Abstract
We report that knocking down the expression of inositol polyphosphate 4-phosphatase type II (INPP4B) in human epithelial cells, like knockdown of PTEN, resulted in enhanced Akt activation, anchorage-independent growth, and enhanced over all motility. In xenograft experiments overexpression of INPP4B resulted in reduced tumor growth. INPP4B preferentially hydrolyzes phosphatidylinositol-3,4-bisphosphate (PI(3,4)P2) with no effect on phosphatidylinositol-3.4.5-triphosphate (PI(3,4,5)P3), suggesting that PI(3,4)P2 and PI(3,4,5)P3 may cooperate in Akt activation and cell transformation. Dual knockdown of INPP4B and PTEN resulted in cellular senescence. Finally, we find loss-of-heterozygosity (LOH) at the INPP4B locus in a majority of basal-like breast cancers as well as in a significant fraction of ovarian cancers, which correlated with lower over all patient survival, suggesting that INPP4B is a tumor suppressor.
Introduction
During the generation and progression of epithelial tumors, oncogenes and tumor suppressor genes cause multiple cell-autonomous alterations resulting in aberrant control of proliferation, apoptosis, angiogenesis, and cellular life span (Hahn and Weinberg, 2002). The cyclic regulation of the mammary gland confers vulnerability to these cancer-promoting processes, and imbalances between proliferation and apoptosis sets the scene for mammary tumorigenesis (Scheid and Woodgett, 2001). The phosphinositide 3-kinase (PI3K)-Akt pathway is deregulated in a wide spectrum of human cancers, and gain or loss-of-function mutations of several components of the pathway lead to neoplastic transformation in various model systems (Engelman et al., 2006; Vivanco and Sawyers, 2002).
In vivo activation of PI3K increases the intracellular amounts of phosphatidylinositol(3,4)bisphosphate (PI(3,4)P2) and phosphatidylinositol(3,4,5) trisphosphate (PI(3,4,5)P3). A variety of signaling proteins, including the Ser/Thr kinases Akt and PDK1 are able to bind to one or both of these PI3K lipid products and are thereby localized to the plasma membrane to initiate cell growth and cell survival pathways. (Plas and Thompson, 2005; Manning and Cantley, 2007).
The termination of signaling downstream of PI3K by degradation of PI(3,4,5)P3 can be mediated by two different types of phosphatases. PTEN dephosphorylates the 3-position of PI(3,4,5)P3 to produce PI(4,5)P2, while SHIP family phosphatases dephosphorylate the 5-position to produce PI(3,4)P2 (Figure 1). Loss of PTEN protein expression or function is frequently observed in human cancers (Vazquez and Sellers, 2000), and deletion of PTEN in mice results in a variety of tumors (Di Cristofano et al., 1998; Kishimoto et al., 2003), indicating that uncontrolled signaling through PI3K contributes to cancer formation and metastasis. In contrast, loss of genes for SHIP family members has not correlated with solid tumor formation, thus the role of these phosphatases in regulating PI3K signaling is complex and controversial. Some studies have indicated that SHIP family members suppress PI3K signaling (Scheid et al., 2002; Sharrard and Maitland, 2007; Sleeman et al., 2005), while other studies suggest that the product of SHIP family members, PI(3,4)P2, is a positive regulator of Akt (Franke et al., 1997; Frech et al., 1997; Klippel et al., 1997; Scheid et al., 2002). The PH-domains of Akt and PDK1 bind both PI(3,4)P2 and PI(3,4,5)P3 in vitro, suggesting these lipids may be redundant in their ability to mediate Akt activation (Cantley, 2002). The possibility that both PI(3,4,5)P3 and PI(3,4)P2 may be necessary for optimal growth signaling downstream of PI3K has not been adequately addressed.
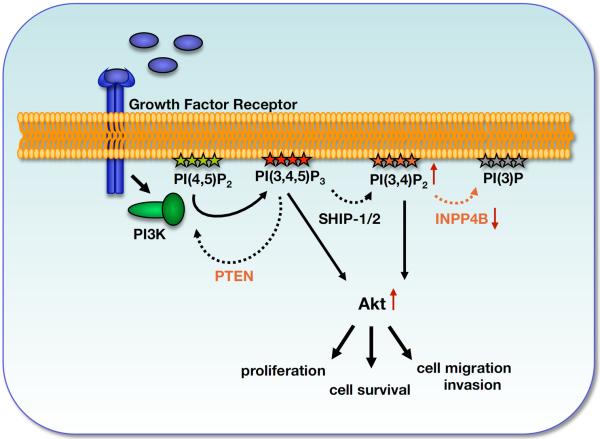
Figure 1. A model indicating the roles of PTEN and INPP4B in regulation of signaling downstream of Class I PI3Ks.
Upon stimulation of Class 1 PI3Ks two major phospholipid pools are generated: PI(3,4,5)P3 and PI(3,4)P2. PTEN hydrolyzes the 3′-phosphate of PI(3,4,5)P3 to terminate PI3K signaling. However, SHIP family members hydrolyze the 5′-phosphate of PI(3,4,5)P3 to generate PI(3,4)P2, which, like PI(3,4,5)P3, can facilitate PDK1-dependent phosphorylation and activation of AKT. INPP4B converts PI(3,4)P2 to PI(3)P. Loss of PTEN or loss of INPP4B results in prolonged activation of Akt, and subsequently in increased cell proliferation, cell migration and invasion.
The inositol polyphosphate 4-phosphatase type II (INPP4B) was initially isolated from rat brain as an enzyme that hydrolyzes the 4-position phosphate of PI(3,4)P2, and to a lesser degree inositol(3,4)bisphosphate (Ins((3,4)P2) and Ins(3,4,5)P3, in vitro (Norris et al., 1997; Norris et al., 1995). The ability of INPP4B to hydrolyze PI(3,4,5)P3 and its effects on phosphoinositides in intact cells was not investigated, thus, the in vivo function of INPP4B remains elusive. However, INPP4B expression was recently shown to be silenced in malignant proerythroblasts. These cells displayed increased phospho-Akt levels, which could be reduced by re-expression of INPP4B (Barnache et al., 2005). In addition, recent studies described a frequently deleted region in breast cancer cell lines and basal-like, high grade breast tumors using high-resolution comparative genomic hybridization analysis (chromosome location 4q31.1-31.21 and chromosome location 4q31.3-q32.1, respectively) comprising 6 genes, including INPP4B (Naylor et al., 2005; Bergamaschi et al., 2006; Chin et al., 2007). Johannsdottir et al. also found LOH in this region in breast cancers (Johannsdottir et al., 2004). Further, INPP4B was identified out of a collection of RNAis to give rise to anchorage-independent growth in human mammary epithelial cells (HMEC) (Westbrook et al., 2005).
These observations raise the possibility that INPP4B might be affecting PI3K signaling in vivo and that it might function as a tumor suppressor. In this study we investigate this possibility with retroviral knockdown of INPP4B in human epithelial cell systems.
Results
Knockdown of INPP4B results in anchorage-independent growth
To investigate the role of INPP4B on cell transformation in the human mammary epithelial cell line HMEC, retroviral hairpin constructs (shRNA) were used to knockdown its expression. An shRNA construct directed against Renilla luciferase (shRNA-Renilla) was used as negative control for the experiments. Two shRNA constructs (shRNA-INPP4B-1 and shRNA-INPP4B-2) were found to effectively reduce expression of INPP4B in several cell lines as judged by decreased mRNA levels (figure 2A). In MCF-7 cells, where INPP4B protein levels were high enough to be detected by Western Blotting, these constructs were shown to reduce INPP4B protein to the same extend (figure 2B). Infection of HMEC cells with either shRNA-INPP4B-1 or shRNA-INPP4B-2 resulted in anchorage-independent growth comparable to that observed with an shRNA directed against PTEN (figure 2C and 2D). Knocking down INPP4B or PTEN also increased the rate of proliferation of HMEC cells and the effect of knocking down INPP4B could be reversed by expression of a knockdown resistant INPP4B, indicating that the shRNA was acting on target (figure 2E).
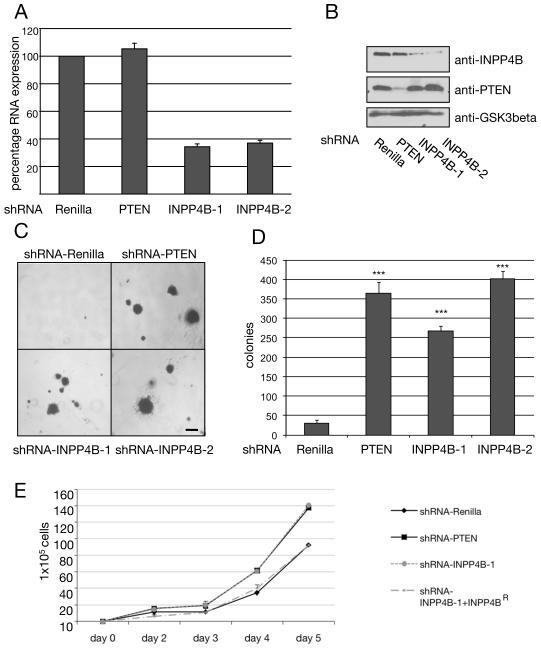
Figure 2. Knockdown of INPP4B results in anchorage-independent growth and increased cell proliferation.
(A) Semi-quantitative RNA-levels of stable HMEC shRNA-INPP4B knockdown colonies grown in anchorage-independent growth assays determined by RT-PCR. Both shRNA hairpins, INPP4B-1 and INPP4B-2, directed against INPP4B resulted in approximately 65% knockdown at the RNA level (n = 3). (B) Knockdown of INPP4B and PTEN in MCF-7 cells. The human mammary epithelial cancer cell line MCF-7 was infected with shRNA hairpins directed against Renilla, PTEN, and INPP4B (‘INPP4B-1’ and ‘INPP4B-2’). Protein levels of INPP4B and PTEN in cells expressing the indicated shRNA were determined by using immunoblotting. Protein loading was shown using antibody directed against GSK3beta. (C) Anchorage-independent growth assay of stable HMEC cell pools infected with retrovirus expressing shRNA directed against Renilla, PTEN, and two hairpin constructs directed against INPP4B (‘INPP4B-1’ and ‘INPP4B-2’). Results are representative of four independent experiments with similar results. Scale bar, 1mm. (D) Quantification of anchorage-independent growth assay of stable HMEC shRNA cell pools (***p < 0.001, n = 3). (E) Proliferation assay of stably infected HMEC shRNA cell pools. Cells from shRNA-Renilla, shRNA-PTEN, shRNA-INPP4B-1 and shRNA-INPP4B+INPP4BR (rescue cell pool expressing knockdown resistant INPP4B) were counted on day 2, 3, 4 and 5 in a Coulter counter. Averages of three independent experiments are represented. All data are shown as mean +/− SD.
INPP4B knockdown results in increased migratory and invasive behavior of human mammary epithelial cells
Migration and invasion are hallmarks of tumor cells with the potential to metastasize to new locations in the body. To analyze the migratory behavior of human mammary epithelial cells, HMEC cells expressing the various shRNA constructs were grown until confluency and a scratch wound was introduced with a pipette tip. Pictures were taken at time points t = 0 hours and t = 8 hours as indicated (figure 3A). Cells expressing shRNA directed against PTEN and INPP4B closed the wound more rapidly than control cells (figure 3A; quantified in figure 3B). The increased rate of wound closure due to INPP4B knockdown was partially rescued by expressing a wild type INPP4B construct that circumvents the knockdown (‘INPP4BR’), but was not affected by expressing the catalytically inactive mutant of INPP4B (‘INPP4B-CD’) (figure 3C – expression of INPP4BR and INPP4B-CD is presented in supplemental figure 1). The PI3K inhibitor, LY294002 also reduced the rate of wound closure in PTEN and INPP4B knockdown cell pools significantly, consistent with the idea that increased cell migration was due to elevated PI(3,4)P2 or PI(3,4,5)P3 levels for INPP4B or PTEN knockdown, repectively (figure 3D).
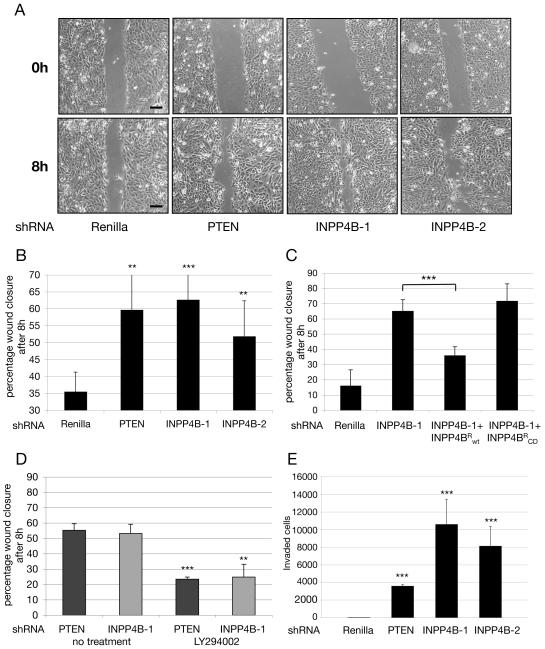
Figure 3. Knockdown of INPP4B results in strong migratory and invasive behavior.
(A) Knocking down INPP4B or PTEN with shRNA enhances wound closure following a scratch of confluent HMEC cells. Cells were photographed at time points t = 0 hours and t = 8 hours after wound introduction. Samples were observed under a light microscope. Scale bars, 50μm. (B) Quantification of wound closure. Displayed are the percentages of wound closure at 8 hours after wound introduction determined using NIH Image J (Averages from 3 experiments like those in part A are presented **p < 0.01 and ***p < 0.001). (C) Expression of an INPP4B sequence (INPP4BR) that circumvents the knockdown of endogenous INPP4B reverses enhanced wound closure induced by INPP4B-1 shRNA. Expression of a catalytically inactive form of INPP4B (INPP4BR-CD) did not suppress wound closure induced by INPP4B-1 shRNA. The experiment was analogous to that in part A (Averages from three experiments are presented, ***p<0.001). (D) The PI3K inhibitor, LY294002 (10 μM), inhibit accelerated wound closure induced by knocking down INPP4B or PTEN in HMEC. The experiments were analogous to those presented in Part A. (Averages from 3 experiments are presented: ***p < 0.001, **p < 0.01). (E) Knocking down the expression of PTEN or INPP4B enhances the ability of MCF-10A cells to cross a matrigel barrier in a Boyden chamber assay. MCF-10A were stably infected with shRNA directed against Renilla, PTEN, and INPP4B (INPP4B-1 and INPP4B-2). Cells were seeded in transwell chambers containing matrigel as a barrier. No stimulus was applied to the bottom chamber, in order to examine non-directional invasive behavior in knockdown cell pools. 24 hours after seeding, invaded cells were fixed, stained and quantified. (Average from three experiments are presented: ***p < 0.001). All data are shown as mean +/− SD.
We also investigated the effect of knocking down INPP4B on the ability of MCF-10A cells to intravasate through a Matrigel layer in a Boyden chamber revealing the invasive nature of shRNA knockdown cell pools. As shown in figure 3E, knocking down INPP4B had a greater effect on promoting migration through Matrigel than knocking down PTEN.
In addition to the enhanced migratory behavior, INPP4B knockdown cells displayed a spindle-like and fibroblastic morphology compared to Renilla or PTEN knockdown controls (supplemental figure 2A), suggestive of an epithelial-mesenchymal transdifferentiation (EMT) (Lee et al., 2006). Semi-quantitative RT-PCR of selected epithelial and mesenchymal marker genes revealed a large increase in N-Cadherin, a decrease in gamma-Catenin, and a subtle decrease in E-Cadherin RNA-levels in INPP4B knockdown cell pools compared to Renilla control cell pools (supplemental figure 2B). Further, we observed a strong increase in the fibroblast marker fibronectin compared to the Renilla knockdown control. These changes were consistent with a more mesenchymal phenotype. In contrast, RNA levels of EMT markers in PTEN knockdown cell pools revealed a significant decrease in N-Cadherin and a strong increase in vimentin levels, but increased E-Cadherin levels and little fibronectin expression (supplemental figure 2C).
INPP4B knockdown in MCF-10A mammary epithelial cells results in distorted acini architecture in three-dimensional culture
Using the MCF-10A three-dimensional culture model of mammary epithelial cells, we examined how knockdown of INPP4B influenced the morphogenesis of polarized epithelial structures. Knockdown of INPP4B, or PTEN in MCF-10A cells resulted in enhanced and dysmorphic growth of acini in three-dimensional cultures (figure 4A, left panels). In parallel, cells were treated with the mTORC1 inhibitor Rapamycin, the PI3K inhibitor LY294002, or with the MEK inhibitor UO126 starting day 1 of culture (figure 4A, middle and right panels). Rapamycin had only a minor effect on dysmorphic growth of INPP4B knockdown acini in three-dimensional assays, and did not have any impact on PTEN knockdown cell pools. LY294002 and UO126 treatment caused a general growth delay of acini formation resulting in smaller spheres compared to untreated cell pools. In INPP4B and PTEN knockdown cell pools, UO126 treatment only delayed the onset of architecturally disturbed acini growth, but LY294002 treatment resulted in almost normal appearance of spheres comparable to Renilla knockdown controls. These data suggest that signals downstream of PI3K/Akt are important for the disturbed architectural growth of INPP4B knockdown spheres.
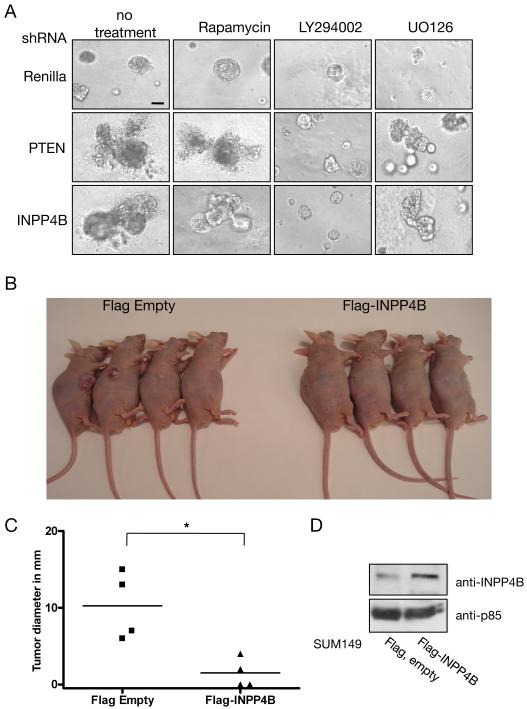
Figure 4. Knocking down INPP4B or PTEN in MCF-10A cells results in dysmorphic cell clusters upon growth in three-dimensional culture, and overexpression of INPP4B suppresses tumor growth in the SUM149 xenograft mouse model.
(A) Stable MCF-10A shRNA cell pools were grown on a matrigel layer under differentiating conditions and observed at day 21 of culture. Left panels display grown, untreated spheres of Renilla, PTEN, and INPP4B knockdown cells. The same stable shRNA cell pools were treated starting day 1 with 1 nM Rapamycin (middle left panels), 10 μM LY294002 (middle right panels), or 10 μM UO126, respectively (right panels). Representative pictures are shown. The experiment was repeated three times with similar results. Scale bar, 50μm. (B) Nude mice were injected with the human invasive ductal carcinoma cell line SUM149 expressing Flag-INPP4B (n=4) or empty Flag control (n=4). Mice were sacrificed 4 weeks post injection and tumor growth and size evaluated. All mice injected with SUM149 cells expressing empty Flag vector showed tumor growth (n=4), while mice injected with SUM149 cells expressing Flag-INPP4B showed reduced tumor growth and size (n=2). (C) Quantification of tumor diameter in xenograft experiment. All mice in the control group (SUM149+Flag Empty) showed tumor growth (n=4). Overexpression of Flag-INPP4B in SUM149 resulted in reduced tumor growth (n=4, p=0.011). (D) Protein levels of INPP4B in SUM149 cells expressing empty Flag vector or Flag-INPP4B were determined by using immunoblotting. Protein loading was shown using antibody directed against p85.
Overexpression of INPP4B in SUM149 cells reduced tumor growth in a xenograft mouse model
The human invasive ductal carcinoma cell line SUM149 was infected with a retroviral vector expressing Flag-INPP4B or Flag-tag, only and the resultant cells were subcutaneously injected into nude mice. The SUM149 cells expressing empty Flag-vector produced much larger tumors as compared to cells expressing Flag-INPP4B (figure 4B, quantified in figure 4C). The total level of INPP4B was approximately 2 to 3 fold higher in the SUM149 cells expressing Flag-INPP4B compared to levels in control SUM149 cells (figure 4D). These data indicate that overexpression of INPP4B in the human mammary tumor cell line SUM149 suppresses tumor growth in a xenograft mouse model.
Effect of INPP4B on cellular phosphoinositides
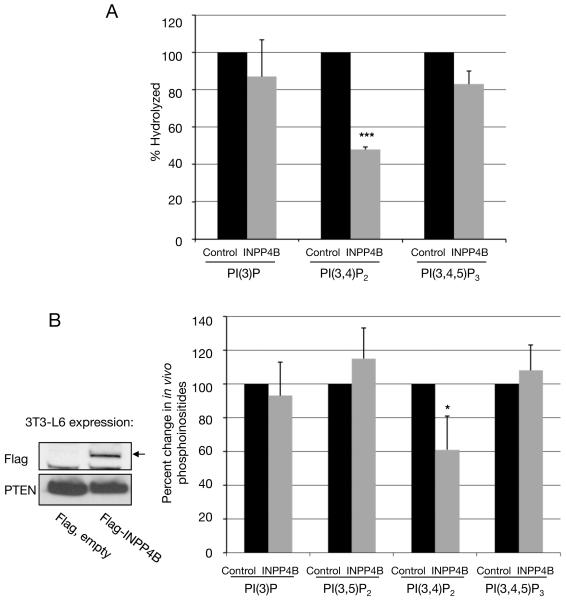
Previous studies have indicated that INPP4B hydrolyzes PI(3,4)P2, however PI(3,4,5)P3 hydrolysis was not examined and the effect of INPP4B on cellular phosphoinositides was not investigated (Norris et al., 1995). In Figure 5A we show that INPP4B prefers PI(3,4)P2 over PI(3,4,5)P3 in in vitro assays. To verify that PI(3,4)P2 is also an in vivo substrate of INPP4B we overexpressed the enzyme in 3T3-L6 myoblast cells, a cell line that produces easily detectable levels of PI(3)P, PI(3,4)P2, PI(3,5)P2 and PI(3,4,5)P3 (Mandl et al., 2007). The cellular level of PI(3,4)P2 in INPP4B expressing cells was reduced significantly. In contrast, the levels of the other lipids, including PI(3,4,5)P3, were not affected (figure 5B). Although one might expect an increase in PI(3)P due to hydrolysis of the 4-position of PI(3,4)P2, PI(3)P is at much higher levels than PI(3,4)P2 and most of this lipid is probably produced by class III PI3K, rather than through hydrolysis of PI(3,4)P2. These findings indicate that the effects of INPP4B knockdown in the mammary epithelial cell system are due to elevated PI(3,4)P2 levels alone, rather than elevated PI(3,4,5)P3 levels, as is the case for PTEN knockdown.
Figure 5. Substrate specificity of INPP4B.
(A) FLAG-tagged INPP4B was immunoprecipitated from 293T cells and incubated with either [32P]-PI(3)P, or [32P]-PI(3,4)P2 or [32P]-PI(3,4,5)P3. As control, the phosphoinositides were incubated with Flag-immunoprecipitate from transfected 293T cells expressing an empty Flag-expression construct. The percent hydrolysis of each lipid was determined by chloroform/methanol/HCL extraction, thin layer chromatography and Phosphoimager analysis. (n=3: ***p < 0.001). (B) Overexpression of human INPP4B causes a reduction in cellular PI(3,4)P2 levels in vivo. 3T3-L6 cells were transiently transfected with empty Flag or Flag-INPP4B constructs and labeled with [32P]-inorganic phosphate. Lipids were extracted, deacylated and the headgroups separated by HPLC. The radioactivity in the glyceroyl-phosphoryl-inositol moieties of each of the D-3 phosphorylated phosphoinositides was then normalized to the total radioactivity in the more abundant phosphoinositides, PI(4)P and PI(4,5)P2 (which did not significantly vary between experiments). The bars indicate per cent changes in these ratios in INPP4B transfected cells compared to control cells. The actual ratios of radioactivity in each lipid to total radioactivity in PI(4)P plus PI(4,5)P2 in the control 3T3-L6 cells were: PI(3)P – 3.2%, PI(3,5)P2 – 0.035%, PI(3,4)P2 – 0.17%, PI(3,4,5)P3 – 0.69%. (Averages from three experiments are presented: *p<0.1). Protein expression of Flag-INPP4B in 3T3-L6 cells was demonstrated by Immunoblotting. Protein loading was shown in using antibody directed against PTEN. (Inset). All data are shown as mean +/− SD.
Knockdown of both INPP4B and PTEN leads to cellular senescence
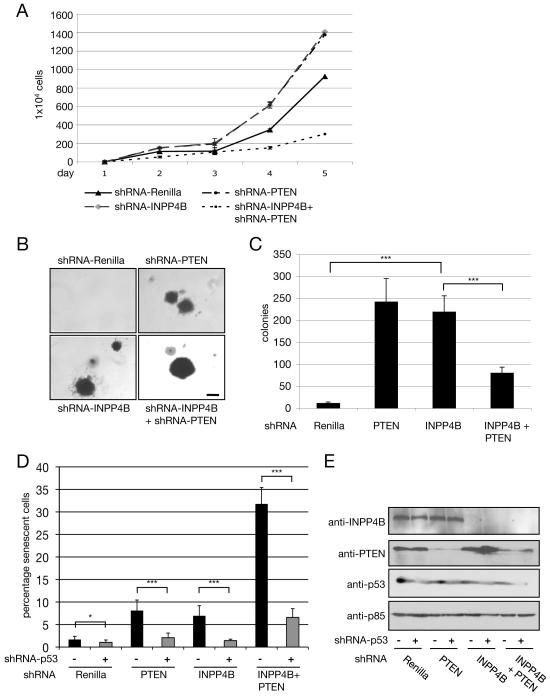
In order to address whether INPP4B and PTEN have redundant functions, HMEC cells were subjected to dual knockdown of INPP4B and PTEN. Unexpectedly, knocking down INPP4B and PTEN simultaneously resulted in decreased cell proliferation compared to Renilla knockdown control (figure 6A), and resulted in fewer anchorage-independent colonies compared to INPP4B or PTEN single knockdown cell pools (figure 6B and 6C). However, the few colonies that formed from the double knockdown cells were larger than those formed from cells with single knockdown of INPP4B or PTEN (figure 6B). These results indicate that loss of both INPP4B and PTEN results in impaired cell growth but that rare events (genetic or epigenetic) occur in the double knockdown cells that allow enhanced growth compared to knockdown of either INPP4B or PTEN alone.
Figure 6. Double knockdown of INPP4B and PTEN in HMEC cells results in increased cellular senescence.
(A) Proliferation assay of stably infected HMEC shRNA cell pools including double knockdown of INPP4B and PTEN. Cells were counted on indicated days. Results are representative of three independent experiments with similar results. (B) Morphology of anchorage-independent growth assays of stable HMEC cell pools infected with retrovirus expressing shRNA directed against Renilla, PTEN, INPP4B, and INPP4B + PTEN. Results are representative of three independent experiments with similar results. Scale bar, 1mm. (C) Quantification of anchorage-independent growth assay of stable HMEC shRNA cell pools (***p<0.001, n=3). (D) SA-beta-Gal activity of stable shRNA HMEC cell pools without or with knockdown of Trp53 was assessed. The error bars indicate the standard deviation of three replicates (n = 3: *p<0.1, ***p<0.001). Data are shown as mean +/− SD. (E) Protein levels of INPP4B, PTEN, and Trp53 in cells expressing the indicated shRNA were determined by using immunoblotting. Protein loading was shown using antibody directed against p85. All data are shown as mean +/− SD.
Since mutations in K-ras, B-raf, PTEN and NF-1 have been shown to trigger cellular senescence in vivo (Chen et al., 2005; Collado et al., 2005)(Braig et al., 2005; Courtois-Cox et al., 2006; Dankort et al., 2007), we investigated the effect of knocking down PTEN or INPP4B on senescence of HMEC cells. In Figure 6D, we show that knocking down expression of PTEN or INPP4B leads to increased cellular senescence, while knocking down both INPP4B and PTEN results in a dramatic increase in cellular senescence. Ras- or PTEN-induced senescence can be bypassed by inactivating the Rb and transformation related protein 53 (Trp53) pathways. Cell senescence in INPP4B knockdown cell pools, as well as in INPP4B and PTEN dual knockdown cell pools could be rescued by knocking down Trp53 using a retroviral hairpin construct (figure 6D). The effect of the shRNAs against INPP4B, PTEN and Trp53 on protein levels is presented in figure 6E. Representative pictures of HMEC knockdown cell pools stained for the senescence marker SA-beta-Gal are shown in supplemental figure 3.
INPP4B knockdown affects the PI3-kinase/Akt signal transduction pathway
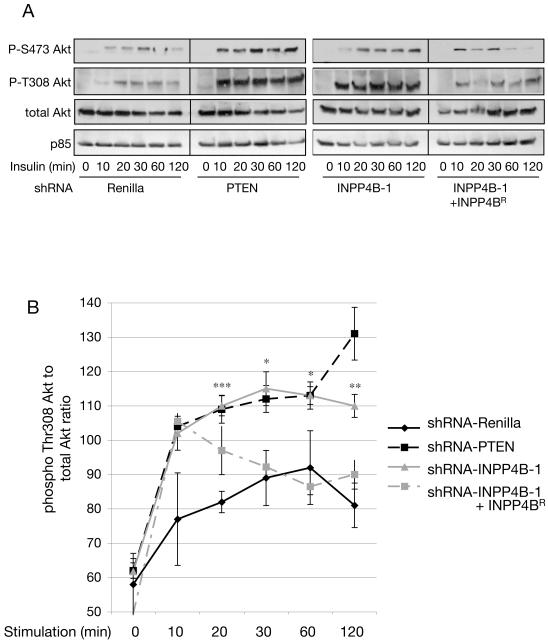
Time course experiments were performed to compare the effects of INPP4B knockdown versus PTEN knockdown on Akt signaling. Knocking down INPP4B or PTEN in HMEC cells had little effect on basal Akt activation (as judged by phosphorylation at Thr-308 or Ser-473) but had a significant effect on the magnitude and duration of insulin-stimulated Akt activation (Fig. 7A and B). This was also reflected in an increase in phosphorylation of Tuberin and GSK3beta at sites known to be dependent on AKT (supplemental figure S4). Expression of a form of INPP4B that circumvents knockdown (INPP4BR) suppressed AKT activation. Dual knockdown of INPP4B and PTEN in HMEC cells caused a more prolonged phosphorylation of AKT at Thr308 than that caused by knocking down either protein alone (supplemental figure 5).
Figure 7. Knocking down the expression of INPP4B increases the duration of AKT activation in response to insulin.
(A) Stable HMEC cell pools expressing shRNA vectors directed against Renilla, PTEN, INPP4B (‘INPP4B-1’) or HMEC cell pools expressing the knockdown resistant INPP4B expression construct in the shRNA-INPP4B-1 cell pool (‘INPP4B-1 + INPP4BR’) were serum-starved and stimulated with 100nM insulin for the indicated time periods. Cell lysates were separated by SDS-PAGE and phospho-Akt detected with specific antibodies in Western blot analysis. INPP4B knockdown cell pools display increased and prolonged phospho-Thr308 and phospho-Ser473 Akt compared to Renilla control knockdown cell pools. Expression of a Flag-tagged INPP4B construct harboring two silent mutations at Ser487 to render it resistant to shRNA knockdown in shRNA-INPP4B-1 cell pools (INPP4BR) reversed prolonged Akt phosphorylation. The results are representative of 4 separate experiments.
(B) Quantification of phospho-Thr308 Akt normalized to total Akt levels (n = 4, *p< 0.1, **p<0.01, ***p<0.001). Quantification of each single time point was determined using NIH ImageJ (n = 4). Data are shown as mean +/− SEM.
Loss of heterozygosity of INPP4B in BRCA1-mutant and basal-like breast tumors
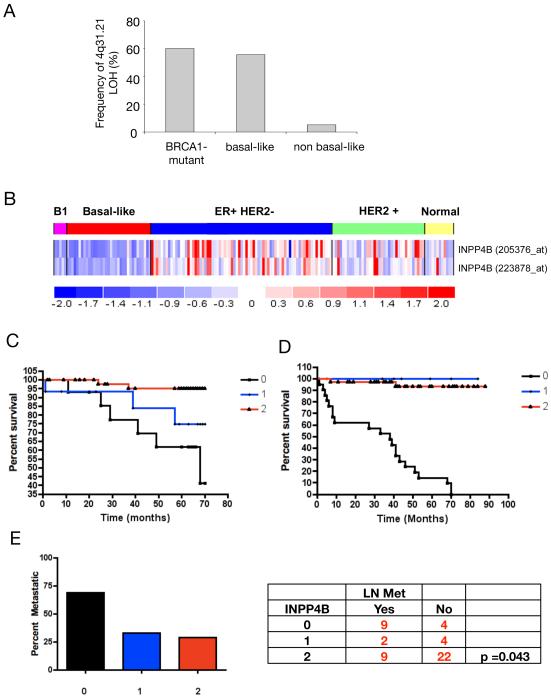
To detect potential genomic alterations of INPP4B in breast cancer, we analyzed a 10K SNP array set of 43 previously published breast cancers (Richardson et al., 2006). Comparison of loss of heterozygosity (LOH) patterns in the major subtypes of high grade breast cancer revealed that allelic loss at 4q31.21 was predominantly seen in the sporadic triple-negative (basal-like) tumors and in tumors arising in patients carrying a BRCA1 germline mutation. Figure 8A presents frequency of allelic loss in percent at chromosome position 4q31.21 for each tumor group. LOH in region 4q31.21 occurred in 60%, 55.6%, and 5% of BRCA1-mutant, sporadic basal-like, and high grade non-basal-like tumors, respectively (p<0.001). INPP4B mRNA expression level determined using two different array probes for INPP4B revealed lower relative expression (progressively darker shades of blue color) in BRCA1-mutant (B1) and sporadic basal-like breast tumors compared to other high-grade breast tumors (figure 8B). The triple negative or basal-like tumor subtype in which LOH and decreased expression of INPP4B was observed, is frequently a tumor subtype noted for increased metastatic potential and poor patient outcome (Sorlie et al., 2001)(Kapp et al., 2006).
Figure 8. Loss of heterozygosity at the INPP4B locus occurs frequently in BRCA1-mutant and basal-like breast cancers and ovarian cancers and correlates with decreased overall patient survival and lymph node involvement.
(A) Frequency of loss of heterozygosity (LOH) in the 4q31 region of Chromosome 4 from 43 high-grade human breast tumors is shown. In the data set, five tumors were carrying germline BRCA1 mutations, 18 additional tumors displayed basal-like features and 20 tumors were non-basal-like. The data come from 10K SNP array and were analyzed using dChip software. LOH of INPP4B, located at 4q31.21, occurred in 60%, 55.6%, and 5% of BRCA1-mutant, basal-like, and non-basal-like tumors, respectively (p<0.001). (B) Differential expression of INPP4B in subtypes of breast cancers analyzed by gene expression array. Sample subsets are indicated across the top as follows: BRCA1 germline mutation-associated tumors (B1, pink), sporadic basal-like tumors (basal-like, red), Luminal predominantly ER positive and HER2 negative tumors (ER+/HER2-, blue), and HER2 positive tumors (HER2+, green), and normal breast samples from mammographic reductions (Normal, yellow). Each column represents an individual sample. Each row represents the relative expression level measured from two different probes to INPP4B. Relative expression levels are represented as follows: mean levels are shown in white, expression levels above mean in progressively darker shades of red, and expression levels below mean in progressively darker shades of blue. The relative expression color scale is shown at the bottom. (C) Percent survival graph over time (in months) for breast cancer tissue stained for INPP4B expression (n = 112). Tissues from breast cancer patients were stained for INPP4B and scored for expression levels (0 = no expression, 1 = low expression, 2 = high expression). Breast cancer patients with no expression of INPP4B in tumor tissue showed significantly shorter over all survival compared to patients with low and high INPP4B expression in breast tumor tissue (p = 0.0018). (D) Percent survival graph over time (in months) for ovarian cancer tissue stained for INPP4B expression (n = 50). Tissues from ovarian cancer patients were stained for INPP4B and scored for protein expression levels (0 = no expression, 1 = low expression, 2 = high expression). Ovarian cancer patients with no expression of INPP4B in tumor tissue showed poor over all survival compared to patients with low or high INPP4B expression (p<0.0001). (E) Ovarian cancer tissues were analyzed for correlation of loss of INPP4B protein expression and lymph node involvement (LN Met). Lymph node metastases were found significantly more frequent in ovarian cancer tissues, which lost INPP4B protein expression (p = 0.043). The graph displays percent of patients with positive lymph nodes over INPP4B expression levels (0 = no expression, 1 = low expression, 2 = high expression).
Probing a different cohort of breast cancer patient tissues comprising mostly subclasses other than BRCA1-mutant and basal-like with an antibody directed against INPP4B, we found significant correlation between loss of INPP4B protein expression and decreased over all patient survival (figure 8C, patient information is summarized in Table S1, examples of scoring patient tissue for INPP4B expression levels by immunohistochemistry is shown in supplemental figure S6). Breast cancer patient tissues were also stained for progesterone receptor (PR), estrogen receptor (ER), and Heregulin2 (Her2)/NEU expression levels in order to indicate hormone dependence of breast tumor tissue (supplemental table 1). These data suggest that loss of INPP4B expression in breast tumor tissues is associated with poor patient outcome.
LOH of INPP4B correlates with decreased survival in ovarian cancer
LOH at 4q31.1-3, the region that includes INPP4B and other genes, has previously been observed in esophageal cancers (Sterian et al., 2006) prompting us to investigate chromosomal locus 4q31.1-3 in a variety of solid cancers. It was found high rates of copy number deletions at 4q31.1-3 in ovarian cancers (39.8%) and melanomas (21.6%) (Supplemental table 2). Copy number loss of INPP4B on chromosomal locus 4q31.1-3 was not observed as focal deletion but involving various genes in addition to INPP4B. Based on these findings, we investigated levels of INPP4B expression in randomly chosen ovarian cancers using IHC and found that, as in breast cancers, low levels of INPP4B correlated with decreased patient survival (Fig. 8D). Also, we found an inverse correlation between INPP4B expression levels and lymph node involvement (Fig. 8E, patient information is summarized in Table S3). Further, copy number analysis of a previously described ovarian tumor cohort revealed that out of 118 ovarian tumors investigated, 37 showed deletion at the INPP4B locus only, 8 tumors showed deletion at the PTEN locus only, and 10 tumors showed deletion at both the INPP4B locus and PTEN locus (n=118, supplemental figure 7A) (Dariush Etemadmoghadam et al., 2009). Since we found high cellular senescence upon loss of both, INPP4B and PTEN, unless Trp53 was also knocked down, we further investigated whether tumors with deletion of either lipid phosphatase (INPP4B or PTEN) or both showed Trp53 mutations. Re-analysis of the described ovarian cohort showed that most tumors harbored mutations in Trp53 whether or not INPP4B or PTEN were deleted (supplemental figure 7B), consistent with previous studies indicating high rates of Trp53 mutations in this disease (Krypuy et al., 2007).
Discussion
Here we present evidence that INPP4B functions as a tumor suppressor. Knocking down the expression of this enzyme in HMEC cells results in anchorage-independent growth and enhanced motility, similar to changes induced in response to knockdown of the PTEN tumor suppressor gene. Knocking down INPP4B in the human breast epithelial cell line, MCF-10A, resulted in dysmorphic growth of acinar structures in three dimensional Matrigel cultures, and increased intravasation of Matrigel in a Boyden chamber assay. These results were also similar to those observed upon knocking down PTEN. Knockdown of INPP4B and knockdown of PTEN both enhanced and prolonged AKT activation in HMEC cells in response to insulin, indicating that these two phosphoinositide phosphatases have similar effects on signaling downstream of PI3K and that they may play similar roles as tumor suppressors.
However, there were also some differences in cellular responses to knocking down INPP4B versus PTEN. Knocking down INPP4B in HMEC cells resulted in a more spindle-like cell structure compared to control cells or cells in which PTEN was knocked down. In addition, while the HMEC cells in which INPP4B was knocked down had enhanced N-cadherin and fibronectin expression, the HMEC cells in which PTEN was knocked down showed decreases in N-cadherin and fibronectin but increases in vimentin and E-cadherin. The MCF-10A cells with INPP4B knockdown also showed a greater ability to invade a Matrigel barrier compared to cells with PTEN knockdown. The similarities and differences observed between loss of PTEN and loss of INPP4B can probably be explained by the different substrate specificities of these two phosphatases. PTEN preferentially dephosphorylates the D-3 position of PI(3,4,5)P3 while here we show that INPP4B preferentially dephosphorylates the D-4 position of PI(3,4)P2 both in vitro and in vivo. While PI(3,4,5)P3 (produced by Class IA PI3Ks) is strongly implicated in cell transformation and in activation of AKT, the in vivo function of PI(3,4)P2 is not yet clear (Figure 1). This lipid can be produced by both class I and II PI3Ks in vitro using PI(4)P as a substrate. It can also be produced in vitro by SHIP family phosphatases using PI(3,4,5)P3 as a substrate. The major pathway for producing PI(3,4)P2 in vivo is not yet clear and may vary from tissue to tissue. But we clearly show that the main substrate of INPP4B is PI(3,4)P2, while PI(3,4,5)P3 is not affected. Interestingly, while some pleckstrin homology (PH) domains, such as those on BTK and Grp1, bind only to PI(3,4,5)P3, other PH domains, such as those on AKT and PDK1, can bind to both PI(3,4,5)P3 and PI(3,4)P2. Yet other PH domains, such as TAPP1 bind only to PI(3,4)P2. Despite apparently redundant abilities of PI(3,4)P2 and PI(3,4,5)P3 to bind to both AKT and PDK1 PH domains and to facilitate PDK1 phosphorylation of AKT in vitro, most in vivo studies have suggested that PI(3,4,5)P3 is more important than PI(3,4)P2 for Akt activation. For example, deletion of SHIP1, a phosphatase that converts PI(3,4,5)P3 to PI(3,4)P2, generally results in enhanced signaling downstream of PI3K and SHIP1 appears to be a tumor suppressor in hematopoietic cells (Gilby et al., 2007; Ong et al., 2007). Yet one study suggested that PI(3,4,5)P3 and PI(3,4)P2 are both needed for full activation of AKT (Scheid et al., 2002). It is also possible that, although PI(3,4,5)P3 and PI(3,4)P2 are redundant for activation of AKT in vivo in some tissues, INPP4B (or related family members) prevents PI(3,4)P2 from achieving the threshold necessary to activate Akt. In any event, our results indicate that knocking down INPP4B results in an increase in sustained AKT activation and cell transformation, similar to that observed upon knocking down PTEN.
Knocking down both INPP4B and PTEN in mammary epithelial cells resulted in cellular senescence. Previous studies had shown that complete loss of PTEN drives cells into senescence in vivo (Chen et al., 2005). Our findings that knocking down both INPP4B and PTEN causes a synergistic increase in senescence compared to knockdown of either message alone is consistent with the idea that superactivation of the PI3K pathway can result in senescence (Nogueira et al., 2008). Consistent with previous studies, senescence could be circumvented by knocking down Trp53. These results suggest that while loss of function of one allele of INPP4B or PTEN will contribute to tumor formation, loss of function of both alleles of INPP4B or PTEN (or of both) would be selected against in epithelial tumors. This may explain why we have not detected point mutations in INPP4B in basal-like breast cancers (Alexander Miron, unpublished data). Recently LOH of PTEN was detected in a significant fraction of basal-like breast cancer, while point mutations in PTEN were not found (Saal et al., 2008). This is presumably due to defects in DNA repair causing chromosomal rearrangements and microcopy number aberrations. The failure to see point mutations in the remaining allele is consistent with the idea that complete loss of PTEN results in senescence unless Trp53 is mutated.
Finally, our results indicate that decreased expression of INPP4B correlates with poor outcome in both breast cancer and ovarian cancer patients. Our results indicate that the PI3K pathway is likely to play a major role in driving this subset of cancers. A number of PI3K inhibitors are now in clinical trials and loss of INPP4B expression may provide a marker for selecting patients who will respond to these drugs.
Experimental Procedures
Cell culture
HMECs expressing hTERT and SV40 LT (TLM-HMECs) were cultured in mammary epithelial growth medium (MEGM, Cambrex) (Zhao et al., 2003). MCF-10A cells were maintained as described (Debnath et al., 2003). 293T and 3T3-L6 cells were grown in DMEM with 10% bovine serum. SUM149PT cell line was provided by S. Ethier and grown as described (http://www.asterand.com/Asterand/human_tissues/149PT.htm). Stable cell lines were generated by transduction with the indicated retroviral supernatants in the presence of 10 μg/ml polybrene, transduced cells were selected for resistance with puromycin (2 μg/ml) or neomycine (250 μg/ml). Anchorage independent proliferation assays were performed as described (Zhao et al., 2003), except cells were suspended in a top layer of 2% low-melting agarose (Seakem) in MEGM. For growth curves, cells were seeded at a densitiy of 1.0 × 104 per well in 6-well plates and cultured in MEGM. Cells were trypsinized and counted at day 0, 1, 2, 3, 4, and 5. Insulin stimulation experiments were performed with 100nM after overnight serum starvation. Used inhibitors concentrations: Wortmannin 50 nM, LY294002 10 μM, Rapamycin 1 nM, UO126 10 μM.
Immunoblotting
Cells were lysed in lysis buffer (20 mM Tris (pH7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 μg/ml Leupeptin, 1 mM DTT. Western blotting was performed with the following antibodies: αPTEN-HRP (Santa Cruz 7974), αP-Ser473Akt (Cell Signaling 9271), αP-Thr308AKT (Cell Signaling 4056), αINPP4B (Santa Cruz 12318), αGSK3beta (Cell Signaling 9315), αp85 (L. Cantley), αAKT(total) (Santa Cruz 8312), αP-Tuberin(Thr1462) (Cell Signaling 3611S), αP-p70S6kinase(Thr389) (Cell Signaling 9206S), αTuberin (Cell Signaling 3612), αP-GSK3alpha/beta(Ser21/9) (Cell Signaling 9331S), αp70S6kinase (Cell Signaling 2708).
Retroviral vectors and virus production
The shRNA hairpin-expressing retroviral vector MSCV-U6miR30-PIGdeltaRI, MSCV-U6miR30-PIGdeltaRI-FF2 (Renilla), and MSCV-U6miR30-PIGdeltaRI-PTEN were a generous gift from Steven Elledge (Harvard University, Boston, MA). Specific shRNA-hairpins directed against human INPP4B-1 (RHS1764-9399376) and INPP4B-2 (RHS1764-9207446) were obtained from OpenBiosystems. Retroviral vector pLXSN was a kind gift from Christopher Carpenter lab (Harvard University, Boston, MA). VSV-pseudotyped retroviruses were produced as previously described (Ory et al., 1996). Specific hairpin directed against human p53 was obtained from Addgene plasmid 10672, and specific hairpin directed against PTEN (hygromycin-resistant) was described in Mandl et al. (Mandl et al., 2007). Human INPP4B originated from the Origin cDNA expression library and was a kind gift of Robin Ketteler (Harvard University, Boston, MA). The rescue construct pLXSN/Flag-INPP4BR harbors two silent mutations at Ser687 to render it resistant to shRNA-INPP4B-1 knockdown. The catalytic-dead pLXSN/Flag-INPP4BR (‘INPP4BR-CD’) construct harbors a Cys842Ala mutation to abolish phosphatase activity as well as two silent mutations at Ser687 to avoid knockdown.
Semi-quantitative RT-PCR
Semi-quantitative SYBR RT-PCR was performed at the Harvard-BIDMC core facility (Victoria Petkova) using the following primers: INPP4B: 5′-GCC GAC CAC ATC ACC ACA G-3′ (sense), 5′-TTT CCG CTC ACA CTT TCC G-3′ (antisense). Beta-actin (control): 5′-AAC CTA ACT TGC GCA GAA AAC-3′ (sense), 5′-TTT ACA CGA AAG AAC TGC TAT C-3′ (antisense). E-cadherin1, type1: 5′-TGC TCT TGC TGT TTC TTC GG-3′ (sense), 5′-TGC CCC ATT CGT TCA AGT AG-3′ (antisense). Fibronectin: 5′-CTG TTA CTG GTT ACA GAG TAA-3′ (sense), 5′-TAG GTC ACC CTG TAC CTG GAA-3′ (antisense). Vimentin: 5′-ACC AGG TCC GTG TCC TCG T-3′ (sense), 5′-CTG CCC AGG CTG TAG GTG-3′ (antisense). Gamma-catenin: 5′-TAC GGC AAC CAG GAG AGC AAG-3′ (sense), 5′-TCT TCA GCA CAC TCT CCA GGC-3′ (antisense). N-cadherin: 5′-GCC CCT CAA GTG TTA CCT CAA-3′ (sense), 5′-AGC CGA GTG ATG GTC CAA TTT-3′ (antisense).
Morphogenesis assays
The 3D culture of MCF-10A cells was performed as previously described (Debnath et al., 2003). Used inhibitor concentrations: Rapamycin (1mM stock), (100mg/ml), UO126 (10mM). The assay media DMEM/F12 supplemented with 3% donor horse serum, 5ng/ml EGF 10 μg/ml insulin, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone and 2% Matrigel) was replaced with media starting on day 1 and re-fed every 4 days thereafter. Where indicated UO126, Rapamycin, or LY294002 were added to cultures at the reported dose on day 1 of 3D culture and replaced every 4 days thereafter. Acini formation was documented with phase contrast pictures.
Matrigel Invasion Assays
Invasion assays were carried out as previously described using Transwell chambers (Corning) with 8μm pore membranes coated with Matrigel. Cells were seeded at a density of 1×104 cells/well in triplicates and allowed to invade through the Matrigel for 24 hours. Growth medium was applied to both sides. Cells that had invaded to the lower surface of the membrane were fixed and stained with TolueneO blue.
SA-beta-Gal Activitiy Assay
SA-beta-Gal staining was performed as described (Dimri et al., 1995), and percentages were assessed by counting at least 300 cells. Cell pools were infected with shRNA-Trp53 and analyzed 3 days later. Cells were stained for 8 hours before evaluation.
In vitro phosphatase assays and in vivo analysis of phosphoinositides
[32P]-PI(3)P, [32P]-PI(3,4)P2 and [32P]-PI(3,4,5)P3 were synthesized by incubating PI, PI(4)P, and PI(4,5)P2 with class Ia PI3K (immunoprecipitated from 293T cells) and γ-[32P]-ATP (Whitman et al., 1987), Each of the radio-labeled lipids was incubated for 3 hrs at 30° with either INPP4B (Flag-immunoprecipitate from 293T cells expressing Flag-INPP4B) or with a control Flag-immunoprecipitate. The lipids were extracted in chloroform/HCl and separated by thin layer chromatography (TLC). The substrates and products from each phosphatase assay were quantified by Phosphoimager and the per cent hydrolysis was calculated. Quantitative HPLC analysis of deacetylated phosphoinositides from [32P]-inorganic phosphate-labeled L6 cells was performed as previously described (Auger et al., 1989). Counts for each single phosphatidylinositol lipid were normalized to combined counts of the major phosphoinositides, PI(4)P, and PI(4,5)P2.
Immunohistochemistry
The study cohort comprised breast and ovarian tumors consecutively ascertained at the Memorial Sloan-Kettering Cancer Center (MSKCC). All biopsies were evaluated at MSKCC, and the histologic diagnosis was based on established standard criteria. Use of tissues were approved by Institutional Review Board (IRB) waivers and the Human Biospecimen Utilization Committee (HBUC). TMAs were stained with INPP4B (goat polyclonal, Santa Cruz Biotechnology, Inc, Santa Cruz, CA Cat No. sc-12318 at 1:150 dilution) and PTEN (mouse monoclonal, clone 6H2.1, Cascade Bioscience Inc, Winchester, MA at 1:75 dilution) were applied after pre-treatment with 10mM citrate buffer, pH 6.0 and microwave. Secondary antibody biotinylated anti-goat made in rabbit (1:500) and ABC (avidin-biotin conjugate) Elite Vectastain (1:25) (Vector Lab, Burlingame, CA) were applied. PTEN was stained and scored as previously published (Min Sup et al., 2008).
Copy number deletion analysis
Copy number loss of INPP4B in tumor tissues were analyzed as described in Lin et al., 2008). Analysis of ovarian tumor tissue for loss of INPP4B and PTEN expression was performed as described (Dariush Etemadmoghadam et al., 2009). Study population consisted of 118 patients diagnosed with epithelial ovarian, primary peritoneal, or fallopian tube cancer. The Australian Ovarian Cancer Study (AOCS) was performed by the Westmead Hospital (n=73, Sidney, Australia). Other samples originated from Haukeland University Hospital of Bergen (Bergen, Norway), and Jikei University (Tokyo, Japan). This project had institutional ethics review board approval at all participating centers. AOCS samples subset was further analyzed for Trp53 mutation as previously described (Krypuy et al., 2007).
Xenograft experiments
SUM149PT cells infected with pLXSN-Flag, empty or pLXSN-Flag-INPP4B were selected for 2 weeks. Male nude mice were injected with 1×106 cells resuspended in 100μl Matrigel at the right flank. Tumors were grown over a time period of 4 weeks and mice subsequently sacrificed. Tumor occurrence and size were evaluated. All animal procedures followed protocols approved by the Animal Care and Use Committee (IACUC), Beth Israel Deaconess Medical Center at Harvard Medical School.
Significance.
Loss of function mutations or deletions of PTEN and activating mutations or amplifications of PIK3CA are amongst the most common events in solid tumors. Here we show that the phosphoinositide phosphatase INPP4B plays a similar role to PTEN in suppressing the PI3K/AKT signaling pathway, and thereby suppressing tumor growth. We also show that INPP4B is frequently deleted in a variety of solid tumors, including the majority of basal-like breast cancers. These results suggest that PI3K pathway inhibitors in current clinical trials may be effective in treating cancers where INPP4B is deleted.
Supplementary Material
Acknowledgments
We would like to thank Thomas Westbrook, Steven Elledge and Mohammed Bentires-Alj for reagents, technical help and discussion concerning the project. We also want to thank Robin Ketteler, Ina Rhee, Jonathan Hurov, and Matt van der Heiden for critical reading of the manuscript. Further we thank Victoria Petkova for the semi-quantitative RT-PCR analysis (core facility).
References
- Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- Barnache S, Le Scolan E, Kosmider O, Denis N, Moreau-Gachelin F. Phosphatidylinositol 4-phosphatase type II is an erythropoietin-responsive gene. Oncogene. 2005 doi: 10.1038/sj.onc.1209187. [DOI] [PubMed] [Google Scholar]
- Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M, Naderi A, Roberts I, Barbosa-Morais NL, Garcia MJ, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. PTEN is essential for embryonic development and tumour suppression. Nat Genet. 19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George G, Getz G, Tothill R, Okamoto A, et al. Integrated Genome-wide DNA Copy Number and Expression Analysis Identifies Distinct Mechanisms of Primary Chemoresistance in Ovarian Carcinomas. Clin Cancer Res. 2009;15:OF1–OF10. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- Gilby DC, Goodeve AC, Winship PR, Valk PJ, Delwel R, Reilly JT. Gene structure, expression profiling and mutation analysis of the tumour suppressor SHIP1 in Caucasian acute myeloid leukaemia. Leukemia. 2007 doi: 10.1038/sj.leu.2404864. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- Johannsdottir HK, Johannesdottir G, Agnarsson BA, Eerola H, Arason A, Johannsson OT, Heikkila P, Egilsson V, Olsson H, Borg A, et al. Deletions on chromosome 4 in sporadic and BRCA mutated tumors and association with pathological variables. Anticancer Res. 2004;24:2681–2687. [PubMed] [Google Scholar]
- Kapp AV, Jeffrey SS, Langerod A, Borresen-Dale AL, Han W, Noh DY, Bukholm IR, Nicolau M, Brown PO, Tibishirani R. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7:231. doi: 10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto H, Hamada K, Saunders M, Backman S, Sasaki T, Nakano T, Mak TW, Suzuki A. Physiological functions of Pten in mouse tissues. Cell Struct Funct. 2003;28:11–21. doi: 10.1247/csf.28.11. [DOI] [PubMed] [Google Scholar]
- Klippel A, Kavanaugh WM, Pot D, Williams LT. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryouy M, Ahmed AA, Etemadmoghadam D, Hyland SJ, Australian Ovarian Cancer Study Group. deFazio A, Fox SB, Brenton JD, et al. High resolution melting for mutation scanning og TP53 exons 5-8. BMC Cancer. 2007;7:168. doi: 10.1186/1471-2407-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, et al. Modeling Genomic Diversity and Tumor Dependency in Malignant Melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl A, Sarkes D, Carricaburu V, Jung V, Rameh L. Serum withdrawal-induced accumulation of phosphoinositide 3-kinase lipids in differentiating 3T3-L6 myoblasts: distinct roles for Ship2 and PTEN. Mol Cell Biol. 2007;27:8098–8112. doi: 10.1128/MCB.00756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSPA-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, Zaks TZ, Weber BL. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen C, Xu P, Chen M, Tonic I, Unterman T, Hay N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cell. 2008;14:458–470. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris FA, Atkins RC, Majerus PW. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. Evidence for conserved alternative splicing in the 4-phosphatase family. J Biol Chem. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- Norris FA, Auethavekiat V, Majerus PW. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J Biol Chem. 1995;270:16128–16133. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- Ong CJ, Ming-Lum A, Nodwell M, Ghanipour A, Yang L, Williams DE, Kim J, Demirjian L, Qasimi P, Ruschmann J, et al. Small-molecule agonists of SHIP1 inhibit the phosphoinositide 3-kinase pathway in hematopoietic cells. Blood. 2007;110:1942–1949. doi: 10.1182/blood-2007-03-079699. [DOI] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DR, Thompson CB. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Saal LH, Gruvberger-Saal SK, Persson C, Lovgren K, Jumppanen M, Staaf J, Jonsson G, Pires MM, Maurer M, Holm K, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid MP, Huber M, Damen JE, Hughes M, Kang V, Neilsen P, Prestwich GD, Krystal G, Duronio V. Phosphatidylinositol (3,4,5)P3 is essential but not sufficient for protein kinase B (PKB) activation; phosphatidylinositol (3,4)P2 is required for PKB phosphorylation at Ser-473: studies using cells from SH2-containing inositol-5-phosphatase knockout mice. J Biol Chem. 2002;277:9027–9035. doi: 10.1074/jbc.M106755200. [DOI] [PubMed] [Google Scholar]
- Scheid MP, Woodgett JR. Phosphatidylinositol 3′ kinase signaling in mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:83–99. doi: 10.1023/a:1009520616247. [DOI] [PubMed] [Google Scholar]
- Sharrard RM, Maitland NJ. Regulation of protein kinase B activity by PTEN and SHIP2 in human prostate-derived cell lines. Cell Signal. 2007;19:129–138. doi: 10.1016/j.cellsig.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Sleeman MW, Wortley KE, Lai KM, Gowen LC, Kintner J, Kline WO, Garcia K, Stitt TN, Yancopoulos GD, Wiegand SJ, Glass DJ. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, can de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterian A, Kan T, Berki AT, Mori Y, Olaru A, Schulmann K, Sato F, Wang S, Paun B, Cai K, et al. Mutational and LOH analyses of the chromosome 4q region in esophageal adenocarcinoma. Oncology. 2006;70:168–172. doi: 10.1159/000094444. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Sellers WR. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim Biophys Acta. 2000;1470:M21–35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. A genetic screen for candidate tumor suppressors identifies. REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Whitman M, Kaplan D, Roberts T, Cantley L. Evidence for two distinct phosphatidylinositol kinases in fibroblasts. Implications for cellular regulation. Biochem J. 1987;247:165–174. doi: 10.1042/bj2470165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM, Hahn WC. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.