Abstract
New gene regulation study tools such as microRNA (miRNA or miR) analysis may provide unique insights into the remarkable ability of the liver to regenerate. In addition, we have previously shown that ursodeoxycholic acid (UDCA) modulates mRNA levels during liver regeneration. Bile acids are also homeotrophic sensors of functional hepatic capacity. The present study was designed to determine whether miRNAs are modulated in rats following 70% partial hepatectomy (PH) and elucidate the role of UDCA in regulating miRNA expression during liver regeneration (LR). Total RNA was isolated from livers harvested at 3–72 h following 70% PH or sham operations, from both 0.4% (wt/wt) UDCA and control diet-fed animals. By using a custom microarray platform we found that several miRNAs are significantly altered after PH by >1.5-fold, including some previously described as modulators of cell proliferation, differentiation, and death. In particular, expression of miR-21 was increased after PH. Functional modulation of miR-21 in primary rat hepatocytes increased cell proliferation and viability. Importantly, UDCA was a strong inducer of miR-21 both during LR and in cultured HepG2 cells. In fact, UDCA feeding appeared to induce a sustained increase of proliferative miRNAs observed at early time points after PH. In conclusion, miRNAs, in particular miR-21, may play a significant role in modulating proliferation and cell cycle progression genes after PH. miR-21 is additionally induced by UDCA in both regenerating rat liver and in vitro, which may represent a new mechanism behind UDCA biological functions.
Keywords: bile acids, liver regeneration, miR-21, miRNAs
the liver displays a remarkable capacity to regenerate after injury, ischemia, and resection. In this process, quiescent liver cells go through a series of well-orchestrated steps within distinct and recognizable stages of priming, cell cycle progression, proliferation, growth, and, finally, arrest (34). Interestingly, bile acids may play a key role during the regenerative phases. A recent study showed that bile acids may regulate liver regeneration (LR) through farnesoid X receptor α and thus be considered members of the growing family of hepatomitogens (16). Through this receptor, bile acids may stimulate FoxM1b, a proliferation-specific transcription factor, known to act in hepatocyte proliferation. Furthermore, we have shown that during rat LR there are significant modifications in the bile acid pool that markedly affect the levels in whole liver tissue (21). Ursodeoxycholic acid (UDCA) and deoxycholic acid (DCA) feeding differentially modulate steady-state mRNA levels in the liver, with profound implications in regeneration. Bile acids are also capable of modulating apoptosis. In the liver, UDCA usually acts as a prosurvival agent, whereas DCA is strongly proapoptotic (7, 9). Interestingly, apoptosis represents a key process in fine tuning the regenerative response (12). Finally, our recent gene expression profiling studies have shown that UDCA and its taurine-conjugate derivative, tauroursodeoxycholic acid (TUDCA) modulate several transcripts that might influence LR, including genes involved in apoptosis, cell cycle control, and proliferation, transcription factors, and proto-oncogenes (7, 8).
microRNAs (miRNAs or miRs) comprise a class of small noncoding RNAs that, in general, negatively modulate gene expression, thus playing diverse regulatory roles during development and cellular homeostasis. Cellular activities such as differentiation, proliferation, and apoptosis are typically regulated by miRNAs. In fact, we have recently shown that miRNAs may play a role in liver apoptosis by acting as a mitochondrial rheostat for gene regulation (23). Nevertheless, although little is known about their involvement in tissue and organ regeneration, recent studies would suggest that miRNAs play a prominent role. For example, during pancreas regeneration miR-15a, -15b, -16, and -195 may be responsible for neurogenin 3 inhibition (18). Changes in miRNA expression at specific phases of the regenerative process have also been observed during wound healing (5). In addition, regeneration is optimized through depletion of miR-133 in amputated zebrafish fins (43). Finally, miRNA-mediated gene regulation may also contribute to the control of homeostasis and regeneration in planarians, free-living flat worms best known for their regenerative capacity (29). Similarly, the list of studies on the role of miRNAs during LR is growing. We have shown that miRNAs are differentially expressed in free, cytoskeleton-bound, and membrane-bound polysome populations during LR (24). Moreover, it was recently shown that miRNAs control hepatocyte proliferation during LR in mice (26, 33).
In this study, we determined miRNA profiles from rat quiescent nonresected liver and regenerating liver, from 3 to 72 h after 70% partial hepatectomy (PH). Importantly, we also examined the effect of UDCA feeding in modulating regeneration-associated miRNA profiles and its potential role in liver repair from injury.
MATERIALS AND METHODS
Animals and diets.
Male Sprague-Dawley rats (Harlan Laboratories, Indianapolis, IN) weighing ∼175 g were maintained on a 12-h light-dark cycle and fed standard laboratory chow ad libitum for 3 days. Animals were then fed a diet of standard laboratory chow supplemented with 0.4% (wt/wt) UDCA or no bile acid addition (control). Triplicate gas chromatography analyses of the individual diets provided accurate concentrations of the bile acid, which closely matched the estimated feed composition. After 2 wk on the supplemented diets, animals were weighed and either 70% PHs or sham operations were performed under isoflurane anesthesia (21). At various times post-PH, the animals were killed by exsanguination under isoflurane anesthesia. The blood was collected for gas chromatography bile acid analysis (21). The liver remnant was removed, rinsed in normal saline, immediately flash frozen in liquid nitrogen, and stored at −80°C.
All animal work was conducted according to protocols submitted to and approved by the Institutional Animal Care and Use Committee at the University of Minnesota. In addition, all animals received humane care in compliance with the institute's guidelines, outlined in Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health (publication no. 86-23, revised 1985).
RNA extraction and processing.
Liver samples were processed for RNA extraction with the TRIZOL Reagent (Invitrogen, Carlsbad, CA). Briefly, the frozen tissue was placed in TRIZOL Reagent and immediately homogenized by using a prechilled mortar and pestle. Samples were then processed for isolation of total RNA, following the manufacturer's instructions. RNA samples typically showed A260/280 ratios between 1.9 and 2.1. RNA integrity was further confirmed by use of denaturing agarose gels.
Preparation of labeled RNA and array hybridization.
Microarray analysis was performed by using a custom microarray platform, highly sensitive, allowing the profiling of even weakly expressed miRNAs (19). Briefly, a miRNA probe set containing ∼1140 oligonucleotides as probes, complementary to Caenorhabditis elegans, Drosophila, zebrafish, mouse, rat, and human miRNAs was purchased from Invitrogen. The set also included a number of internal and negative control probes. Oligonucleotides were printed in quadruple on Corning GAPSII-coated slides in the Microarray Facility at the University of Minnesota. For RNA labeling, 25 μg of total RNA was ligated to 0.5 μg of a synthetic linker, pCU-DY547 (Dharmacon, Lafayette, CO). To control the hybridization process, reference DNA oligonucleotides complementary to a subset of mammalian miRNAs were combined and labeled with a ULYSIS Alexa Fluor 647 Kit (Invitrogen). Labeled RNAs and DNAs were then mixed and hybridized to microarray slides (38). Finally, slides were scanned with a ScanArray 5000 machine (Perkin Elmer, Waltham, MA).
Data filtering and analysis.
Microarray images were processed with Bluefuse software (BlueGnome, Cambridge, UK) to quantify pixel intensities. Individual spots on the slides were further inspected to exclude abnormal spots from subsequent calculations. To determine the changes in miRNA expression between sham- and PH-operated animals and/or control and UDCA-fed animals, Bluefuse raw data were analyzed via GeneSpringGX v 10.0 (Agilent Technologies, Santa Clara, CA). Samples were first normalized relative to 28s rRNA and baseline corrected to the median of all samples. Replicate data were consolidated into groups based on feeding (control and UDCA), treatment (sham and PH), and time (3, 6, 12, 18, 24, 36, 48, and 72 h) following PH and were organized by using the hierarchical clustering and analysis of variance (ANOVA) functions in the GeneSpring software. Hierarchical clustering was done by use of the clustering function (condition tree) and Euclidean correlation as a distance metric. Two-way ANOVA analysis and asymptotic P value computation without any error correction on the samples was performed to search for miRNAs which varied most prominently across the different groups. P value cutoff was set to 0.05. Only changes ≥50% in at least one of the time points for each sample were considered significant.
Northern blot analysis.
Approximately 20 μg of RNAs from each of the different time point samples were run on a 7 M urea/10% polyacrylamide gel and transferred to a Hybond-N+ membrane (GE Healthcare, Fairfield, CT) for Northern blot detection of miRNAs (19). The membrane was then probed with an end-labeled oligonucleotide complementary to the miRNA of interest and analyzed using a PhosphorImager (GE Healthcare). Sequences of the oligonucleotide end-labeled probes were miR-21, 5′-GCTAGTCAACATCAGTCTGATA-3′; and miR-19b, 5′-TCAGTTTTGCATGGATTTGCACA-3′.
Quantitative RT-PCR.
Real-time RT-PCR was performed using an Applied Biosystems 7300 System (Applied Biosystems, Foster City, CA), to quantitate and confirm the expression of selected miRNAs that were identified by microarray analysis. For each sample and miRNA tested, the RT reaction was performed using the TaqMan MicroRNA Reverse Transcription kit from Applied Biosystems. U64 snRNA was used as the normalization control for rat livers and primary hepatocyte cultures, whereas RNU6B served as the normalization control for HepG2 cells. Each RT reaction contained 0.66 ng/μl of total RNA, 1× stem-loop RT specific primer, 1× reaction buffer, 0.25 mM each of dNTPs, 3.33 U/μl Multiscribe RT enzyme, and 0.25 U/μl RNase inhibitor. The 15-μl reactions were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. The PCR reaction was performed by use of a standard TaqMan PCR kit protocol (Applied Biosystems). Briefly, following the RT step, 1.33 μl of the RT reaction were combined with 1 μl of a TaqMan MicroRNA Assay (20×; forward primer, reverse primer, and probe) and 17.67 μl of TaqMan Universal PCR Master Mix, No AmpErase UNG in a 20-μl final volume. The reactions were incubated in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, and were run in triplicate. The relative amounts of miRNAs were determined by the 2−ΔΔCt method, where ΔΔCt = (Cttarget − CtU64) sample − (Cttarget − CtU64) calibrator.
Cell cultures and treatments.
Primary rat hepatocytes were isolated from male Sprague-Dawley rats (100–150 g) by collagenase perfusion (25). After isolation, hepatocytes were resuspended in complete William's E medium and plated on Primaria tissue culture dishes (BD Biosciences, San Jose, CA) at 5 × 103 cells/cm2. The cells were maintained at 37°C in a humidified atmosphere of 5% CO2 for 4 h, to allow attachment. Plates were then incubated in William's E medium containing 10% heat-inactivated fetal bovine serum (Invitrogen) and when indicated treated with 100 μM UDCA or DCA (Sigma-Aldrich, St. Louis, MO), or no addition (control), for 4, 16, 24, 40, and 48 h before harvest for RNA isolation. In parallel studies, HepG2 cells were grown in Dulbecco's modified Eagle's Medium (Invitrogen) supplemented with 10% serum, 1% nonessential amino acids, 100 units/ml penicillin, 100 U/ml streptomycin, and 0.25 U/ml amphotericin B. Cells were plated at 5 × 103 cells/cm2 and maintained at 37°C in a humidified atmosphere of 5% CO2 for 12 h. UDCA or DCA were then added to cells for 2, 4, 6, 24, and 48 h before harvest for RNA isolation.
For functional analyses, primary rat hepatocytes were transfected 4 h after plating with a specific miR-21 precursor (Pre-miR-21; Applied Biosystems) or a Pre-miR negative control at 60 nM, by using the siPORT NeoFX transfection agent (Applied Biosystems). Alternatively, miR-21 silencing was performed by transfecting hepatocytes with a miR-21-specific inhibitor (anti-miRNA-21), in parallel with an anti-miRNA negative control (Applied Biosystems). To further validate the experimental model, cells were cotransfected with a miR-21 sensor comprising two sequences complementary to mature miR-21 (pGL3-miR-21 sensor) or with a pGL3-control plasmid (Promega, Madison, WI). pRL-SV40 (Promega) was used for transfection normalization. Cells were incubated with 100 μM UDCA or DCA, 4 h after transfections. The hepatocytes were harvested at 24 and 48 h posttransfection and processed for miR-21 expression analysis and cell viability assays, respectively.
Evaluation of cell proliferation and death.
At the indicated times, general cell death was evaluated by the LDH assay kit (Sigma). LDH activity was evaluated in cell culture media, using a Bio-Rad microplate reader model 680 (Bio-Rad, Hercules, CA). Cell proliferation was evaluated with the CellTiter96 AQueous Non-Radioactive Cell Proliferation Assay (Promega), using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS). Finally, cells were processed for luciferase assay and transfection efficiency normalization.
Luciferase activity.
At the indicated times, firefly and Renilla luciferase activities were measured by use of the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used as a transfection normalization control.
RESULTS
Comprehensive miRNA expression analysis after partial hepatectomy in rats.
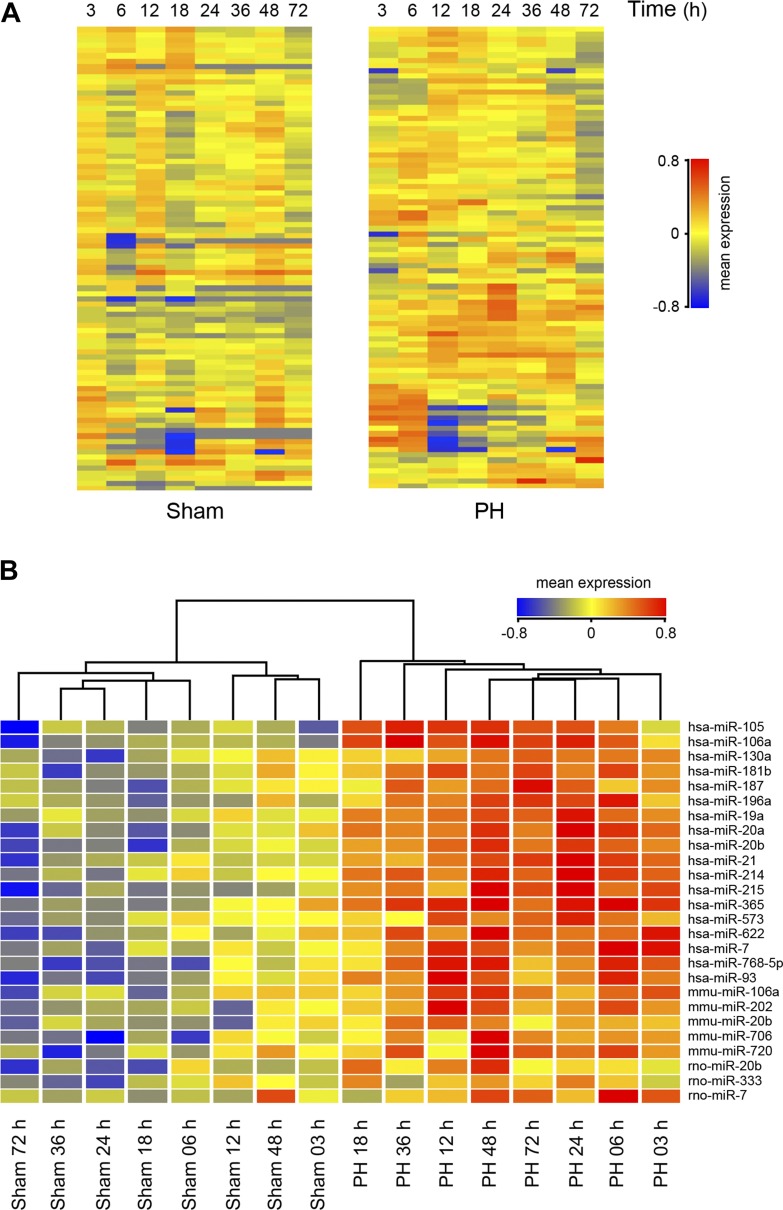
Using a custom microarray platform, we analyzed expression levels of hundreds of mammalian (human, mouse, and rat) miRNA probes, from 3 to 72 h post-PH in rats, during which time critical proliferation and cell division signals are activated. A clearly distinct miRNA microarray profile was observed between sham- and PH-operated animals, from 3 to 72 h after surgery (Fig. 1A). Supervised hierarchical clustering analysis, using Pearson correlation, showed that sham-operated animals clustered together and separately from the PH-operated rats (Fig. 1B). This subset of 26 miRNAs was found to exhibit >1.5-fold alterations in expression levels following PH, in at least one of the time points (Table 1). Although significant changes were identified as upregulation of miRNA expression levels, a few miRNAs displayed a negative trend. Validated targets for identified miRNAs included several cell cycle-, growth-, and proliferation-related genes, which may play a role during LR. These were retrieved from miRecords (http://mirecords.umn.edu/miRecords), a resource for animal miRNA-target interactions developed at the University of Minnesota.
Fig. 1.
Differentially expressed liver microRNAs (miRNAs or miRs) in partial hepatectomy (PH)- vs. sham-operated rats, from 3 to 72 h after PH. A: distribution of 88 distinct miRNAs in the regenerating liver. These miRNA probes represent the transcripts that gave the strongest normalized hybridization signals in all sampled populations. Overall, miRNA expression was increased in PH samples. B: hierarchical clustering of miRNA genes whose expression was significantly altered by >1.5-fold in at least 1 time point after PH. Each row represents an individual gene and each column represents 3 RNA liver samples from 3 different animals. Results are presented to highlight the clustering analysis, with the most distant groups on the left and right extremes of the hierarchical tree. The relative abundance of individual miRNAs in different time point populations is presented as heat maps, with the lowest and highest intensity values in blue and red, respectively. The scale is shown at right, with the relative abundance in arbitrary units.
Table 1.
Modulation of miRNA expression profiles during liver regeneration
| Time after PH, h |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | 3 | 6 | 12 | 18 | 24 | 36 | 48 | 72 | Validated targets |
| hsa-miR-105 | 1.26 | 1.42 | 1.23 | 1.34 | 2.10 | 1.53 | 1.17 | 1.72 | |
| hsa-miR-106a | 1.33 | 1.51 | 1.25 | 1.69 | 2.11 | 1.35 | 1.60 | 1.76 | VEGFA, RB1, AML1, APP |
| hsa-miR-130a | 1.46 | 1.35 | 1.53 | 1.40 | 1.71 | 2.25 | 1.88 | 2.14 | TAC1, CSF1, MAFB, MEOX2, HOXA5 |
| hsa-miR-181b | 1.08 | 0.98 | 1.57 | 2.08 | 1.58 | 1.26 | 1.94 | 1.57 | GLUR2, VSLN1, TCL1A, AICDA |
| hsa-miR-187 | 1.34 | 2.14 | 1.65 | 1.55 | 1.89 | 1.62 | 1.31 | 1.75 | |
| hsa-miR-196a | 1.23 | 1.83 | 1.64 | 1.44 | 1.68 | 2.09 | 1.13 | 1.72 | IKK2, ANXA1, HOXA7, HOXD8, HOXC7, HOXB8 |
| hsa-miR-19a | 1.27 | 1.69 | 2.50 | 1.36 | 1.52 | 1.52 | 1.44 | 1.61 | PTEN |
| hsa-miR-20a | 1.22 | 2.06 | 1.42 | 1.97 | 2.40 | 1.36 | 1.71 | 2.07 | VEGFA, AML1, E2F1, CCND1 |
| hsa-miR-20b | 1.42 | 1.68 | 1.53 | 1.95 | 2.06 | 1.70 | 1.65 | 2.05 | VEGFA |
| hsa-miR-21 | 1.55 | 1.66 | 1.62 | 2.22 | 2.10 | 2.10 | 2.13 | 2.70 | 27 validated targets |
| hsa-miR-214 | 1.03 | 1.51 | 2.02 | 1.32 | 1.47 | 1.51 | 1.26 | 1.40 | PTEN |
| hsa-miR-215 | 1.31 | 1.64 | 1.39 | 1.54 | 1.26 | 1.38 | 1.40 | 2.01 | |
| hsa-miR-365 | 1.76 | 2.13 | 1.55 | 1.11 | 1.97 | 1.60 | 1.66 | 1.69 | |
| hsa-miR-573 | 1.25 | 1.32 | 1.31 | 1.43 | 1.90 | 1.80 | 1.31 | 2.00 | |
| hsa-miR-622 | 1.01 | 1.22 | 1.00 | 1.50 | 2.02 | 1.16 | 1.25 | 1.42 | |
| hsa-miR-7 | 1.38 | 1.97 | 0.89 | 1.24 | 2.17 | 1.77 | 1.81 | 1.75 | PAK1, RAF1, IRS1, IRS2, EGFR |
| hsa-miR-768-5p | 1.20 | 2.04 | 2.28 | 1.77 | 1.97 | 1.70 | 1.57 | 1.84 | |
| hsa-miR-93 | 1.15 | 1.25 | 1.54 | 1.17 | 2.12 | 1.24 | 1.29 | 1.96 | VEGFA, E2F1, p21 |
| mmu-miR-106a | 1.64 | 1.72 | 1.53 | 1.73 | 2.23 | 1.88 | 2.17 | 2.57 | MYLIP, ARID4B |
| mmu-miR-202 | 1.26 | 1.53 | 1.38 | 1.07 | 1.41 | 3.78 | 1.35 | 1.61 | |
| mmu-miR-20b | 1.53 | 1.47 | 1.54 | 1.39 | 2.24 | 1.47 | 1.66 | 2.47 | MYLIP, ARID4B |
| mmu-miR-706 | 1.29 | 2.22 | 1.67 | 1.81 | 2.24 | 1.95 | 1.78 | 1.83 | |
| mmu-miR-720 | 1.51 | 2.20 | 1.41 | 1.07 | 1.29 | 1.37 | 1.01 | 1.61 | |
| rno-miR-20b | 1.43 | 1.39 | 1.46 | 1.42 | 2.74 | 1.69 | 1.63 | 1.91 | |
| rno-miR-333 | 1.36 | 2.48 | 1.72 | 1.43 | 1.89 | 2.20 | 1.97 | 1.52 | |
| rno-miR-7 | 1.23 | 1.93 | 0.93 | 1.18 | 1.83 | 1.74 | 1.49 | 1.70 | |
List of microRNAs (miRNAs, miRs) expressed differentially in partial hepatectomy (PH) animals compared with sham animals. Mean expression values were determined following global normalization and statistical analysis as described in materials and methods. AICDA, activation-induced cytidine deaminase; AML1, runt-related transcription factor 1; ANXA1, annexin A1; APP, amyloid precursor protein; ARID4B, AT rich interactive domain 4B (Rbp1 like); CCND1, cyclin D1; CSF1, colony stimulating factor 1; E2F1, E2F transcription factor 1; EGFR, epidermal growth factor receptor; GLUR2, glutamate receptor ionotropic AMPA 2; HOXA5; homeobox A5; HOXA7, homeobox A7; HOXB8, homeobox B8; HOXC8, homeobox C8; HOXD8, homeobox D8; IKK2, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta; IRS1, insulin receptor substrate 1; IRS2, insulin receptor substrate 2; MAFB, v-maf musculoaponeurotic fibrosarcoma oncogene homolog B; MEOX2, mesenchyme homeobox 2; MYLIP, myosin regulatory light chain interacting protein; p21, cyclin-dependent kinase inhibitor 1A; PAK1, p21 protein (Cdc42/Rac)-activated kinase 1; PTEN, phosphatase and tensin homolog (mutated in multiple advanced cancers 1); RAF1, v-raf-1 murine leukemia viraloncogene homolog 1; RB1, retinoblastoma 1; TAC1, tachykinin precursor 1; TCL1A, T-cell leukemia/lymphoma 1A; VEGFA, vascular endothelial growth factor A; VSLN1, visinin-like 1.
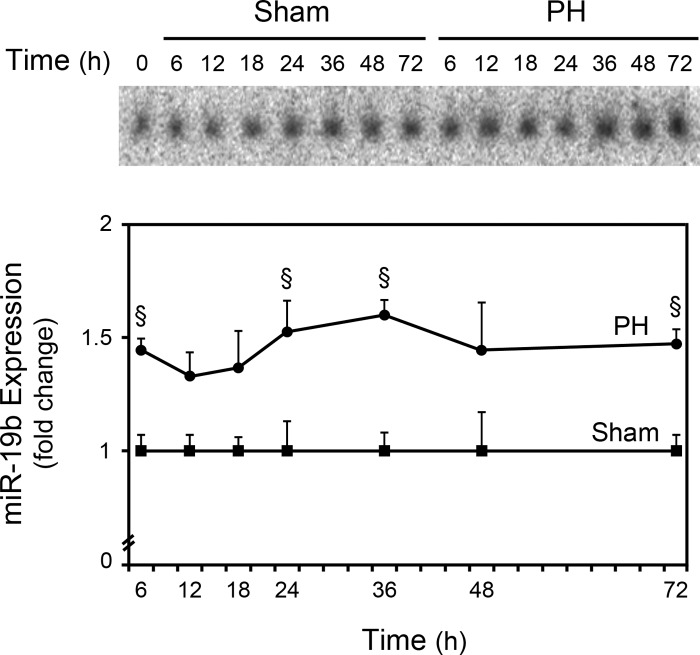
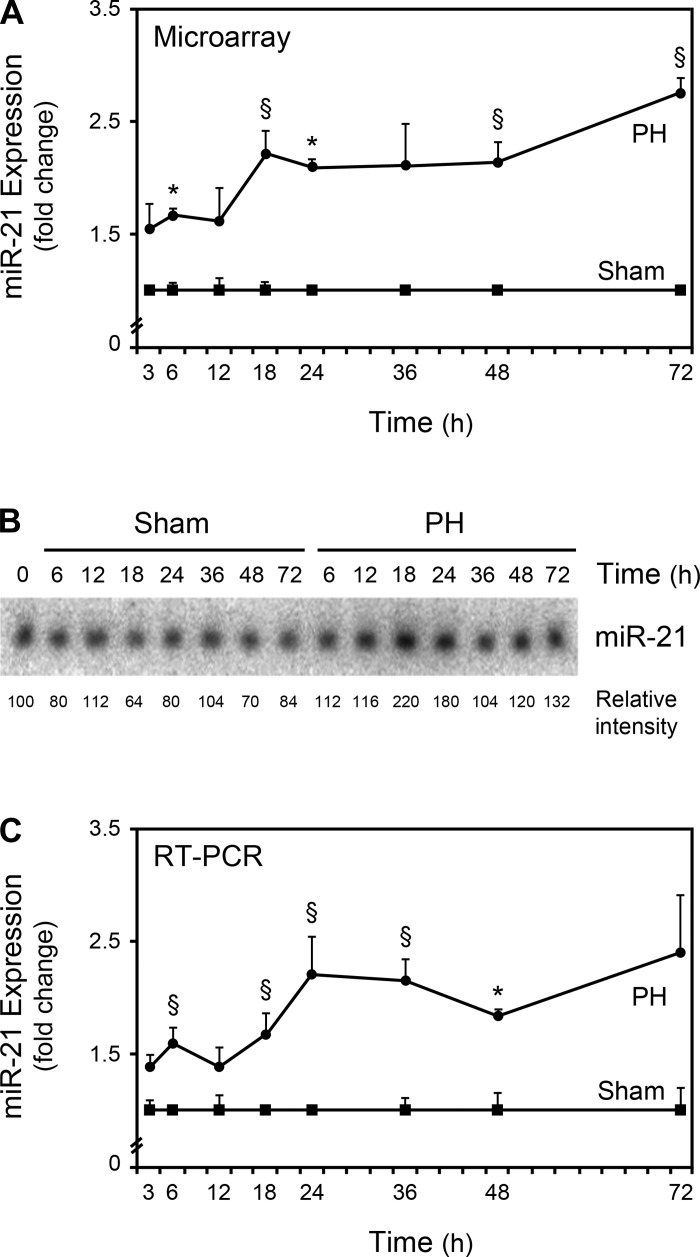
To validate the microarray data, we performed Northern blot and real-time RT-PCR analyses for selected miRNAs. Increased expression of miR-19, belonging to the miR-17–92 cluster, were correlated and in agreement with the microarray results (Fig. 2). miR-21 was found to be significantly increased after PH by microarray analysis (Fig. 3A) and may contribute to liver regeneration through Pellino-1 (26) and Btg2 (33). Furthermore, miR-21 was recently shown to be transcriptionally activated by activator protein 1 (AP-1), signal transducer and activator of transcription-3 (Stat3), and transforming growth factor β (30). Interestingly, all of these proteins play an important role in the initiation of hepatic regeneration (20) and, in this regard, the transcriptional activation of miR-21 may, in fact, constitute an important mechanism of action during LR. The augmented expression of miR-21 was also demonstrated by Northern blot analysis (Fig. 3B). Finally, real-time RT-PCR further confirmed these results (Fig. 3C), demonstrating that our miRNA microarray platform provided a reliable method to measure miRNA expression levels.
Fig. 2.
Northern blot analysis of miRNA-19b. Northern blots were performed as described in materials and methods. Blots were normalized to 5S rRNA and results are expressed as mean ± SE fold change for 3 different animals. A representative Northern blot is shown. §P < 0.05 from the corresponding controls.
Fig. 3.
miR-21 expression is increased after PH. Expression of miR-21 from 3 to 72 h after PH was evaluated by microarray (A), Northern blot (B), and real-time RT-PCR (C) analyses as described in materials and methods. A representative Northern blot is shown. Results are expressed as mean ± SE fold change for 3 different animals. *P < 0.01 and §P < 0.05 from the corresponding controls.
Functional role of mir-21 in liver cells.
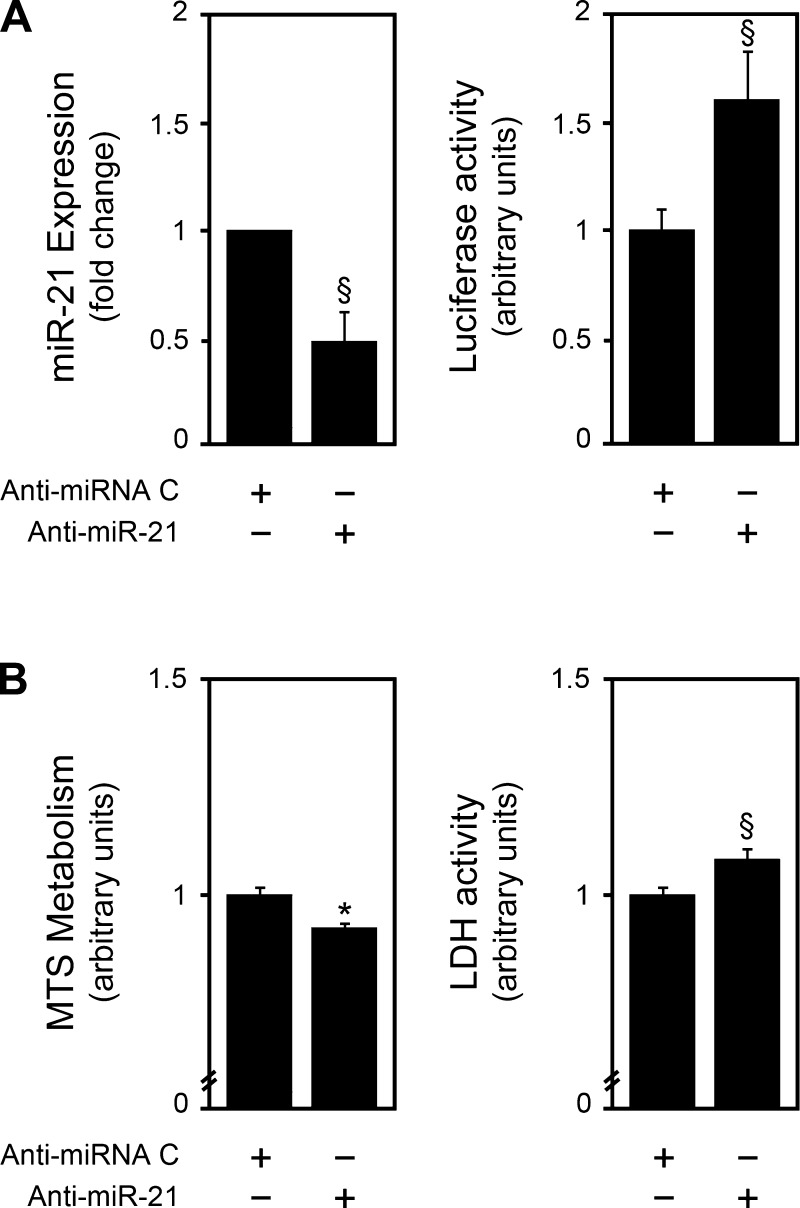
miR-21 is a well-known modulator of cell proliferation and viability in many different cell types. In fact, regardless of which genes are targeted, miR-21 expression is considered to be a proproliferative signal. We characterized the functional effect of miR-21 in primary rat hepatocytes by performing inhibition studies using an anti-miR-21 (Fig. 4A). In accordance with other models, anti-miR-21 decreased cell proliferation (P < 0.01) and viability (P < 0.05) (Fig. 4B).
Fig. 4.
miR-21 modulates cell proliferation and viability in primary rat hepatocytes. Primary rat hepatocytes were transfected with a miR-21 inhibitor as described in materials and methods. miR-21 silencing (A) was assessed by real-time RT-PCR (left) or luciferase activity (right) assays. Cell proliferation and viability (B) were assessed by the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) metabolism (left) and lactate dehydrogenase (LDH) activity (right) assays, respectively. Results are expressed as means ± SE from at least 4 different experiments. §P < 0.05 and *P < 0.01 from the corresponding controls.
Effects of UDCA on miRNA expression profile after PH in rats.
Our previous results have shown that bile acids, including UDCA, are potent modulators of proliferation-related transcripts (8), particularly during LR (21). In addition, UDCA was shown to double the proliferative response of the liver, compared with controls, after PH (3). Therefore, rats were maintained on standard rat chow supplemented with 0.4% (wt/wt) UDCA or no addition for 14 days before PH, and its effects on miRNA expression were assessed 36 h after PH. UDCA plasma levels were used to confirm enrichment in the hepatic portal system. In fact, plasma levels of UDCA increased ∼20-fold in rats that were fed the UDCA diet for 2 wk compared with standard chow diet (P < 0.05). Furthermore, UDCA became the predominant bile acid species, representing ∼50% of the total bile acid pool, consistent with previous studies (21).
Unsupervised clustering analysis showed a significant partition between control- and UDCA-diet-fed animals. In fact, the modulation of the miRNA expression profiles with UDCA feeding was striking (Table 2). We aimed to specifically identify differential miRNA expression, by comparisons of mean expression levels between 1) all UDCA-fed and non-UDCA-fed animal samples, 2) sham samples of the UDCA- and control diet-fed animals, 3) PH samples of the UDCA- and control diet-fed animals, and 4) sham- and PH-liver samples of the UDCA-fed animals.
Table 2.
Effect of UDCA on miRNA expression profiles during liver regeneration
| miRNA | UDCA vs. Control | UDCA Sham vs. Control Sham | UDCA PH vs. Control PH | UDCA PH vs. UDCA Sham | Validated Targets |
|---|---|---|---|---|---|
| hsa-miR-106a | 1.52 | VEGFA, RB1, AML1, APP | |||
| hsa-miR-143 | 1.54 | 1.50 | MAPK7, KRAS | ||
| hsa-miR-181d | 2.25 | ||||
| hsa-miR-191 | 1.82 | ||||
| hsa-miR-199b | 0.47 | ||||
| hsa-miR-20a | 1.51 | VEGFA, AML1, E2F1, CCND1 | |||
| hsa-miR-21 | 1.81 | 2.43 | 27 validated targets | ||
| hsa-miR-24 | 1.99 | NOTCH1, p38, p16, ACVR1B, DHFR | |||
| hsa-miR-302d | 2.65 | 3.59 | VEGFA, TRPS1, MBLN2, KLF13 | ||
| hsa-miR-31 | 1.49 | ||||
| hsa-miR-342 | 0.50 | ||||
| hsa-miR-4255p | 2.59 | ||||
| hsa-miR-451 | 0.45 | ABCB1 | |||
| hsa-miR-513 | 1.91 | 1.76 | 2.13 | ||
| hsa-miR-539 | 2.65 | ||||
| hsa-miR-552 | 0.48 | ||||
| hsa-miR-568 | 0.31 | 0.32 | |||
| hsa-miR-594 | 1.80 | ||||
| hsa-miR-603 | 0.41 | ||||
| hsa-miR-620 | 0.45 | ||||
| hsa-miR-638 | 2.90 | ||||
| hsa-miR-7 | 2.45 | PAK1, RAF1, IRS1, IRS2, EGFR | |||
| mmu-miR-362 | 0.50 | ||||
| mmu-miR-451 | 0.42 | ||||
| mmu-miR-674* | 1.74 | ||||
| mmu-miR-685 | 0.50 | ||||
| mmu-miR-691 | 0.35 | ||||
| mmu-miR-708 | 0.26 | ||||
| mmu-miR-720 | 4.94 | ||||
| mmu-miR-744 | 1.53 | ||||
| mmu-miR-760 | 2.04 | 1.90 | |||
| mmu-miR-761 | 0.49 | ||||
| mmu-miR-762 | 1.92 | ||||
| mmu-miR-764 | 0.50 | ||||
| 3p | |||||
| rno-miR-329 | 0.48 | 0.44 | |||
| rno-miR-346 | 1.76 | ||||
| rno-miR-350 | 0.50 | 0.50 | |||
| rno-miR-382* | 0.38 | CAPN8, CNTN4, SEPT3, TAGLN, VIM | |||
| rno-miR-7 | 2.31 |
List of miRNAs differentially expressed at 36 h after PH between ursodeoxycholic acid (UDCA)-fed animals and control diet-fed animals, UDCA-fed Sham animals and control-diet Sham animals, UDCA-fed PH animals and control-diet PH animals, and UDCA-fed PH and Sham animals. Mean expression values were determined following global normalization and statistical analysis as described in materials and methods. ABCB1, ATP-binding cassette, sub-family B (MDR/TAP), member 1; ACVR1B, activin A receptor, type IB; AML1, runt-related transcription factor 1; APP, amyloid precursor protein; CAPN8, calpain 8; CCND1, cyclin D1; CNTN4, contactin 4; DHFR, dihydrofolate reductase; E2F1, E2F transcription factor 1; EGFR, epidermal growth factor receptor; IRS1, insulin receptor substrate 1; IRS2, insulin receptor substrate 2; KLF13, Kruppel-like factor 13; KRAS, v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; MAPK7, mitogen-activated protein kinase 7; MBLN2, muscleblind-like 2; NOTCH1, Notch homolog 1; p16, cyclin-dependent kinase inhibitor 2A; p38, mitogen-activated protein kinase 14; PAK1, p21 protein (Cdc42/Rac)-activated kinase 1; RAF1, v-raf-1 murine leukemia viraloncogene homolog 1; RB1, retinoblastoma 1; SEPT3, septin 3; TAGLN, transgelin; TRPS1, trichorhinophalangeal syndrome I; VEGFA, vascular endothelial growth factor A; VIM, vimentin.
A large subset of miRNAs was affected by UDCA feeding, independently of PH. When comparing the two feeding groups, the results showed that UDCA significantly changed the expression of 24 miRNAs, by more than 1.5-fold. Approximately half of these changes were positive, with UDCA reducing the expression of 13 miRNAs (Table 2). When comparing sham samples between UDCA- and control diet-fed animals, we found nine significantly modulated miRNAs, of which only three were downregulated. These results indicate that the previously identified transcriptional role of UDCA (8) is also affecting miRNAs in the normal liver. In fact, and interestingly, UDCA was shown to be a potent modulator of miR-21 during LR; after PH, miR-21 expression was increased by ∼80% in the UDCA-fed group when compared with the control diet-fed animals (Table 2). In addition, UDCA also slightly increased miR-21 expression in sham animals, compared with control diet-fed animals. Of note, most of the miRNAs found to be modulated at 36 h post-PH in the UDCA-fed animals had already been identified as significantly increased, but at earlier time points, in the control diet-fed animals (Table 1), and these included miRs-21, -106a, -20a, and -7. More importantly, their expression levels at 36 h after PH in the UDCA-fed animals were ∼20% on average higher and were statistically significant. Therefore, these results indicate that the UDCA feeding was allowing for a prolonged expression of these particular miRNAs. In sum, it appears that UDCA is a strong modulator of miRNAs in the liver and, particularly, miR-21.
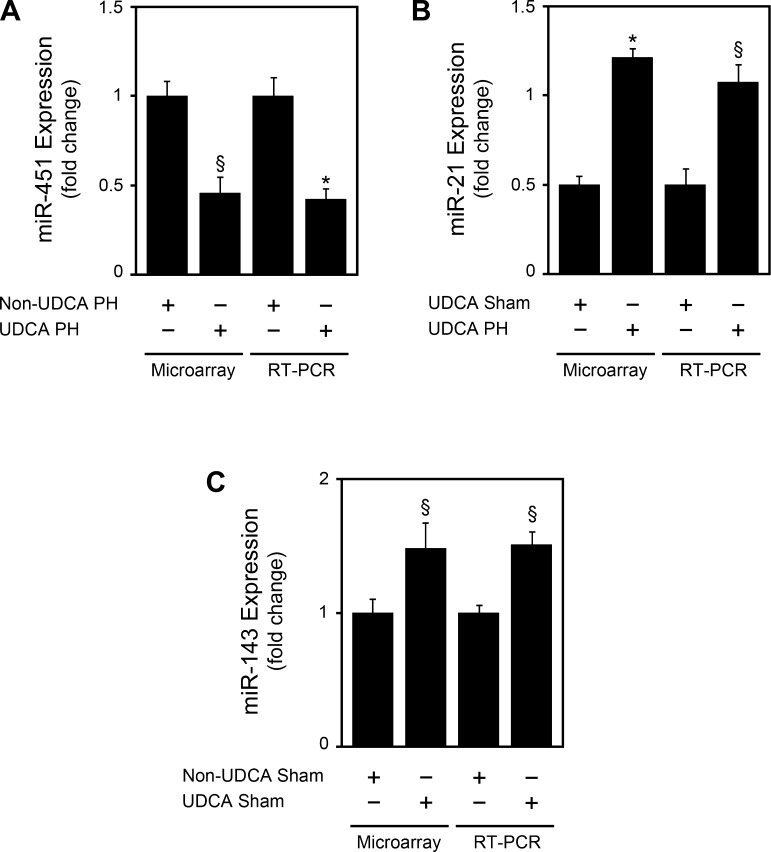
A subset of miRNAs modulated by UDCA in the different comparisons, namely miRs-21, -143, and -451, were chosen to further validate miRNA microarray analysis. Real-time RT-PCR analysis accurately confirmed the modulation by UDCA in all cases (Fig. 5).
Fig. 5.
Ursodeoxycholic acid (UDCA) modulates miR-451, -21, and -143 expression in accordance with microarray results. miRNA expression was evaluated by real-time RT-PCR as described in materials and methods. A: miR-451 expression in UDCA diet-fed animals at 36 h after PH, compared with control diet PH animals. B: miR-21 expression in UDCA-fed animals at 36 h after PH, compared with sham animals. C: miR-143 expression in sham UDCA-diet-fed animals, at 36 h after PH, compared with control diet sham animals. Results are expressed as means ± SE of 3 different animals. *P < 0.01 and §P < 0.05 from the corresponding controls.
Modulation of UDCA cellular effects by mir-21.
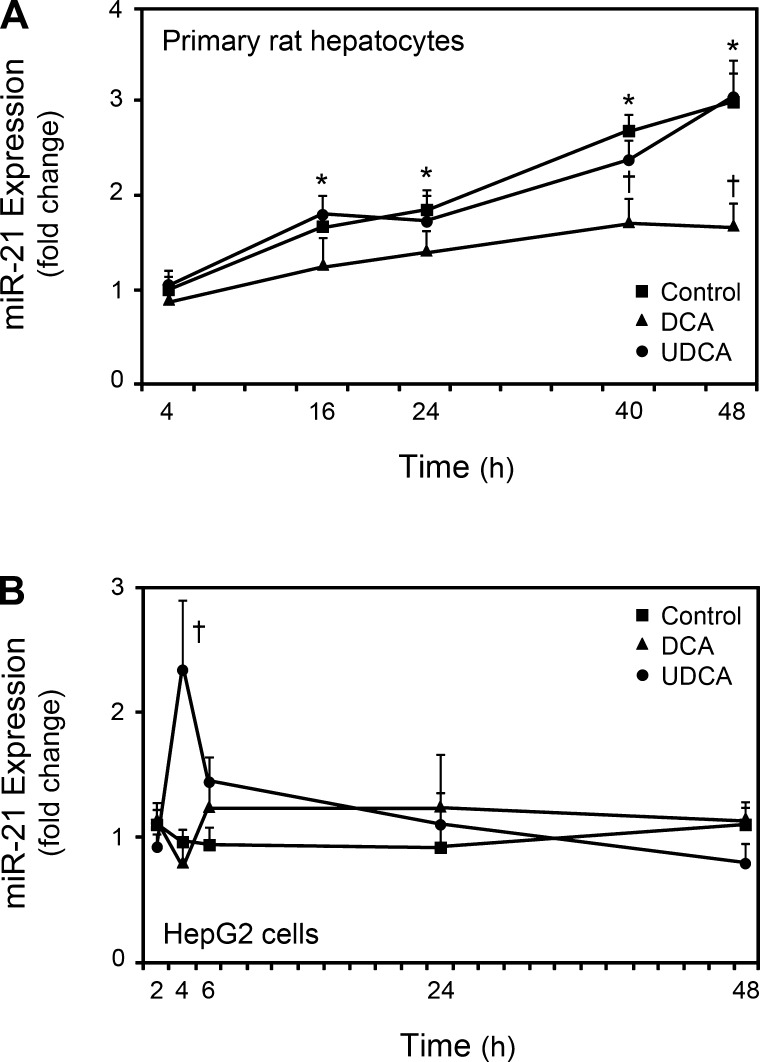
Given the relevance of miR-21, we analyzed the modulation of its expression by UDCA in primary rat hepatocytes and HepG2 cells. In addition, cells were also incubated with DCA for comparison to UDCA. miR-21 expression continuously increased throughout the time course in primary rat hepatocytes (Fig. 6A). Interestingly, although UDCA failed to modulate the expression of miR-21 in primary rat hepatocytes, it significantly induced miR-21 levels in HepG2 cells, at 4 h after incubation (P < 0.05) (Fig. 6B). In turn, DCA significantly inhibited miR-21 expression in primary rat hepatocytes, but not in HepG2 cells, and particularly at 40 and 48 h after incubation (P < 0.05). These results demonstrated a differential regulation of miR-21 by two distinct bile acid species. In particular, the failure of UDCA to increase miR-21 levels in primary rat hepatocytes may be related to their limited life span and progressive loss of liver-specific functions, including proliferation potential in vitro (11). During this dedifferentiation, a dramatic and progressive change in the hepatocyte gene expression patterns occurs, leading to physiological and morphological adaptations. Increased miR-21 levels at later time points, in control cells, may represent one of such adaptations. Ultimately, it may also halt the induction of miR-21 by UDCA. In proliferating HepG2 cells, this induction appears to be rapid and transient. In contrast, UDCA-induced miR-21 expression in vivo showed prolonged elevation (data not shown), perhaps reflecting the continuous administration of UDCA in the diet, or a specific response to the array of molecular stimuli caused by the PH. Finally, UDCA feeding alters the bile acid pool (21), which may also account for the differential effects of UDCA in vivo and in vitro.
Fig. 6.
UDCA and deoxycholic acid (DCA) modulate miR-21 expression in proliferating HepG2 cells and primary rat hepatocytes, respectively. Cells were incubated with 100 μM UDCA, or DCA, or no addition (control) for 4, 16, 24, 40, and 48 h in primary rat hepatocytes (A), or 2, 4, 6, 24, and 48 h in HepG2 cells (B). Total RNA was obtained for real-time RT-PCR analysis as described in materials and methods. Results are expressed as means ± SE for at least 3 different experiments. *P < 0.01 from control 4 h and †P < 0.05 from the respective control.
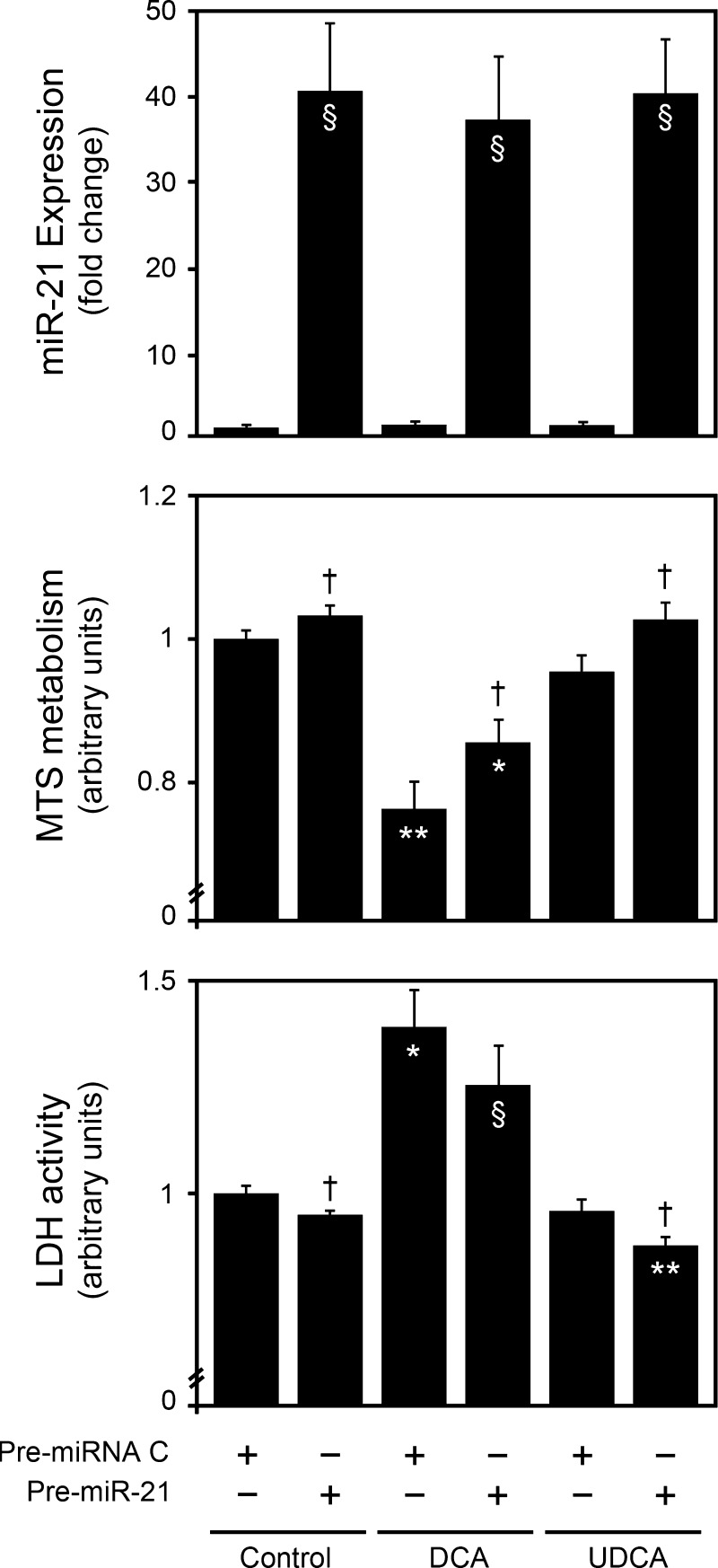
To further explore the notion that the cellular effects of bile acids are dependent on miR-21, we performed overexpression studies using a miR-21 precursor (Fig. 7, top). In agreement with the miR-21 inhibition studies (Fig. 4), pre-miR-21 overexpression alone slightly increased cell proliferation and viability (P < 0.05) (Fig. 7, middle and bottom). In pre-miRNA control treated cells, DCA strongly inhibited cell proliferation (P < 0.001) and viability (P < 0.01). These results are in accordance with our previous findings (7). Interestingly, miR-21 overexpression significantly increased the proproliferative and antiapoptotic effects of UDCA, while inhibiting the loss of cell proliferation and viability by DCA (P < 0.05). More importantly, cell proliferation after miR-21 overexpression was further increased by ∼4- and 2.5-fold in cells treated with DCA and UDCA, respectively, compared with untreated cells (Fig. 7, middle panel). Likewise, cell viability was also further increased by ∼2- and 1.5-fold under the same conditions (Fig. 7, bottom).
Fig. 7.
Bile acids modulate cell proliferation and viability in primary rat hepatocytes, in part, through miR-21. Primary rat hepatocytes were transfected with a miR-21 precursor and incubated with 100 μM UDCA, or DCA, or no addition (control) as indicated in materials and methods. miR-21 overexpression was assessed by real-time RT-PCR (top). Cell proliferation and viability were assessed by the MTS metabolism (middle) and LDH activity (bottom) assays, respectively. Results are expressed as means ± SE from at least 3 different experiments. §P < 0.05, *P < 0.01 and **P < 0.001 from the no-bile-acid pre-miRNA control. †P < 0.05 from the corresponding pre-miRNA controls.
These findings suggested that miR-21 is specifically integrated in the complex network of genes and pathways regulating cell proliferation and viability by bile acids. This might also explain the fact that modulation of miR-21 alone does not dramatically shift the ability of UDCA and DCA to modulate cell proliferation and viability. In fact, miRNAs appear to fine tune cellular functions or act in association with other miRNAs, integrating specific miRNA regulatory networks. Still, UDCA-induced miR-21 appears to contribute to its prosurvival effects. The mechanisms by which bile acids and, in particular, UDCA modulate miR-21 expression are as yet unknown. Indirect mechanisms are likely, as we have previously identified p53 as a key molecular target of UDCA (1, 7, 32), and a recent report revealed that p53 can modulate miRNA processing (37). Nevertheless, UDCA is also a powerful transcriptional modulator (8), and the possibility that bile acids directly regulate miRNA gene expression should also be considered.
DISCUSSION
In this study, we have shown that LR is associated with a specific miRNA pattern, which may play an important role in modulating proliferation and cell cycle progression genes after PH. Furthermore, UDCA modulates miRNA expression, in particular miR-21, in both regenerating rat liver and liver-derived cell cultures.
Many studies on gene expression using microarray analysis have shown that hundreds of genes are likely to be involved in the initial phases of LR (36). It is also well established that posttranscriptional regulation of gene expression is an important level of control during LR (22). In fact, similarly to immediate-early genes, which are induced in a protein synthesis-independent fashion by preexisting transcription factors in hepatocytes, miRNAs could also represent a key mechanism to rapidly optimize gene expression after PH. Still, the abnormal expression of miRNAs, usually in >10- or 100-fold change differences, has already been causally attributed to the development of cancer, including hepatocellular carcinoma (14). In the present report, however, miRNAs modulated during LR seem to fall in a different category, with expression differences averaging less than twofold and therefore possibly functioning to fine tune and stabilize expression profiles.
It is interesting that the modulated miRNAs targeted both pro- and anti-proliferation-related molecules. This is in agreement with a recent report in which miRNA deficiency was shown to result in both hepatocyte apoptosis and proliferation (15). miRs -19a, -21, and -214 for instance were found to be significantly upregulated after PH. They have all been shown to target the phosphatase and tensin homolog deleted on chromosome ten (PTEN), a negative regulator of the PI3K/Akt survival pathway. Because activation of the PI3K/Akt pathway plays a critical role in the early regenerative response of the liver after resection (17), these miRNAs may be, at least in part, responsible for a greater PI3K/Akt activation. miR-21, in particular, has recently been shown to promote cellular proliferation during mouse liver regeneration by targeting Btg2 (26). Thus, regardless of its targets, miR-21 has been described as a proliferative factor in several biological systems. Here we show that it also specifically increases proliferation of primary rat hepatocytes, while decreasing cell death, further supporting a strong role for miR-21 during LR. Still, several other miRNAs might also be as important; miRs -106a, -20a, -20b, and -93 have been validated as modulators of the vascular endothelial growth factor (VEGF). VEGF levels peak 48 h after PH, thus playing an important role in LR, in part, through its effects on neovascularization (4). Since miR-106a, -20a, -20b, and -93 levels are increased more than twofold at 36 h after PH and only increase again 72 h after PH, it may be that they contribute to the rapid and coordinated expression of VEGF. Interestingly, all of these later miRNAs are encoded by the miR-17–92 cluster and its paralogs, typically associated with critical pathways regulating cellular life and death decisions during development and in malignancy (27). In fact, both this cluster and miR-21 have recently been shown to possess differential expression in hepatitis B and C virus infection and their progression to hepatocellular carcinoma (39).
It was recently shown that Stat3 is a potent inducer of the miR-17–92 cluster (6). Stat3 activity is rapidly induced by cytokines such as tumor necrosis factor-α and interleukin-l and -6 after PH. In fact, Stat3 DNA-binding activity is typically increased within 30 min following PH and peaks at more than 30-fold at 3 h (10). Therefore, the rapid activation of miR-17–92 cluster after PH observed in this study may represent an event directly downstream of Stat3 activation, linking the transcriptional to the posttranscriptional control of LR. In fact, the E2F family of transcription factors are among the best characterized targets of the miR-17–92 cluster (40) and have been extensively studied regarding their crucial role during LR. Alternatively, the miR-17–92 cluster may also be activated by p53 (41) or c-Myc (28), two modulators of E2F1 expression known to be significantly activated during early LR (13, 23), providing further support for a transcriptional activation of miRNAs early during LR.
UDCA feeding appears to be shifting the liver miRNA expression patterns, both before and after PH. Interestingly, most changes appear to favor a proproliferative environment. Comparing the miRNA expression profiles between UDCA- and control diet-fed animals, at 36 h after PH, miR-451 showed a more than twofold downregulation by UDCA. This decrease may reflect a greater proliferation potential of hepatocytes induced by UDCA after PH, since miR-451 was recently shown to inhibit cell proliferation (2). More importantly, UDCA is allowing for a greater expression of miRNAs belonging to the miR-17–92 cluster and miR-21, which may account for its role during LR. In particular, miR-21 expression after PH in UDCA-fed rats was twofold increased compared with PH control-diet animals. miR-21 was also differentially modulated by distinct bile acids in vitro. Proapoptotic DCA inhibited miR-21 expression in primary rat hepatocytes and was less cytotoxic after miR-21 overexpression. In turn, under identical conditions, UDCA increased cell proliferation and reduced cell death. Therefore, although not yet fully characterized, UDCA modulation of miR-21 may contribute to the unique ability of the liver to regenerate after PH.
Interestingly, the miR-21 target programmed cell death 4 (PDCD4) is a powerful inhibitor of the AP-1 transcription factor (42), which has recently been shown to control LR through a molecular pathway involving p53, p21, and the p38 mitogen-activated protein kinase (35). This novel link underscores the importance of these factors in regulating the ability of the liver to repair itself following stress responses such as PH and/or liver injury.
Curiously, bile acids are strong modulators of AP-1 activity; UDCA has been shown to inhibit bile acid-induced activation of AP-1 (31). Therefore, the increase of miR-21 expression potentiated by either UDCA or PH itself may also contribute to LR through PDCD4. Particularly, the offsetting effects of bile acids on the rate and extent of LR (3, 21) may be coupled with miR-21 and PDCD4 modulation, in addition to PDCD4 targets. Furthermore, our previous microarray gene expression studies have shown that UDCA and TUDCA are potent modulators of several LR-related transcripts. These include LR p53-related gene and several insulin-like growth factor genes (8), which might also be associated with the miR-21/PDCD4 pathway during LR. Finally, it was recently demonstrated that during LR in mice miR-21 directly inhibits Btg2, a cell cycle inhibitor that prevents activation of FoxM1 (26). In turn, during LR in mice, cell cycle progression is controlled by bile acid-dependent mechanisms that also activate FoxM1 (16). Therefore, the upregulation of miR-21 by UDCA may constitute a novel mechanism for bile acids to activate this DNA synthesis transcription factor.
Collectively, these studies provide important information regarding the regulation of LR via miRNAs and its modulation by UDCA signaling. A further understanding of LR and potential endogenous modulators may enable us to improve the regenerative response following resection and/or diseases that require transplantation. Furthermore, it may ultimately provide new therapies to induce the patient's own liver to regenerate.
GRANTS
This research was supported by National Institutes of Health American Recovery and Reinvestment Act grant R01 DK081865-01 (to C. J. Steer) and grant PTDC/SAU-OSM/102099/2008 (to R. E. Castro), PhD fellowship SFRH/BD/60521/2009 (to D. M. S. Ferreira), and postdoctoral fellowship SFRH/BPD/65212/2009 (to P. M. Borralho) from Fundação para a Ciência e a Tecnologia, Lisbon, Portugal.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Amaral JD, Castro RE, Solá S, Steer CJ, Rodrigues CM. p53 is a key molecular target of ursodeoxycholic acid in regulating apoptosis. J Biol Chem 282: 34250–34259, 2007. [DOI] [PubMed] [Google Scholar]
- 2. Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, Jimenez P, Rodriguez J, Garcia-Foncillas J. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res 15: 2281–2290, 2009. [DOI] [PubMed] [Google Scholar]
- 3. Barone M, Francavilla A, Polimeno L, Ierardi E, Romanelli D, Berloco P, Di Leo A, Panella C. Modulation of rat hepatocyte proliferation by bile salts: in vitro and in vivo studies. Hepatology 23: 1159–1166, 1996. [DOI] [PubMed] [Google Scholar]
- 4. Bockhorn M, Goralski M, Prokofiev D, Dammann P, Grunewald P, Trippler M, Biglarnia A, Kamler M, Niehues EM, Frilling A, Broelsch CE, Schlaak JF. VEGF is important for early liver regeneration after partial hepatectomy. J Surg Res 138: 291–299, 2007. [DOI] [PubMed] [Google Scholar]
- 5. Bostjancic E, Glavac D. Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatovenerol Alp Panonica Adriat 17: 95–102, 2008. [PubMed] [Google Scholar]
- 6. Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, Ulrich S, Speich R, Huber LC. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res 104: 1184–1191, 2009. [DOI] [PubMed] [Google Scholar]
- 7. Castro RE, Amaral JD, Solá S, Kren BT, Steer CJ, Rodrigues CM. Differential regulation of cyclin D1 and cell death by bile acids in primary rat hepatocytes. Am J Physiol Gastrointest Liver Physiol 293: G327–G334, 2007. [DOI] [PubMed] [Google Scholar]
- 8. Castro RE, Solá S, Ma X, Ramalho RM, Kren BT, Steer CJ, Rodrigues CM. A distinct microarray gene expression profile in primary rat hepatocytes incubated with ursodeoxycholic acid. J Hepatol 42: 897–906, 2005. [DOI] [PubMed] [Google Scholar]
- 9. Castro RE, Steer CJ, Rodrigues CM. Bile acids as modulators of apoptosis. In: Hepatotoxicity: From Genomics to In Vitro and In Vivo Models, edited by Sahu SC. Chichester, UK: Wiley, 2007, p. 391–419. [Google Scholar]
- 10. Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology 21: 1443–1449, 1995. [PubMed] [Google Scholar]
- 11. Elaut G, Henkens T, Papeleu P, Snykers S, Vinken M, Vanhaecke T, Rogiers V. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab 7: 629–660, 2006. [DOI] [PubMed] [Google Scholar]
- 12. Fan G, Kren BT, Steer CJ. Regulation of apoptosis-associated genes in the regenerating liver. Semin Liver Dis 18: 123–140, 1998. [DOI] [PubMed] [Google Scholar]
- 13. Fausto N, Mead JE, Braun L, Thompson NL, Panzica M, Goyette M, Bell GI, Shank PR. Proto-oncogene expression and growth factors during liver regeneration. Symp Fundam Cancer Res 39: 69–86, 1986. [PubMed] [Google Scholar]
- 14. Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med 12: 2189–2204, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology 49: 618–626, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 312: 233–236, 2006. [DOI] [PubMed] [Google Scholar]
- 17. Jackson LN, Larson SD, Silva SR, Rychahou PG, Chen LA, Qiu S, Rajaraman S, Evers BM. PI3K/Akt activation is critical for early hepatic regeneration after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol 294: G1401–G1410, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol 311: 603–612, 2007. [DOI] [PubMed] [Google Scholar]
- 19. Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 29: 2394–2399, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kountouras J, Boura P, Lygidakis NJ. Liver regeneration after hepatectomy. Hepatogastroenterology 48: 556–562, 2001. [PubMed] [Google Scholar]
- 21. Kren BT, Rodrigues CM, Setchell KD, Steer CJ. Modulation of steady-state messenger RNA levels in the regenerating rat liver with bile acid feeding. Liver Transpl 7: 321–334, 2001. [DOI] [PubMed] [Google Scholar]
- 22. Kren BT, Steer CJ. Posttranscriptional regulation of gene expression in liver regeneration: role of mRNA stability. FASEB J 10: 559–573, 1996. [DOI] [PubMed] [Google Scholar]
- 23. Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol 6: 65–72, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kren BT, Wong PY, Shiota A, Zhang X, Zeng Y, Steer CJ. Polysome trafficking of transcripts and microRNAs in regenerating liver after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol 297: G1181–G1192, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mariash CN, Seelig S, Schwartz HL, Oppenheimer JH. Rapid synergistic interaction between thyroid hormone and carbohydrate on mRNAS14 induction. J Biol Chem 261: 9583–9586, 1986. [PubMed] [Google Scholar]
- 26. Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol 298: G535–G541, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell 133: 217–222, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843, 2005. [DOI] [PubMed] [Google Scholar]
- 29. Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the Planarian Schmidtea mediterranea: a model system for stem cell biology. RNA 12: 1640–1649, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 37: 918–925, 2009. [DOI] [PubMed] [Google Scholar]
- 31. Shah SA, Volkov Y, Arfin Q, Abdel-Latif MM, Kelleher D. Ursodeoxycholic acid inhibits interleukin 1 beta and deoxycholic acid-induced activation of NF-kappaB and AP-1 in human colon cancer cells. Int J Cancer 118: 532–539, 2006. [DOI] [PubMed] [Google Scholar]
- 32. Solá S, Ma X, Castro RE, Kren BT, Steer CJ, Rodrigues CM. Ursodeoxycholic acid modulates E2F-1 and p53 expression through a caspase-independent mechanism in transforming growth factor beta1-induced apoptosis of rat hepatocytes. J Biol Chem 278: 48831–48838, 2003. [DOI] [PubMed] [Google Scholar]
- 33. Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH, Frandsen NM, Willenbring H. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 51: 1735–1743, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steer CJ. Liver regeneration. FASEB J 9: 1396–1400, 1995. [DOI] [PubMed] [Google Scholar]
- 35. Stepniak E, Ricci R, Eferl R, Sumara G, Sumara I, Rath M, Hui L, Wagner EF. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev 20: 2306–2314, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci USA 99: 11181–11186, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature 460: 529–533, 2009. [DOI] [PubMed] [Google Scholar]
- 38. Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods 1: 47–53, 2004. [DOI] [PubMed] [Google Scholar]
- 39. Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 49: 1098–1112, 2009. [DOI] [PubMed] [Google Scholar]
- 40. Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282: 2130–2134, 2007. [DOI] [PubMed] [Google Scholar]
- 41. Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH. Repression of the miR-17–92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28: 2719–2732, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene 20: 669–676, 2001. [DOI] [PubMed] [Google Scholar]
- 43. Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev 22: 728–733, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]