Abstract
Background
Ferredoxin-NADP(H) reductase (FNR) from Pisum sativum and Flavodoxin (Fld) from Anabaena PCC 7119 have been reported to protect a variety of cells and organisms from oxidative insults. In this work, these two proteins were expressed in mitochondria of Cos-7 cells and tested for their efficacy to protect these cells from oxidative stress in vitro.
Principal Findings
Cos-7/pFNR and Cos-7/pFld cell lines expressing FNR and Fld, respectively, showed a significantly higher resistance to 24 h exposure to 300–600 µM hydrogen peroxide measured by LDH retention, MTT reduction, malondialdehyde (MDA) levels and lipid peroxide (LPO; FOX assay) levels. However, FNR and Fld did not exhibit any protection at shorter incubation times (2 h and 4 h) to 4 mM hydrogen peroxide or to a 48 h exposure to 300 µM methyl viologen. We found enhanced methyl viologen damage exerted by FNR that may be due to depletion of NADPH pools through NADPH-MV diaphorase activity as previously observed for other overexpressed enzymes.
Significance
The results presented are a first report of antioxidant function of these heterologous enzymes of vegetal and cyanobacterial origin in mammalian cells.
Introduction
Ferredoxin-NADP(H) reductases (FNRs; EC 1.18.1.2) constitute a family of hydrophilic, monomeric enzymes that contain non-covalently bound flavin adenine dinucleotide (FAD) as a prosthetic group. These ubiquitous flavoenzymes are present in animals, plants, parasites and prokaryotes, where they catalyze the reversible exchange of electrons between two molecules of a variety of obligatory one-electron carriers, such as ferredoxins (Fds) and flavodoxins (Flds), and a single molecule of NADP(H) [1]. Although FNRs are present in all kingdoms, plant isoforms are 200–500 times more active than their animal or prokaryote counterparts [1], property that could explain their notable competency as antioxidants in bacterial models [2], [3]. FNRs have demonstrated to protect proteobacteria and cyanobacteria from oxidative stress [2]–[5]. Pisum sativum (pea) FNR accomplishes functional complementation of mutant Escherichia coli defective for mrvA gene that are unable to grow aerobically in the presence of the radical propagator methyl viologen (MV: 1,1′-dimethyl-4,4′-bipyridinium dichloride) [4]. This complementation is strictly dependent on accumulation of functional transgenic FNR [4]. The NADP(H) dependent activities of the reductase were necessary and sufficient for detoxification, without participation of either Fd or Fld in the process [2]. Transgenic tobacco plants expressing knocked down levels of FNR are abnormally prone to photo-oxidative injury [6]. When grown under autotrophic conditions for 3 weeks, specimens with 20–40% extant reductase underwent leaf bleaching, elevated malondialdehyde (MDA) levels and membrane damage [6].
Given these antecedents, we hypothesized that pea FNR could be beneficial in the protection of mammalian cells from the oxidative injury produced by cold ischemia/reperfusion. In a previous work [7], we applied FNR gene transfection to isolated hepatocytes preserved in University of Wisconsin solution and then transplanted into recipient animals. We observed that, after cold preservation, transplanted hepatocytes expressing pea FNR were found in the parenchyma of recipient rat livers in quantities 20–50 times greater than controls and were devoid of the surrounding inflammatory infiltrates normally present when hepatocyte engraftment occurs. These results were indicative of an advantage of FNR-expressing cells during implantation. Two facts were already known: 1- donor hepatocytes show a burst of reactive oxygen species (ROS) soon after transplantation (cold ischemia-reperfusion injury) [8]–[12]; and 2- the obstruction of sinusoids produced by donor hepatocyte arrival through portal circulation recruits Kuppfer cells that exert their local action by releasing of ROS [13], [14]. Consequently, it seemed that FNR transfected hepatocytes could overcome these challenges better and engraft more effectively in the receptor liver than control cells [7]. Since our original hypothesis was that an advantage of FNR-expressing hepatocytes should be due to an antioxidant effect of this transgene, we became interested to assess whether pea FNR protects mammalian cells from oxidative insults in a less complex in vitro model that allowed a direct challenge to oxidants and non-ambiguous interpretation of the results.
Flavodoxins, on the other hand, are small acidic proteins that transfer electrons at low potentials and contain a non-covalently bound flavin mononucleotide (FMN) as their sole redox center. They act in various electron transport systems as functional analogs of Fds. Despite the fact that they have been found only in bacteria and algae, they share similarity with a number of protein domains of both prokaryote and eukaryote origin [15]. In cyanobacteria and enterobacteria, Fld levels increase several fold on exposure to MV and other superoxide (O2 •-)-propagating compounds [5], [16], [17], and overexpression of the flavoprotein in E. coli leads to augmented tolerance toward various sources of oxidative stress [16]. In transgenic tobacco plants, expressing Anabaena Fld in a constitutive fashion, this protein has exerted a notable protection against photo-oxidative and hydric stresses [18]. As in the case of FNR, Fld seems a good candidate for exploration of its protective antioxidative properties in new systems as mammalian cells are.
In this work we attempted the evaluation of the capacity of P. sativum FNR and Anabaena PCC 7119 Fld to protect Cos-7 cells from oxidative stress challenges in vitro. We found that both FNR and Fld protected against hydrogen peroxide (H2O2) after 24 hours of exposure. Surprisingly, we did not observe protection towards MV by neither of these transgenic proteins.
These results reveal antioxidant functions of plant FNR and cyanobacterial Fld in a mammalian cell line opening the opportunity to examine a myriad of applications in diseases where reactive oxygen species (ROS) are know to play a role in etiology (cancer, Alzheimer) and other clinical situations such as transplantation where the involvement of ROS in primary organ failure has been recognized.
Materials and Methods
Genes and plasmids
A full length cDNA (1385 nt; GenBank Accession Number X12446) of Pisum sativum FNR gene was cloned from a pea library [19]. Anabaena Fld gene sequence (540 nt) [20] was obtained by PCR cloning from genomic DNA (GenBank Accession Number S68006). A full length cDNA (1898 nt; GenBank Accession Number D49920) coding for mouse ferredoxin reductase (Fdxr) gene was cloned from a mouse kidney library [21].
To construct pFNR (Figure S1) plasmids p-FR7-1 [21] and pCV105 [22] were used to obtain the sequences of mitochondrial localization signal (MLS) of mouse Fdxr and mature portion of pea FNR genes, respectively. These sequences were fused in frame and inserted into commercial plasmid pcDNA3 (Invitrogen V79020) between the Hind III and Eco RI sites of its multiple cloning site.
To generate plasmid pFld (Figure S2) the sequence of Fld gene from Anabaena PCC 7119 [20], in plasmid pFlavo, was amplified by PCR introducing Sac I and Eco RI restriction sites flanking the open reading frame, fused in frame with MLS of mouse Fdxr gene [21], and inserted into pcDNA3.
pFNR and pFld sequences were checked in a Hitachi 3100 Avant Genetic Analyzer (Applied Biosystems) using the Big Dye kit (Applied Biosystems 4336697 and 4336768) and Avant Data Collection Software (Applied Biosystems) (Figure S3).
Cell culture and transfections
Cos-7 cells (ECACC 87021302) were cultured in DMEM/High glucose (Sigma D6546) supplemented with 2 mM L-glutamine (EuroClone EC B3000D) and 10% fetal bovine serum (FBS, Sigma F7524) in a CO2 incubator at 37°C. Transfections were done using Lipofectamine 2000 (Invitrogen 11668-027) following manufacturer's instructions. Transfectant cells were selected with 0.50 mg/ml G418 (Sigma A1720) for 15 days until complete detachment of mock-transfected cells. The G418 concentration for selection was established by performing a killing curve on Cos-7 cells for 2 weeks. To conduct the experiments, all lines were used within 20 passage numbers to reduce variations due to cell line characteristic alterations.
Evaluation of transgene expression and cellular localization
For RT-Real Time PCR total cellular RNA was isolated using TRI-REAGENT (Sigma T9424) and then treated with DNAse. Quantification and quality evaluation were performed spectrophotometrically and integrity was assessed in agarose-formaldehyde gels. RNA was retrotranscribed into cDNA with the commercial kit iScript (Bio-Rad 170-8890). Primers (FNR FP: 5′TGGTTTGGCATGGCTCTTCC3′; FNR RP: 5′ATCGTTTACTTGCTCTCTGCTTAC3′; Fld FP: 5′TTGATTATTGGCTGTCCTACTTGG3′; Fld RP: 5′CTGCGTAACCTATTTGGTCACC3′) were designed with the aid of Beacon Designer 2.0 (PREMIER Biosoft International). Real Time PCR was performed using iQ SYBR Green Supermix (Bio-Rad 170-8882) in Gene Amp PCR System 2400 thermocycler (Perkin-Elmer) controlled by i-Cycler IQ program (Bio-Rad). Negative controls using non-retrotranscribed RNA were included and the amplification products were checked in agarose gels after Real Time-PCR completion.
The sub-cellular localization of the transgenic proteins was verified. Total proteins were extracted with 20 mM Tris/HCl pH 7.50, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM Na3VO4, 1 mM PMSF and 10 µl/ml protease inhibitor cocktail (Calbiochem 539137), directly from the culture plate. Cytosolic, mitochondrial and nuclear protein fractions were obtained by a differential centrifugation protocol. Briefly, cells (approximately 30×106 cells) were detached from culture surface, mechanically disrupted by several passages trough a 25G syringe avoiding foam formation and debris were eliminated by low speed centrifugation (500 g). Nuclear and mitochondrial enriched fractions were obtained centrifuging at 2,500 g and 10,000 g, respectively, and were further washed and purified by more stringent centrifugation speeds. Cytosolic fraction was cleared by high speed centrifugation. Enrichment and cross-contamination of fractions were determined measuring the activities of the classical organelle marker enzymes: citrate synthase, NADH:ubiquinone oxidoreductase, lactate deshydrogenase, glucose-6-phosphatase, catalase and acid phosphatase as previously reported [23]–[25]. Fractions were probed for transgenic protein presence by standard western blotting (WB) using rabbit anti-FNR and rabbit anti-Fld antibodies and polyclonal swine anti-rabbit IgG/HRP (DakoCytomation P0217). Immunofluorescence (IF) co-localization studies were performed by labeling with the same primary antibodies and fluorescein anti-rabbit IgG (Vector FI-1000); and Mitotracker Red 580 (Molecular Probes M22425) for mitochondrial staining. Images were taken with an inverted confocal microscope (Nikon C1/Eclipse TE-2000-E2). Pearson's Correlation (Rr) and Overlap (R) Coefficients were also calculated (EZ-C1 3.70 software, Nikon).
Diaphorase activity for FNR was measured as the reduction of ferricyanide in a reaction mixture (1 ml) containing 50 mM Tris-HCl pH 8.50, 5 mM MgCl2, 3 mM glucose-6-phosphate, 300 µM NADP+, 1 U glucose-6-phosphate dehydrogenase and 1 mM potassium ferricyanide. The decrease in absorption at λ = 420 nm was monitored at 30°C using an ε420 = 1 mM−1 cm−1 [26].
Oxidative stress induction
Cells were plated 24 h before treatment at a density of 25,000 cells/cm2 and then exposed to 0–4 mM H2O2 (Sigma H1009) for 2 and 4 h, and to 0–600 µM H2O2 for 24 h in complete medium. For MV (Aldrich 85617-7) incubation cells were plated 24 h before treatment at a density of 12,500 cells/cm2. Cells were then exposed to 0–300 µM MV in complete medium for 48 h.
Cell viability and metabolic function
LDH activity was determined by measuring NADH oxidation at λ = 340 nm in a reaction mixture containing 0.6 mM pyruvate and 0.2 mM NADH in 50 mM KPO4 pH 7.50. ΔA/min was monitored for 3 min at 37°C.
For MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide) reduction assay cell monolayers were washed with PBS and 0.50 mg/ml MTT (Sigma M2128) in culture medium was added and incubated for 2 h in a CO2 incubator. After incubation, insoluble violet formazan crystals were dissolved with 3% SDS and 0.03 N HCl in 70% isopropanol and absorbance at λ = 570 nm was read.
Oxidative damage to lipids
Fe3+/xylenol orange (XO: 1-methyl-1-phenylethylhydroperoxide sodium salt) complex formation due to lipid peroxides present in the sample (FOX assay) was determined [27], [28]. Monolayers were washed and incubated with 100 µM XO (Aldrich 513296), 250 µM Fe (NH4)2 (SO4)2 and 110 mM HClO4 for 30 min at room temperature. Absorbance was then read at λ = 560 nm.
Malondialdehyde (MDA) quantification was performed in an HPLC system using a C18 column. Mobil phase consisted in 30 mM KH2PO4 pH 4.00/methanol (65/35% v/v) at a flow rate of 1.5 ml/min [29]. UV detection was set at λ = 254 nm. As standard, TEP (1,1,3,3-tetraethoxypropane, Sigma T9889) was diluted in 0.1 M HCl, boiled for 5 min and diluted to 50 µM MDA (ε = 13700 M−1 cm−1 at λ = 254 nm). Samples and standard were diluted (1/20) in cold HClO4 (final concentration 25 mM), filtered and injected immediately into the column (20 µl). Culture supernatants were used as it was shown that MDA is almost completely released to culture medium within 1 h after production [30].
Data analysis
All the data were expressed as percentage of change with respect each cell line not exposed to H2O2 or MV (control cultures, 100%). Results are expressed as mean ± standard deviations (SD). Experiments were performed in triplicate and its number (n) is reported in the figures. Groups of data were compared by analysis of variance (ANOVA) followed by Tukey post test. Statistical analyses were performed by InStat 3.05 (GraphPad Software Inc.) free software.
Results
Cell lines
Three new cell lines were generated by transfection with pcDNA3, pFNR and pFld plasmids and named Cos-7/pcDNA3, Cos-7/pFNR and Cos-7/pFld, respectively. They showed conserved epithelial cell morphology and no evident changes in shape or size (cellular or nuclear) with respect to original Cos-7 cells. Rate of growth was also similar in the absence of G418 antibiotic but to some extent slower in its presence (Figure S4). Based on this evidence, the results obtained have been normalized to controls performed with the same cell line.
Evaluation of transgene expression and cellular localization
We confirmed by RT-PCR that Cos-7 and Cos-7/pcDNA3 do not express the analyzed messengers, Cos-7/pFNR express only pea FNR and Cos-7/pFld express only Anabaena Fld. These results were confirmed and expanded by WB and IF were the cellular localization of FNR in Cos-7/pFNR and Fld in Cos-7/pFld was assessed (Figure S5). For the FNR detection in WB two positive bands were seen in Cos-7/pFNR total extract and mitochondrial fraction lanes; no bands were revealed in the lanes corresponding to total extracts of Cos-7, Cos-7/pcDNA3 and Cos-7/pFld or cytoplasmic and nuclear fractions of Cos-7/pFNR cells (Figure S5 A). The major band showed a Mr of 35.76 kDa identical to the expected one (35.34 kDa) for the processed form of the enzyme after cleavage of the MLS at the predicted site. The minor band presented a Mr of 43.23 kDa somewhat higher than calculated for the entire protein (39.72 kDa). Co-localization estimations gave Rr = [0.571-0.846] and R = [0.977-0.985] (n = 10 cells for each of 2 independent staining analyses) pointing to a high degree of overlap between mitochondria and FNR signals (Figure S5 B). Similar studies were performed for Fld. Again, positive bands in WB were only observed for total extract and mitochondrial fraction of Cos-7/pFld while all other lines and fractions resulted negative (Figure S5 A). Also in this case two bands were found: a major band of 33.24 kDa (higher than 24.84 kDa expected for the whole protein) and a minor one of 19.87 kDa very similar in Mr to the predicted one (20.46 kDa) for the processed protein lacking the MLS. By IF (Figure S5 B) overlap coefficients Rr = [0.602-0.741] and R = [0.973-0.982] were found (n = 10 cells for each of 2 independent staining analyses) indicating elevated co-localization between Fld and mitochondria.
Cos-7/pFNR total diaphorase activity was almost twice of that of Cos-7/pcDNA3 (49.6±1.6 mU/mg prot vs. 27.1±4.3 mUI/mg prot, p<0.0001).
Hydrogen peroxide cytotoxicity
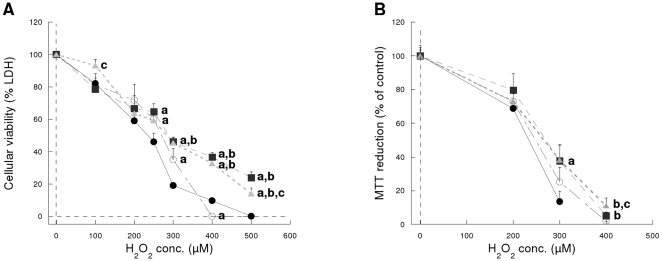
Figure 1 shows the effects of 24 h incubation with H2O2 on the cell viability and MTT test. Both FNR and Fld showed a clear protective effect (p<0.0001 by ANOVA) at H2O2 concentrations higher than 300 µM as evidenced by the difference with Cos-7 and Cos-7/pcDNA3 control cells. No protection was observed at 2 and 4 h incubation time even thought at the highest H2O2 concentration tested (4 mM), cell viability was reduced to 40% and 10%, respectively (Figure S6).
Figure 1. Cytotoxicity after 24 h exposure to H2O2.
(A) Cellular viability measure as LDH retention (n = 4 experiments, in triplicate). (B) Cellular metabolism measured as the reduction of MTT (n = 12 experiments, in triplicate). (•) Cos-7 cells, (○) Cos-7/pcDNA3 cells (▪), Cos-7/pFNR cells and (▴) Cos-7/pFld cells. a different from Cos-7 (p<0.05), b different from Cos-7/pcDNA3 (p<0.05), c different from Cos-7/pFNR (p<0.05).
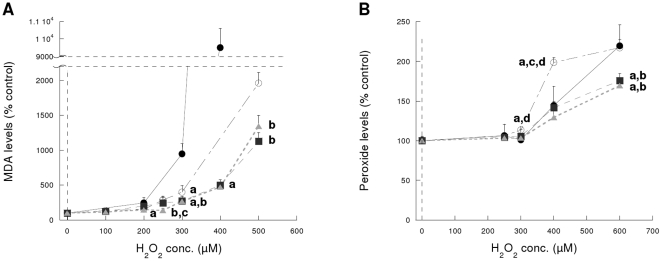
With regard to the measurement of the oxidative damage to lipids, a similar protection was observed after an exposure of 24 h (Figure 2) since both MDA and lipoperoxides levels were significantly lower (p<0.001 by ANOVA) than in control lines. After 4 h incubation lipid peroxide levels (FOX assay) did not differ from untreated controls or between cell lines (not shown).
Figure 2. Oxidative damage to lipids after 24 h exposure to H2O2.
(A) MDA levels. (B) Peroxide levels (FOX assay). (•) Cos-7 cells, (○) Cos-7/pcDNA3 cells, (▪) Cos-7/pFNR cells and (▴) Cos-7/pFld cells. a different from Cos-7 (p<0.05), b different from Cos-7/pcDNA3 (p<0.05), c different from Cos-7/pFNR (p<0.05), d different from Cos-7/pFld (p<0.05). n = 4 experiments, in triplicate.
Methyl viologen cytotoxicity
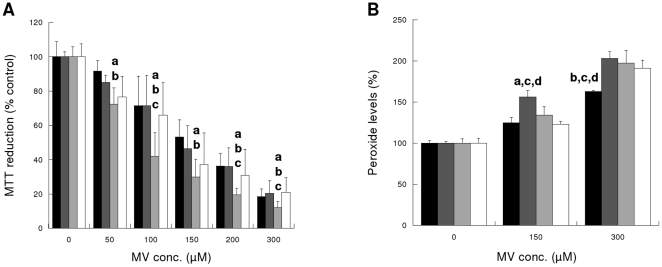
We could not find any protection exerted by FNR or Fld against MV injury as measured by MTT reduction and FOX assay (Figures 3A and B, respectively). Moreover, Cos-7/pFNR resulted to be more susceptible to injury than the other cell lines (Figure 3A).
Figure 3. Cytotoxicity after 48 h exposure to MV.
(A) Cellular metabolism (MTT reduction). (B) Lipid peroxidation levels (FOX assay). Black bars: Cos-7 cells, dark grey bars: Cos-7/pcDNA3 cells, light grey bars: Cos-7/pFNR cells and white bars: Cos-7/pFld cells. a different from Cos-7 (p<0.05), b different from Cos-7/pcDNA3 (p<0.05), c different from Cos-7/pFNR (p<0.05), d different from Cos-7/pFld (p<0.05). n = 4 experiments, in triplicate.
Discussion
In this work we investigated the capacity of P. sativum FNR and Anabaena Fld to protect Cos-7 cells from oxidative stress induced by hydrogen peroxide and methyl viologen.
Previous work performed by various groups demonstrated the successful interactions and electron transfer of the components of hybrid systems as Anabaena FNR and bovine adrenodoxin [31] and bovine adrenodoxin reductase and Anabaena PCC 7119 Fld [32]. It was also shown that NADPH-Fld reductase/Fld from E. coli can function as electron donors to bovine P450c17 to the same proportional extent (hydrolase to lyase ratio) as does P450 reductase [33]. NADPH-cytochrome P-450 oxidoreductase gene has most probably arisen through the fusion of the ancestral genes of Fld and FNR [15]. All these evidences prompted us to undertake the present work to test the hypothesis that heterologous proteins originating from plant and cyanobacteria would still be functional in Cos-7 cells.
Both proteins were directed to mitochondria via the fusion to their sequence of a MLS of mouse ferredoxin reductase. A number of works reported that proteins can be successfully directed to mitochondria by just fusing their N-terminal ends to a MLS [34]–[37]. Mouse Fdxr MLS was recognized by Cos-7 cells where it effectively directed both FNR and Fld to mitochondria. For FNR the MLS seems to be processed correctly as shown by WB analysis were we found a major band corresponding to the size of the mature protein and with only a small proportion remaining as the full-length, non-processed form. This is different for Cos-7/pFld cells where the proportion of the processed form was smaller indicating that the MLS have been cleaved less effectively. This can be explained taking into account that Fld does not posses a localization signal in its native form while pea FNR complete gene bears a chloroplast localization signal making its mature portion sequence appropriate for enzymatic digestion after transport. The exact position at which the cleavages of FNR and Fld are accomplished by Cos-7 cells is still to be determined.
Mitochondria have clearly been recognized as the most important physiological source of ROS being, at the same time, a cellular target for free radical-mediated damage as described for a series of diseases and during aging. Mitochondria are also the origin place of signaling molecules that command cell cycle, proliferation and apoptosis. Conservation of the mitochondrial DNA and respiratory chain components integrity is of most importance as they are the main source of cell energy production through oxidative phosphorylation. If antioxidant defense mechanisms are compromised or overwhelmed for any reason, a disease can arise as for example neurodegenerative disorders, cancer or aging [38]. In mammalian cell mitochondria, H2O2 provokes the disruption of the permeability transition with the consequent induction of apoptosis [39]. The main defense system against H2O2 is constituted by the glutathione redox cycle and Glutathione Peroxidase activities [39] together with Mn-SOD and the reductants NADH2 and UQH2. Recently, it has been demonstrated that MV accumulates in mitochondria through a carrier-mediated transport which requires intact membrane potential and, once inside, increases intramitochondrial O2 •- production [40]. Reduction and autooxidation of MV outside the mitochondria seemed to be negligible [40]. In mammalian mitochondria, different from other organisms, MV is mainly reduced by Complex I by reverse electron transport [40]. For these reasons, we considered mitochondria an appropriate cellular site to express FNR and Fld as a first approximation to examine their actions.
After 2 and 4 h exposure to H2O2 at concentrations above 1.5 mM, we observed a marked lost of metabolic function as measured by MTT reduction (Figure S6 A) for all cell lines with no protection from FNR and Fld. The lack of protection is probably due to the high concentration of H2O2 used which has been reported to induce a rapid death by necrosis [41]. These H2O2 concentrations were chosen because they allowed to measure a reduction of MTT detoxification at the short-time exposures studied (2 and 4 h).
Cellular viability (LDH release) and metabolism (MTT test) were protected by both FNR and Fld towards exposure to hydrogen peroxide for 24 h at concentrations of 300 µM and above (Figure 1). It is known that cells can display different responses to cytotoxicity injuries going from quiescence to apoptosis and necrosis, with different degrees in between. It has been reported that at H2O2 concentrations of 250–400 µM cells enter a “permanently growth-arrested state”, sometimes confused with cell death in toxicity studies, while at higher concentrations (0.5–1.0 mM) they will enter the apoptotic pathway [41]. The range of concentrations at which FNR and Fld display protection in this work overlap both situations described and thereby it is possible that the two phenomena are occurring with cell death gaining importance at the higher concentrations tested.
Hydrogen peroxide preferentially partitions into lipid and hydrophobic cores of proteins. The oxidative biomarker hypothesis proposes the theory that measurement of levels of oxidized biomolecules can provide an index of the levels of oxidant to which a biological system has been exposed [42]. Cells expressing FNR and Fld showed reduced oxidative damage of lipids (Figure 2) indicating that the protection is probably exerted through an antioxidative action towards H2O2. Even though a mechanism of protection through repair can not be discarded and it has been observed for iron-sulfur centers of hydro-lyases in FNR-overexpressing bacterial cells after oxidative stress induction [43], FNR and Fld are not repairing enzymes per se. However, it is possible that some repairing pathways are working more efficiently in cells expressing FNR and Fld because they are more protected by the actions of these two proteins. Moreover, even though low H2O2 concentrations (120–150 µM) are known to be rapidly metabolized by cells and provoke a reversible cellular arrest that last only 2 to 4 h [41], Kang et al. [44] have found several fold increased ROS levels in their Cos-7-pcDNA3.0 cells (equivalent to Cos-7/pcDNA3 in this work) after 24 h incubation with 250 and 500 µM H2O2. This observation reinforces the hypothesis that oxidative stress persists 24 h after induction by hydrogen peroxide.
MDA levels were much higher for Cos-7 than Cos-7/pcDNA3 cells. This difference is not really surprising as Cos-7 cell line has a faster rate of growth than all the transfected lines when grown in the presence of G418 (Figure S4). Faster growing cells are metabolically more active and, as a consequence, more susceptible to damages by exposure to cytotoxic compounds. Given this observation, we considered important to include both lines (Cos-7 and Cos-7/pcDNA3) as controls in this study to be able to discriminate between the effects of the expression of FNR and Fld per se (when compared with Cos-7/pcDNA3) and including other factors of the process of transfection and selection (when compared with Cos-7). Both controls are necessary to allow comparison in the case transfection or transduction systems different from the one used here were employed.
The protection from hydrogen peroxide-induced damage resulted of similar magnitude for FNR and Fld. This finding was not expected a priori because FNR and Fld are enzymes with different properties regarding kinetics and, most probably, specificity of interactions with endogenous cellular components. We have measured the diaphorase activity of Cos-7/pFNR and found that it doubles that of the control line Cos-7/pcDNA3 supporting the idea that FNR is functional and elevates basal levels of this enzymatic activity. For Fld we do not have an estimation of the expression of this protein.
MTT reduction and LPO (FOX assay) were analyzed under MV induced injury. Cells demonstrated a major reduction in viability/growth (20% of controls after 48 h incubation with 300 µM MV) and almost duplication of LPO content (Figure 3) but we could not see any protection in cells expressing FNR and Fld. These findings were surprising since both FNR and Fld have shown to protect a number of eukaryote and prokaryote organisms from MV exposure [2], [4], [5], [45]–[52]. Shimizu et al. [53] showed protection from MV damage (50–300 µM for 48 h) in Cos-7 cells overexpressing oxidized protein hydrolase. This finding does not exclude, however, that changes associated with the overexpression of FNR and/or Fld may result in the lack of protection we observed.
Other authors using MV to induce oxidative stress have observed that protective enzymes work better if their expression is directed to mitochondria when compared with cytosolic localization [54], [55]. MLS in Cos-7/pFld has not been efficiently excised but this fact did not avoid protection from H2O2. Anyway, if non-processed MLS-Fld displays lower or no function, it is possible that the achieved levels of Fld activity are insufficient to cope with MV challenge. Regarding this point van Leeuwen et al. [56] have reported that the reaction of MV radicals with Fld increases with Fld concentration and decreases when the concentration of oxidized viologen raises. In our studies we could be in the presence of low Fld (unprocessed protein) and high oxidized MV levels (high MV concentration imposed) explaining our observations.
Moreover, Figure 3A shows reduced viabilities for Cos-7/pFNR compared to the other cell lines for all the MV concentrations tested. In this sense, it has been reported that NADPH-MV diaphorase reaction of overexpressed FNR towards MV can enhance redox cycling of this compound with the consequent depletion of NADPH and augmentation of oxidative damage [4], [52]. Elroy-Stein et al. [57] reported an increased MV-mediated cytotoxity and enhancement of LPO in cells overexpressing Cu/Zn-SOD. They showed that clones possessing the highest transgene activities were less resistant to MV and that even the most resistant clones (expressing lower levels of SOD) were gradually deteriorating in the presence of low concentrations of MV for extended periods (50 µM for 48 h) [57].
In this work we have demonstrated that P. sativum FNR and Anabaena PCC 7119 Fld are able to reduce the damages caused by H2O2 but not MV in Cos-7 cells in vitro. This is a first study performed to evaluate these two genes as candidates for future applications in gene therapy such as amelioration of diseases and transplantation conditions for which ROS participation have been established.
Supporting Information
A) Circular map of pFNR showing the principal attributes of the plasmid B) Schematic representation of hybrid FNR gene between restriction sites Hind III (red) and EcoR I (yellow) of pFNR. In green: translation initiation (ATG) and termination (TAA) codons; in blue: limit between mouse Fdrx mitochondrial localization signal (MLS) and mature portion of Pisum sativum FNR (FNR). C) Fusion site sequence. In red: part of Fdxr MLS (last 2 codons); in green: residual mouse Fdrx mature portion (4 codons); in black: an additional codon created by the fusion; in grey: residual P. sativum FNR chloroplastic localization signal (1 codon) and in blue: part of P. sativum FNR mature portion (first 2 codons). Underlined in black: Sac I restriction site.
(0.03 MB PDF)
A) Circular map of pFld showing the principal attributes of the plasmid. B) Schematic representation of hybrid Fld gene between restriction sites Hind III (red) and EcoR I (yellow) of pFld. In green: translation initiation (ATG) and termination (TAA) codons; in blue: limit between mouse Fdrx mitochondrial localization signal (MLS) and Anabaena Fld (Fld). C) Fusion site sequence. In red: part of Fdxr MLS (last two (2) codons); in green: residual mouse Fdrx mature portion (4 codons); in black: additional codons created by the fusion (4 codons) and in blue: part of Anabaena Fld (ATG, first codon). Underlined in black: Sac I restriction site.
(0.02 MB PDF)
A) Sequence of hybrid FNR gene between restriction sites Hind III (red) and EcoR I (yellow) of pFNR. In green: translation initiation (ATG) and stop (TAA) codons. In blue: Sac I restriction site. B) Sequence of hybrid Fld gene between restriction sites Hind III (red) and EcoR I (yellow) of pFld. In green: translation initiation (ATG) and stop (TAA) codons. In blue: Sac I restriction site.
(0.01 MB PDF)
Rate of growth of cell lines. Grown in culture medium without G418: Balck: Cos-7; dark grey: Cos-7/pcDNA3; light grey: Cos-7/pFNR; white: Cos-7/pFld. Grown in culture medium with G418: oblique dashed: Cos-7/pcDNA3; horizontal dashed: Cos-7/pFNR; oblique squared: Cos-7/pFld. ANOVA results: * different from cells cultured in the presence of G418; # different from all other cell lines. At 72 h (not marked in the figure to avoid confusion): 1) Cos-7/pFNR +G418 is also different from Cos-7/pcDNA3 -G418 and Cos-7/pFld +G418; and 2) Cos-7/pFNR -G418 is also different from Cos-7/pFld -G418.
(0.01 MB PDF)
A- Western blot analyses. FNR: detection using antibody developed against P. sativum FNR (C7: total extract from Cos-7; C7/p3: total extract from Cos-7/pcDNA3; C7/pFNR: total extract from Cos-7/pFNR; C7/pFld: total extract from Cos-7/pFld; C: cytoplasmic fraction of Cos-7/pFNR cells; M: mitochondrial franction of Cos-7/pFNR cells; N: nuclear fraction of Cos-7/pFNR cells). Fld: detection using antibody developed against Anabaena Fld (C7: total extract from Cos-7; C7/p3: total extract from Cos-7/pcDNA3; C7/pFNR: total extract from Cos-7/pFNR; C7/pFld: total extract from Cos-7/pFld; C: cytoplasmic fraction of Cos-7/pFld cells; M: mitochondrial franction of Cos-7/pFld cells; N: nuclear fractions of Cos-7/pFld cells). Positive band positions are pointed with red arrows (43.23 and 35.76 kDa for FNR; and 33.24 and 19.87 kDa for Fld). Positions of molecular weight markers are marked with blue arrows and their weights in kDa are indicated. B- Typical merges of the photomicrographs taken for co-localization studies. Staining: nuclei in blue (Hoechst 33258); mitochondria in red (MitoTracker Red); transgenic protein in green (fluorescein conjugated specific antibody).
(0.48 MB PDF)
Hydrogen peroxide induced cytotoxicity after (A) 2 h exposure and (B) 4 h exposure. Black: Cos-7; dark grey: Cos-7/pcDNA3; light grey: Cos-7/pFNR and white: Cos-7/pFld cells. n = 6 experiments in triplicate.
(0.01 MB PDF)
Acknowledgments
We are grate to Dra. Adriana Krapp, Dr. Eduardo Ceccarelli and Dr. Nestor Carrillo from Instituto de Biología de Rosario (IBR, CONICET, Rosario, Argentina) for their useful help in cloning techniques to construct the plasmids used and the generous gift of the primary antibodies against FNR and Fld. For the fluorescence co-localization studies we have utilized the services of the confocal microscopy facility of IBR/IFISE (CONICET, Rosario, Argentina). Dra. Mediavilla and Dr. Guibert are members of CONICET (National Scientific and Technology Council), Argentina.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by StartUp Grant AREA Science Park/Centro Studi Fegato to Dra. Mediavilla and a postdoctoral fellowship CONICET/Sistema Cientifíco de Trieste to Dra. Mediavilla. Dra. Mediavilla and Dr. Guibert are members of CONICET (National Scientific and Technology Council), Argentina. Italian Liver Foundation (legge26/00) to Professor Tiribelli. Grant PICT N° 703 "Préstamo BID" from Agencia Nacional de Promoción Científica y Tecnológica, Argentina. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carrillo N, Ceccarelli EA. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur J Biochem. 2003;270:1900–1915. doi: 10.1046/j.1432-1033.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 2.Krapp AR, Tognetti VB, Carrillo N, Acevedo A. The role of ferredoxin-NADP+ reductase in the concerted cell defense against oxidative damage – studies using Escherichia coli mutants and cloned plant genes. Eur J Biochem. 1997;249:556–563. doi: 10.1111/j.1432-1033.1997.00556.x. [DOI] [PubMed] [Google Scholar]
- 3.Pomposiello PJ, Demple B. Identification of SoxS-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182:23–29. doi: 10.1128/jb.182.1.23-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krapp AR, Carrillo N. Functional complementation of the mvrA mutation of Escherichia coli by plant ferredoxin-NADP+ oxidoreductase. Arch Biochem Biophys. 1995;317:215–221. doi: 10.1006/abbi.1995.1156. [DOI] [PubMed] [Google Scholar]
- 5.Yousef N, Pistorius EK, Michel KP. Comparative analysis of idiA and isiA transcription under iron starvation and oxidative stress in Synechococcus elongatus PCC 7942 wild-type and selected mutants. Arch Microbiol. 2003;180:471–483. doi: 10.1007/s00203-003-0618-4. [DOI] [PubMed] [Google Scholar]
- 6.Palatnik JF, Tognetti VB, Poli HO, Rodriguez RE, Blanco N, et al. Transgenic tobacco plants expressing antisense ferredoxin-NADP(H) reductase transcripts display increased susceptibility to photo-oxidative damage. Plant J. 2003;35:332–341. doi: 10.1046/j.1365-313x.2003.01809.x. [DOI] [PubMed] [Google Scholar]
- 7.Mediavilla MG, Krapp A, Carrillo N, Rodriguez JV, Tiribelli C, et al. Efficient cold transfection of pea ferredoxin-NADP(H) oxidoreductase into rat hepatocytes. J Gene Med. 2006;8:306–313. doi: 10.1002/jgm.847. [DOI] [PubMed] [Google Scholar]
- 8.Rauen U, Polzar B, Stephan H, Mannherz HG, de GH. Cold-induced apoptosis in cultured hepatocytes and liver endothelial cells: mediation by reactive oxygen species. FASEB J. 1999;13:155–168. doi: 10.1096/fasebj.13.1.155. [DOI] [PubMed] [Google Scholar]
- 9.Kerkweg U, Li T, de GH, Rauen U. Cold-induced apoptosis of rat liver cells in University of Wisconsin solution: the central role of chelatable iron. Hepatology. 2002;35:560–567. doi: 10.1053/jhep.2002.31869. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez JV, Mamprin ME, Guibert EE, Labadie G. Protective effect of glutathione (GSH) over glutathione monoethyl-ester (GSH-E) on cold preservation of isolated rat liver cells. Cell Transplant. 1995;4:245–251. doi: 10.1177/096368979500400208. [DOI] [PubMed] [Google Scholar]
- 11.Mamprin ME, Guibert EE, Rodriguez JV. Glutathione synthesis during the rewarming of rat hepatocytes preserved in the University of Wisconsin solution. Cryobiology. 2001;43:310–315. doi: 10.1006/cryo.2001.2364. [DOI] [PubMed] [Google Scholar]
- 12.Mamprin ME, Guibert EE, Rodriguez JV. Glutathione content during the rinsing and rewarming process of rat hepatocytes preserved in University of Wisconsin solution. Cryobiology. 2000;40:270–276. doi: 10.1006/cryo.2000.2242. [DOI] [PubMed] [Google Scholar]
- 13.Bilzer M, Jaeschke H, Vollmar AM, Paumgartner G, Gerbes AL. Prevention of Kupffer cell-induced oxidant injury in rat liver by atrial natriuretic peptide. Am J Physiol. 1999;276:G1137–G1144. doi: 10.1152/ajpgi.1999.276.5.G1137. [DOI] [PubMed] [Google Scholar]
- 14.Joseph B, Malhi H, Bhargava KK, Palestro CJ, McCuskey RS, et al. Kupffer cells participate in early clearance of syngeneic hepatocytes transplanted in the rat liver. Gastroenterology. 2002;123:1677–1685. doi: 10.1053/gast.2002.36592. [DOI] [PubMed] [Google Scholar]
- 15.Porter TD, Kasper CB. NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry. 1986;25:1682–1687. doi: 10.1021/bi00355a036. [DOI] [PubMed] [Google Scholar]
- 16.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh AK, Li H, Sherman LA. Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiol Plant. 2004;120:27–35. doi: 10.1111/j.0031-9317.2004.0232.x. [DOI] [PubMed] [Google Scholar]
- 18.Tognetti VB, Palatnik JF, Fillat MF, Melzer M, Hajirezaei MR, et al. Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. Plant Cell. 2006;18:2035–2050. doi: 10.1105/tpc.106.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman BJ, Gray JC. Characterisation of a full-length cDNA clone for pea ferredoxin-NAPD+ reductase. Plant Mol Biol. 1988;10:511–520. doi: 10.1007/BF00033606. [DOI] [PubMed] [Google Scholar]
- 20.Fillat MF, Borrias WE, Weisbeek PJ. Isolation and overexpression in Escherichia coli of the flavodoxin gene from Anabaena PCC 7119. Biochem J. 1991;280(Pt 1):187–191. doi: 10.1042/bj2800187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh S, Iemura O, Yamada E, Yoshimura T, Tsujikawa K, et al. cDNA cloning of mouse ferredoxin reductase from kidney. Biochim Biophys Acta. 1995;1264:159–162. doi: 10.1016/0167-4781(95)00172-d. [DOI] [PubMed] [Google Scholar]
- 22.Ceccarelli EA, Viale AM, Krapp AR, Carrillo N. Expression, assembly, and processing of an active plant ferredoxin-NADP+ oxidoreductase and its precursor protein in Escherichia coli. J Biol Chem. 1991;266:14283–14287. [PubMed] [Google Scholar]
- 23.Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- 24.Baudhuin P. Isolation of rat liver peroxisomes. Methods Enzymol. 1974;31:356–368. doi: 10.1016/0076-6879(74)31039-7. [DOI] [PubMed] [Google Scholar]
- 25.Trouet A. Isolation of modified liver lysosomes. Methods Enzymol. 1974;31:323–329. doi: 10.1016/0076-6879(74)31034-8. [DOI] [PubMed] [Google Scholar]
- 26.Serra EC, Carrillo N, Krapp AR, Ceccarelli EA. One-step purification of plant ferredoxin-NADP+ oxidoreductase expressed in Escherichia coli as fusion with glutathione S-transferase. Protein Expr Purif. 1993;4:539–546. doi: 10.1006/prep.1993.1071. [DOI] [PubMed] [Google Scholar]
- 27.Hermes-Lima M, Willmore WG, Storey KB. Quantification of lipid peroxidation in tissue extracts based on Fe(III)xylenol orange complex formation. Free Radic Biol Med. 1995;19:271–280. doi: 10.1016/0891-5849(95)00020-x. [DOI] [PubMed] [Google Scholar]
- 28.Wolff SP. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods in Enzymology. 1994;233:182–189. [Google Scholar]
- 29.Karatas F, Karatepe M, Baysar A. Determination of free malondialdehyde in human serum by high-performance liquid chromatography. Anal Biochem. 2002;311:76–79. doi: 10.1016/s0003-2697(02)00387-1. [DOI] [PubMed] [Google Scholar]
- 30.Morel I, Lescoat G, Cillard J, Pasdeloup N, Brissot P, et al. Kinetic evaluation of free malondialdehyde and enzyme leakage as indices of iron damage in rat hepatocyte cultures. Involvement of free radicals. Biochem Pharmacol. 1990;39:1647–1655. doi: 10.1016/0006-2952(90)90107-v. [DOI] [PubMed] [Google Scholar]
- 31.Faro M, Schiffler B, Heinz A, Nogues I, Medina M, et al. Insights into the design of a hybrid system between Anabaena ferredoxin-NADP+ reductase and bovine adrenodoxin. Eur J Biochem. 2003;270:726–735. doi: 10.1046/j.1432-1033.2003.03433.x. [DOI] [PubMed] [Google Scholar]
- 32.Zollner A, Nogues I, Heinz A, Medina M, Gomez-Moreno C, et al. Analysis of the interaction of a hybrid system consisting of bovine adrenodoxin reductase and flavodoxin from the cyanobacterium Anabaena PCC 7119. Bioelectrochemistry. 2004;63:61–65. doi: 10.1016/j.bioelechem.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins CM, Waterman MR. NADPH-flavodoxin reductase and flavodoxin from Escherichia coli: characteristics as a soluble microsomal P450 reductase. Biochemistry. 1998;37:6106–6113. doi: 10.1021/bi973076p. [DOI] [PubMed] [Google Scholar]
- 34.Horwich AL, Kalousek F, Mellman I, Rosenberg LE. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO J. 1985;4:1129–1135. doi: 10.1002/j.1460-2075.1985.tb03750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurt EC, Pesold-Hurt B, Schatz G. The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett. 1984;178:306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- 36.Hurt EC, Muller U, Schatz G. The first twelve amino acids of a yeast mitochondrial outer membrane protein can direct a nuclear-coded cytochrome oxidase subunit to the mitochondrial inner membrane. EMBO J. 1985;4:3509–3518. doi: 10.1002/j.1460-2075.1985.tb04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurt EC, Pesold-Hurt B, Suda K, Oppliger W, Schatz G. The first twelve amino acids (less than half of the pre-sequence) of an imported mitochondrial protein can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO J. 1985;4:2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai J, Rodriguez AM, Melendez JA, Cederbaum AI. Overexpression of catalase in cytosolic or mitochondrial compartment protects HepG2 cells against oxidative injury. J Biol Chem. 1999;274:26217–26224. doi: 10.1074/jbc.274.37.26217. [DOI] [PubMed] [Google Scholar]
- 40.Cocheme HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 41.Davies KJA. The broad spectrum of responses to oxidative stress in proliferating cells. In: Hutzinger O, editor. The handbook of environmental chemistry. Springer Berlin/Heilderberg; 2005. pp. 63–75. [Google Scholar]
- 42.Griffiths HR, Moller L, Bartosz G, Bast A, Bertoni-Freddari C, et al. Biomarkers. Mol Aspects Med. 2002;23:101–208. doi: 10.1016/s0098-2997(02)00017-1. [DOI] [PubMed] [Google Scholar]
- 43.Giro M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152:1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- 44.Kang HJ, Hong SM, Kim BC, Park EH, Ahn K, et al. Effects of heterologous expression of thioredoxin reductase on the level of reactive oxygen species in COS-7 cells. Mol Cells. 2006;22:113–118. [PubMed] [Google Scholar]
- 45.Krapp AR, Rodriguez RE, Poli HO, Paladini DH, Palatnik JF, et al. The flavoenzyme ferredoxin (flavodoxin)-NADP(H) reductase modulates NADP(H) homeostasis during the soxRS response of Escherichia coli. J Bacteriol. 2002;184:1474–1480. doi: 10.1128/JB.184.5.1474-1480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palatnik JF, Valle EM, Carrillo N. Oxidative stress causes ferredoxin-NADP+ reductase solubilization from the thylakoid membranes in methyl viologen-treated plants. Plant Physiol. 1997;115:1721–1727. doi: 10.1104/pp.115.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez RE, Krapp AR, Carrillo N. The mvrA locus of Escherichia coli does not encode a ferredoxin-NADP+ reductase. Microbiology. 1998;144(Pt 9):2375–2376. doi: 10.1099/00221287-144-9-2375. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez RE, Lodeyro A, Poli HO, Zurbriggen M, Peisker M, et al. Transgenic tobacco plants overexpressing chloroplastic ferredoxin-NADP(H) reductase display normal rates of photosynthesis and increased tolerance to oxidative stress. Plant Physiol. 2007;143:639–649. doi: 10.1104/pp.106.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zurbriggen MD, Tognetti VB, Fillat MF, Hajirezaei MR, Valle EM, et al. Combating stress with flavodoxin: a promising route for crop improvement. Trends Biotechnol. 2008;26:531–537. doi: 10.1016/j.tibtech.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Gaudu P, Weiss B. Flavodoxin mutants of Escherichia coli K-12. J Bacteriol. 2000;182:1788–1793. doi: 10.1128/jb.182.7.1788-1793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girardini JE, Dissous C, Serra E. Schistosoma mansoni ferredoxin NADP(H) oxidoreductase and its role in detoxification. Mol Biochem Parasitol. 2002;124:37–45. doi: 10.1016/s0166-6851(02)00158-5. [DOI] [PubMed] [Google Scholar]
- 52.Liochev SI, Hausladen A, Beyer WF, Jr, Fridovich I. NADPH: ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci U S A. 1994;91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu K, Fujino T, Ando K, Hayakawa M, Yasuda H, et al. Overexpression of oxidized protein hydrolase protects COS-7 cells from oxidative stress-induced inhibition of cell growth and survival. Biochem Biophys Res Commun. 2003;304:766–771. doi: 10.1016/s0006-291x(03)00657-0. [DOI] [PubMed] [Google Scholar]
- 54.Mockett RJ, Bayne AC, Kwong LK, Orr WC, Sohal RS. Ectopic expression of catalase in Drosophila mitochondria increases stress resistance but not longevity. Free Radic Biol Med. 2003;34:207–217. doi: 10.1016/s0891-5849(02)01190-5. [DOI] [PubMed] [Google Scholar]
- 55.Tien Nguyen-nhu N, Knoops B. Mitochondrial and cytosolic expression of human peroxiredoxin 5 in Saccharomyces cerevisiae protect yeast cells from oxidative stress induced by paraquat. FEBS Lett. 2003;544:148–152. doi: 10.1016/s0014-5793(03)00493-9. [DOI] [PubMed] [Google Scholar]
- 56.van Leeuwen JW, van DC, Veeger C. A pulse-radiolysis study of the reduction of flavodoxin from Megasphaera elsdenii by viologen radicals. A conformational change as a possible regulating mechanism. Eur J Biochem. 1983;135:601–607. doi: 10.1111/j.1432-1033.1983.tb07694.x. [DOI] [PubMed] [Google Scholar]
- 57.Elroy-Stein O, Bernstein Y, Groner Y. Overproduction of human Cu/Zn-superoxide dismutase in transfected cells: extenuation of paraquat-mediated cytotoxicity and enhancement of lipid peroxidation. EMBO J. 1986;5:615–622. doi: 10.1002/j.1460-2075.1986.tb04255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Circular map of pFNR showing the principal attributes of the plasmid B) Schematic representation of hybrid FNR gene between restriction sites Hind III (red) and EcoR I (yellow) of pFNR. In green: translation initiation (ATG) and termination (TAA) codons; in blue: limit between mouse Fdrx mitochondrial localization signal (MLS) and mature portion of Pisum sativum FNR (FNR). C) Fusion site sequence. In red: part of Fdxr MLS (last 2 codons); in green: residual mouse Fdrx mature portion (4 codons); in black: an additional codon created by the fusion; in grey: residual P. sativum FNR chloroplastic localization signal (1 codon) and in blue: part of P. sativum FNR mature portion (first 2 codons). Underlined in black: Sac I restriction site.
(0.03 MB PDF)
A) Circular map of pFld showing the principal attributes of the plasmid. B) Schematic representation of hybrid Fld gene between restriction sites Hind III (red) and EcoR I (yellow) of pFld. In green: translation initiation (ATG) and termination (TAA) codons; in blue: limit between mouse Fdrx mitochondrial localization signal (MLS) and Anabaena Fld (Fld). C) Fusion site sequence. In red: part of Fdxr MLS (last two (2) codons); in green: residual mouse Fdrx mature portion (4 codons); in black: additional codons created by the fusion (4 codons) and in blue: part of Anabaena Fld (ATG, first codon). Underlined in black: Sac I restriction site.
(0.02 MB PDF)
A) Sequence of hybrid FNR gene between restriction sites Hind III (red) and EcoR I (yellow) of pFNR. In green: translation initiation (ATG) and stop (TAA) codons. In blue: Sac I restriction site. B) Sequence of hybrid Fld gene between restriction sites Hind III (red) and EcoR I (yellow) of pFld. In green: translation initiation (ATG) and stop (TAA) codons. In blue: Sac I restriction site.
(0.01 MB PDF)
Rate of growth of cell lines. Grown in culture medium without G418: Balck: Cos-7; dark grey: Cos-7/pcDNA3; light grey: Cos-7/pFNR; white: Cos-7/pFld. Grown in culture medium with G418: oblique dashed: Cos-7/pcDNA3; horizontal dashed: Cos-7/pFNR; oblique squared: Cos-7/pFld. ANOVA results: * different from cells cultured in the presence of G418; # different from all other cell lines. At 72 h (not marked in the figure to avoid confusion): 1) Cos-7/pFNR +G418 is also different from Cos-7/pcDNA3 -G418 and Cos-7/pFld +G418; and 2) Cos-7/pFNR -G418 is also different from Cos-7/pFld -G418.
(0.01 MB PDF)
A- Western blot analyses. FNR: detection using antibody developed against P. sativum FNR (C7: total extract from Cos-7; C7/p3: total extract from Cos-7/pcDNA3; C7/pFNR: total extract from Cos-7/pFNR; C7/pFld: total extract from Cos-7/pFld; C: cytoplasmic fraction of Cos-7/pFNR cells; M: mitochondrial franction of Cos-7/pFNR cells; N: nuclear fraction of Cos-7/pFNR cells). Fld: detection using antibody developed against Anabaena Fld (C7: total extract from Cos-7; C7/p3: total extract from Cos-7/pcDNA3; C7/pFNR: total extract from Cos-7/pFNR; C7/pFld: total extract from Cos-7/pFld; C: cytoplasmic fraction of Cos-7/pFld cells; M: mitochondrial franction of Cos-7/pFld cells; N: nuclear fractions of Cos-7/pFld cells). Positive band positions are pointed with red arrows (43.23 and 35.76 kDa for FNR; and 33.24 and 19.87 kDa for Fld). Positions of molecular weight markers are marked with blue arrows and their weights in kDa are indicated. B- Typical merges of the photomicrographs taken for co-localization studies. Staining: nuclei in blue (Hoechst 33258); mitochondria in red (MitoTracker Red); transgenic protein in green (fluorescein conjugated specific antibody).
(0.48 MB PDF)
Hydrogen peroxide induced cytotoxicity after (A) 2 h exposure and (B) 4 h exposure. Black: Cos-7; dark grey: Cos-7/pcDNA3; light grey: Cos-7/pFNR and white: Cos-7/pFld cells. n = 6 experiments in triplicate.
(0.01 MB PDF)