Abstract
The Ndc80 complex is a key site of regulated kinetochore-microtubule attachment, but the molecular mechanism underlying its function remains unknown. Here we present a subnanometer resolution cryo-electron microscopy reconstruction of the human Ndc80 complex bound to microtubules, sufficient for precise docking of crystal structures of the component proteins. We find that Ndc80 binds the microtubule with a tubulin monomer repeat, recognizing α- and β-tubulin at both intra- and inter-dimer interfaces in a manner that is sensitive to tubulin conformation. Furthermore, Ndc80 complexes self-associate along protofilaments via interactions mediated by the amino-terminal tail of the Ndc80 protein, the site of phospho-regulation by the Aurora B kinase. Ndc80's mode of interaction with the microtubule and its oligomerization suggest a mechanism by which Aurora B could regulate the stability of load-bearing Ndc80-microtubule attachments.
The Ndc80 complex is a member of the conserved KMN kinetochore network, which also includes the KNL-1 and Mis12 complexes1. The Ndc80 complex is the key site for kinetochore-microtubule attachment1,2,3 and a landing pad for the spindle assembly checkpoint4,5,6. Although extensively characterized genetically7,8 and biochemically1,2,3,9,10, the mechanisms by which the Ndc80 complex effects and coordinates these activities remain elusive.
The complex is an elongated, 57 nm heterotetramer composed of Ndc80, Nuf2, Spc24, and Spc25, each having a globular domain connected to a coiled-coil that mediates dimerization: Spc24 with Spc25, and Ndc80 with Nuf29,10,11,12. Tetramerization via the dimerized coiled-coils9,10 results in a dumbbell architecture, with the Spc24–25 globular head at one end mediating kinetochore association1,3, and the Ndc80-Nuf2 head at the other mediating microtubule binding1,2,3,12. The Ndc80-Nuf2 coiled-coil contains a break which makes the rod-like complex highly bendable13.
Crystallographic structures of both globular head domains have been obtained2,11, as well as that of a chimeric version of the human complex containing a minimal coiled-coil, where Ndc80 was fused to Spc25 and Nuf2 to Spc2412. This 17 nm “bonsai” complex retained microtubule binding and kinetochore localization. Both Ndc80 and Nuf2 contain a Calponin Homology Domain (CHD), which is also present in other microtubule binding proteins14,15. The unstructured, positively charged 80 amino-acid N-terminal tail of the Ndc80 protein is required for high-affinity microtubule binding2,3,12,16,17, likely by interaction with the acidic C-terminal tails of tubulin, or E-hooks. This region of the Ndc80 protein, the site of phospho-regulation by the Aurora B kinase3,12,18, is absent from all crystal structures. The key questions of how the Ndc80 complex binds and remains attached to microtubules (MTs) during MT depolymerization and how this attachment is regulated during mitosis remain unanswered.
Ndc80 binds tubulin with a novel monomeric repeat
We used cryo-EM to obtain a structure of the Ndc80 bonsai complex12 including the N-terminal tail bound to MTs. We employed an implementation19 of the Iterative Helical Real Space Reconstruction method20, which utilizes reference-free classification to sort helical segments based on symmetry and sample quality (Supplementary Fig. 1–2). Class averages show densities protruding from the microtubule (Supplementary Fig. 1b, top right) with the chevron-like orientation also reported for the C. elegans complex1,21. Power spectra of class averages show layer lines at 1/40 Å−1 (and subsequent orders), corresponding to the spacing of the tubulin monomer (Supplementary Fig. 1b, bottom right), but lack the 1/80 Å−1 layer line typically observed for microtubule-binding proteins which recognize the tubulin heterodimer22,23,24. This result suggested that Ndc80 binds to each tubulin monomer.
A helical reconstruction of the C. elegans complex suggested alternating binding of the two CHDs to strong and weak sites present in each tubulin dimer21. In order to determine the arrangement of the Ndc80 complex on the MT lattice without imposing any symmetry or averaging, we obtained tomographic reconstructions of negatively-stained MTs saturated with Ndc80 bonsai (Supplementary Fig. 1c). Single volume slices allow us to visualize individual tubulin monomers and bound Ndc80 molecules, which are found with a 40 Å spacing within the thickness of a single protofilament. Thus, the human Ndc80 complex binds both α- and β-tubulin monomers.
Microtubule site recognition of the Ndc80 complex
Using a microtubule as a starting reference (Supplementary Fig. 3), we obtained a reconstruction of the Ndc80-bound microtubule at 8.6 Å resolution (FSC 0.143 criterion, Supplementary Fig. 4a), allowing us to visualize secondary-structure (Fig. 1a; Supplementary Fig. 3–4). The asymmetric units of reconstructed 13 and 14 protofilament microtubules were essentially identical (Supplementary Fig. 5). Docking of the crystal structures of Ndc80 bonsai lacking the N-terminus (Bonsai ΔN)12 and tubulin25 allowed us to build a pseudo-atomic model of this interface (Fig. 1b). The excellent correspondence between our map and the crystal structure of Bonsai ΔN argue against major rearrangements in the globular domains of the complex upon microtubule binding.
Figure 1. Structure of the Ndc80-microtubule interface.
a, End on (from the plus-end) and side views of the microtubule-Ndc80 complex cryo-EM reconstruction (tubulin, green; Ndc80-Nuf2 head, blue; disordered Spc24–25 head, red). b, Orthogonal views of docked crystal structures (Ndc80, blue; gold-Nuf2 gold; green-tubulin). In ball and stick: ordered residues adjacent to the N-terminus of Bonsai ΔN (magenta), ordered residues in tubulin preceding the E-hooks (red). The region of the map occupied by Nuf2 (right panel) is at higher radius and thus is of lower resolution. c, Orthogonal views of the positive difference density (magenta) between the cryo-EM reconstruction and the docked crystal structures, contoured at 2.5 σ. The dotted lines indicate a proposed path for the Ndc80 N-terminus.
The surface of the Ndc80 complex binding the MT lattice is minimal, and includes the C-terminal end of helix G and the short helices B and F in the Ndc80 protein. This “toe” recognizes a site between two tubulin monomers, a “toe-print” present at both intra- and inter-dimer interfaces composed of the short helix H11' in one tubulin subunit and the loop connecting H8 with S7 in the other (Fig. 2a, purple and orange, respectively). Superposition of the crystal structures of α- and β-tubulin and multiple-sequence alignments show the toe-print to be highly conserved between the two tubulin monomers (Fig. 2a, bottom), consistent with the ability of the Ndc80 complex to recognize them both. The end of the tubulin crystal structure marking the beginning of the disordered E-hook of tubulin (Fig. 2a, red), required for high-affinity Ndc80 binding1,12, is adjacent to the toe-print, from where we propose it extends and acts as a second, distinct binding site.
Figure 2. The Ndc80 toe-print is a tubulin conformation sensor.
a, Superposition of intra-dimer and inter-dimer interfaces, viewed from the outside of the microtubule (α-tubulin, green; β-tubulin, blue; ordered residues adjacent to the E-hooks, red; conserved toe-print segments, purple and orange). Consensus sequences are indicated, with deviations between the monomers in parenthesis. b, SDS-PAGE of co-sedimentation assays with straight (taxol) and curved (vinblastine) tubulin polymers with the indicated Ndc80 bonsai constructs. S: supernatant, P: pellet. [Tubulin monomer] = 6 μM, [Ndc80 bonsai] = 0.5 μM. c, Quantification of b. Error bars represent ± s.d., n = 3. d, SDS-PAGE of sedimentation assays after cold-induced depolymerization of dynamic microtubules in the presence and absence of Ndc80 bonsai. e, Negative-stain EM of Ndc80-induced cold-stable microtubules and straight tubulin sheets. Scale bar, 50 nm.
The interface we observe is largely in agreement with mutagenesis analysis of Ndc80 residues important for binding (e.g. K123, K166, H176)12. Interestingly, the Nuf2 CHD is not in contact with the surface of the microtubule, yet mutations in this region also disrupt binding12. The positively charged surface of this domain is approximately 15 Å from the E-hook of a laterally adjacent tubulin monomer and thus could be engaged in an electrostatic interaction.
The Ndc80 toe is a tubulin conformation sensor
During MT disassembly protofilaments bend outwards by kinking of tubulin at intra-and inter-dimer interfaces. In spite of its small size, the toe-print spans the proposed hinge point between tubulin monomers and could thus be disrupted by tubulin bending26 (Fig. 2a). We therefore hypothesized that Ndc80's toe acts as a sensor of the conformational state of tubulin and that the complex would bind preferentially to straight protofilaments.
Using co-sedimentation assays we investigated the ability of Ndc80 bonsai to bind vinblastine-induced tubulin spirals (Supplementary Fig. 6), a polymer analogous to the peels observed at microtubule ends27. Because spirals retain the E-hooks, which make a major contribution to binding affinity, we expected the interaction to be reduced, not eliminated. Indeed, we observed a modest but statistically significant (P < 0.05, Student's t-test) reduction in affinity for this form of tubulin versus the straight microtubule conformation (Fig. 2b top, 2c). We next sought to delineate the relative contributions of the toe versus the N-terminus of Ndc80 to tubulin binding. N-terminus-7D, a phosphomimetic construct of the 7 Aurora B phosphorylation sites confirmed in vitro12 (Supplementary Fig. 7), showed significantly reduced affinity towards the straight microtubule conformation and negligible affinity towards the vinblastine-induced, bent conformation, reflecting the increased relative contribution of the toe-print interaction to affinity when the N-terminus-E-hook interaction is impaired (Fig. 3b–c). This result is consistent with a bipartite binding mechanism, with the Ndc80 N-terminus providing affinity without conformation sensitivity, and the toe providing a smaller contribution to affinity that is exquisitely sensitive to tubulin conformation.
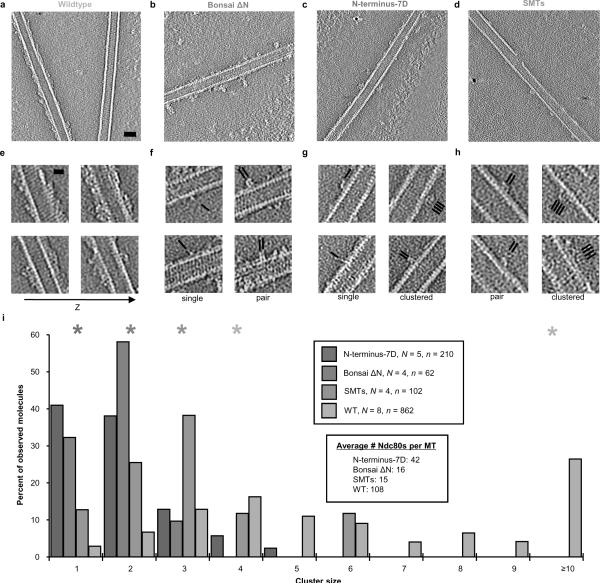
Figure 3. Cluster formation requires the N-terminus of the Ndc80 protein.
a–d, Central slices of tomograms of microtubule-bound Ndc80 constructs under subsaturating conditions. Ndc80:tubulin monomer ration was 1:2 for wild-type and SMTs and 2:1 for bonsai N-terminus mutants. SMTs: subtilisin-cleaved microtubules. Scale bar, 25 nm (black dots = gold fiducials). e, Serial slices of the wild-type reconstruction 4.5 nm apart show cooperative binding and cluster formation only along (not between) protofilaments. Scale bar, 10 nm. f–h, Selected views of clusters in Bonsai ΔN, N-terminus-7D, and SMTs reconstructions, respectively. Black lines indicate position and orientation of Ndc80 molecules. i, Quantification of cluster size populations. N = number of reconstructed MT segments, n = total number of Ndc80 complexes observed. Asterisks indicate the most probable cluster size for each of the populations (wild-type has two peaks). See Supplementary Table 1 for pair-wise statistical comparison of these distributions.
We next investigated the effect of the complex's small bias in affinity towards straight tubulin. We polymerized tubulin into dynamic microtubules, then initiated depolymerization by cooling in the presence and absence of Ndc80 bonsai. Cold-stable microtubules and straight tubulin sheets were observed only in the presence of Ndc80 (Fig. 3d–e). Together, these results are consistent with Ndc80 favoring a straight tubulin conformation, and with this specificity being mediated by the toe. Our studies also indicate that Ndc80 has a stabilizing effect on microtubules.
Ndc80 self-assembles on microtubules
In our cryo-EM reconstruction connections appear between Ndc80 complexes along protofilaments (Fig. 1 a–b), supporting and explaining the observed cooperative binding of the complex to MTs12,17. We collected tomograms of negatively-stained microtubules with non-saturating amounts of Ndc80 complex to test for the presence of Ndc80-Ndc80 interactions under more physiological conditions. We found that the complex forms clusters along protofilaments that retain tubulin monomer spacing (Fig. 3a, e; Supplementary Fig. 8). Decoration was heterogeneous, with some microtubules approaching saturation and others almost undecorated (Fig. 3a, Supplementary Figure 9a), a direct manifestation of cooperativity. We do not observe ordered Ndc80 self-association in the absence of microtubules (Supplementary Fig. 10). Analysis of the pooled distributions of cluster number and size indicates that it is most probable for a complex to be found in a cluster of 4 molecules or in large clusters of more than ten molecules (Fig. 3i, Supplementary Fig. 9b). Given the reported number of 6–8 Ndc80s per microtubule at the kinetochore, the larger clusters are probably not physiologically relevant28,29. Our data is therefore consistent with the formation of two or three clusters per kinetochore MT.
Analysis of our docking results shows densities not accounted for by the crystals structures. We attempted to visualize these regions by calculating a difference-map between the experimental density map and the docked crystal structures (Fig. 1c; Supplementary Fig. 11). The map shows positive density peaks adjacent to the C-terminal end of the tubulin crystal structure, corresponding to the extension of the H12 helices in each monomer. Stabilization of these residues upon Ndc80 binding is consistent with this region of tubulin contributing to affinity1,12. Small extra density is also observed by the toe-print. The largest difference density is between the globular domain of Nuf2 and the αH–αI helical hairpin of Ndc80, with a series of small peaks running along the groove between two longitudinally adjacent complexes, connecting this region with the N-terminal end of the Bonsai ΔN crystal structure.
The most parsimonious explanation of this path of density is that it corresponds to the N-terminus of Ndc80. We propose that the N-terminus mediates weak contacts between the globular heads of Ndc80 and Nuf2 before making a strong contact, corresponding to the major density peak. It then could bind the C-terminus of tubulin (necessary for high-affinity), likely in an adjacent protofilament. The total mass (80 residues) of the N-terminus is not accounted for by these densities, which probably correspond to ordered points of contact formed by this mostly unstructured polypeptide.
Ndc80's N-terminus mediates regulated self-assembly
In agreement with this proposal, we found that Bonsai ΔN was deficient in cluster formation (Fig. 3b, f, i). The presence of some clusters of two complexes suggests that the N-terminus is the major but not sole molecular determinant of clustering. We found that N-terminus-7D showed a clustering phenotype similar to Bonsai ΔN, with a cluster size of 1 molecule being the most probable despite a higher average surface density, and a few clusters of moderate size (Fig. 3c, g, i). We therefore conclude that the N-terminus of Ndc80 mediates Ndc80-Ndc80 interactions, as previously speculated12, and that phosphorylation at Aurora B sites is capable of modulating this binding.
Next, we dissected the N-terminus's differential contributions to affinity and cooperativity. Subtilisin cleavage of the E-hooks from tubulin significantly reduces affinity of Ndc80 for MTs1,12,17. As expected, after this treatment we found a significantly lower average surface density of Ndc80 compared to un-cleaved MTs (Fig. 3d, h, i). If the N-terminus of Ndc80 were only contributing indirectly to cluster formation by increasing microtubule-binding affinity, the cluster size distribution for this condition should be similar to the N-terminus mutants. Instead, we observe a most probable cluster size of 3 molecules, despite an average surface density lower than that of N-terminus-7D (Fig. 3i). This observation strongly supports our hypothesis that the Ndc80 N-terminus forms specific intermolecular interactions between complexes on the MT surface. It also supports cluster formation occuring after MT binding, since the average cluster size on subtilisin-cleaved MTs is smaller than on uncleaved MTs.
Role of Ndc80 clusters in mitosis
It was recently demonstrated that Aurora B's activity is governed by spatial localization rather than directly by tension30, confirming previous proposals31,32. Based on this finding, we envision a scenario where Ndc80 complexes begin phosphorylated at an unattached kinetochore (Fig. 4a)33. When the kinetochore encounters a microtubule, individual Ndc80 heads would initially bind it through a low-affinity interaction. The bound heads would escape the Aurora B phosphorylation zone provided that intrakinetochore stretching increased as microtubule binding by additional heads allowed the site to come under tension34. This could occur via straightening of the flexible Ndc80 complex itself13 (Fig. 4a) or by the stretching of a compliant link in the inner kinetochore35. As the phosphorylation zone is cleared, phosphatase activity (possibly KMN network-localized Protein Phosphatase 136) would dephosphorylate the N-terminus of Ndc80, resulting in the formation of high-affinity Ndc80 clusters.
Figure 4. Proposed models of attachment maturation and biased diffusion.
a, A cartoon illustrating the phospho-regulated formation of Ndc80 clusters in vivo concomitant with stable kinetochore-mirotubule attachment (colors are as Fig. 1b). In this schematic the assembly of the cluster biased diffusion machinery is regulated by the spatial localization of Aurora B and a counterbalancing phosphatase rather than directly by tension. b, A cartoon diagramming the proposed biased diffusion process for coupling chromosome movement to microtubule depolymerization via the Ndc80 complex. Colors are as in Fig. 1b, except the Ndc80-Nuf2 head is shown in gold and unidentified filaments are shown in grey.
Once assembled, the cluster arrangement is consistent with Ndc80 maintaining load-bearing attachments via biased diffusion37 (Fig. 4b), as recently proposed from functional studies38. An alternative model, based on shuffling, is also consistent with our data (Supplementary Discussion, Supplementary Fig. 12). Our results support a biased-diffusion model with significant differences from previous studies presuming either a continuous sleeve37 or sites uncoupled from the subunit spacing of the microtubule38. We propose that a set of identical, coupled binding sites assemble to form linear arrays along protofilaments that match the longitudinal subunit spacing of the MT, only after the kinetochore comes under tension. Consistent with this hypothesis, outer kinetochore rearrangement upon microtubule interaction has been observed in vivo39.
A cluster could diffuse on the microtubule lattice, but its diffusion would become biased at a microtuble end. Thus, a shrinking microtubule would pull the attached chromatid polewards. Cluster diffusion should be facilitated by the 40 Å rather than 80 Å spacing of the Ndc80 complexes, with a shorter distance to the transition state between binding sites during diffusion. Thus, monomer binding may have evolved to enable a biased-diffusion mechanism with sufficiently rapid kinetics (40 Å is the smallest step a MT-binding element with a specific footprint can take longitudinally along the microtubule). Our finding that the Ndc80 toe serves as a tubulin conformation sensor suggests MT subunit loss is not required to bias diffusion: the curving of protofilaments at a depolymerizing end would suffice40. Depolymerizing ends have been observed at metaphase kinetochores in vivo, apparently stabilized by attached filaments of unknown identity (Fig. 4b)41.
While the KMN network and the Ndc80 complex are conserved from yeast to humans, the mechanism of microtubule attachment may not be. We propose that fungal Ndc80 has diverged to act as a coupler to the Dam1 complex, and our structural results, coupled with conservation analysis, suggest a possible binding site for Dam1 on microtubule-bound Ndc80 (Supplementary Discussion, Supplementary Fig. 13).
We have used cryo-EM and molecular docking to define the interface at the heart of metazoan kinetochore-microtubule attachment. Our studies also demonstrate the microtubule-mediated self-assembly of the Ndc80 complex, which directly involves the Aurora B-regulated N-terminus of the Ndc80 protein.
Methods Summary
Cryo-electron microscopy and helical reconstruction
Taxol-stabilized microtubules were decorated with Ndc80 bonsai after application to glow-discharged C-flat grids (Protochips), then plunge-frozen in ethane slush. Images were collected on KODAK SO-163 film with a Tecnai F20 electron microscope operating at 200kV at a nominal magnification of 50,000× with dose of 15 electrons/Å2. Micrographs were digitized with a Nikon Super CoolScan 8000 scanner with a step size of 6.35 μm. Image processing and projection-matching alignment was carried out using programs from the EMAN, IMAGIC, and SPIDER packages, and final refinement and CTF correction was performed with a version of FREALIGN adapted for this work to implement helical symmetry. Visualization and molecular docking was performed with UCSF Chimera. Amplitude-weighted difference maps were calculated using the program DIFFMAP (available at: http://emlab.rose2.brandeis.edu/software). References for all image analysis and visualization software can be found in the Methods section.
Tubulin co-sedimentation assays
Tubulin polymerized with or without conformation-stabilizing drugs was mixed with Ndc80 bonsai, then layered on to a 50% glycerol cushion supplemented with additives and pelleted by ultracentrifugation in a Beckman TLA100 rotor at 90,000 RPM for 10 minutes12. Supernatant and pellet fractions were analyzed by SDS-PAGE.
Electron tomography
Samples were prepared on C-flat grids augmented with a continuous carbon layer, and stained with uranyl formate. Tilt series were collected on 2k×2k CCD cameras using a JEOL3100 microscope operating at 300 kV with the SerialEM package or a Phillips CM200 microscope operating at 200 kV with Digital Micrograph (Gatan Inc.). Processing was carried out with programs from the EMAN, SPIDER, and IMOD software packages. References for all image analysis software can be found in the Methods section.
Methods
Mutagenesis
Ndc80 bonsai point mutations were generated using standard procedures for site-directed mutagenesis. The N-terminus-7D mutant was generated as follows: Synonymous mutations were performed which destroyed the StuI restriction site at bp 81–86 of the Ndc80-Spc25 protein coding sequence and subsequently reintroduced it at bp 250–255. An NdeI restriction site was also created 5' to the beginning of the coding sequence. This construct thus features a swappable N-terminus cassette. A sequence coding for the 7 in vitro verified12 Aurora B phosphorylation sites mutated to aspartic acid (S4D, S8D, S15D, S44D, S55D, S62D, S69D) was synthesized (GENEART) and cloned NdeI-StuI into this construct using standard protocols.
Biochemical sample preparation
Ndc80 bonsai proteins were purified as described12. 10mg/ml bovine brain tubulin (Cytoskeleton Inc.) was polymerized in CB1 buffer (80mM PIPES pH 6.8, 1mM EGTA, 1mM MgCl2, 1mM GTP, 10% glycerol) for 15 minutes at 37°C before addition of 160μM taxol, followed by 30–60 minutes of further incubation. We have found this procedure primarily produces 13 and 14 protofilament microtubules. Microtubules were pelleted at 17,000 RCF in a tabletop microcentrifuge for 20 min., then resuspended in room temperature EM Buffer (80 mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 0.05% Nonidet P-40) supplemented with 160 μM taxol. Tubulin concentration was assayed by A280 after depolymerization on ice with 50mM CaCl2.
Ndc80 bonsai at 2 mg/ml was rapidly thawed before storage on ice, diluted 1:1 with EM Buffer, then desalted into EM Buffer using a Zeba Spin Desalting Column (Pierce). The removal of salt resulted in a rapid precipitation of the sample, which was clarified by ultracentrifugation in a Beckman TLA100 rotor for 20 minutes at 40,000 RPM at 4°C. The sample was then warmed to room temperature for approximately 15 minutes prior to EM grid preparation, which resulted in a second round of precipitation/self-association. Analysis of the sample at this stage by negative-stain EM did not reveal any ordered assemblies (Supplementary Fig. 10). The sample was once again clarified for 3 min. at 17,000 RCF in a tabletop microcentrifuge. Protein concentration was estimated with the Coomasie Plus Protein Assay Reagent (Pierce) using Bovine Serum Albumin (BSA) as a reference. It is notable that the amount of precipitation was dependent on the state of the N-terminal domain: the Bonsai ΔN and N-terminus-7D constructs were significantly more stable under low-salt conditions.
Cryo-sample preparation
C-flat grids (Protochips) were glow-discharged using an Edwards Carbon Evaporator. Taxol-stabilized microtubules were diluted to 0.25 mg/ml in EM Buffer supplemented with 20 μM taxol, and 2 μl was applied to the grid in the humidity chamber of a Vitrobot (Maastricht Instruments). After 1 minute, 4 μl of ~0.7 mg/ml Ndc80 bonsai was added, corresponding to a molar ratio of ~2:1 Ndc80:tubulin monomer, and incubated for 1 minute. The grid was then briefly blotted before a second 4 μl addition of Ndc80 and 1 minute incubation. 2 μl was then removed from the grid, which was then blotted for 2 seconds and plunged into ethane slush. This protocol is essentially similar to that employed for visualization of the C. elegans complex21, with minor modification.
Cryo-electron Microscopy
100 micrographs were collected on KODAK SO-163 film with a Tecnai F20 electron microscope operating at 200 kV at a nominal magnification of 50,000X with dose of 15 electrons/Å2 between 1.2 μm and 2.2 μm underfocus. Micrographs were digitized with a Nikon Super CoolScan 8000 scanner with a step size of 6.35 μm. After digitization the power spectra of carbon present in each image was examined, and images lacking Thon rings to at least 8 Å were excluded.
Image processing and IHRSR
We carried out helical processing essentially as described19, with some modifications. Unless otherwise indicated, all processing steps were implemented with the SPIDER package42. The pixel size of the digitized micrographs was calibrated to be 1.24 Å by examination of the power spectrum of samples of Tobacco Mosaic Virus (TMV) imaged under identical conditions. CTF parameters were estimated for each image using the program CTFFIND343. Images were divided into three approximate defocus groups (1.2 μm, 1.7 μm, 2.2 μm). Segments 768 pixels long were extracted from the micrographs with the helix option in BOXER44, using 90% overlap20. Pixel intensities were normalized, and large-scale gradients in intensity due to variations in ice thickness were subtracted. A two-dimensional wiener filter was then applied to each segment, with the intention of aiding the detection of highly ordered segments, which should diffract to high-resolution. Subsequent experience has demonstrated that this step is not necessary for robust classification based on helical quality, and may in fact be detrimental in cases with a low signal-to-noise ratio in the low spatial frequency regime, as amplification of high-resolution spatial frequency components in raw images drives alignment based on noise rather than signal. The segments were then masked such that each image contained approximately 2 turns of the 1-start helix, corresponding to 78 asymmetric units, and decimated three-fold to a pixel size of 3.72 Å.
We then subjected the data to reference-free two-dimensional classification as described19. This method successfully sorts segments based on protofilament number and helical quality, as is apparent in Supplementary Fig. 2. Since microtubules can incorporate varying numbers of protofilaments in vitro45, and segments vary in their quality, this method is used as an alternative to assessing each individual microtubule by manual inspection of moiré patterns and power spectra. Out of an initial set of 10,253 segments, only 1,475 were members of classes which corresponded to well-ordered 13 protofilament microtubules and were selected for further processing.
Segments from defocus groups were combined prior to IHRSR with SPIDER. Using a naked microtubule as a reference, the reconstruction converged after 10 rounds of refinement. Particles with cross-correlation scores more than one standard deviation below the mean were excluded, corresponding to approximately 30% of the data. The final refined helical parameters correspond to a rise of 42.7 Å per tubulin monomer. This is slightly greater than the reported value of 41.7 Å for taxol-stabilized microtubules46, but could result from an error in our pixel size estimation since TMV was not observed in situ. The full three-start symmetry of the microtubule was applied after refinement was complete by real-space averaging. The resolution of the reconstruction at this stage was limited to approximately 20 Å (bronze reconstruction, Supplementary Fig 3), as filtering the volume at higher resolution did not reveal any additional features.
This limit in resolution was found to be at least partially due to limited pixel sampling. With no further alignment of the particles, simply reducing the decimation factor of the original CTF-corrected data from three-fold to two-fold (2.48 Å/pixel) resulted in a discrete jump in resolution, to approximately 12–15 Å resolution (silver reconstruction, Supplementary Fig 3). The mask on each particle was removed at this stage, increasing the number of asymmetric units incorporated into the reconstruction, since not all segments incorporated into the reconstruction were from adjacent positions in microtubule filaments. Approximately 50,000 asymmetric units were incorporated at this stage, and secondary structure elements began to be visualized when a B-factor of −450 Å2 was applied with the program bfactor (available at: http://emlab.rose2.brandeis.edu/software) using the cosine edge mask option with a radius of 8.5 Å. This model was used as the input for final refinement in Frealign.
Final Refinement with Frealign
We used the computer program Frealign47 to refine our reconstruction. Frealign was originally designed to work with single particle images. To work with helical particles, we implemented a helical symmetry operator (symmetry operator H in Frealign) and followed a reconstruction algorithm described previously48. Briefly, the stack of overlapping sections of the helical particles is processed as a conventional single particle stack except for the following differences: Masking of the helical sections is done using a rectangular mask that is aligned with the helical axis of the particle in each section. Using the helical symmetry operator, a set of Euler angles and shifts producing equivalent views for each section can be calculated. Using this set of alignment parameters, each section is inserted multiple times into the reconstruction. The number of insertions is matched to the degree of overlap between helical sections such that it corresponds to the number of symmetry-related subunits contained in the non-overlapping part of each section. In the case of the Ndc80-decorated microtubules, the unique, non-overlapping area of the filament was chosen to contain approximately one helical turn of 13 subunits. In addition to this, the 3-start helical symmetry of the 13 protofilament microtubule increased the number of unique subunits per segment 3-fold, to 39. Therefore, in the present case, each helical segment is inserted into the 3D reconstruction 39 times using Euler angles and shifts that represent equivalent views of the particle. The final reconstruction is masked using a cylindrical mask with user-defined radius, and the helical symmetry is imposed in real space (option BEAUTIFY in Frealign) to remove the small density gradient along the helical axis due to the reconstruction algorithm48.
For the refinement of Euler angles and shifts for each helical section, the reference projections from the helical reference are masked with a soft-edged mask to reset the image density to background level in a 10% margin near the edges of the image. This is necessary to avoid truncation of reference projections at the edges when applying the small shifts typically observed during alignment. Furthermore, the refinement is carried out using the weighted correlation coefficient49, but without the use of the absolute value. For helical structures, the low-resolution correlation terms are essentially insensitive to shifts along the helical axis and, therefore, if the absolute value is used, the high-resolution layer lines may be rendered out of phase during alignment. Finally, the Euler angles describing in-plane and out-of-plane alignments of each section are restrained as previously described50, but using separate statistics for each segment. No restraints were used for the shifts. The final reconstruction was scaled to the same amplitude profile as the input reconstruction, and is shown in gold in Supplementary Fig. 3.
Difference Map Calculation
The experimental map was segmented, and an appropriate number of Ndc80 bonsai and tubulin crystal structures were docked into the map to approximately account for the total mass with UCSF Chimera51. These crystal structures were converted into SPIDER volume format using the CP FROM PDB command, then filtered to 8 Å resolution. Although this procedure retained the orientation of the crystal structures relative to each other, this volume had to be re-docked into the experimental map, which was accomplished using a cross-correlation search with SPIDER. The amplitude-weighted difference map was calculated using the program DIFFMAP (available at: http://emlab.rose2.brandeis.edu/diffmap). Amplitude weighting was critical for a meaningful comparison, as the amplitudes of the volume derived from the crystal structures were down-weighted several hundred fold in all resolution shells.
Tubulin co-sedimentation assays
For biochemical assays Ndc80 bonsai proteins were desalted into Binding Buffer (80 mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 5% sucrose). We found that this buffer increased the solubility of the protein in the absence of salt while allowing for accurate determination of protein concentration by A280. The phenyl group of Nonidet P-40 results in significant absorbance of EM Buffer at 280 nm, which is refractory to reproducible protein concentration measurement by this method. The presence of sucrose significantly reduces contrast in the electron microscope, and thus Binding Buffer was not used for any of the imaging experiments shown. In control imaging experiments we have found no differences in Ndc80-microtubule interactions or tubulin polymer behavior between the two buffers (data not shown).
To generate vinblastine spirals, we diluted tubulin stored in CB1 buffer to 3 mg/ml in Binding Buffer supplemented with 1 mM vinblastine sulfate (Sigma) at room temperature. After two hours, robust formation of spiral aggregates was observed (Supplementary Fig. 6).
Taxol microtubules or vinblastine spirals (6 μM tubulin monomer) and Ndc80 bonsai proteins (0.5 μM) were mixed in a 50 μl reaction volume and incubated at room temperature for 15 minutes in the presence of 20 μM taxol or 1 mM vinblastine. The binding reactions were layered on to a 100 μl 50% glycerol cushion containing buffer components and the appropriate drug and polymers were pelleted by ultracentrifugation at 90,000 R.P.M. in a Beckman TLA 100 rotor for 10 minutes at 25°C, essentially as described12. Supernatant and pellet fractions were collected and precipitated in 90% ethanol at −20°C for 16 hours prior to analysis by SDS-PAGE. Gels were stained with Flamingo Fluorescent Gel Stain (Bio-Rad), and imaged with a Typhoon Trio (GE Healthcare). Apparent slight degradation of the Ndc80 complex was observed in the presence of 1 mM vinblastine sulfate (Fig. 2b), but this did not cause spurious pelleting of the complex in the absence of tubulin. Quantification was performed with ImageJ52
To test for cold stabilization, 10 mg/ml tubulin in CB1 buffer was polymerized for 45 minutes at 37°C. A binding reaction containing Ndc80 bonsai in Binding Buffer was set up on ice, and then heated to 37°C for 1 minute. Dynamic microtubules were then added to this reaction for a final concentration of 20 μM tubulin and 3.3 μM Ndc80 and incubated at 37°C for 10 minutes. The reaction was then shifted to ice for 30 minutes, and subsequently analyzed by pelleting assay as above, except ultracentrifugation was performed at 4°C.
To analyze the outcomes of the described experiments by negative stain EM, we repeated them substituting EM Buffer for Binding Buffer. Samples were prepared on continuous carbon grids, stained with 2% uranyl acetate, and imaged on a 1k×1k CCD camera with a Tecnai T12 electron microscope operating at 120kV between approximately 1 μm and 2 μm underfocus.
Negative stain sample preparation for tomography
Samples were prepared as described for cryo-EM, except that the grids were augmented with a layer of thin carbon and treated with gold fiducial markers (British BioCell International) prior to sample application. To achieve sub-saturating binding, wild-type Ndc80 bonsai was diluted to 0.15 mg/ml and only a single application was performed, corresponding to a 1:2 Ndc80:tubulin monomer ratio. To generate SMTs, taxol-stabilized microtubules were digested with 1.5 μM subtilisin A (Calbiochem) in EM Buffer for 30 minutes at 37°C. The reaction was stopped with 2 mM PMSF, and the SMTs were pelleted and resuspended in EM Buffer prior to sample preparation.
To achieve saturation in the case of wild-type and to observe binding in the case of Bonsai ΔN and N-terminus-7D, double-application of 0.6–0.7 mg/ml Ndc80 was performed, corresponding to a 2:1 Ndc80:tubulin monomer ratio. Samples were stained with 2% uranyl formate.
Electron Tomography and Cluster Quantification
The tomograms displayed in Figure 1 were derived from tilt series collected on a 2k×2k CCD camera from −60° to 60° using a JEOL 3100 microscope operating at 300 kV at approximately 2 μm underfocus with a nominal magnification of 40,000×. The acquisition was performed semi-automatically using a version of SerialEM (available at: http://bio3d.colorado.edu) adapted for operating JEOL microscopes. Raw tomographic image stacks were aligned either manually with the eTomo suite of progams, or automatically using the software RAPTOR53. Tomographic reconstructions were constructed using the eTomo suite of programs and visualized using the IMOD software package54.
The tomograms displayed in Figure 3 were derived from tilt series collected from −65° to 65° on a 2k×2k CCD camera using a Phillips CM200 microscope operating at 200 kV at approximately 2.5 μm underfocus at a nominal magnification of 39,000×. Images were filtered to 25–30 Å, before the first phase-inversion of the contrast-transfer function, and decimated 2-fold prior to reconstruction as described above.
Cluster quantification was performed by manual inspection of the reconstructions. The raw data are shown in Supplementary Fig. 9, and consist of number of counts versus cluster size. Clusters ≥10 molecules in size were ignored in the case of the wild-type, as we believe they are not physiologically relevant and would bias comparisons between the distributions towards spurious difference. Pair-wise comparisons were performed using Welch's t-test55, which is appropriate for samples featuring both different numbers of observations and possibly unequal variances (which the unbiased estimator of the variance of the distributions suggested in this case), and p-values are shown in Supplementary Table 1. In all but one of the cases p < 0.0015, suggesting that the data derived from each of the conditions shown in Fig. 3 and Supplementary Fig. 9 do indeed sample different distributions. The exception was N-terminus-7D versus Bonsai ΔN, where we find a probability of 0.61 of sampling the same distribution. This supports our assertion that these two mutants phenocopy each other.
Conservation Analysis and Structure Alignments
We performed multiple-sequence alignments using CLUSTALW256, and mapped this analysis on to the Ndc80 bonsai crystal structure using the CONSURF server57. The alignments included 48 fungi sequences and 39 metazoan sequences in the case of Ndc80 and 52 fungi sequences and 22 metazoan sequences in the case of Nuf2, and are available on request. α- and β-tubulin sequences from six representative organisms (S. cerevisiae, C. elegans, D. melongaster, X. laevis, B. taurus, and H. sapiens) were compared. Since the Ndc80 bonsai crystal structure is of the human complex, fungal sequences had to be threaded on to the structure by alignment with the human sequence. Since we observe a similar pattern of conservation at the toe-print interface, we believe this procedure was successful. All super-positions of crystal structures were performed with the MatchMaker function in UCSF Chimera51.
Supplementary Material
Acknowledgements
We are grateful to Ken Downing for supporting the work carried out by D.A.B., to Claudio Ciferri for his knowledge and advice on the Ndc80 complex and critical reading of the manuscript, and to Patricia Grob and Slaton Lipscomb for EM and computer support, respectively. We would also like to acknowledge Dieter Typke and Bob Glaeser for advice on data collection, and Chuck Sindelar for discussion of data processing strategies. This work was funded by a grant from the National Institute of General Medical Sciences (E.N). E.N. and N.G. are Howard Hughes Medical Institute Investigators.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 3.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 4.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 5.DeLuca JG, et al. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 6.Kemmler S, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. Embo J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wigge PA, et al. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciferri C, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 11.Wei RR, et al. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HW, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 19.Ramey VH, Wang HW, Nogales E. Ab initio reconstruction of helical samples with heterogeneity, disorder and coexisting symmetries. J Struct Biol. 2009;167:97–105. doi: 10.1016/j.jsb.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci U S A. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoenger A, Gross H. Structural investigations into microtubule-MAP complexes. Methods Cell Biol. 2008;84:425–444. doi: 10.1016/S0091-679X(07)84014-3. [DOI] [PubMed] [Google Scholar]
- 24.des Georges A, et al. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat Struct Mol Biol. 2008;15:1102–1108. doi: 10.1038/nsmb.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 26.Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson L, Jordan MA, Morse A, Margolis RL. Interaction of vinblastine with steady-state microtubules in vitro. J Mol Biol. 1982;159:125–149. doi: 10.1016/0022-2836(82)90035-3. [DOI] [PubMed] [Google Scholar]
- 28.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joglekar AP, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 32.Andrews PD, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 33.Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- 41.McIntosh JR, et al. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 43.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 44.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 45.Wade RH, Chretien D, Job D. Characterization of microtubule protofilament numbers. How does the surface lattice accommodate? J Mol Biol. 1990;212:775–786. doi: 10.1016/0022-2836(90)90236-F. [DOI] [PubMed] [Google Scholar]
- 46.Arnal I, Wade RH. How does taxol stabilize microtubules? Curr Biol. 1995;5:900–908. doi: 10.1016/s0960-9822(95)00180-1. [DOI] [PubMed] [Google Scholar]
- 47.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Sachse C, et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol. 2007;371:812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Chen JZ, et al. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 53.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161:260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 55.Welch BL. The generalisation of student's problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 56.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Landau M, et al. Consurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.