and orientations of substituent groups. (47) For example, in residue (a) of the disialoside, values for anti-orientations of 11.7 ± 0.7 Hz, 9.4 ± 0.8 Hz and 10.4 ± 0.5 Hz for the H3ax-H4, H4-H5 and H5-H6 protons, respectively, computed from the MD simulation were in good agreement with the experimental values of 12.2 ± 0.1 Hz, 10.2 ± 0.1 Hz and 10.2 ± 0.1 Hz, respectively. A value of 4.6 ± 1.2 Hz was computed for the gauche H3eq-H4 proton pair from the MD data, which also showed good agreement with the experimental value of 4.4 ± 0.4 Hz. A complete list of the pyranosyl 3JHHvalues is presented in Table S1.These J-values indicated that the pyranosyl rings remained entirely in the 2C5 conformation although the ring torsion angles displayed modest oscillations of approximately (±15°). Therefore, the conformational properties of the methyl α-(2,8)-di- and trisialosides were characterized here by monitoring the time-dependent evolution or populations of the φ, ψ, ω8 and ω7 angles (see Figure 1) during the MD and REMD simulations.
In order to ascertain whether the results from the present simulations were consistent with experiment, and so could be relied on to shed some light on the conformational properties of the strain B haptens, inter-proton distances and scalar 3JHH-coupling constants were computed from the MD trajectories and compared with experimental data determined herein.
3.1.1 Methyl α-(2,8)-disialoside (a-b-OMe)
The experimental NMR coupling constants and those computed from the MD simulations of I are presented in Table 4. It can be seen that the values derived from the MD and REMD simulations were statistically indistinguishable, and the back-calculated 3JHH values showed good overall agreement with the experimental coupling constants. In terms of conformational properties, the small experimental 3JbH6-bH7 and 3JbH7-bH8 values of <1.0 and 1.5 Hz, respectively, for these sets of protons in the internal reducing Neu5Ac residue, were consistent with a ±gauche orientation, due to the symmetry of the Karplus relationship. (47) The precise rotamer assignment required the utilization of nOe data, Table 5. The strong nOes measured between the bH8-bH6 and bH8-bH7 protons, confirmed the −gauche and +gauche conformations for the bH6-bC6-bC7-bH7 and bH7-bC7-bC8-bH8 dihedral angles, respectively, which corresponded to a +gauche orientation for both the ω7 (bO6-bC6-bC7-bO7) and ω8 (bO7-bC7-bC8-bO8) angles. In the non-reducing terminal residue, the small magnitude of the 3JaH6-aH7 coupling constant (1.4 Hz) for the glyceryl side chain indicated a +gauche conformation for the ω7 (aO6-aC6-aC7-aO7) angle. In contrast, the large 3JaH7-aH8 value of 9.5 Hz for this residue suggested an anti orientation for these protons, which corresponded to an anti-orientation for this ω8 (aO7-aC7-aC8-aO8) torsion angle. These results are similar to those reported previously and indicate that the orientation of ω8 depends heavily on whether O8 is glycosylated or free. (9, 46) The MD and REMD simulations gave similar nOe values, which compared well with the experimentally-derived distances, Table 5. The inter-glycosidic nOe distances were largely determined by the linkage dihedral angles; therefore the fact that the MD simulations were sampling experimentally consistent torsion space was also supported by the good agreement between the experimental and the MD-derived inter-glycosidic nOe distances. For example the nOe distances measured between the aH3ax–bH8 protons was 2.5 ± 0.8 Å for both the MD and REMD, respectively, which may be compared with the experimental value of 3.0 ± 0.03 Å. The experimentally-derived nOe distance between the aH3eq–bH8 protons was 4.0 ± 0.04 Å, compared to 3.8 ± 0.5 Å for both the MD and REMD simulations.
Table 4.
NMR and computed 3JHH coupling constants (Hz) for inter-residue torsion angles in fragments of NmB sialobioside, α-Neu5Ac-(2-8)-α-Neu5Ac-OMe (a-b).
| Angle | Linkage | Spins | NMR | MD (100 ns)a | REMD (20 ns)a,b |
|---|---|---|---|---|---|
| ω7 | Terminal | aH6-aH7 | 1.4 ± 0.1 | 1.0 ± 0.8 | 1.0 ± 0.8 |
| Internal | bH6-bH7 | < 1.0 | 1.2 ± 0.9 | 1.2 ± 0.9 | |
| ω8 | Terminal | aH7-aH8 | 9.5 ± 0.9 | 7.9 ± 0.6 | 8.1 ± 0.6 |
| Internal | bH7-bH8 | 1.5 ± 0.2 | 2.0 ± 0.7 | 2.0 ± 0.7 | |
| ω9 | Terminal (free rotor) | aH8-aH9R | 6.1 ± 0.6 | 6.7 ± 1.5 | 5.9 ± 1.5 |
| aH8-aH9S | 2.4 ± 0.2 | 3.7 ± 0.9 | 3.6 ± 0.9 | ||
| Internal (free rotor) | bH8-bH9R | 6.1 ± 0.6 | 7.3 ± 1.6 | 7.4 ± 1.7 | |
| bH8-bH9S | 4.1 ± 0.4 | 2.9 ± 1.3 | 2.9 ± 1.3 | ||
Conformation-defining intra-residue J-couplings are shown in bold.
REMD simulations were performed at 8 temperatures for a total of 160 ns.
Table 5.
Experimental (ISPA) NOE distances (Å) and average inter-proton distances from MD and REMD simulations for fragments of NmB sialobioside, α-Neu5Ac-(2-8)-α-Neu5Ac-OMe (a-b).
| Spins | NMR | MD 100 ns | REMD 20ns | Spins | NMR | MD (100ns)a | REMD (20ns)a,b |
|---|---|---|---|---|---|---|---|
| aH3ax – aH3eq | 1.8 ± 0.02 | 1.7 ± 0.1 | 1.7 ± 0.1 | bH8 – bH6 | 2.9 ± 0.03 | 2.5 ± 0.2 | 2.5 ± 0.2 |
| aH3ax – aH4 | 3.4 ±0.04 | 3.0 ± 0.1 | 3.0 ± 0.1 | bH8 – bH7 | 2.5 ± 0.03 | 2.6 ± 0.2 | 2.6 ± 0.2 |
| aH3ax – aH5 | 2.8 ± 0.03 | 2.6 ± 0.2 | 2.7 ± 0.2 | aH3ax – bH8 | 3.0 ± 0.03 | 2.5 ± 0.8 | 2.5 ± 0.9 |
| aH3eq – aH4 | 2.8 ± 0.03 | 2.4 ± 0.1 | 2.4 ± 0.1 | aH3eq – bH8 | 4.0 ± 0.04 | 3.8 ± 0.5 | 3.8 ± 0.5 |
| bH3ax – bH4 | 3.2 ± 0.04 | 3.0 ± 0.1 | 3.0 ± 0.1 | bH5 – bNMe | 4.3 ± 0.05 | 4.5 ± 0.1 | 4.5 ± 0.1 |
| bH3ax – bH5 | 3.0 ± 0.04 | 2.6 ± 0.2 | 2.6 ± 0.2 | bH9R – bH8 | 2.5 ± 0.03 | 2.8 ± 0.3 | 2.8 ± 0.3 |
Conformation-defining intra- and inter-residue nOe’s are shown in bold.
REMD simulations were performed at 8 temperatures for a total of 160 ns.
The distances are averaged over the MD trajectories as <1/R6>(−1/6)
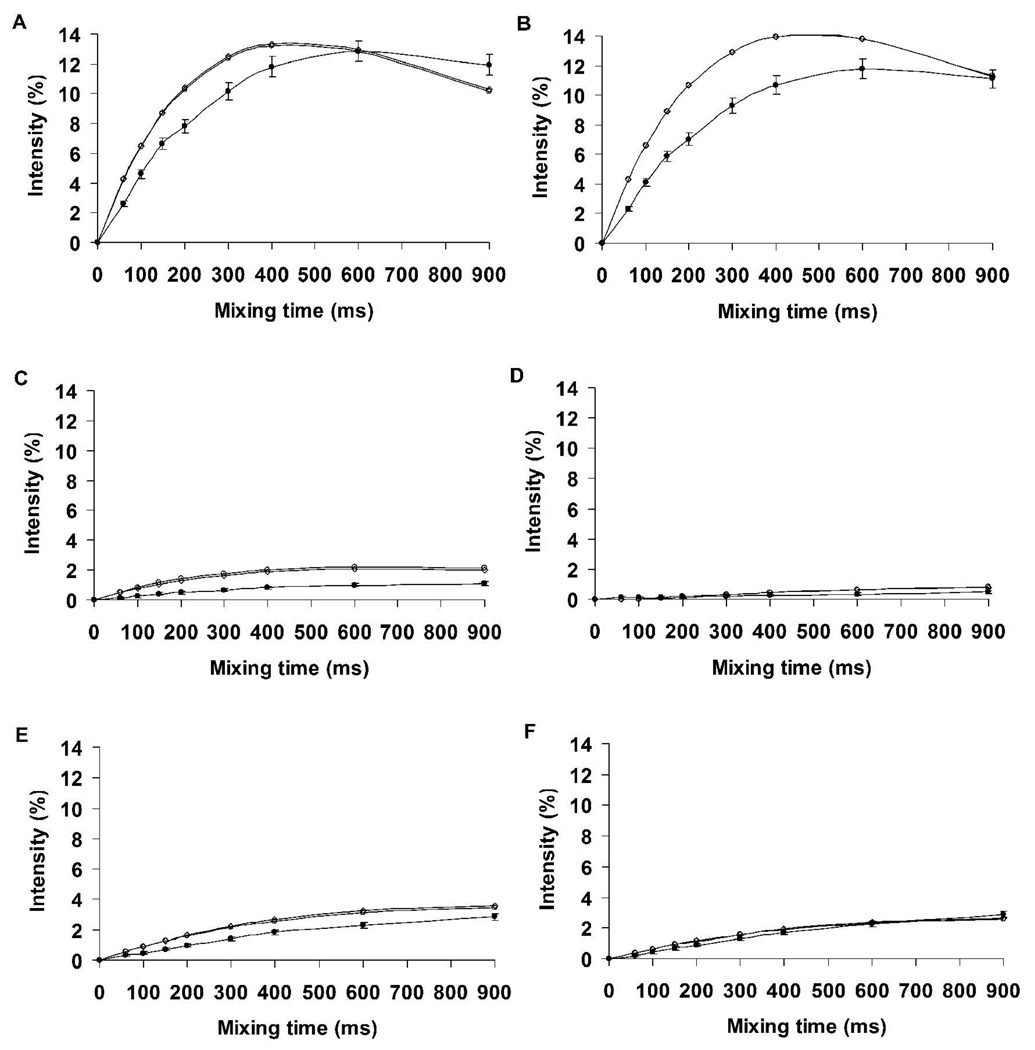
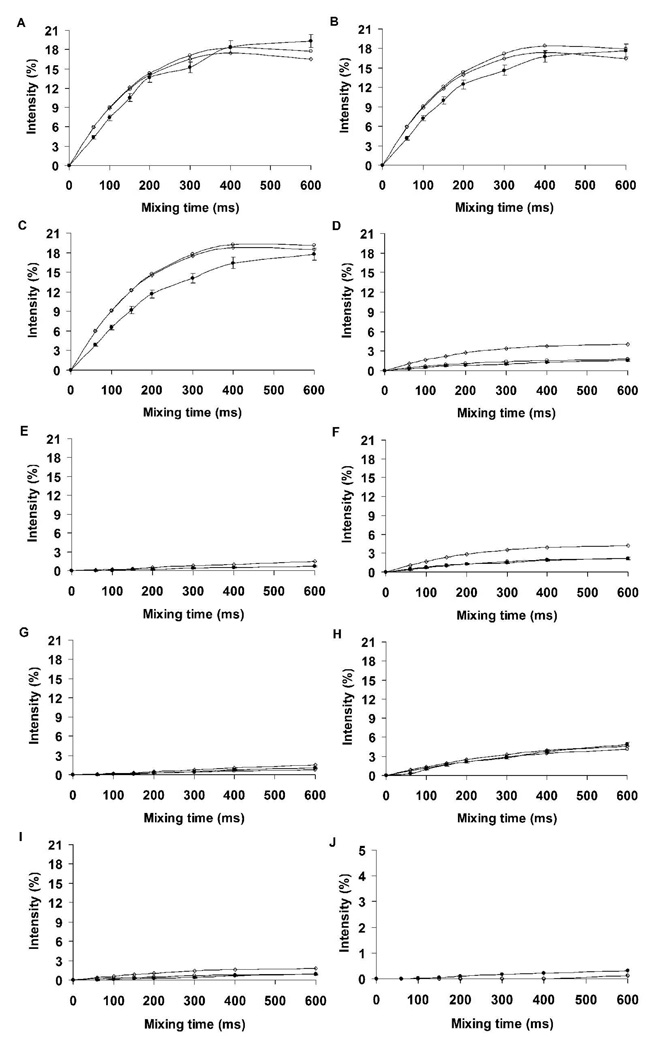
To determine the fit between the experimental and theoretical nOe intensities in I, nOe build-up curves were computed employing a complete relaxation matrix analysis. A single overall correlation time of 0.5 ns for all pairs of protons resulted in good agreement between the experimental and theoretical data, Figure 6. Notably, the theoretical data gave rise to nOe intensities that were in quantitative agreement was excellent for the conformationally significant proton pairs (aH3ax-bH8, aH3eq-bH8, bH8-bH6 and bH8-bH7) over the entire range of mixing times.
Figure 6.
Experimental and theoretical nOe build-up curves of proton pairs in disialoside (a-b-OMe). A, aH3ax-aH3eq; B, bH3ax-bH3eq; C, aH3ax-bH8; D, aH3eq-bH8; E, bH8-bH6; F, bH8- bH7. A correlation time of 0.5 ns was employed in the theoretical predictions. Experiment ●; Conventional MD ○; REMD ◊
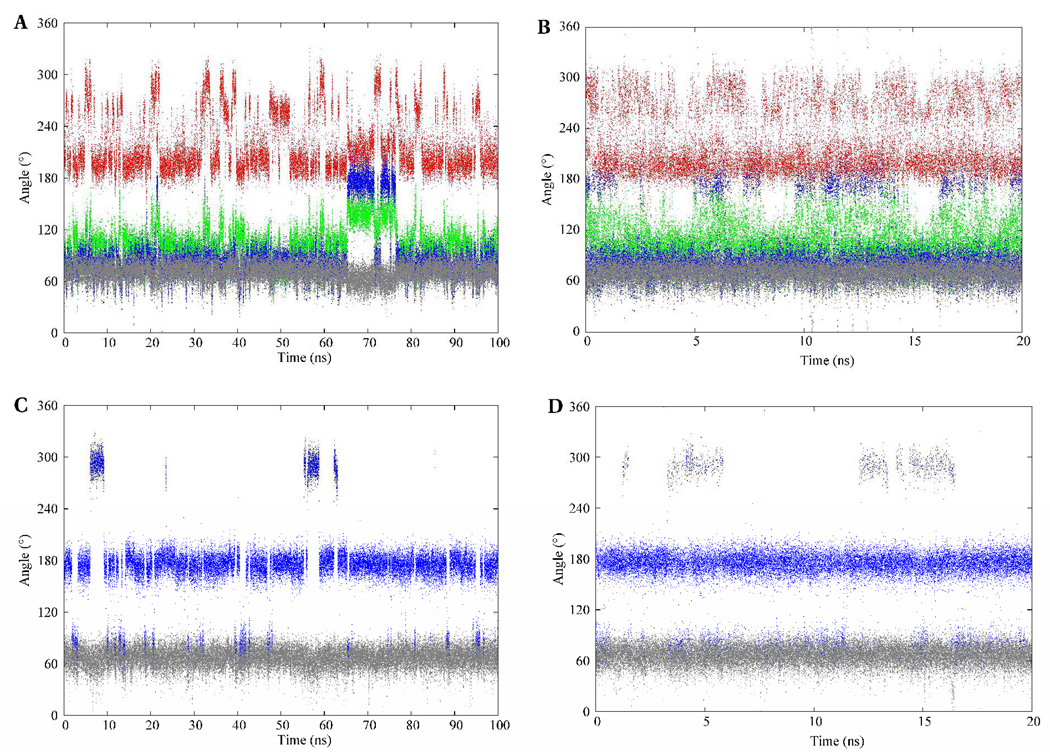
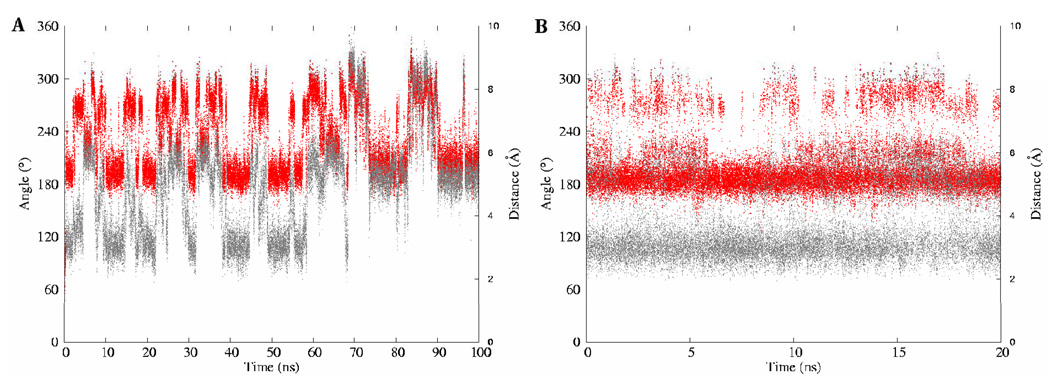
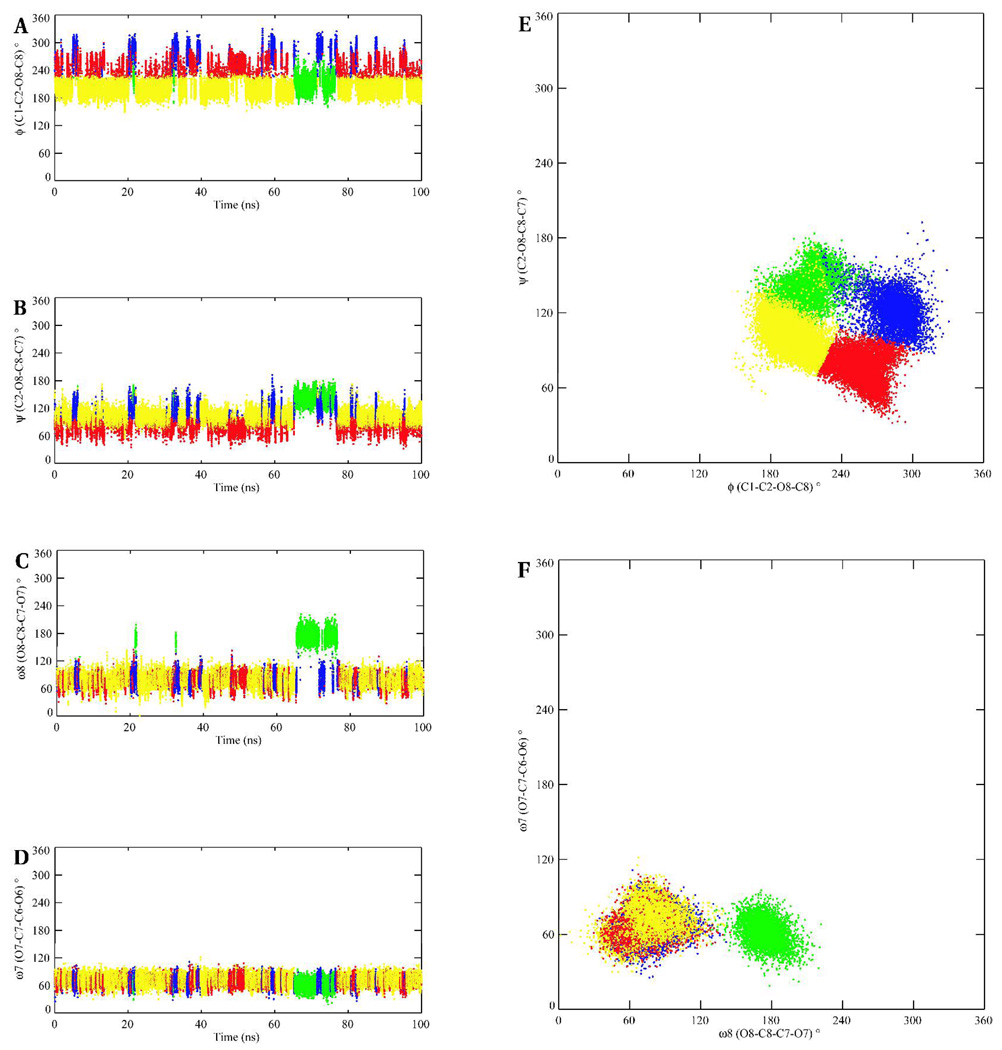
The conformational itineraries of the glyceryl exocyclic dihedral angles from the 100-ns MD simulation of I are presented in Figure 7. Despite the potential for the glycosidic linkage to populate a wide range of conformations, the data from the MD and REMD simulations suggested that well-defined subsets of conformations were preferentially adopted. The ω7 angle predominantly sampled the +gauche rotamer in both Neu5Ac residues. In contrast, the ω8 angle exhibited a preference for several staggered rotamers depending on whether the O8 atom was involved in a glycosidic bond. Consistent with the experimental vicinal 3JH7-H8 coupling constants, the ω8 angle was predicted to exist predominantly in the anti orientation when the O8 atom was not involved in a glycosidic bond, but preferred the gauche orientation otherwise. The predominance of the anti conformation when O8 is non-glycosidically linked, has been observed in X-ray crystal structures (48) and inferred from solution-phase NMR (9, 46, 49) data. Two intra-residue hydrogen bonds, between the oxygen atoms of the carboxylate group with the hydroxyl group at C8, and between the carbonyl group of the acetamido functionality and the hydroxyl group at C7, seem to be the origin of this rotamer’s stability. (49) However, the magnitude of this stabilization is not clear, since a similar anti orientation for the ω8 angle has been observed in the X-ray crystal structures of the β-anomer, (48) in which the formation of the intra-residue hydrogen bond to the carboxylate group is impossible.
Figure 7.
The conformational variations of the exocyclic glyceryl dihedral angles of the methyl α-(2,8)-disialoside. φ – red; ψ – green; ω8 – blue; ω7 – gray. The A–B and C–D panels depict dihedral angle variations of the glyceryl region involved and not involved in glycosidic bond formation, respectively. The left and right columns represent trajectories from the conventional MD and REMD simulations, respectively.
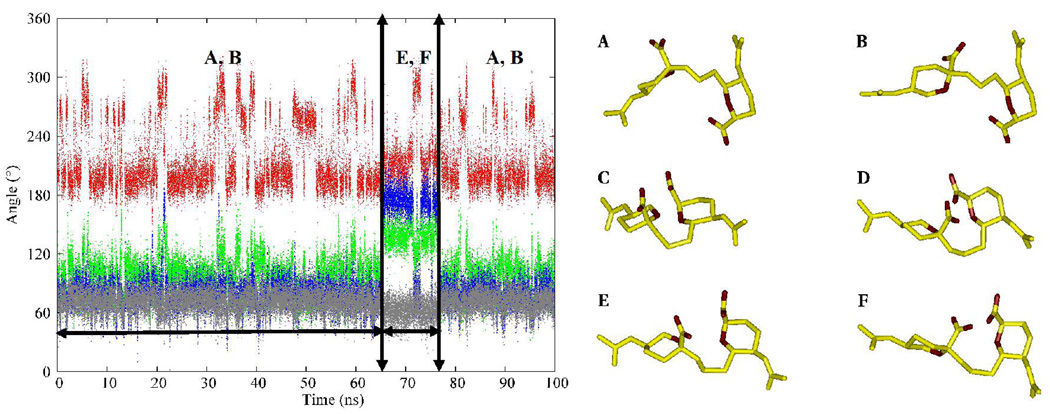
The φ-angle in Neu5Ac residues is known to populate two conformations, −gauche and anti. (49, 50) There are no conclusive experimental data on the solution conformational properties of the ψ angle; however, potential energy calculations suggest that the ψ angle oscillates between 105–130°. (9) Over the course of the MD simulations, correlated motions were observed between the conformational itineraries of the ω8, and φ and ψ dihedral angles of the glycosidic linkage region. During the first 65 ns, the ω8 angle predominantly populated the +gauche rotamer (78 ± 13°), the φ angle displayed transitions between its two solution preferred conformations, (−161 ± 14°, −91 ± 17°), while the ψ angle adopted a value of 98 ± 18°. After 65 ns the ω8 angle effected a transition to the anti rotamer. During the time interval that the ω8-anti rotamer persisted, if the φ angle adopted either the −gauche (−91 ± 17°) or anti (−161 ± 14°) conformations then a +gauche ψ (98 ± 18°) would introduce unfavorable electrostatic repulsions due to the close proximity of the carboxyl groups and ring oxygen atoms on two adjacent Neu5Ac residues, Figure 8. Presumably to alleviate these interactions, the ψ angle flips to the anti rotamer, (142 ± 12°). In general, the preferred conformational preferences of the exocyclic dihedral angles observed during the MD simulations are in accordance with previous reports from experimental NMR studies of the respective angles. (9, 46, 50) The majority of the transitional correlations between the ω8, and φ and ψ angles were also observed in the REMD simulation. Comparisons of the relative distributions of the rotamers of the exocyclic glyceryl side chain computed from experimental J-values, as well as the MD and REMD simulations are shown in Table 6. For the conformationally significant dihedral angles, particularly ω7 and ω8, the theoretical populations showed good agreement with experiment. Only a qualitative agreement was observed between the theoretical and experimental populations for the freely rotating ω9 angle.
Figure 8.
Possible repulsive interactions between the charged carboxyl groups and the O6 ring oxygen atoms on two adjacent residues. (φ, ψ, ω8) = (−161, 98, 78) and (−91, 98, 78) in A and B, respectively, denoting the solution-preferred structures during the first 65 ns of the simulation. (φ, ψ, ω8) = (−161, 98, 175) and (−91, 98, 175) in C and D, respectively, represent unfavorable interactions that would occur if the ω8 angle adopted the anti rotamer while the ψ angle remained in the +gauche (98°) state. To alleviate the repulsive interactions, the ψ angle makes a transition to the anti rotamer, E and F with (φ, ψ, ω8) = (−161, 142, 175) and (−91, 142, 175), respectively.
Table 6.
NMR-based and computed populations (%) for inter-residue torsion angles in fragments of NmB disialoside, α-Neu5Ac-(2-8)-α-Neu5Ac-OMe (a-b).
| Angle | Linkage | NMRa, b gauche/trans/-gauche |
MD (100ns) gauche/trans/-gauche |
REMD (20ns)dgauche/trans/-gauche |
|---|---|---|---|---|
| ω7 | Terminal a | 100/0/0 | 100/0/0 | 100/0/0 |
| ω7 | Internal b | 100/0/0 | 100/0/0 | 100/0/0 |
| ω8 | Terminal a | 0/100c/0 | 13/80/7 | 13/83/4 |
| ω 8 | Internal b | 96 ± 3/3± 3/0 | 90/10/0 | 90/10/0 |
| ω 9 | Terminal a | 54 ± 8 /0/50 ± 12 | 61/18/21 | 52/16/32 |
| ω9 | Internal b | 55 ± 8/24 ± 9/20 ± 15 | 73/2/25 | 75/4/21 |
Range is consistent with the estimated error in the experimental J-values, only one symmetric solution is presented.
NMR populations derived by employing limiting J-values from rotational isomeric states computed from conventional MD (the REMD simulations did not give significantly different limiting J-values).
Estimated based on the magnitude of the J-value.
REMD simulations were performed at 8 temperatures for a total of 160 ns.
3.1.2 Methyl α-(2,8)-trisialoside (a-b-c-OMe)
Methyl α-(2,8)-trisialoside (II) was included in this study to investigate the effects of multiple Neu5Ac residues on the conformational properties of the 2,8-linkage. To the best of our knowledge only two experimental studies have been carried out on the trisaccharide hapten. (9, 46) In those studies the trisaccharide fragment terminated in the β-configuration, limiting the relevance of those data. The vicinal homonuclear 3JHH-coupling constants in II, determined experimentally and those from our MD simulations, are presented in Table 7. The observed 3JH6-H7 constants for all the Neu5Ac residues from the experiment were less than 1.5 Hz, which indicated a gauche relationship between protons H6 and H7. The average 3JH6-H7 values computed from the MD-simulation were also of low magnitude, and were in excellent agreement with the experimental values. As in the case of the disaccharide, the large experimental 3JaH7-aH8 value of 9.6 Hz indicates that these protons are predominantly in the anti orientation for the non-reducing terminal residue. The MD-computed value for 3JaH7-aH8 (7.7 ± 0.6 Hz) also suggested a predominantly anti orientation, which upon further analysis was confirmed to be 78% anti and 22% gauche. In contrast to the large 3JaH7-aH8 values observed in the non-reducing terminal residue, the 3JH7-H8 values for the internal (b) and reducing-internal (c) residues, in which O8 is involved in glycosidic bonds, were less than 4.0 Hz, indicating a conformational change to the gauche orientation. That a gauche rotamer was preferred for the a-b and b-c linkages, respectively, was also demonstrated by the relatively small magnitude of the 3JH7-H8 coupling constants observed in the MD data (2.1 ± 0.7 and 2.1 ± 0.8 for the MD and REMD simulations, respectively). In addition, the small-magnitude of the 3JH7-H8 values for the residues b and c imply similarities between the conformational properties of the ω8 angle in both the a-b and b-c linkages. In previous experimental NMR studies of the Neu5Ac trisaccharide (9, 46) the anomeric center of the reducing-terminal Neu5Ac residues was in the β-configuration, which led to the conclusion that the conformational properties of the ω8 angle in the two linkage regions differed. (46) Here, for the all α-configuration, we see that this is not the case. Good agreement was also obtained between the experimental and MD-computed values for the 3JH8-H9R and 3JH8-H9S couplings; however, the rotameric preferences of the C8-C9 bond are not directly pertinent to the overall 3D conformational properties of the CPS.
Table 7.
Conformationally sensitive NMR and computed 3JHH coupling constants (Hz) for inter-residue torsion angles in fragments of NmB trisialoside, α-Neu5Ac-(2-8)-α-Neu5Ac-(2-8)-α-Neu5Ac-OMe (a-b-c).
| Angle | Linkage | Spins | NMR | MD (100 ns)a | REMD(20ns)a |
|---|---|---|---|---|---|
| ω7 | Terminal | aH6-aH7 | 1.5 ± 0.2 | 1.0 ± 0.8 | 1.0 ± 0.8 |
| Internal | bH6-bH7 | < 1.0 | 0.9 ± 0.8 | 0.9 ± 0.7 | |
| Internal | cH6-cH7 | < 1.0 | 1.1 ± 0.9 | 1.3 ± 1.0 | |
| ω8 | Terminal | aH7-aH8 | 9.6 ± 1.0 | 7.7 ± 0.6 | 8.1 ± 0.5 |
| Internal | bH7-bH8 | < 4.0 | 3.6 ± 0.9 | 1.9 ± 0.9 | |
| Internal | cH7-cH8 | < 4.0 | 2.1 ± 0.7 | 2.1 ± 0.8 | |
REMD simulations were performed at 8 temperatures for a total of 160 ns.
nOe distances were computed from the MD and REMD simulations of the trimer and compared with the available experimental data, Table 8. Unlike the case of the dimer, the agreement with the experimental values in the trisaccharide was only qualitative, suggesting a need for longer sampling time. The nOe values from the REMD simulation were in better agreement relative to the MD simulation, except for one case, bH3eq–cH7. It should be noted that a very weak (<1%) long-range nOe was observed between protons H9 of the internal residue (b) and H7 of the reducing terminal residue (c), Figure 5. The ISPA-derived experimental distance between these protons was 3.1 ± 0.06 Å. Given that the bH9 protons were not unambiguously identified, the center of mass of the bH9R and bH9S protons was employed to determine the bH9-cH7 distance from the MD trajectories. Inter-proton distances of 3.0 ± 1.4 and 3.5 ± 2.1 Å were obtained from the REMD and MD data, respectively. Further investigations revealed that the bH9-cH7 distance was highly sensitive to the φ-angle of the b-c linkage, Figure 9. In particular, these protons were within the range of a detectable nOe (<3.5 Å) only when the φ-angle adopted the anti rotamer. Since the MD data displayed a mixture of both the anti and gauche rotamers for this angle, the reproduction of the bH9-cH7 distance can be utilized as an indicator of whether a simulation is correctly sampling the φ-angle rotamers in polysialic acids. The average distances between protons bH9 and cH7, computed from the first 70 ns of the conventional MD simulation for the periods when the φ-angle is anti (52%) or −gauche (48%) were 3.3 ± 1.8 Å and 5.3 ± 1.5 Å, respectively. Using these values the experimental distance of 3.1 Å equates to a ratio of anti:−gauche rotamers of approximately 100:0. The MD simulation therefore underestimates the population of the anti rotamer, leading to a slightly long value for the bH9-cH7 distance. Despite the marked improvement of the nOe-consistent population during the REMD simulation, these observations suggest a possible need for improvement in the force field parameterization for the φ-angle in Neu5Ac. Nonetheless, the better performance of the REMD simulation indicates that enhanced-sampling methods can be effectively employed in studying the conformational properties of larger fragments of polysaccharides. Theoretical and experimental nOe build-up intensities in II, computed with the utilization of a complete relaxation matrix are shown in Figure 10. The subtle variations in the bH9-cH7 nOe notwithstanding, a correlation time of 0.55 ns resulted in excellent overall agreement between the experimental and theoretical data for all pairs of protons. Rotamer populations computed from the experimental J-values and directly from the MD simulations are shown in Table 9. In general, the agreement with experiment was excellent with the exception of the ω8 populations of the internal linkages, for which the theoretical data, nevertheless, correctly ranked the rotameric preferences.
Table 8.
Experimental (ISPA) NOE distances (Å) and average inter-proton distances from MD and REMD simulations for fragments of NmB trisialoside, α-Neu5Ac-(2–8)-α-Neu5Ac-(2-8)-α-Neu5Ac-OMe (a-b-c).
| Spins | NMR | MD (100 ns)a | REMD (20 ns)a, b | Spins | NMR | MD (100 ns)a | REMD (20 ns)a, b |
|---|---|---|---|---|---|---|---|
| bH3ax – cH8 | 2.6 ± 0.06 | 2.6 ± 1.0 | 2.3 ± 0.8 | bH6 – bH8 | 2.3 ± 0.05 | 2.5 ± 0.4 | 2.4 ± 0.2 |
| bH3eq – cH8 | 3.5 ± 0.08 | 3.9 ± 0.5 | 3.6 ± 0.5 | aH3ax – bH8 | 2.8 ± 0.07 | 2.7 ± 1.1 | 2.3 ± 0.8 |
| bH3eq – cH7 | 3.9 ± 0.09 | 4.6 ± 0.4 | 5.0 ± 0.5 | aH3eq – bH8 | 3.4 ± 0.08 | 4.0 ± 0.5 | 3.6 ± 0.4 |
| bH6 – bH7 | 2.2 ± 0.05 | 2.4 ± 0.1 | 2.4 ± 0.1 | bH9 – cH7c | 3.1 ± 0.06 | 3.5 ± 2.1 | 3.0 ± 1.4 |
Conformationally insensitive nOe’s for the trisialoside were similar to the disialoside values, and are omitted.
REMD simulations were performed at 8 temperatures for a total of 160 ns.
Given that the bH9 protons were not unambiguously identified, the center of mass of the bH9R and bH9S protons was employed to determine the bH9-cH7 distance from the MD trajectories.
The distances are averaged over the MD trajectories as <1/R6>(−1/6)
Figure 9.
Correlation between the bH9-cH7 inter-proton distance (gray) and the rotameric properties of the φ-angle (red) of the b-c linkage in the trisaccharide (a-b-c-OMe). Traditional MD (A); Replica exchange MD (B).
Figure 10.
Experimental and theoretical nOe build-up curves for proton-pairs in the trisaccharide. A, aH3ax-aH3eq; B, bH3ax-bH3eq; C, cH3ax-cH83eq; D, aH3ax-bH8; E, aH3eq-bH8; F, bH3ax-cH8; G, bH3eq-cH8; H, bH8-bH6; I, bH9-cH7; J, bH3e-cH7. A correlation time of 0.55 ns was employed in predicting the theoretical nOe intensities. Experiment ●; Conventional MD ○; REMD ◊
Table 9.
NMR-based and computed populations (%) for inter-residue torsion angles in fragments of NmB trisialoside, α-Neu5Ac-(2-8)-α-Neu5Ac-(2-8)-α-Neu5Ac-OMe (a-b-c).
| Angle | Linkage | NMRa, b gauche/trans/-gauche |
MD(100 ns) gauche/trans/-gauche |
REMD (20 ns)d gauche/trans/-gauche |
|---|---|---|---|---|
| ω7 | Terminal a | 100/0/0 | 100/0/0 | 100/0/0 |
| ω7 | Internal b | 100/0/0 | 100/0/0 | 100/0/0 |
| ω7 | Internal c | 100/0/0 | 100/0/0 | 100/0/0 |
| ω8 | Terminal a | 0/100c/0 | 15/74/7 | 14/82/4 |
| ω8 | Internal b | 60 ± 9 / 25 ± 3 / 0 | 70 / 30 / 0 | 92/8/0 |
| ω8 | Internal c | 66 ± 5 / 31 ± 9 / 0 | 89 / 11 / 0 | 82/10/8 |
Range is consistent with the estimated error in the experimental J-values, only one symmetric solution is presented.
NMR populations derived by employing limiting J-values from rotational isomeric states computed from conventional MD (the REMD simulations did not give significantly different limiting J-values).
Estimated based on the magnitude of the J-value.
REMD simulations were performed at 8 temperatures for a total of 160 ns.
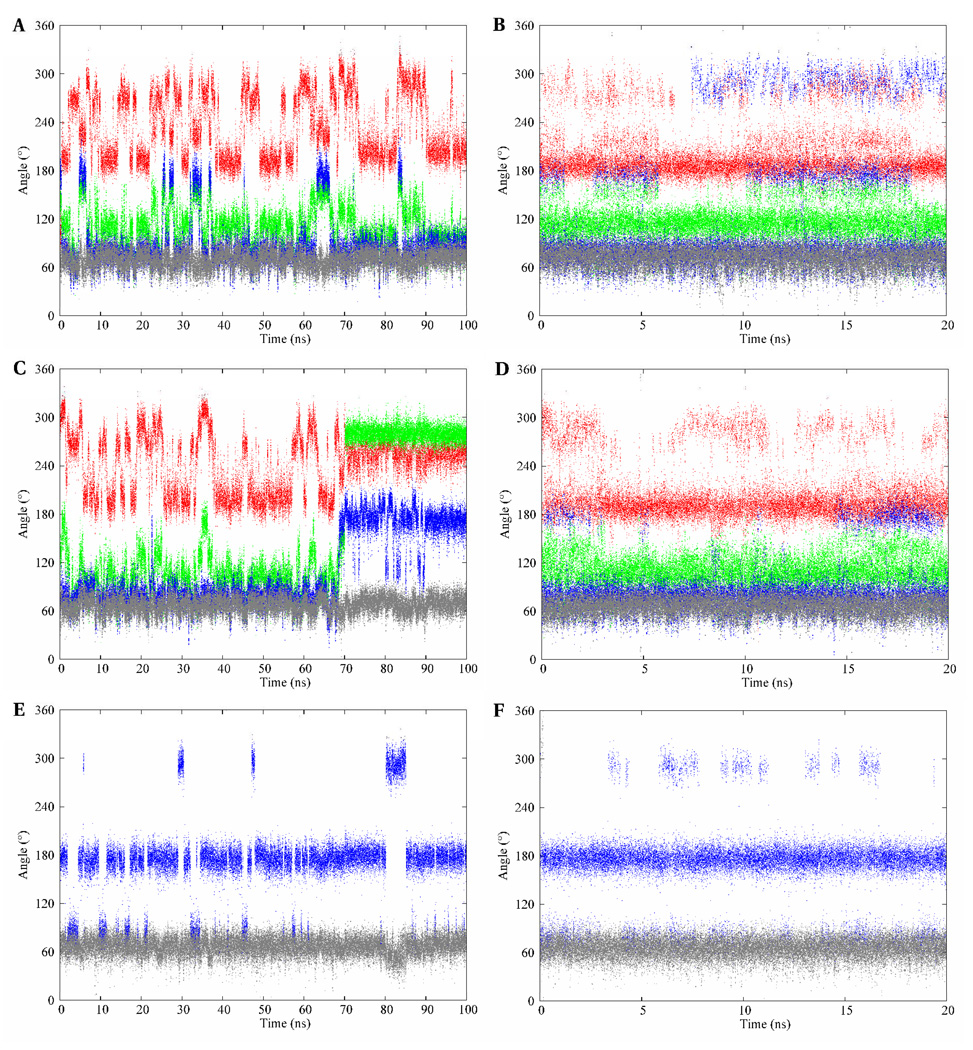
The time series for the rotational angles of the glyceryl side chain in II over the 100-ns simulation are presented in Figure 11. In all three Neu5Ac residues the ω7 angle populated only the +gauche rotamer. The ω8 angle predominantly adopted the anti conformation in the glyceryl side chain of the non-reducing terminal residue, while in the internal and reducing-terminal residues the +gauche rotamer was preferred. Over the course of the simulation, the ψ angle oscillated around 101 ± 30° and 109 ± 27° for the a-b and b-c linkages, respectively, while the φ angle frequently populated the anti and −gauche rotamers in both linkage regions. As in the case of the linkage properties of I, correlated motions were observed between the ω8, and φ and ψ dihedral angles, which were strongly evident for the terminal (a-b) linkage in II. During the first 70 ns, the ω8 angle predominantly populated the +gauche (76 ± 12°) rotamer and the φ angle displayed transitions between its two solution preferred conformations (−159 ± 14° and −84 ± 19°). After 70 ns the ω8 dihedral angle effected a transition to the anti (174 ± 15°) rotamer. The presumed unfavorable electrostatic repulsions, discussed earlier in the disaccharide, again resulted in the ψ and φ angles effecting conformational transitions to their respective −gauche rotamers, −80 ± 8° and −104 ± 14°, respectively. However, unlike in the case of the disaccharide, these rotamers persisted for the remainder of the simulation. Despite the plausible explanation for the existence of these rotamers observed during the last 30 ns of the conventional MD simulation, we sought to determine whether they represented experimentally relevant conformations.
Figure 11.
The conformational variations of the exocyclic glyceryl dihedral angles of the methyl α-(2,8)-trisialoside. φ – red; ψ – green; ω8 – blue; ω7 – gray. The top, middle and bottom panels represent trajectories from the reducing, internal, and terminal glyceryl side chains, respectively. The left and right columns represent the conventional MD and REMD simulations, respectively.
Therefore, J-values and inter-proton distances were computed pre- and post-conformational transition of the ω8 angle, the first 70 and last 30 ns, respectively. A closer observation of the J-values computed from the conventional MD and REMD simulations indicated that the J-values were statistically similar except for the 3JbH7-bH8 values, where the φ, ψ and ω8 transitions occurred. The value computed from the conventional MD simulation was about a factor of two (3.6 ± 0.9 Hz) greater than that determined from the REMD simulation (1.9 ± 0.9 Hz). The higher value may be due to contributions from the anti rotamers of ω8. The J-values computed before and after the conformational transition are presented in Table 10. These values are statistically similar and in good agreement with the experimental data except the 3JbH7-bH8 values that are 1.4 ± 0.8 Hz and 8.7 ± 0.1 Hz for the first 70 and last 30 ns, respectively. The large value of 8.7 ± 0.1 Hz suggests that the last 30 ns may not be entirely experimentally relevant. However, the value of 1.4 ± 0.8 Hz is still small relative to the experimental value of < 4.0 Hz, suggesting that the conformational properties of the ω8 angle may be described by contributions from both the +gauche and anti rotamers, with a predominance of the +gauche rotamer.
Table 10.
NMR and computed 3JHH coupling constants (Hz) for inter-residue torsion angles in fragments of NmB trisialoside during the first 70 ns and last 30 ns of the conventional MD simulation.
| Pre-conformational transition (the first 70 ns) | ||||
|---|---|---|---|---|
| Angle | Linkage | Spins | NMR | MD |
| ω7 | Terminal | aH6-aH7 | 1.5 ± 0.2 | 1.0 ± 0.8 |
| Internal | bH6-bH7 | < 1.0 | 0.9 ± 0.7 | |
| Internal | cH6-cH7 | < 1.0 | 1.1 ± 0.9 | |
| ω8 | Terminal | aH7-aH8 | 9.6 ± 1.0 | 7.6 ± 0.6 |
| Internal | bH7-bH8 | < 4.0 | 1.4 ± 0.8 | |
| Internal | cH7-cH8 | < 4.0 | 2.4 ± 0.7 | |
| Post-conformational transition (the last 30 ns) | ||||
| Angle | Linkage | Spins | NMR | MD |
| ω7 | Terminal | aH6-aH7 | 1.5 ± 0.2 | 1.0 ± 0.8 |
| Internal | bH6-bH7 | < 1.0 | 1.0 ± 0.8 | |
| Internal | cH6-cH7 | < 1.0 | 1.2 ± 0.9 | |
| ω8 | Terminal | aH7-aH8 | 9.6 ± 1.0 | 7.8 ± 0.6 |
| Internal | bH7-bH8 | < 4.0 | 8.7 ± 0.1 | |
| Internal | cH7-cH8 | < 4.0 | 1.4 ± 0.7 | |
The inter-proton distances prior to and after the conformational transitions are presented in Table 11. It should be noted that the inter-proton distances were statistically indistinguishable in the b-c linkage for both segments of the trajectory. However, notable differences were observed in the a-b linkage. For example, the aH3ax-bH8 distance during the first 70 ns was 2.6 ± 1.0 Å, which compared well with the experimental value of 2.8 ± 0.07 Å, relative to the distance of 4.1 ± 0.2 Å computed during the last 30 ns. The bH6-bH8 and aH3eq-bH8 were also in better agreement with the experimental data during the first 70 ns of the simulation compared to the last 30 ns. Coupled with the J-values, these inter-proton distances indicate that during the last 30 ns of the conventional MD simulation experimentally non-relevant conformations were sampled.
Table 11.
Experimental (ISPA) NOE distances (Å) and average inter-proton distances from MD and REMD simulations for fragments of NmB trisialoside, during the first 70 ns and last 30 ns of the conventional MD simulation.
| Pre-conformational transition (the first 70 ns) | |||||
|---|---|---|---|---|---|
| Spins | NMR | MD | Spins | NMR | MD |
| bH3ax – cH8 | 2.6 ± 0.06 | 2.7 ± 1.0 | bH6 – bH8 | 2.3 ± 0.05 | 2.4 ± 0.2 |
| bH3eq – cH8 | 3.5 ± 0.08 | 3.9 ± 0.5 | aH3ax – bH8 | 2.8 ± 0.07 | 2.6 ± 1.0 |
| bH3eq – cH7 | 3.9 ± 0.09 | 4.6 ± 0.4 | aH3eq – bH8 | 3.4 ± 0.08 | 3.9 ± 0.5 |
| bH6 – bH7 | 2.2 ± 0.05 | 2.4 ± 0.1 | bH9 – cH7 | 3.1 ± 0.06 | 3.3 ± 1.8 |
| Post-conformational transition (the last 30 ns) | |||||
| Spins | NMR | MD | Spins | NMR | MD |
| bH3ax – cH8 | 2.6 ± 0.06 | 2.6 ± 1.0 | bH6 – bH8 | 2.3 ± 0.05 | 2.9 ± 0.3 |
| bH3eq – cH8 | 3.5 ± 0.08 | 3.8 ± 0.4 | aH3ax – bH8 | 2.8 ± 0.07 | 4.1 ± 0.2 |
| bH3eq – cH7 | 3.9 ± 0.09 | 4.6 ± 0.4 | aH3eq – bH8 | 3.4 ± 0.08 | 4.5 ± 0.1 |
| bH6 – bH7 | 2.2 ± 0.05 | 2.4 ± 0.1 | bH9 – cH7 | 3.1 ± 0.06 | 5.3 ± 1.5 |
The distances are averaged over the MD trajectories as <1/R6>(−1/6)
4. Conformational clustering
In order to identify unique global conformational states and correlated internal rotations, the trajectories of I were subjected to a clustering analysis. (40) Another key attribute of such an analysis is the accessibility of pertinent information about the interconversion pathways between conformational states, which cannot be easily discerned from traditional torsion angle scatter plots or from time dependent trajectory representations of dihedral angles. The conformer families were generated on the basis of inter-residue dihedral angles: φ, ψ, ω8 and ω7.
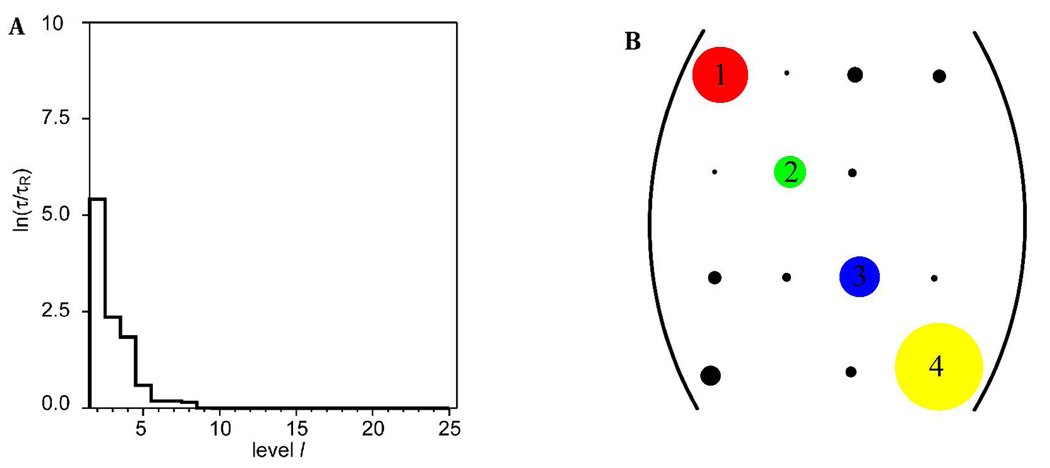
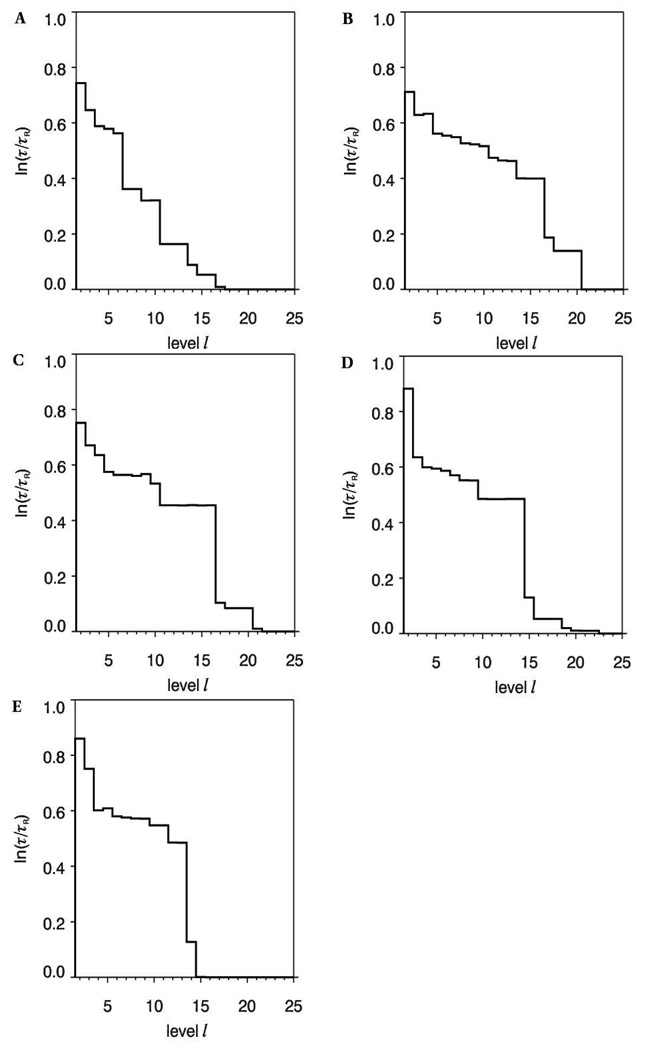
A logarithmic plot of the fastest transition time scales τl (relative to the time scale of the 25 dimensional model τ25) for the hierarchy of Markov states obtained from the trajectory of I is shown in Figure 12A. The largest changes in time scale suggest a four-state model is adequate to describe the conformational space. The representative values of the glycosidic linkages of the four clusters present in the four-state model, and the abundance of each state, are presented in Table 12. Two states differing in more one than dihedral angle will effect transitions through correlated or concerted changes, while states with differences in a single angle undergo interconversions through isolated transitions. Transition probabilities were generally low in the case of strongly coupled or correlated transitions between two or more torsion angles, which may be ascribed to the overall additive barrier to interconversion. Examination of the four-state transition matrix (Figure 12) indicates that state 1 (red) undergoes less frequent transitions to state 3 (blue) than to state 4 (yellow). The principal difference between states 1 and 3 lies in the ψ angles with values of 71 and 128°, respectively. States 1 and 4 differ in the (φ, ψ) domain with values of (−97°, 71) and (−167°, 106°), respectively. Given that there are two concerted transitions in the 1–4 pathway, it might be expected that the 1–3 pathway would be preferred. However, the transitions between states 1 and 4 occur with higher probability. This discrepancy is explained in terms of the low barrier for the transitions between the −gauche and anti rotamers of the φ angle involved in the 1–4 route. State 2 (green) is characterized by an anti (173°) orientation of the ω8 angle, and by an increase in the ψ angle (143°). This is an isolated state, as indicated by the extremely low transition frequency (Figure 12), which is accessed essentially only from state 3 (blue). The reason for its isolation arises in the need for simultaneous changes in ω8 and ψ, to compensate for the electrostatic repulsions that would otherwise result from a single transition in ω8 (Figure 8). The 1–2 pathway involves three concerted transitions (φ, ψ and ω8) and would be disfavored, compared to two concerted changes (φ and ω8) and (ψ and ω8) in the 2–3 and 2–4 routes, respectively. The 2–3 pathway is more probable compared to the 2–4 pathway, because the 2–3 route involves the φ angle oscillating frequently between the −gauche and anti rotamers. Pathways 3–4 and 1–4 both involve changes in the (φ, ψ) domain. However, the 3–4 route occurs with a lower probability. It must be the case that the 3–4 pathway is strongly competing with another pathway. Further examination indicated that state 3 also frequently interchanges with state 1, the (1–3) route involving only the ψ angle. In terms of populations, which are indicated by the number of partition volumes in Table 12, the isolated state 2 accounted for 8% of the conformer distribution. States 1 and 3 accounted for 16 and 12%, respectively, while state 4 was the dominant state, at 64%.
Figure 12.
Plot of the relaxation time-scale, lnτ/τR, between the various hierarchic Markov states identified from the MD-generated trajectories of the methyl α-(2,8)-disialoside, A. A graphical representation of the transition matrix between the Markov states when a four-state model is employed to describe the molecular conformational distribution of the dimer, B. The diameters of the diagonal peaks represent the populations of each state and cross-peaks denote populations of states and transition probabilities, respectively. The sizes of the cross-peaks are related to the additive energy barrier to interconversion.
Table 12.
The conformations (°) of representative members of the four clusters in the four-state Markov model in the methyl α-(2-8)-disialoside
| State | φ(C1C2O8C8) | ψ(C2O8C8C7 | ω8(O8C8C7O7) | ω7(O7C7C6O6) | Populations from partitions volumes (%)a |
|---|---|---|---|---|---|
| 1 (red) | −97 | 71 | 73 | 65 | 16 |
| 2 (green) | −142 | 143 | 173 | 62 | 8 |
| 3 (blue) | −82 | 128 | 78 | 67 | 12 |
| 4 (yellow) | −167 | 106 | 74 | 69 | 64 |
Provides an estimate of what fraction of the data set belongs to a particular conformation. A total of 25 partition volumes were employed.
Plots of the time evolution of the glycosidic angles of the 4-state model during the simulation, and scatter plots of the (φ, ψ) and (ω8, ω7) dihedral angles are presented in Figure 10. During the first 65 ns and the last 20 ns of the simulation interconversions occurred predominantly between states 1 and 4, with much lower sampling of states 2 and 3, respectively. However, within the 65–80 ns interval, states 2 and 3 were preferentially populated. It should be noted that the major pathway from state 2 was via state 3. The scatter plots, Figure 13, indicate that the (φ, ψ)-space is clearly separated into four states for the 4-state model, while the (ω8, ω7) space showed an overlap of states 1, 3 and 4.
Figure 13.
The conformational transition pathway of the four plausible clusters of methyl α- (2,8)-disialoside over the course of the MD simulation, left column. A, B, C and D represent the pathways of the φ, ψ, ω8 and ω7 angles, respectively. The scatter plots of the exocyclic dihedral angles, right column: E – (φ, ψ) and F – (ω8, ω7). The figures employ the color-coding of Figure 12 to indicate the various conformations.
Effects of chemical group modifications
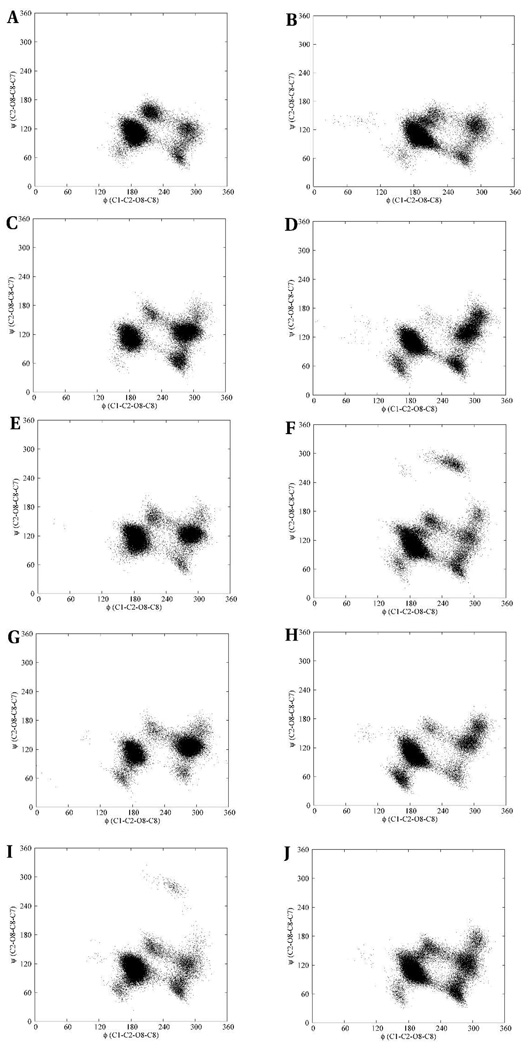
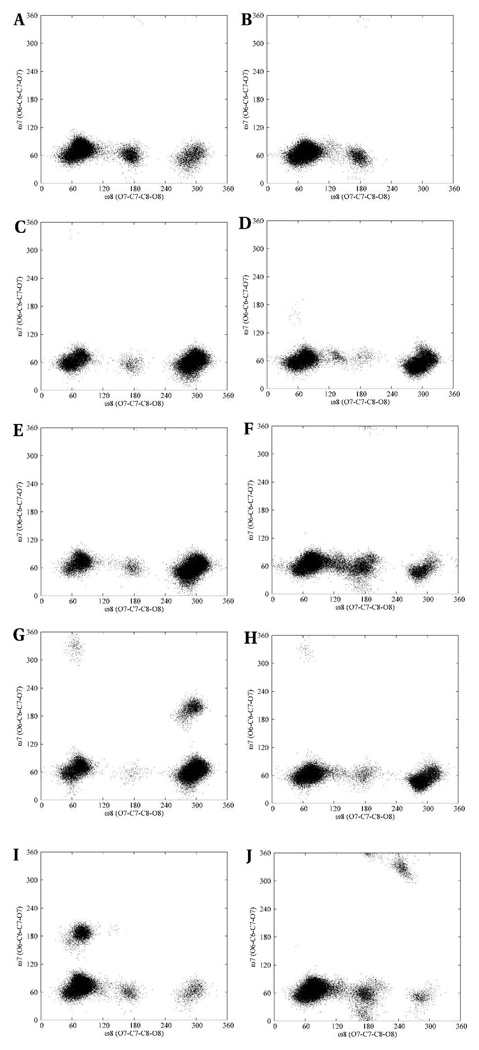
The effects of chemical group modifications on the conformational properties of II were determined by comparing the (φ, ψ) and (ω8, ω7) scatter plots of the linkage dihedral angles with those of the N-Gc, N-Pr, N-Prop and N-But analogs. The multiple conformational distributions in the (φ, ψ) scatter plots indicate a significant degree of flexibility in the trisaccharide hapten and its analogs, Figure 14. For any given trisaccharide the φ and ψ angles for both the reducing and non-reducing terminal residues displayed similar distributions with the exception that the N-Pr and N-But analogs occupied additional (φ, ψ)-spaces in the non-reducing and reducing linkage regions, respectively. Despite these differences in the number of conformations between the reducing and non-reducing linkage regions in each analog, subsets of the phase-space were common to both linkage regions, albeit to varying extents. The (φ, ψ)-space of the native trimer, compared to all the analogs, exhibited similar conformational properties in terms of the number and conformation of the states, except for the N-Pr derivative, which displayed a significant population of a conformation with (φ, ψ)-values both between −120° and −60°. The (ω8, ω7)-space showed less flexibility in all cases, and considerable phase-space overlaps were seen in all the linkages of the trisaccharides, Figure 15. Amongst all the linkages the (ω8, ω7)-space showed more rigidity primarily due to the restricted flexibility of the ω7 angle. However, in the N-Prop and N-But analogs the ω7 angle populated isolated families with (ω8, ω7) values of about (−300, 180) and (60, 180), respectively. These additional conformational families were unexpected, given the preference for ω7 to adopt a +gauche conformation in the native hapten.
Figure 14.
The (φ, ψ) conformational space of the native N-acetyl α-(2,8)-trisialoside and its analogs. The reducing and non-reducing linkages are on the left and right columns, respectively. N-acetyl (A, B), N-glycolyl (C, D), N-propionyl (E, F), N-propyl (G, H) and N-butanoyl (I, J).
Figure 15.
(ω8, ω7) scatter plots of the conformational space of the native N-acetyl α-(2,8)- trisialoside and its analogs. The reducing and non-reducing linkages are on the left and right columns, respectively. N-acetyl (A, B), N-glycolyl (C, D), N-propionyl (E, F), N-propyl (G, H) and N-butanoyl (I, J)
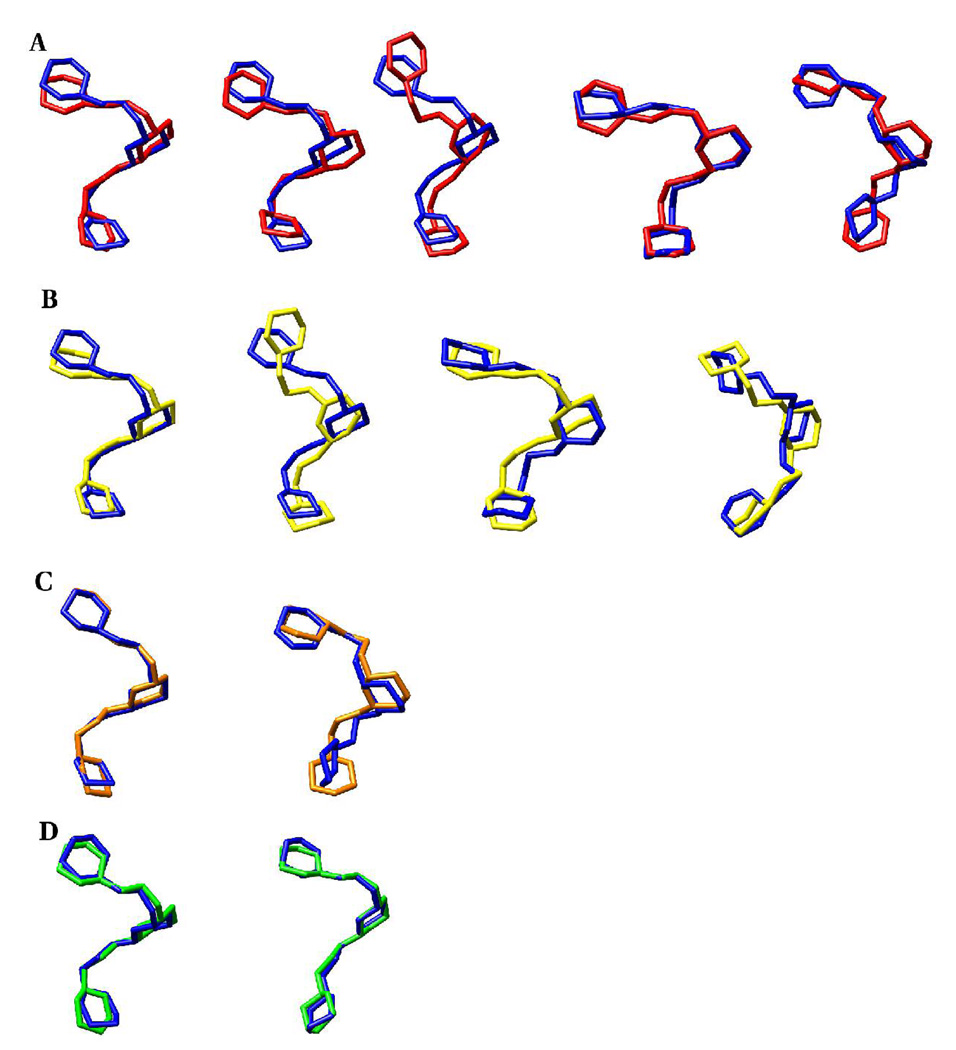
In general the chemical group modifications resulted in similarities in the conformational families in the trisaccharides, although there were conformational states unique to some systems. To provide a structural interpretation for this observation, representative structures of conformational states identified via the Bayesian statistical method were generated. Plots of the relaxation time scales determined for each trajectory, Figure 16, indicated that the following number of states could adequately describe the conformational properties of each trisaccharide: II (10 states), N-Gc (16 states), N-Pr (16 states), N-prop (14 states) and N-But (13 states). The representative conformations of the most populated states of the trisaccharide fragments are presented in Table 13. Based on dihedral angle matches, conformer family A of the N-Ac trimer will have the highest similarity with conformer family A of the N-Gc, N-Pr, N-Prop and N-But analogs, respectively. It is also evident that conformer family B of II and N-But trimers are close structural matches. However, these angle-matching comparisons may fail to identify certain global structural similarities. In particular, MD simulations have indicated that oligosaccharides may display similar overall topologies even though their internal linkages possess different dihedral angles, (51) which are due to correlated transitions between the dihedral angles in the linkages. Therefore, the conformational similarities between the representative structures of the most populated clusters of the analogs and the native trisaccharide were also determined by RMSD comparisons. This approach revealed further structural matches that were not identified initially based on angle-similarities, Table 14 and Figure 17. For example, the A and J conformers of II and the N-Gc analog, respectively, differ in the φ and ω8 angles of the b-c linkage. Despite these differences, they share very similar overall structures, Figure 17. The relative abundance of the states, Table 14, was employed to determine the fraction of the states displaying similar overall topologies. In the case of II and the N-Gc trisialosides, 24% of the NGc conformers (A, C, J) exhibited similar backbone topologies as the A conformer of II (36%).Another 8% of the conformers of II (C), could be mapped to 12% of the N-Gc conformers (C), while 8% of the conformers of II (E), were similar to 8% of the N-Gc conformers (I). Such comparisons were extended to all the other analogs (Table 14).
Figure 16.
Plot of the relaxation time scale, ln(τ/τR) between the hierarchic Markov states identified from the REMD trajectories of II (A), N-Gc (B), N-Prop (C), N-Pr (D) and N-But (E)
Table 13.
Linkage torsion angles of representative members of the predominant clusters populated during the REMD simulations of the native trimer and its N-acetyl substituted analogs (a-b-c-OMe)
| Conformer | a-b | a-b | a-b | a-b | b-c | b-c | b-c | b-c |
|---|---|---|---|---|---|---|---|---|
| φ(C1C2O8C8) | Ψ(C2O8C8C7) | ω8(O8C8C7O7) | ω7(O7C7C6O6) | φ(C1C2O8C8) | Ψ(C2O8C8C7) | ω8(O8C8C7O7) | ω7(O7C7C6O6) | |
| N-acetyl | ||||||||
| A (36%) | −170 | 106 | 78 | 70 | −174 | 115 | 78 | 75 |
| B (8%) | −73 | 132 | 77 | 62 | −174 | 112 | 76 | 74 |
| C (12%) | −159 | 103 | 76 | 65 | −82 | 96 | 76 | 67 |
| D (12%) | −173 | 108 | 73 | 67 | −131 | 147 | −176 | 59 |
| E (8%) | 61 | 121 | 143 | 62 | −164 | 128 | −87 | 65 |
| F (8%) | −157 | 141 | 179 | 54 | −178 | 100 | 87 | 75 |
| N-glycolyl | ||||||||
| A (8%) | −175 | 109 | 65 | 59 | 178 | 115 | −67 | 60 |
| C (8%) | −177 | 90 | 92 | 67 | −115 | 81 | 36 | 60 |
| G (8%) | −64 | 139 | −49 | 59 | −178 | 109 | −73 | 61 |
| H (16%) | −179 | 112 | −67 | 54 | −76 | 125 | −62 | 63 |
| I (8%) | −175 | 109 | 65 | 59 | 178 | 115 | −67 | 60 |
| J (8%) | −171 | 106 | 73 | 65 | −72 | 123 | −67 | 61 |
| K (8%) | −73 | 137 | −93 | 59 | −94 | 131 | −93 | 60 |
| N-propionyl | ||||||||
| A (12%) | −173 | 106 | 76 | 69 | −172 | 114 | 74 | 72 |
| L (8%) | −174 | 133 | 153 | 53 | −175 | 109 | −67 | 60 |
| M (8%) | −169 | 104 | 81 | 71 | −76 | 120 | −70 | 59 |
| N (20%) | −172 | 108 | 73 | 65 | −179 | 121 | −71 | 56 |
| O (8%) | −177 | 113 | −68 | 54 | −73 | 126 | −65 | 62 |
| N-propyl | ||||||||
| A (12%) | −169 | 101 | 78 | 65 | −176 | 112 | 74 | 72 |
| P (12%) | −160 | 108 | −8 | 64 | −81 | 127 | 96 | 62 |
| Q (8%) | −170 | 101 | 77 | 61 | −179 | 117 | −67 | 61 |
| R (16%) | −170 | 101 | 75 | 65 | −75 | 125 | −64 | 62 |
| S (12%) | −177 | 113 | −74 | 47 | −73 | 128 | −61 | 64 |
| T (8%) | −176 | 113 | −75 | 47 | −71 | 132 | −68 | −164 |
| N-butanoyl | ||||||||
| A (32%) | −172 | 113 | 75 | 66 | −173 | 113 | 77 | 76 |
| B (8%) | −74 | 118 | 77 | 64 | −171 | 112 | 76 | 76 |
| U (12%) | −85 | 112 | 131 | 46 | −176 | 96 | 77 | 160 |
| V (8%) | −94 | 71 | 69 | 62 | −174 | 110 | 75 | 74 |
| W (8%) | −174 | 72 | 60 | 57 | −158 | 109 | 53 | 74 |
Table 14.
Backbonea RMSD (Å) comparison between the representative conformations of the native trimer and those of the analogs, populated during the REMD simulations
| N-acetyl | ||||||
|---|---|---|---|---|---|---|
| N-glycolyl | Ab | B | C | D | E | F |
| A | 0.79 | 1.14 | 1.25 | 1.66 | 1.87 | 1.46 |
| C | 1.00 | 1.30 | 0.69 | 1.29 | 1.51 | 1.37 |
| G | 2.81 | 2.53 | 2.28 | 2.20 | 2.14 | 2.35 |
| H | 1.83 | 1.75 | 1.90 | 2.16 | 2.22 | 1.73 |
| I | 1.27 | 1.46 | 1.38 | 1.10 | 1.26 | 1.67 |
| J | 1.39 | 1.76 | 1.85 | 1.92 | 2.09 | 1.94 |
| K | 2.01 | 1.98 | 1.83 | 1.90 | 1.70 | 1.75 |
| N-propionyl | ||||||
| A | 0.72 | 1.19 | 1.21 | 1.61 | 1.85 | 1.45 |
| L | 1.85 | 1.67 | 1.40 | 1.17 | 1.25 | 1.31 |
| M | 1.50 | 1.88 | 1.91 | 1.90 | 2.06 | 2.00 |
| N | 1.28 | 1.49 | 1.31 | 0.98 | 1.18 | 1.65 |
| O | 1.80 | 1.78 | 1.90 | 2.13 | 2.17 | 1.73 |
| N-propyl | ||||||
| A | 0.49 | 1.29 | 1.61 | 1.91 | 2.03 | 1.58 |
| P | 1.86 | 1.78 | 1.32 | 1.22 | 1.67 | 2.15 |
| Q | 1.28 | 1.60 | 1.42 | 1.01 | 1.17 | 1.77 |
| R | 1.52 | 1.92 | 1.90 | 1.82 | 1.94 | 2.01 |
| S | 1.86 | 1.78 | 1.93 | 2.12 | 2.11 | 1.68 |
| T | 2.07 | 1.95 | 2.07 | 2.16 | 2.24 | 1.82 |
| N-butanoyl | ||||||
| A | 0.48 | 1.00 | 1.18 | 1.60 | 1.83 | 1.40 |
| B | 1.17 | 0.35 | 1.23 | 1.67 | 2.02 | 1.23 |
| U | 1.22 | 1.10 | 1.63 | 1.80 | 2.04 | 1.37 |
| V | 0.87 | 0.92 | 1.39 | 1.75 | 2.13 | 1.59 |
| W | 0.84 | 1.40 | 1.55 | 1.86 | 2.08 | 1.84 |
Pyranose ring and glycosidic linkage atoms.
Highlighted entries indicate the best structural matches (see also Figure 14).
Figure 17.
Backbone (ring and glycosidic linkage atoms) superposition of the representative members from the most populated clusters during the REMD simulations of the native and Nacetyl analogs of the methyl α-(2,8)-trisialoside
A. N-acetyl (blue) and N-glycolyl (red): A-A, A-C, A-J, C-C, E-I
B. N-acetyl (blue) and N-propionyl (yellow): A-A, A-M, C-A, E-N
C. N-acetyl (blue) and N-propyl (orange): A-A, E-Q
D. N-acetyl (blue) and N-butanoyl (green): A-A, B-B
These topological similarities suggest that a common core conformational epitope is present in the NmB CPS and its analogs. These observations may be utilized to rationalize the results from efforts to enhance the immunogenicity of the NmB CPS by chemically modifying the N-acetyl moiety. (11, 14, 18–22) In those studies, both IgM (20) and IgG (14, 18, 19) classes of antibodies, respectively, were utilized. Attempts to characterize the epitopes of the N-propionylated CPS suggested that the IgG mAbs recognized both short and extended conformational epitopes, (14) and a subset of the mAbs cross-reacted with human PSA and encapsulated NmB. (14) In another study (20) IgM mAbs cross-reacted equally with the native NmB CPS and its derivatives, while different reactivities were noted among other species. In that study NmB CPS specific IgMs were bactericidal and safe when tested in mice. These studies indicate that low affinity IgMs and fully matured IgGs can show cross-reactivity between the native and modified NmB CPS. Regardless of the immunoglobulin class, the observed cross-reactivity or lack thereof may be rationalized in terms of interactions determined by the global (core) configuration of the polysaccharide, and those dependent on local interactions with pendant or exocyclic groups. Despite the observed similarities in backbone topologies, Figure 17, the exocyclic groups are displayed uniquely in each trisaccharide, Figure 18.
Figure 18.
The display of key exocyclic groups relative to the backbone (ring and glycosidic linkage atoms) conformations of the representative cluster members of the native methyl α-(2,8)-trisialoside and its analogs.
A. N-acetyl (blue) and N-glycolyl (red): A-A, A-C, A-J, C-C, E-I
B. N-acetyl (blue) and N-propionyl (yellow): A-A, A-M, C-A, E-N
C. N-acetyl (blue) and N-propyl (orange): A-A, E-Q
D. N-acetyl (blue) and N-butanoyl (green): A-A, B-B
When cross-reactivity is observed among the altered CPS analogs the elicited mAbs are recognizing a common topology, Figure 17, whose fine specificities do not likely depend on the N-acyl groups or on strong interactions with them. On the other hand, the absence of cross-reactivity either arises from significant variations in the overall 3D shape (unlikely based on the data presented here) or from incompatibilities with the presentation of pendant groups in the CPS. An antibody that has evolved to recognize the non-native N-acyl derivatives, such as mAb 13D9, may not be able to make optimal contacts when the native CPS is encountered, despite the presence of a common conformational backbone. This is particularly evident in the comparison between the native structure and the synthetic N-Pr or N-But analogs, Figure 18 and Figure 19. The smaller size of the N-acetyl group may prevent it from interacting effectively with a binding pocket deep enough to accommodate the larger N-Pr and N-But groups. This would explain the observation that although mAb 13D9 requires an extended epitope, as in the native CPS, it is unable to recognize the native CPS. (23) It was reported previously that absence of cross-reactivity between the native CPS and the synthetic N-Pr and N-But polysaccharides could be due to conformational changes in the CPS induced by the modifications. (20) In contrast, the MD simulations performed in this work suggest there is significant overlap in the conformational space of the native and chemically modified saccharides, observations that are in accord with experimental NMR data. (11)
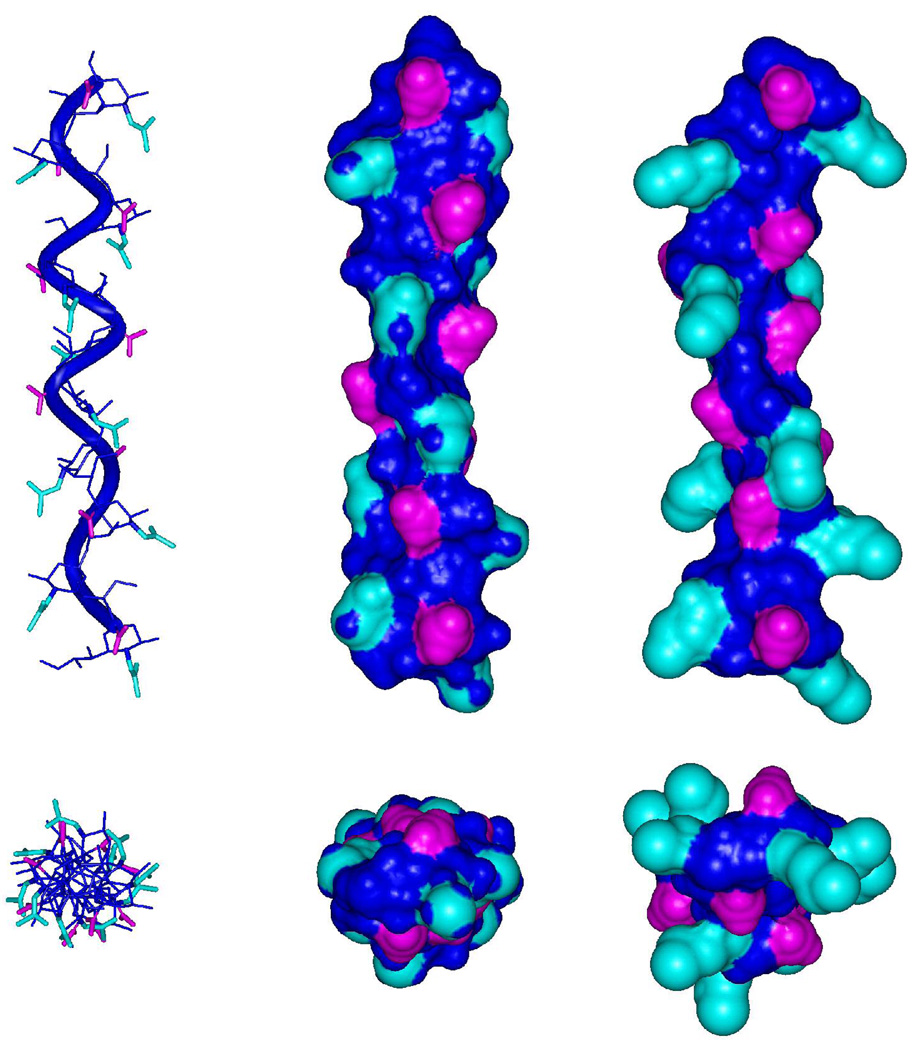
Figure 19.
Longitudinal and axial views of decasaccharide helical structures generated by propagating representative conformational states derived from the REMD simulations of the native trisaccharide (left and middle panels) and the N-butanoyl analog (right panel). The helical repeat is eight residues in each helix, shown to overlap in the axial views. Cyan, N-acyl groups; Pink, carboxyl groups; Blue, all other atoms.
To illustrate the key structural properties pertinent to antibody recognition, decasaccharide helices were generated from structures selected from the conformational states identified during the REMD simulations of II and the N-But analog. The building blocks were selected to be consistent with the experimental nOe and J-value data. Conformational states whose φ-angles adopted the anti rotamer were selected because this rotamer preference led to the reproduction of the bH9-cH7 distance. States were also required to display the +gauche rotamers of ω8 and ω7, consistent with a strong nOe between H8-H6, and low magnitude 3J-values between H8-H7 and H7-H6. Based on these criteria, state A was selected for both II and its N-But analog. As expected, the nOe distances computed from these states are consistent with the NMR data, Table 15. The helical parameters, n (the number of residues per turn) and h (vertical rise per residue in Å), were calculated based on the method described by Miyazawa et al. (52) Vectors consisting of the glycosidic oxygen atoms (O8) were employed to describe virtual bonds in each decasaccharide. Using these vectors distances, angles and dihedral angles for the central residues (residues 4 to 7) were computed for each decasaccharide and employed to determine n and h. The (n, h) values were (2.2, 4.5 Å) and (2.3, 5.2 Å) for the A state of II and its N-But analog, respectively. It should be noted that in solution, these polymers are dynamic and interconvert between states, and these idealized helices are not representative of the overall structure. Nonetheless, the helices generated here, Figure 19, are representative of the local epitopes that are presented to antibodies. The outward projection of the N-acyl groups from the helical axis is evident, suggesting that these groups may be crucial for antibody recognition. In these helices the N-acetyl and N-butanoyl groups constitute 33% and 46% of their respective surfaces. Consequently, an antibody whose specificity involves the N-butanoyl moiety would be expected to have much lower affinity for the N-acetyl analog. In each case, the carboxylate groups are also presented on the helical surfaces, although in the N-But analog these groups are markedly less accessible.
Table 15.
Experimental (ISPA) nOe distances (Å), inter-proton distances and 3J-values from the N-acetyl and Nbutanoyl trisaccharide (a-b-c-OMe) conformer families employed to generate the decasaccharide helices
| Experimental (ISPA) nOe and MD-derived distances (Å) | |||||||
|---|---|---|---|---|---|---|---|
| Spins | NMR (native) | N-acetyl | N-butanoyl | Spins | NMR (native) | N-acetyl | N-butanoyl |
| bH3ax – cH8 | 2.6 ± 0.06 | 2.2 ± 0.2 | 2.3 ± 0.2 | bH6 – bH8 | 2.3 ± 0.05 | 2.4 ± 0.2 | 2.4 ± 0.2 |
| bH3eq – cH8 | 3.5 ± 0.08 | 3.5 ± 0.2 | 3.6 ± 0.2 | aH3ax – bH8 | 2.8 ± 0.07 | 2.2 ± 0.2 | 2.3 ± 0.2 |
| bH3eq – cH7 | 3.9 ± 0.09 | 5.1 ± 0.2 | 5.1 ± 0.2 | aH3eq – bH8 | 3.4 ± 0.08 | 3.6 ± 0.2 | 3.6 ± 0.2 |
| bH6 – bH7 | 2.2 ± 0.05 | 2.4 ± 0.1 | 2.4 ± 0.1 | bH9 – cH7 | 3.1 ± 0.07 | 2.8 ± 0.4 | 2.9 ± 0.4 |
| Experimental and MD-derived 3J-values | |||||||
| Angle | Linkage | Spins | NMR (native) | N-acetyl | N-butanoyl | ||
| ω7 | Internal | bH6-bH7 | < 1.0 | 1.0 ± 0.7 | 1.1 ± 0.9 | ||
| Internal | cH6-cH7 | < 1.0 | 1.5 ± 1.0 | 1.6 ± 1.0 | |||
| ω8 | Internal | bH7-bH8 | < 4.0 | 1.2 ± 0.7 | 1.2 ± 0.7 | ||
| Internal | cH7-cH8 | < 4.0 | 1.2 ± 0.8 | 1.1 ± 0.6 | |||
REMD simulations were performed at 8 temperatures for a total of 160 ns.
The distances are averaged over the MD trajectories as <1/R6>(−1/6)
Two salient points emerge from all these considerations: cross-reactivity arises between the N-acyl analogs when overall backbone conformation is conserved and when the immune response has not matured to recognize the acyl moiety as an immunodominant epitope. Therefore, these simulations suggest that alternative chemical modifications may be sought, subject to the constraints that the integrity of the N-acetyl moiety, and the backbone core conformations, are not compromised. These modifications could involve the introduction of small hydrophobic groups at positions that are hydrophilic in the native structure, and thus lees immunogenic. To the best of our knowledge, no such derivatizations have been performed with the NmB CPS. However, within the accuracy limitations of the force field and MD simulations, it is now feasible to predict the effect of chemical modifications on the CPS structure and to identify the structural similarities between native and synthetic immunogens.
Conclusion
Because of the absence of a protective vaccine against NmB, and the postulated importance of a conformational epitope in mediating the antigenicity and bactericidal activity related to this CPS, considerable attention has been given to characterizing its 3D shape and dynamics (9, 11, 18, 25, 26, 46). In addition, the weak immunogenicities of both the proteinconjugated and non-conjugated forms of the NmB CPS have resulted in alternative vaccine design attempts that typically involve chemical group modifications of the acyl groups (11, 14, 18, 21, 22) or the carboxylate group. (11)
As an initial step in quantifying the antigenicities of native and chemically modified NmB CPS, conformational properties of di- and trisaccharide haptens have been determined by MD simulations and confirmed by NMR spectroscopy. That the MD simulations identified experimentally relevant conformations was demonstrated by the good agreement between the experimental and theoretical NMR observables. In addition, a Bayesian statistical analysis (40) was employed to identify conformational states from the trajectories. To the best of our knowledge this is the first time such an analysis has been applied to investigate carbohydrate conformational properties. With the preliminary agreement demonstrated here between the MD data and the experimental data for the native haptens, the study was extended to shed light on the effects of chemical group modifications on epitope size and fine structure. These theoretical simulations present a unique opportunity to generate atomic level insight into the conformational space of native and synthetic CPS analogs. It is also suggested that efforts employed in the quest for more immunogenic carbohydrate vaccines that can mimic NmB should consider chemical group modifications that do not perturb the N-acetyl moieties or the global conformation. MD simulations provide a much needed tool for the rational design of such synthetic vaccines, and provide a solid basis for the interpretation of the diverse and complex data from immunological studies.
Supplementary Material
Pyranose ring vicinal proton-proton coupling constants of methyl α-(2,8)-di/trisialoside. This material is available free of charge via the internet at http://pubs.acs.org
Acknowledgments
The authors thank the National Institutes of Health (grants RR05357 and GM55230) for funding. Computational time was provided in part by a grant (#9607) from the Environmental Molecular Sciences Laboratory (EMSL) at Pacific Northwest National Laboratory (PNNL). V.S. thanks the Boehringer Ingelheim Fonds for a Ph.D scholarship. RJW thanks H. J. Jennings (NRC, Canada) and J. B. Robbins (NIH) for useful discussions.
Abbreviations
- Nm
Neisseria meningitidis
- NmB
Neisseria meningitidis serogroup B
- NmC
Neisseria meningitidis serogroup C
- mAb
monoclonal antibody
- ISPA
isolated spin-pair approximation
- PSA
polysialic acid
- Neu5Ac
α-5-N-acetylneuraminic acid
- CPS
capsular polysaccharide
- MD
molecular dynamics
- REMD
replica exchange molecular dynamics
- E. coli
Escherichia coli
- N-Gc
N-glycolyl
- N-Pr
N-propionyl
- N-Prop
N-propyl
- N-But
N-butanoyl
References
- 1.Ferreiros CM, Gomez JA, Criado MT. Prevention of meningococcal disease: present and future. Rev. Med. Microbiol. 1998;9:29–37. [Google Scholar]
- 2.Tikhomirov E, Santamaria M, Esteves K. Meningococcal disease: public health burden and control. World Health Statistics Quarterly. 1997;50:170–177. [PubMed] [Google Scholar]
- 3.Tsang RSW, Zollinger WD. Serological specificities of murine hybridoma monoclonal antibodies against Neisseria meningitidis serogroups B, C, Y and W135 and evaluation of their usefulness as serogrouping reagents by indirect whole-cell enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 2005;12:152–156. doi: 10.1128/CDLI.12.1.152-156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardotti A, Averani G, Berti F, Berti S, Galli C, Giannini S, Fabbri B, Proietti D, Ravenscroft N, Ricci S. Size determination of bacterial capsular oligosaccharides used to prepare conjugate vaccines against Neisseria meningitidis groups Y and W135. Vaccine. 2005;23:1887–1899. doi: 10.1016/j.vaccine.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee AK, Jennings HJ, Kenny CP, Martin A, Smith ICP. Structural determination of polysaccharide antigens of Neisseria meningitidis serogroup Y, serogroup W-135, and serogroup BO. Can. J. Biochem. 1976;54:1–8. doi: 10.1139/o76-001. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee AK, Jennings HJ, Kenny CP, Martin A, Smith ICP. Structural determination of sialic acid polysaccharide antigens of Neisseria meningitidis serogroup B and serogroup C with C-13 nuclear magnetic resonance. J. Biol. Chem. 1975;250:1926–1932. [PubMed] [Google Scholar]
- 7.Bundle DR, Smith ICP, Jennings HJ. Determination of the structure and conformation of bacterial polysaccharides by carbon 13 nuclear magnetic resonance. Studies on group-specific antigens of Neisseria meningitidis serogroups A and X. J. Biol. Chem. 1974;249:2275–2281. [PubMed] [Google Scholar]
- 8.Kabat EA, Liao J, Osserman Ef, Gamian A, Michon F, Jennings HJ. The epitope associated with the binding of the capsular polysaccharide of the group B meningococcus and of Escherichia coli K1 to a human monoclonal macroglobulin, IGMNOV. J. Exp. Med. 1988;168:699–711. doi: 10.1084/jem.168.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisson JR, Baumann H, Imberty A, Perez S, Jennings HJ. Helical epitope of the group B meningococcal α-(2-8)-linked sialic acid polysaccharide. Biochemistry. 1992;31:4996–5004. doi: 10.1021/bi00136a012. [DOI] [PubMed] [Google Scholar]
- 10.Jennings HJ, Roy R, Michon F. Determinant specificities of the group B and group C polysaccharides of Neisseria meningitidis. J. Immunol. 1985;134:2651–2657. [PubMed] [Google Scholar]
- 11.Baumann H, Brisson JR, Michon F, Pon R, Jennings HJ. Comparison of the conformation of the epitope of α-(2-8) polysialic acid with its reduced and N-acyl derivatives. Biochemistry. 1993;32:4007–4013. doi: 10.1021/bi00066a022. [DOI] [PubMed] [Google Scholar]
- 12.Weintraub A. Immunology of bacterial polysaccharide antigens. Carbohydr. Res. 2003;338:2539–2547. doi: 10.1016/j.carres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2:355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 14.Granoff DM, Bartoloni A, Ricci S, Gallo E, Rosa D, Ravenscroft N, Guarnieri V, Seid RC, Shan A, Usinger WR, Tan S, McHugh YE, Moe GR. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic aicd. J. Immunol. 1998;160:5028–5036. [PubMed] [Google Scholar]
- 15.Devi SJN, Robbins JB, Schneerson R. Antibodies to poly[(2-8)-α-Nacetylneuraminic acid] and poly[(2-9)-α-N-acetylneuraminc acid] are elicited by immunization of mice with Escherichia coli K92 conjugates: Potential vaccines for groups B and C meningococci and E. coli K1. Proc. Natl. Acad. Sci. USA. 1991;88:7175–7179. doi: 10.1073/pnas.88.16.7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lifely MR, Gilbert AS, Moreno C. Sialic acid polysaccharide antigens of Neisseria meningitidis and Escherichia coli. Esterification between adjacent residues. Carbohydr. Res. 1981;94:193–203. doi: 10.1016/s0008-6215(00)80717-x. [DOI] [PubMed] [Google Scholar]
- 17.Azurmendi HF, Vionnet J, Wrightson L, Trinh LB, Shiloach J, Freedberg DI. Extracellular structure of polysialic acid explored by on cell solution NMR. Proc. Natl. Acad. Sci. USA. 2007;104:11557–11561. doi: 10.1073/pnas.0704404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans SV, Sigurskjold BW, Jennings HJ, Brisson JR, To R, Tse WC, Altman E, Frosch M, Weisgerber C, Kratzin HD, Klebert S, Vaesen M, Bittersuermann D, Rose DR, Young NM, Bundle DR. Evidence for the extended helical nature of polysaccharide epitopes. The 2.8Å resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for α-(2-8)-polysialic acid. Biochemistry. 1995;34:6737–6744. doi: 10.1021/bi00020a019. [DOI] [PubMed] [Google Scholar]
- 19.Jennings HJ, Roy R, Gamian A. Induction of meningococcal group B polysaccharide-specific IgG antibodies in mice by using an N-propionylated B polysaccharide-tetanus toxoid conjugate vaccine. J. Immunol. 1986;137:1708–1713. [PubMed] [Google Scholar]
- 20.Lifely MR, Esdaile J. Specificity of the immune response to the group B polysaccharide of Neisseria meningitidis. Immunology. 1991;74:490–496. [PMC free article] [PubMed] [Google Scholar]
- 21.Pon RA, Khieu NH, Yang QL, Brisson JR, Jennings HJ. Serological and conformational properties of E. coli K92 capsular polysaccharide and its N-propionylated derivative both illustrate that induced antibody does not recognize extended epitopes of polysialic acid: Implications for a comprehensive conjugate vaccine against groups B and C N. meningitidis. Can. J. Chem. 2002;80:1055–1063. [Google Scholar]
- 22.Pon RA, Lussier M, Yang QL, Jennings HJ. N-propionylated group B meningococcal polysaccharide mimics a unique bactericidal capsular epitope in group B Neisseria meningitidis. J. Exp. Med. 1997;185:1929–1938. doi: 10.1084/jem.185.11.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pon RA, Biggs NJ, Jennings HJ. Polysialic acid bioengineering of neuronal cells by N-acyl sialic acid precursor treatment. Glycobiology. 2007;17:249–260. doi: 10.1093/glycob/cwl075. [DOI] [PubMed] [Google Scholar]
- 24.Lifely MR, Lindon JC, Williams JM, Moreno C. Structural and conformational features of the Escherichia coli K92 capsular polysaccharide. Carbohydr. Res. 1985;143:191–205. doi: 10.1016/s0008-6215(00)90708-0. [DOI] [PubMed] [Google Scholar]
- 25.Henderson TJ, Venable RM, Egan W. Conformational flexibility of the group B meningococcal polysaccharide in solution. J. Am. Chem. Soc. 2003;125:2930–2939. doi: 10.1021/ja0210087. [DOI] [PubMed] [Google Scholar]
- 26.Venable RM, Bizik F, Henderson TJ, Egan W. Molecular dynamics simulations of an α-(2->8)-linked sialic acid tetramer in vacuum and solvent. J. Mol. Struct. (Theochem) 1997;395:375–388. [Google Scholar]
- 27.Cumming DA, Carver JP. Virtual and Solution Conformations of Oligosaccharides. Biochemistry. 1987;26:6664–6676. doi: 10.1021/bi00395a016. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Outeirino J, Kadirvelraj R, Woods RJ. Structural elucidation of type III group B Streptococcus capsular polysaccharide using molecular dynamics simulations: the role of sialic acid. Carbohydr. Res. 2005;340:1007–1018. doi: 10.1016/j.carres.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Vasudevan SV, Balaji PV. Molecular dynamics of alpha2-8 linked disialoside: Conformational analysis and implications for binding to proteins. Biopolymers. 2002;63:168–180. doi: 10.1002/bip.10019. [DOI] [PubMed] [Google Scholar]
- 30.Kirschner KN, Woods RJ. Solvent interactions determine carbohydrate conformation. Proc. Natl. Acad. Sci. USA. 2001;98:10541–10545. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Outeirino J, Kirschner KN, Thobhani S, Woods RJ. Reconciling solvent effects on rotamer populations in carbohydrates - A joint MD and NMR analysis. Can. J. Chem. 2006;84:569–579. doi: 10.1139/v06-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirschner KN, Yongye AB, Gonzalez-Outeirino J, Tschampel SM, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yongye AB, Foley BL, Woods RJ. On achieving experimental accuracy from molecular dynamics simulations of flexible molecules: aqueous glycerol. J. Phys. Chem. A. 2008;112:2634–2639. doi: 10.1021/jp710544s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Case DADTA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Lou R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister P, Caldwell JW, Ross WS, Kollman PA. AMBER 8. San Francisco, CA: University of California; 2004. [Google Scholar]
- 35.Jorgensen WL, Swenson CJ. Optimised intermolecular potential functions for amides and peptides. Structure and properties of liquid amides. J. Am. Chem. Soc. 1985;107:569–578. [Google Scholar]
- 36.Darden T, York D, Pederson L. Particle mesh Ewald: A N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 37.van Gunsteren WF, Berendsen HJC. Algorithms for macromolecular dynamics and constraint dynamics. Mol. Phys. 1977;34:1311–1327. [Google Scholar]
- 38.Okur A, Wickstrom L, Layten M, Geney R, Song K, Hornak V, Simmerling C. Improved efficiency of replica exchange simulations through use of a hybrid explicit/implicit solvation model. J. Chem. Theory Comput. 2006;2:420–433. doi: 10.1021/ct050196z. [DOI] [PubMed] [Google Scholar]
- 39.Rathore N, Chopra M, J. de Pablo J. Optimal allocation of replicas in parallel tempering simulations. J. Chem. Phys. 2005;122:024111. doi: 10.1063/1.1831273. [DOI] [PubMed] [Google Scholar]
- 40.Schultheis V, Hirschberger T, Carstens H, Tavan P. Extracting Markov models of peptide conformational dynamics from simulation data. J. Chem. Theory Comput. 2005;1:515–526. doi: 10.1021/ct050020x. [DOI] [PubMed] [Google Scholar]
- 41.Kloppenburg M, Tavan P. Deterministic annealing for density estimation by multivariate normal mixtures. Phys. Rev. E. 1997;55:R2089–R2092. [Google Scholar]
- 42.James TL, Leu HE. CORMA. San Francisco: University of California; 1999. [Google Scholar]
- 43.Thrippleton MJ, Keeler J. Elimination of zero-quantum interference in two-dimensional NMR spectra. Angew. Chem. Int. Edit. 2003;42:3938–3941. doi: 10.1002/anie.200351947. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - A Multidimensional Spectral Processing System Based On Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.NMRView. NMRView Software. One Moon, Scientific, Inc.; [Google Scholar]
- 46.Michon F, Brisson JR, Jennings HJ. Conformational differences between linear α-(2-8)-linked homosialooligosaccharides and the epitope of the group B meningococcal polysaccharide. Biochemistry. 1987;26:8399–8405. doi: 10.1021/bi00399a055. [DOI] [PubMed] [Google Scholar]
- 47.Haasnoot CAG, Deleeuw F, Altona C. Relationship between Proton-Proton NMR Coupling-Constants and Substituent Electronegativities .I. An empirical generalization of the Karplus Equation. Tetrahedron. 1981;36:2783–2792. [Google Scholar]
- 48.Kooijman H, Kroonbatenburg LMJ, Kroon J, Breg JN, Deboer JL. Structure of α-D-N-acetyl-1-O-methylneuraminic acid methyl ester. Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 1990;46:407–410. [Google Scholar]
- 49.Poppe L, Stuike-Prill S, Meyer B, van Halbeek H. The solution conformation of sialyl-α-(2,6)-lactose studied by modern NMR techniques and Monte Carlo simulation. J. Biomol. NMR. 1992;2:109–136. doi: 10.1007/BF01875524. [DOI] [PubMed] [Google Scholar]
- 50.Breg J, Kroon-Batenburg MJL, Strecker G, Montreuil J, Vliegenthart JFG. Conformational analysis of the sialyl-α-(2-3/6) N-acetyllactosamine structural element occurring in glycoproteins, by two-dimensional nOe 1H-NMR spectroscopy in combination with energy calculations by hard-sphere exo-anomeric and molecular mechanics force-field with hydrogen-bonding potential. Eur. J. Biochem. 1989;178:727–739. doi: 10.1111/j.1432-1033.1989.tb14504.x. [DOI] [PubMed] [Google Scholar]
- 51.Woods RJ, Pathiaseril A, Wormald MR, Edge CJ, Dwek RA. The high degree of internal flexibility observed for an oligomannose oligosaccharide does not alter the overall topology of the molecule. Eur. J. Biochem. 1998;158:372–386. doi: 10.1046/j.1432-1327.1998.2580372.x. [DOI] [PubMed] [Google Scholar]
- 52.Sugeta H, Miyazawa T. General method for calculating helical parameters of polymer chains from bond lengths, bond angles and internal rotation angles. Biopolymers. 1967;5:673–679. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pyranose ring vicinal proton-proton coupling constants of methyl α-(2,8)-di/trisialoside. This material is available free of charge via the internet at http://pubs.acs.org