Abstract
The daily timing of when PERIOD (PER) proteins translocate from the cytoplasm to the nucleus is a critical step in clock mechanisms underpinning circadian rhythms in animals. Numerous lines of evidence indicate that phosphorylation plays a prominent role in regulating various aspects of PER function and metabolism, including changes in its daily stability and subcellular distribution. In this report, we show that phosphorylation of serine 661 (Ser661) by a proline-directed kinase(s) is a key phospho-signal on the Drosophila PER protein (dPER) that regulates the timing of its nuclear accumulation. Mutations that block phosphorylation at Ser661 do not affect dPER stability but delay its nuclear entry in key pacemaker neurons, yielding longer behavioral rhythms. Intriguingly, abolishing phosphorylation at Ser661 also attenuates the extent of dPER hyperphosphorylation in vivo, suggesting the phosphorylated state of Ser661 regulates phosphorylation at other sites on dPER. Indeed, we identify Ser657 as a site that is phosphorylated by the glycogen synthase kinase GSK-3β (SHAGGY; SGG) in a manner dependent on priming at Ser661. Although not as dramatic as mutating Ser661, mutations that abolish phosphorylation at Ser657 also lead to longer behavioral periods, suggesting that a multi-kinase hierarchical phosphorylation module regulates the timing of dPER nuclear entry. Together with evidence in mammalian systems, our findings implicate proline-directed kinases in clock mechanisms and suggest that PER proteins are key downstream targets of lithium therapy, a potent inhibitor of GSK-3β used to treat manic depression, a disorder associated with clock malfunction in humans.
Introduction
Phosphorylation programs have emerged as important regulatory schemes common to circadian (≅24 h) pacemakers that help drive daily changes in the stabilities, subcellular localizations, protein–protein interactions, and activities of central “clock” proteins (for review, see Bae and Edery, 2006; Gallego and Virshup, 2007). A conserved feature of circadian clocks in animals is that PERIOD (PER) proteins undergo daily changes in phosphorylation, modifications that are likely to be the principal biochemical “state variable” driving the pace of these clocks. Studies in Drosophila melanogaster have been instrumental in our understanding of clock mechanisms in general and mammalian ones in particular.
The D. melanogaster intracellular clock mechanism is most generally represented as two interconnected feedback loops with overlaying posttranslational regulatory circuits (for review, see Hardin, 2005). Prominent players in the first or “major” loop are PER [herein referred to as Drosophila PER (dPER)], TIMELESS (TIM), CLOCK (dCLK), and CYCLE (CYC) (homolog of mammalian BMAL1). dCLK and CYC are transcription factors of the basic helix–loop–helix/PAS (Per–Arnt–Sim) superfamily that heterodimerize to stimulate the daily transcription of dper and tim, in addition to other clock and downstream genes. dPER plays a pivotal role in driving cyclical gene expression by undergoing daily translocation from the cytoplasm to the nucleus in which it functions as a critical nexus in the phase-specific inhibition of dCLK–CYC transcriptional activity. Kinases are key players controlling when in a daily cycle dPER engages in autoinhibition by regulating its stability, timing of nuclear entry, duration in the nucleus, and possibly repressor potency (for review, see Bae and Edery, 2006).
How phosphorylation modulates the timing of dPER nuclear entry/accumulation is not clear. dPER is progressively phosphorylated in a time-of-day-specific manner, appearing as newly synthesized hypophosphorylated isoforms in the cytoplasm during the late day/early night to exclusively hyperphosphorylated variants in the nucleus during the late night/early morning that are rapidly degraded (Edery et al., 1994). Three kinases, DOUBLETIME (DBT) [homolog of mammalian casein kinase Iδ/ε (CK1δ/ε)], CK2 (formerly known as casein kinase 2), and SHAGGY (SGG) [Drosophila homolog of glycogen synthase kinase-3β (GSK-3β)] have been implicated in regulating the nuclear localization of dPER (Bao et al., 2001; Martinek et al., 2001; Lin et al., 2002, 2005; Akten et al., 2003; Cyran et al., 2005). Unlike DBT and CK2, it is thought that the effects of SGG on dPER might be rather indirect via TIM (Martinek et al., 2001). Herein we identify Ser661 as a key phospho-signal regulating the timing of dPER nuclear entry. Phosphorylation at Ser661 enables subsequent phosphorylation at Ser657 by SGG. Together, the results implicate a novel role for proline-directed kinases in clock mechanisms and raise the intriguing possibility that PER proteins are key targets of lithium, which is a potent inhibitor of GSK-3β and used in the treatment of manic depression, a disorder linked to altered clock function in humans (for review, see McClung, 2007).
Materials and Methods
Transgenic flies.
To generate transgenic flies carrying dper mutations, we used the previously characterized vector that contains a 13.2 kb dper genomic fragment tagged with an hemagglutinin (HA) epitope and multiple histidine residues (10XHis) at the C terminal (13.2per+–HAHis) (Lee et al., 1998). A XbaI–BamHI subfragment of this vector (including sequences encoding amino acids 1–870 of dPER) was subcloned into pGEM7 vector (Promega). This was used as the template for site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene). Mutated dper regions were confirmed by DNA sequencing and used to replace the corresponding fragment in the 13.2per+–HAHis plasmid. Transgenic flies were generated by Genetics Services using standard P element-mediated transformation techniques and w1118per+ (referred to as either wper+ or w) embryos as hosts. For each construct, several independent germ-line transformants in the wper+ background were obtained, yielding; p{dper}, p{dper/S661A}, and p{dper/S657A}. In addition, the transgenes were crossed into a wper01 genetic background (kindly provided by P. Hardin, Texas A & M University, College Station, TX), as described previously (Kim et al., 2007). All flies were routinely reared at room temperature (22–25°C) and maintained in vials or bottles containing standard agar–cornmeal–sugar–yeast–tegosept media.
Locomotor activity.
Locomotor activity was continuously monitored and recorded in either 15 or 30 min bins by placing individual adult male flies (3- to 7-d-old males) in glass tubes and using a Trikinetics system as described previously (Kim et al., 2007). Briefly, flies were kept in incubators at the indicated temperature (18°, 25°, or 29°C) and entrained for at least three cycles of 12 h light/dark cycle (LD) [in which Zeitgeber time (ZT) 0 is the start of the light period], followed by at least 7 d in constant-dark conditions (DD) for determination of free-running period. Cool white fluorescent light (∼2000 lux) was used during LD, and the temperature did not vary by >0.5°C between the light and dark periods. Data analysis was done on a Macintosh computer with the FaasX software (kindly provided by F. Rouyer, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). Rhythmic flies were defined by χ2 periodogram analysis with the following settings: power ≥20, width ≥2, and number of peaks ≤3. Values for individual flies were pooled to obtain an average value for each independent line analyzed (see Table 2) (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). For each construct, locomotor activity rhythms were measured from at least two independent lines in the wper01 and wper+ genetic backgrounds, which are representative of behavioral results obtained with other independent transgenic lines (data not shown). The wild-type control dper-containing transgene [wper01;per+–HAHis (line termed M16); more simply referred to here as per0;p{dper} (M16)] was described previously (Ko et al., 2007).
Table 2.
Locomotor activity rhythms of dper mutants and control transgenic fliesa
| Genotypeb | Period (h ± SEM) | Powerc | Rhythmicity (%)d | Total fliese |
|---|---|---|---|---|
| w1118 | 23.7 ± 0.1 | 46.9 | 93.3 | 15 |
| wper0;p{dper} (M16) | 23.4 ± 0.1 | 87.9 | 93.8 | 16 |
| wper0;p{dper} (M12) | 24.2 ± 0.1 | 108.9 | 100 | 16 |
| wper0;p{dper/S661A} (F9) | 25.4 ± 0.7 | 56.6 | 100 | 17 |
| wper0;p{dper/S661A} (F25) | 25.7 ± 0.1 | 157.5 | 100 | 12 |
| w1118;p{dper/S661A} (F9) | 23.8 ± 0.1 | 68.1 | 78.6 | 14 |
| w1118;p{dper/S661A} (F25) | 24.1 ± 0.1 | 47.7 | 85.7 | 14 |
| wper0;p{dper/S661D} (F7) | 25.4 ± 0.1 | 125.9 | 100 | 17 |
| wper0;p{dper/S661D} (F35) | 25.1 ± 0.1 | 128 | 94.4 | 18 |
| w1118;p{dper/S661D} (F7) | 23.5 ± 0.1 | 44.2 | 81.3 | 17 |
| w1118;p{dper/S661D} (F35) | 23.5 ± 0.2 | 26.1 | 50 | 8 |
| wper0;p{dper/S657A} (M11) | 24.6 ± 0.1 | 158 | 100 | 15 |
| wper0;p{dper/S657A} (M34) | 24.6 ± 0.1 | 172.7 | 100 | 14 |
aFlies were kept at 25°C and exposed to 4 d of LD cycles, followed by 7 d of DD.
bIndependent transgenic lines are designated by the numbers in parentheses.
cPower is a measure of the strength or amplitude of the rhythm in arbitrary units.
dPercentage of flies showing locomotor rhythms with a power value of ≥10 and a width value of ≥2.
eTotal number of flies that survived until the end of the experimental period.
Plasmids for S2 cell expression and site-directed mutagenesis.
Plasmids expressing dper–V5–His fragments of various lengths (supplemental Fig. S1, available at www.jneurosci.org as supplemental material) were a generous gift from Dr. Reppert (University of Massachusetts Medical School, Worcester, MA) and were described previously (Chang and Reppert, 2003). The pAct–dper--V5–His (Kim et al., 2007) and pMT–dbt–V5–His (Ko et al., 2002) plasmids used in this study were described previously. To generate either full-length or truncated versions of dper containing point mutations (e.g., S661A, P662A), we used the previously described pAct–per–V5–His vector (Ko et al., 2002) in combination with the QuickChange Site-Directed Mutagenesis kit (Stratagene), and the final construct was confirmed by DNA sequencing. We also generated a construct whereby expression of the open reading frame (ORF) for sgg was placed under the control of the metallothionein promoter (pMT). This was done by extracting total RNA from Drosophila Schneider S2 cells and using reverse transcription (RT)-PCR in the presence of appropriate primers that also contained suitable restriction enzyme sites to facilitate subcloning the sgg ORF into the pMT–V5–His vector (Invitrogen). Final constructs were confirmed by sequencing before use.
Cell culture-based assays.
S2 cells and Drosophila Expression System (DES) expression medium were obtained from Invitrogen, and transient transfections were performed using either cellfectin (Invitrogen) or effectene (Qiagen) according to the instructions of the manufacturers. For each transient transfection, 0.8 μg of different dper containing plasmids and 0.2 μg of pMT–dbt–V5–His or empty control pMT–V5–His plasmids were used. Expression of dbt or sgg was induced by adding 500 μm CuSO4 to the culture media 36 h after transfection, and cells were collected at the indicated times after induction. When indicated, the proteasome inhibitor MG132 (50 μm; Sigma) or cycloheximide (10 μg/ml; Sigma) were added to the media 20 h after dbt induction, and cells were collected 4 h later.
To measure dCLK-mediated transactivation, we used the well characterized reporter-based assay (Darlington et al., 1998), with the modifications described previously (Kim and Edery, 2006). Briefly, S2 cells were transfected with 2 ng of a pMT–dClk vector either alone or mixed with various amounts of one of the following; pAct–dper+, pAct–dper(S661A), or, pAct–dper(S657A). One day after transfection, 500 μm CuSO4 (final) was added to the media, cells were collected 24 h after inducing dClk expression, and luciferase (Luc) activity was measured.
Phosphorylation site mapping.
Hygromycin-resistant stable S2 cell lines expressing pMT–3XFLAG–His–dper alone or with pMT–sgg were established for dPER purification. Phosphorylation site mapping using mass spectrometry and data analysis were performed as described previously (Schlosser et al., 2005; Vanselow et al., 2006; Chiu et al., 2008).
Immunoblotting.
For immunoblotting of proteins produced in S2 cells, the cells were washed with PBS, cell-free extracts were prepared, and immunoblotting of target proteins was performed as described previously (Ko et al., 2002). Briefly, cells were homogenized in EB1 solution (10 mm HEPES, pH 7.5, 5 mm Tris-HCl, pH 7.5, 50 mm KCl, 10% glycerol, 2.5 mm EDTA, 5 mm DTT, 0.2% Triton X-100, and 2.5 mm NaF), with the addition of a protease inhibitor cocktail at the concentration as recommended by the manufacturer (Roche). In the case of extracts generated from fly material, representative lines [wild-type control, (M16); S661A(F25), S661D(M33), and S657A(F1)] were selected for biochemical analysis, and immunoblotting was performed essentially as described previously (Lee et al., 1998). Briefly, flies were collected by freezing at the indicated times in LD or DD, heads were isolated, and extracts were prepared using EB1 buffer as above. Extracts were resolved by SDS-PAGE and immunoblotted essentially as described previously (Ko et al., 2007). Gels differing in polyacrylamide concentration were used to resolve different proteins [i.e., 6% for dPER; 10.5% for dPER(560–1034)]. Primary antibodies were used at the following dilutions: mouse anti-V5 (Invitrogen), 1:5000; rat anti-HA (3F10; Roche), 1:1000; and rabbit anti-phospho-Ser661 (Rb1; see below), 1:500. Appropriate HRP-conjugated IgG secondary antibodies were used at a 1:1000 dilution (GE Healthcare).
Immunoprecipitation and phosphatase treatment.
Cell-free extracts, either from S2 cells or fly heads, were prepared using EB2 buffer (10 mm HEPES, pH 7.5, 5 mm Tris-HCl, pH 7.5, 50 mm KCl, 10% glycerol, 1 mm EDTA, 1 mm DTT, and 0.1% Triton X-100), with the addition of a protease inhibitor cocktail (Roche), as indicated above. To the extracts, 20 μl of anti-HA agarose (Sigma) or anti-V5 agarose (Sigma) resins were added depending on the sought after target protein and incubated with gentle rotation for 3–5 h at 4°C. Beads were collected using light centrifugation. Phosphatase treatment of immune complexes was performed as described previously (Lee et al., 1998; Kim et al., 2007). Briefly, resins containing dPER-bound immune complexes were divided into three equal aliquots. To one aliquot, vanadate was added to a final concentration of 40 mm, followed by the addition of 200 U of λ-protein phosphatase (New England Biolabs); a second aliquot was treated in an identical manner except that vanadate was omitted, and no addition was made to the final aliquot. Beads containing the immune complexes were mixed with 30 μl of 1× SDS-PAGE sample buffer and incubated for 5 min at 100°C, and the resulting supernatants were resolved by immunoblotting using 6 or 10.5% polyacrylamide gels, as indicated in the legends to figures.
Confocal imaging of adult brains.
Whole mounts of adult brains were prepared and imaged as described previously (Ko et al., 2007). Briefly, adult flies were dissected in ice-cold PBS, and at least five to seven brains were analyzed for each time point. Heads were cut open, fixed in 4% paraformaldehyde for 30 min, and washed with PBS containing 1% Triton X-100. Brains were dissected out, incubated with a blocking solution containing PBT solution (PBS containing 0.5% Triton X-100) with 10% horse serum, and incubated for 30 min to a few hours. Primary antibodies were directly added to the blocking solution and incubated for 1 d at 4°C. The following antibodies and final dilutions were used: (1) anti-HA antibody (3F10; Roche) at 1:100 and (2) anti-pigment dispersing factor (PDF) antibody (C7) at 1:200 (Cyran et al., 2005). Subsequently, the brains were briefly washed with PBT, blocking solution containing secondary antibodies was added, and the brains were incubated overnight at 4°C. The secondary antibodies used were Alexa Fluor 488-conjugated anti-rat IgG (Invitrogen) or tetramethylrhodamine isothiocyanate-conjugated anti-mouse IgG (Sigma), both at a final dilution of 1:200. After several washes with PBT, brains were transferred onto slides and mounted with Vectashield (Vector Laboratories) on a coverslip. Confocal images were obtained with a Leica SP2 confocal microscope and processed with LCS Lite software (Leica) and Photoshop (Adobe Systems).
Generation of dPER phospho-specific S661 antibodies.
Affinity-purified dPER anti-phospho-S661 antibodies were purchased from Proteintech Group. Briefly, two rabbits were immunized with a 15 aa peptide (amino acid 655–GASGPMpSPVHEGSGG–amino acid 669, in which p indicates phosphate, and numbering is based on the full-length dPER sequence) that was conjugated to the carrier keyhole limpet hemocyanin. Antisera was subjected to affinity purification on a resin containing the phospho-peptide, yielding anti-phospho-S661 antibodies [(dPER/pS661)–Rb1 and (dPER/pS661)–Rb2]. Specificity for phosphorylated S661 was confirmed by treating dPER with phosphatases and probing mutant versions of dPER (Fig. 4D) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material).
Figure 4.
S661 is phosphorylated in flies and modulates the hyperphosphorylation of dPER but does not have significant effects on its overall daily levels. A–D, Adult flies of the indicated genotype in the wper0 genetic background (top of panels) were collected at the indicated time during the fourth day of LD (A, C, D) or first day of DD (B). A–C, Head extracts were prepared and analyzed by immunoblotting using anti-HA antibodies. CT, Circadian time. D, Head extracts were subjected to immunoprecipitation using anti-HA antibodies. A portion of the recovered immune complexes was analyzed by immunoblotting in the presence of either anti-HA (top panel) or S661 phospho-specific (anti-pS661; bottom panel) antibodies. For lanes 7 and 8, immune complexes were first treated with phosphatase before analysis by immunoblotting; note that, whereas phosphatase treatment abolishes the ability of the anti-pS661 antibodies to detect dPER (bottom panel, compare lanes 7 and 8 with 1 and 5, respectively), it still reacts with the anti-HA antibody (top panel, lanes 7 and 8). Each experiment was done at least three times, and representative examples are shown.
In vitro kinase assay.
For in vitro kinase assays, S2 cells were transiently transfected with the plasmid pAct–dper(560–1034)–V5 using cellfectin and incubated for 48 h. Cell-free extracts were prepared using EB2 and dPER(560–1034)–V5 affinity purified using anti-V5 agarose, as described above. Immune complexes were first treated with 200 U of λ-phosphatase (New England Biolabs) for 30 min. This was followed by extensive washing with EB2, equilibration in the appropriate kinase reaction buffer, and subsequent subjection to in vitro kinase assays with the indicated commercially available kinase(s) (New England Biolabs) and suggested reaction buffer(s) according to the protocols of the supplier. Briefly, anti-V5 agarose beads with bound dPER(560–1034) were mixed with 50 μl of kinase reaction buffer containing 500 U of the indicated recombinant kinase, 200 μm ATP, and 10 μCi of [γ-32P]ATP, followed by incubation with occasional mild mixing at 30°C for 30 min. For assays in which two kinases were sequentially added, each kinase reaction was done as above in the presence of “cold” and 32P-radiolabeled ATP, except that, after the first incubation with kinase, the beads were extensively washed and reequilibrated in the appropriate kinase reaction buffer for the second kinase. Reactions were terminated by the addition of 2× SDS-PAGE sample buffer and resolved on 10.5% polyacrylamide gels. Subsequently, gels were dried, radioactive bands were visualized using a Typhoon 9400 Imager, and the intensity was quantified using Imagequant software (Molecular Dynamics).
RT-PCR-based assay to measure relative
dper RNA levels. The relative levels of dper mRNA in fly head extracts were measured by semiquantitative RT-PCR as described previously (Majercak et al., 2004). Briefly, vials containing ∼100 young (2–6 d old) adult flies were placed in controlled environmental chambers at 25°C, exposed to at least four LD cycles, and collected by freezing at selected times during LD or the first day of DD. For each time point, total RNA was extracted from ∼30 μl of fly heads using TriReagent (Sigma) and following the recommended procedure of the manufacturer. Approximately 2 μg of total RNA was incubated in a final volume of 20 μl, and reverse transcription was performed using oligo-dT20 as a primer and the ThermoScript RT-PCR kit from Invitrogen, according to the recommended procedure of the manufacturer. A 2 μl aliquot of the reaction was further processed by PCR in a final volume of 50 μl using the dper-specific primers P7197 and P6869, as described previously (Majercak et al., 2004). To control for sample-to-sample differences in total RNA, we also included primers for the noncycling mRNA encoding for cap binding protein 20 (Majercak et al., 2004).
Results
Identification of Ser661 as a phosphorylation site on dPER; effects on global DBT-mediated phosphorylation but not stability
We previously described a simplified system using cultured Drosophila Schneider (S2) cells that can mimic the DBT-dependent progressive phosphorylation of dPER and subsequent targeting of highly phosphorylated isoforms by the F-box protein SLIMB (Drosophila homolog of mammalian β-TrCP) for rapid degradation by the proteasome (Ko et al., 2002; Ko and Edery, 2005). In this system, S2 cells (which do not have an endogenously operating functional clock) are transfected with plasmids containing the dper and dbt open reading frames. Expression of dbt is controlled by the copper-inducible pMT, whereas the constitutive actin5C promoter (pAct) drives expression of epitope-tagged versions of dper. After induction of dbt, full-length dPER undergoes progressive increases in phosphorylation that are readily detected as temporal decreases in electrophoretic mobility (Fig. 1D, compare lanes 1, 3). Importantly, daily changes in the electrophoretic mobility of dPER are mostly or solely attributable to phosphorylation (Edery et al., 1994; Ko et al., 2002). Thus, in both cultured cells and flies, changes in the electrophoretic mobility of dPER serve as a strong indicator of differential phosphorylation.
Figure 1.
Phosphorylation of S661 on dPER, which is highly conserved in Drosophila, regulates the extent of DBT-mediated hyperphosphorylation but not the stability of dPER. A, Sequence alignment of dPER proteins from different Drosophila species (** identifies the conserved Ser–Pro pair; S661–P662 in D. melanogaster). Sequences were aligned using the program ClustalX (www.clustal.org) and processed with GeneDoc (www.nrbsc.org/gfx/genedoc) for better visualization. Amino acids that are identical in all the species analyzed are highlighted in black, whereas those showing less conservation are indicated in gray. Note that the conserved Ser–Pro pair falls in a relatively nonconserved region of dPER. B, C, S2 cells were transiently transfected with pAct–dper(560–1034)–V5–His plasmid that contained either the wild-type (WT) control version (B, lane 1; C) or the S661A mutant (B, lane 2). The wild-type dPER(560–1034) protein exhibits at least two mobility isoforms (indicated at left, arrowheads). C, Immunoprecipitated dPER(560–1034) was mock treated (lane 1) or treated with λ-phosphatase (λPPase) in the absence (lane 2) or presence (lane 3) of the phosphatase inhibitor Na3VO4. D, E, S2 cells were transiently cotransfected with pMT–dbt–V5–His and plasmids containing full-length versions of either wild-type dper (WT) or the S661A mutant (SA). Cells were collected at the indicated times after dbt induction (top) and dPER–V5 analyzed either directly by immunoblotting (D, lanes 1–6; E, lanes 1–4) or after immunoprecipitation and treatment with λ-phosphatase (D, lanes 7, 8). Where indicated, the proteasome inhibitor MG132 (50 μm final) and the translation inhibitor cycloheximide (CHX; 10 μg/ml final) were added to the media at 20 h after dbt induction, and cells were collected 4 h later. Each experiment was done at least three times, and representative examples are shown.
In one approach to map phosphorylation sites, we analyzed a range of dPER deletion mutants using this system (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). We observed that several centrally located dPER fragments [e.g., dPER(513–892) and dPER(560–1034)] exhibit substantial DBT-mediated decreases in electrophoretic mobility (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), indicating that there are multiple phosphorylation sites between amino acids 560–892. Although DBT stimulates the degradation of full-length dPER (Kloss et al., 1998; Price et al., 1998; Ko et al., 2002), highly phosphorylated dPER(513–892) or dPER(560–1034) remain stable (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), in agreement with our recent findings showing that N-terminal phosphorylation sites (e.g., Ser47) are critical for SLIMB binding (Chiu et al., 2008). As an initial attempt to identify physiologically relevant phosphorylation sites between amino acids 560–892, we compared dPER sequences from several Drosophila species. We were intrigued by a highly conserved Ser–Pro pair beginning at amino acid 661 of D. melanogaster PER because it was buried in a nonconserved region (Fig. 1A) and raised the possibility of proline-directed kinases in the Drosophila clockworks.
We first sought to determine whether Ser661 is endogenously phosphorylated in S2 cells by comparing the electrophoretic mobilities of control dPER and a mutant version in which Ser661 was replaced by Ala (S661A). To maximize the ability to discern phosphorylation-dependent electrophoretic mobility differences, we evaluated the effects of mutating S661 within the context of smaller fragments of dPER. Indeed, we observed an additional slower mobility isoform of dPER(560–1034) that was missing in the S661A version [dPER(560–1034)/S661A] (Fig. 1B). Furthermore, of the two mobility isoforms of dPER(560–1034), the fastest comigrates with the mutant dPER(560–1034)/S661A (Fig. 1B). That the slower migrating species is attributable to phosphorylation was confirmed by treating purified dPER(560–1034) with phosphatase (Fig. 1C). These results are consistent with our recent findings using mass spectrometry in which we showed that S661 on full-length dPER expressed in S2 cells is phosphorylated by endogenous kinases (Chiu et al., 2008) (Table 1). The intensity of the slower migrating dPER(560–1034) species is approximately threefold greater compared with the faster migrating isoform (Fig. 1B, lane 1), suggesting that in our S2 culture system the majority of dPER proteins contain a phosphate at S661.
Table 1.
Mapping phosphorylation sites on dPER produced in Drosophila S2 cells
| dPERa,b,c | dPER + SGGd |
|---|---|
| S93, [T96, S97]e,f | |
| S149, S151,g S153 | S149, S151 |
| S164, S169, S174áPñh | |
| S596áPñ | T583, S585, S596áPñ |
| T610áPñ | T610áPñ |
| S661áPñ | S657, S661áPñ |
| [S826, S828]f | |
| T883áPñ | T883áPñ |
| S981áPñ, [T980, T983, S985]f | S981áPñ, [T980, T983, S985áPñ]f |
| S1130áPñ | |
| S1185, S1187áPñ | |
| [S1204, T1205, S1206]f |
aStable cell line expressing dper under the control of pMT inducible promoter.
bdPER residues phosphorylated by endogenous kinase(s) in S2 cells.
cdPER phosphorylation sites data taken from Chiu et al. (2008).
dStable cell line expressing dper and sgg under the control of pMT inducible promoter.
eAmino acids are numbered according to sequence of dPER(1–1224), GenBank accession number P07663.
fOne of the sites in brackets is phosphorylated.
gS151 and S153 were identified previously as putative CK2 phosphorylation sites (Lin et al., 2005).
háPñ denotes residues that are next to a proline.
Having established that Ser661 is endogenously phosphorylated in S2 cells, we sought to determine whether phosphorylation at this site has noticeable effects on the stability and/or global phosphorylation of full-length dPER after induction of DBT. As reported previously, under our experimental conditions, dPER exhibits extensive phosphorylation and substantial degradation by 24 h after dbt induction (Fig. 1D, lanes 1, 3) (Ko et al., 2002; Ko and Edery, 2005; Chiu et al., 2008). Although DBT evoked increases in the global phosphorylation of dPER/S661A, the slowest mobility isoforms detected were below those observed for the control dPER (Fig. 1D, lanes 3, 4). This was the case even in the presence of MG132 to block SLIMB-dependent degradation (Ko et al., 2002) (Fig. 1D, lanes 5, 6) and when we added cycloheximide to inhibit translation and “chase” preexisting dPER isoforms toward hyperphosphorylation (Fig. 1E, lanes 3, 4). These results indicate that the inability to detect the slowest mobility isoforms normally observed with control dPER is not because these variants are extremely unstable in the case of dPER/S661A. Although we have not established a direct relationship between the electrophoretic mobility of dPER and the number of sites phosphorylated, it is highly likely that the extent of DBT-mediated phosphorylation of dPER is attenuated in the S661A mutant.
To better compare the levels of dPER and dPER/S661A after dbt induction, we immunoprecipitated dPER proteins and treated the samples with phosphatase to collapse the different electrophoretic mobility isoforms to a more uniformly sharp band. It is clear that the S661A mutation has little to no effect on DBT-mediated degradation (Fig. 1D, lanes 7, 8). This is consistent with our recent study showing that the main DBT-mediated phosphorylation signals governing the stability of dPER are found in the N-terminal first 100 aa and that global phosphorylation by DBT is not tightly linked to dPER stability (Chiu et al., 2008). After phosphatase treatment, both dPER and dPER/S661A had indistinguishable electrophoretic mobilities (Fig. 1D, lanes 7, 8), indicating that the slight mobility differences in the absence of induced DBT (Fig, 1D, lanes 1, 2) are attributable to differential phosphorylation by endogenous kinases (Fig. 1B). Thus, although blocking phosphorylation at S661 appears to reduce DBT-mediated global phosphorylation of dPER, it does not play a significant role in regulating the stability of dPER. Similar results were also obtained when S661 was replaced with Asp or Glu (data not shown), suggesting that in this context the negatively charged amino acids do not function as phospho-mimetics (see below).
Phosphorylation at S661 primes additional phosphorylation at S657 by SGG
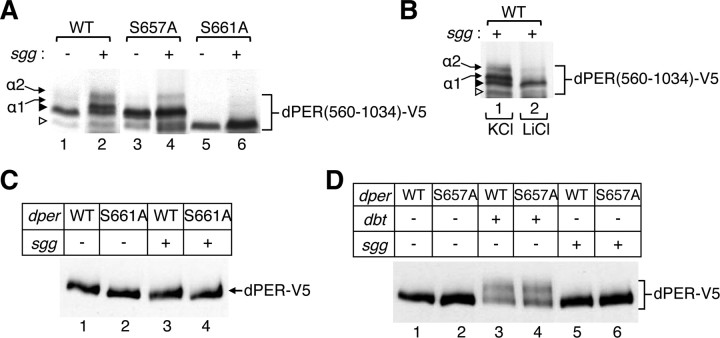
Based on the possibility that phosphorylation of S661 might affect phosphorylation at other sites, we were intrigued by a nearby Ser residue (S657 in D. melanogaster) that is conserved in some but not all Drosophila dPERs (Fig. 1A). Phosphorylation at S661 generates a good consensus for a GSK-3β/SGG site at S657, because this kinase usually requires a priming phosphate located at n + 4 (where n is the site of phosphorylation by GSK-3β/SGG) (Cohen and Frame, 2001). As above, we used the dPER(560-1034) fragment to maximize observing phosphorylation-dependent changes via electrophoretic mobility changes. The induction of recombinant SGG leads to time-dependent decreases in the electrophoretic mobility of dPER(560–1034), resulting in the appearance of at least two slower mobility variants (Fig. 2A, compare lanes 1, 2; herein termed α1 and α2). Treatment of cells with lithium, an inhibitor of GSK-3β/SGG (Cohen and Frame, 2001), but not potassium blocks the appearance of the slower-mobility dPER(560–1034) isoforms observed after induction of SGG (Fig. 2B). However, lithium treatment did not block the appearance of the dPER(560–1034) mobility isoform associated with S661 phosphorylation (Fig. 2B, filled arrowhead; and data not shown). These results indicate that SGG is unlikely to be the S661 kinase, although there are examples in which GSK-3β phosphorylates Ser/Thr residues with an upstream flanking Pro (Li et al., 1998; Kim and Kimmel, 2000; Kannan and Neuwald, 2004). The SGG-dependent phospho-isoforms were abolished or severely reduced when assaying the S661A mutant (Fig. 2A, compare lanes 2, 6), indicating that both the α1 and α2 species are dependent on priming at S661. Also, α2 but not α1 is detectable when probing the S657A mutant (Fig. 2A, compare lanes 2, 4), strongly suggesting that the α1 species is attributable to S657 phosphorylation and that there are multiple SGG-dependent sites on dPER. Studies using in vitro approaches described below (Fig. 3) are consistent with phosphorylation at S661 priming the ability of GSK-3/SGG to phosphorylate S657.
Figure 2.
GSK-3β/SGG phosphorylates S657 and likely other sites on dPER in a hierarchical manner that depends on previous phosphorylation of S661. A, S2 cells were transiently cotransfected with the indicated versions of pAct--dper(560–1034)–V5 [wild type (WT); S657A or S661A] and pMT–sgg–V5–His. Cells were collected either just before inducing recombinant sgg (−) or 24 h after sgg induction (+). Recombinant dPER was visualized by immunoblotting in the presence of anti-V5 antibodies. Exogenous expression of sgg leads to the appearance on several novel mobility isoforms of wild-type dPER(560–1034), herein termed α1 and α2 (arrows, left). Also indicated are the S661 phosphorylated (filled arrowhead) and S661 nonphosphorylated (open arrowhead) dPER(560–1034) mobility isoforms observed in the absence of exogenously expressed kinases (see Fig. 1B). Note that the S657A mutant does not block S661 phosphorylation (lane 4), and, in the presence of recombinant SGG, only the α2 isoform is detected (lane 4). In the case of the S661A mutant, both the α1 and α2 isoforms are not detected (lane 6). B, Detection of the SGG-mediated α1 and α2 dPER(560–1034) isoforms is blocked in the presence of the GSK-3β/SGG inhibitor LiCl (10 mm, final; lane 2) but not KCl (10 mm, final; lane 1). Recombinant sgg was induced for 24 h. C, D, S2 cells were transiently cotransfected with plasmids containing different versions of full-length dper–V5–His [wild type (WT); S657A or S661A] and either pMT–sgg–V5–His or pMT–dbt–V5–His, as indicated (−, +). Cells were collected 24 h after induction of dbt or sgg, and dPER–V5 was visualized by immunoblotting. Each experiment was done at least three times, and representative examples are shown.
Figure 3.
Phosphorylation of S661 by MAPK can prime additional phosphorylation at S657 by GSK-3β. A, B, Wild-type (WT), S661A, or S657A versions of dPER(560–1034)–V5–His were immunoprecipitated from extracts prepared from S2 cells using anti-V5 agarose and pretreated with λ-phosphatase for 30 min, followed by extensive washing and equilibration in MAPK kinase assay buffer. Subsequently, the immune complexes were incubated with (+) or without (−) 500 U of ERK2. After this incubation, immune complexes were washed with extraction buffer, equilibrated in GSK-3β buffer, and incubated with (+) or without (−) 500 U of GSK-3β. Phosphorylation was detected by adding [γ-32P]ATP to the in vitro kinase assays (see Materials and Methods), and radiolabeled bands were visualized by PAGE and autoradiography. The different phospho-isoforms are indicated (arrows, right of panels) and whether Ser657 or Ser661 are phosphorylated (bold). Note that ERK2 does not require previous phosphorylation to phosphorylate S661 of dPER (A, lane 3), an event that can prime subsequent phosphorylation by GSK-3β (A, lane 4). Addition of GSK-3β by itself does not lead to noticeable mobility shift in dPER(560–1034) (A, compare lanes 1 and 2). Addition of ERK2 does not shift the mobility of the S661A mutant (B, compare lanes 1 and 3), and the addition of GSK-3β after ERK2 does not evoke an additional shift in the electrophoretic mobility of either the S661A mutant (B, compare lanes 3 and 4) or S657A mutant (B, compare lanes 5 and 6). Each experiment was done at least three times, and representative examples are shown.
Induction of SGG did not lead to detectable changes in the electrophoretic mobility of full-length dPER nor did it affect its stability (Fig. 2C, lanes 1, 3). Similar results were also obtained for the dPER/S661A and dPER/S657A mutants (Fig. 2C,D). Also, after induction of DBT, the electrophoretic mobilities of dPER/S657A and wild-type dPER were indistinguishable (Fig. 2D, lanes 3, 4), unlike dPER/S661A (Fig. 1D,E). Together, the results obtained in cultured cells indicate that (1) phosphorylation at S657 or S661 on dPER has little to no effects on dPER stability, (2) SGG has no noticeable effects on dPER levels, (3) phosphorylation of dPER by SGG is less extensive compared with DBT and/or the sites phosphorylated by SGG evoke small changes in the electrophoretic mobility of full-length dPER, and (4) the phosphorylated status of S661 appears to have more global effects on dPER phosphorylation compared with that of S657, consistent with its role as a priming site for SGG-mediated phosphorylation of S657 and other sites (Fig. 2A).
It is not clear why detectable levels of S657 phosphorylation require the induction of exogenous SGG (Fig. 2A). If most of Ser661 is endogenously phosphorylated (Fig. 1B), why is endogenous SGG not sufficient to maintain S657 in a mostly phosphorylated state? Although this is not clear, a similar situation has been noted for DBT; despite the presence of endogenous dbt expression in naive S2 cells, ectopic dbt expression is required to observe substantial DBT-mediated phosphorylation of dPER (Ko et al., 2002). A likely reason is that the levels/activity of certain endogenous kinases that target dPER are not sufficient to overcome the strong counterbalance from endogenous protein phosphatases (Sathyanarayanan et al., 2004).
The Ser residue at position 661 is followed by a proline residue, indicating that S661 is phosphorylated by a “Pro-directed” kinase. These kinases are part of the CMGC family of kinases that include several subfamilies, such as mitogen-activated protein kinases (MAPK), cyclin-dependent kinases (CDKs), and dual-specificity tyrosine-regulated kinases (DYRKs) (Kannan and Neuwald, 2004). It is unlikely that the kinase phosphorylating S661 would be from outside the CMGC family because the flanking Pro residue on the substrate greatly constrains the kinds of kinases that can recognize and phosphorylate the amino-proximal Ser/Thr acceptor site (Kannan and Neuwald, 2004). Indeed, replacing P662 with other residues completely abolished phosphorylation of dPER(560–1034) at the S661 site in cultured cells (supplemental Fig. S4C, available at www.jneurosci.org as supplemental material).
To further investigate the phosphorylation of dPER by SGG, we used a mass spectrometric approach (Schlosser et al., 2005; Vanselow et al., 2006) that we used previously to identify DBT-dependent phosphorylation sites on dPER (Chiu et al., 2008). Stable S2 cell lines were generated that coexpress inducible versions of Flag-tagged dPER and untagged SGG. This was followed by proteolytic digestion, phosphopeptide enrichment, and mass spectrometry of affinity-purified dPER–FLAG. We identified ∼20 phosphorylation sites in the dPER + SGG sample, of which S657 was one of 11 sites not identified in the dPER alone sample (Table 1). Although several of the phospho-sites identified in the dPER + SGG sample are identical to those detected previously in dPER + DBT samples (Table 1) (Chiu et al., 2008), phospho-S657 was only observed in the dPER + SGG sample. Likewise, in the dPER + SGG sample, we did not observe phosphorylation of S47, which is phosphorylated by DBT and critical for generating a SLIMB binding region on dPER (Chiu et al., 2008). Thus, it is likely that robust phosphorylation at S47 and S657 is highly dependent on DBT and SGG, respectively, and these phosphorylation events can occur independent of each other.
Nonetheless, more quantitative mass spectrometry procedures would have to be undertaken to try to achieve reliable comparisons between samples. For example, some of the overlap in phospho-sites between the dPER + SGG and dPER + DBT samples might be “false positives” attributable to sample-to-sample variation in detection of sub-stoichiometric phosphorylation by endogenous DBT and/or other kinases. Thus, from our mass spectrometry data, we cannot determine with any degree of certainty what other phospho-sites on dPER besides S657 (in which we have other lines of evidence) might be phosphorylated by SGG in a manner dependent on priming at S661, as suggested from our biochemical results (Figs. 1D, 2A). Ongoing work is aimed at better establishing the physiological roles of the cumulative phospho-sites we identified by mass spectrometry.
To obtain more direct evidence that phosphorylation at S661 can prime subsequent phosphorylation at S657 by SGG, purified dPER(560–1034) was pretreated with λ-phosphatase and subsequently incubated with several commercially available kinases in the presence of [γ-32P]ATP. Although we did not do an extensive interrogation of CMGC kinases, the classic MAP kinase ERK2 (extracellular signal-regulated kinase) evoked a mobility shift in dPER(560–1034) that is consistent with phosphorylation at S661 (Fig. 3A, compare lanes 1, 3). Several control experiments, including an S661A mutant and the use of phospho-specific antibodies (see below), confirmed that ERK2 can phosphorylate S661 (Fig. 3B, compare lanes 1, 3).
In a second set of experiments, phosphatase-treated dPER(560–1034) was first incubated with ERK2, extensively washed, and subsequently incubated with GSK-3β (the mammalian homolog of SGG). The addition of GSK-3β shifted the mobility of the ERK2-phosphorylated dPER(560–1034)–V5–His to a slightly slower isoform (Fig. 3A, compare lanes 3, 4; B, compare lanes 1, 2), remarkably similar to the α1 variant observed in cultured cells (Fig. 2A). When singly added, GSK-3β does not lead to a noticeable mobility shift in dPER(560–1034) (Fig. 3A, compare lanes 1, 2). Also, GSK-3β did not shift the mobility of either an S657A or S661A mutant, whereas the ability of ERK2 to phosphorylate S661 was not affected by blocking phosphorylation at S657 (Fig. 3B). That the fast migrating dPER(560–1034) isoform is still radiolabeled in the absence of exogenous kinases (Fig. 3A, lane 1) or when using the S661A version (Fig. 3B, lanes 3, 4) is likely attributable to phosphorylation by copurifying endogenous kinases at other sites besides S657 and S661 that do not lead to detectable electrophoretic mobility shifts. Thus, our in vitro analysis is consistent with a model whereby S661 is first phosphorylated by a Pro-directed kinase(s), an event that can act to prime additional phosphorylation at S657 by SGG/GSK-3β. Unfortunately, initial attempts using cultured cells and flies were not conclusive in identifying the in vivo kinase(s) phosphorylating S661 of dPER, and ongoing experiments are aimed toward that goal.
Blocking phosphorylation of S661/S657 leads to longer periods
To evaluate the physiological significance of our findings, we generated transgenic flies whereby S661 was replaced with Ala (S661A) or Asp (S661D). In addition, we also analyzed an S657A mutant. The different mutations were engineered into a previously characterized vector that contains a 13.2 kb dper genomic fragment and also includes an HA epitope tag and a stretch of His moieties (10xHis) at the C terminus of the dper open reading frame, facilitating purification and surveillance of the recombinant protein (Lee et al., 1998; Kim et al., 2007). We assayed the function of the transgenes in the arrhythmic per-null wper01 (Konopka and Benzer, 1971), as well as wper+ genetic backgrounds. Flies were subjected to standard entraining conditions of 12 h light/dark cycles (in which ZT0 is defined as the time when the light phase begins) at 25°C, followed by at least 1 week in DD to determine their free-running periods.
As shown previously, transgenic wper01 flies harboring wild-type versions of the transgene [wper01; per+–HAHis; herein more simply referred to as per0;p{dper}] manifest robust locomotor activity rhythms with normal ∼24 h periods (Table 2(Kim et al., 2007; Ko et al., 2007; Chiu et al., 2008). Intriguingly, flies expressing the S661A mutation [per0;p{dper/S661A}] exhibit free-running periods that are ∼2 h longer compared with control transgenics (Table 2). The S661D mutation also lengthened periods by ∼2 h, consistent with results obtained in transfected S2 cells whereby both the S661A and S661D mutants have similar effects in attenuating the extent of DBT-mediated hyperphosphorylation (Fig. 1 and data not shown). When p{dper/S661A} or p{dper/S661D} were evaluated in a per+ background, the period lengthening effects were abolished, indicating that wild-type dPER is dominant in the presence of either mutant allele. This is somewhat unusual because most period-altering mutant alleles of dper are semi-dominant (Hall, 2003). The S657A mutant flies also exhibit longer periods but only ∼1 h. The more severe behavioral phenotype in p{dper/S661A} flies could be attributable to the role of S661 in regulating phosphorylation at multiple sites on dPER (Figs. 1, 2). The timing of evening activity in daily LD cycles is delayed for both the p{dper/S661A} and p{dper/S657A} flies, consistent with a longer endogenous clock period (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Finally, we also determined the effect of ambient temperature on behavioral periods for the p{dper/S661A} flies. A hallmark feature of circadian clocks is that they maintain relatively stable periods over a broad range of temperatures, a property termed temperature compensation (Pittendrigh, 1954). In contrast, the p{dper/S661A} flies manifest progressively longer periods as temperature increases (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), suggesting that the biochemical lesion underlying the altered pace of the mutant clock is exacerbated by heat.
dPER/S661A manifests less extensive hyperphosphorylation in flies
Based on the more severe behavioral phenotype in p{dper/S661A} compared with p{dper/S657A} flies, we mostly focused on characterizing the clockworks in the former mutant. To analyze the effects of the S661A mutation on dPER biochemical rhythms, we collected per0;p{dper/S661A} flies at different times in LD and DD, prepared head extracts, and detected dPER protein using immunoblotting. Although there is a clear rhythm in the levels of the dPER(S661A) protein, the temporal phosphorylation program is less extensive compared with that observed in the control transgenics (Fig. 4A). This is readily observed when comparing dPER mobilities at ZT4, a time in the daily cycle when the slowest mobility isoforms of wild-type dPER are detected (Fig. 4A, top panel, lanes 1, 2; C). Whereas the mobility of control dPER decreases between ZT23.5 and ZT4, that of dPER(S661A) is apparently “stuck” at the overall global phosphorylated state normally attained by ZT23.5 (Fig. 4C). The inability of dPER(S661A) to reach the same degree of hyperphosphorylation as control dPER also occurred under constant-dark conditions, indicating that this is a bona fide circadian difference in dPER protein dynamics (Fig. 4B, bottom panel, lanes 1, 7). These results are remarkably similar to results obtained in S2 cells using recombinant dPER (Fig. 1D), supporting the notion that S661 is a key phospho-signal regulating phosphorylation at multiple sites on dPER. We did not observe reproducible differences in the extent of hyperphosphorylation for the S657A mutant compared with control dPER (data not shown), consistent with findings obtained in cultured cells (Fig. 2D).
To confirm that S661 is phosphorylated in flies, we generated phospho-specific polyclonal antibodies that recognize phosphorylated S661 (anti-pS661). Several lines of evidence demonstrate the high specificity of our anti-pS661 antibody for phosphorylated Ser661 (Fig. 4D) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). For example, phosphatase treatment of dPER expressed in either flies or S2 cells abolished detection by the anti-pS661 antibody (Fig. 4D, bottom panel) (supplemental Fig. S4A, available at www.jneurosci.org as supplemental material). In addition, the anti-pS661 antibody only recognizes the upper band of the dPER(560–1034) protein produced in S2 cells (supplemental Fig. S4A, available at www.jneurosci.org as supplemental material), consistent with our mutational analysis indicating that this slower migrating band is attributable to phosphorylation at S661 (Fig. 1B). Likewise, the S661A mutant is not detected by the anti-pS661 antibody (supplemental Fig. S4B, available at www.jneurosci.org as supplemental material). A similar result was also observed when evaluating the P662A mutant (supplemental Fig. S4C, available at www.jneurosci.org as supplemental material), providing additional evidence that Ser661 is phosphorylated by a Pro-directed kinase. In this regard, S661 is still phosphorylated when probing the dPER(ΔdPDBD) internal deletion mutant (supplemental Fig. S4D, available at www.jneurosci.org as supplemental material), which eliminates the ability of DBT to stably bind and hyperphosphorylate dPER (Kim et al., 2007; Nawathean et al., 2007), further indicating that phosphorylation of S661 is not dependent on DBT function.
When probing extracts prepared from control p{dper} flies collected at different times in a daily cycle, the relative staining intensity of S661 phosphorylated dPER [dPER(pS661)] was similar to that of total dPER, suggesting that phosphorylation at S661 is constitutive and not temporally regulated (Fig. 4D, compare top and bottom panels). Because S661 is endogenously phosphorylated in S2 cells and is not dependent on DBT (Fig. 1B, Table 1) (supplemental Fig. S4D, available at www.jneurosci.org as supplemental material), the combined results suggest that phosphorylation at S661 occurs in a stoichiometric manner on dPER with little to no steady-state removal in flies. This is different from our recent findings in which we showed that the DBT-mediated phosphorylation of Ser47, a key phospho-signal for SLIMB binding, is temporally restricted to the late day/early night (Chiu et al., 2008). Unfortunately, we were not able to generate a high-quality antibody that specifically detects phosphorylated S657.
In addition to dPER levels, the abundance of dper mRNA fluctuates reaching peak levels in the early night and trough values in the late night/early morning (Hardin et al., 1990) (Fig. 5). The daily cycle in dper mRNA levels from per0;p{dper/S661A} mutant flies is similar to the wild-type situation in both LD and DD (Fig. 5), except that the levels of dper RNA in the S661A mutant flies are slightly higher, especially during the daily accumulation phase in LD. The quasi-normal dper RNA rhythm in per0;p{dper/S661A} mutant flies suggests that the transcriptional repressor activity of dPER/S661A is not significantly altered. As an initial strategy to address this issue, we used the standard S2 cell-based assay and measured the ability of exogenously supplied dPER to inhibit dCLK-driven expression of a luciferase reporter placed downstream of an E-box containing circadian regulatory element (Darlington et al., 1998; Chang and Reppert, 2003). Over a wide range of dper-containing plasmid amounts used in the transfection, dPER and dPER/S661A have similar abilities to inhibit dCLK-driven expression (supplemental Fig. S5A, available at www.jneurosci.org as supplemental material). The dPER/S657A mutant also showed similar repressor capability as the wild-type control (supplemental Fig. S5B, available at www.jneurosci.org as supplemental material). We attempted to evaluate the effects of the S661A and S657A mutants in the presence of exogenously supplied kinases (e.g., DBT and SGG) or inhibitors (e.g., lithium ions), but the results were not easily interpreted because kinases have direct effects on the trans-activation potential of dCLK–CYC (data not shown) (Kim and Edery, 2006; Weber et al., 2006). Nonetheless, our combined results indicate that phosphorylation at either S661 or S657 has little effect on dPER stability or its ability to engage in feedback inhibition of dCLK–CYC-driven transcription.
Figure 5.
Cycling of dper mRNA levels in p{dper/S661A} flies. A, B, Transgenic flies expressing either a wild-type (WT; wper0;p{dper}) or the S661A mutant (wper0;p{dper/S661A}) version of dper were collected at the indicated times during either the fourth day of LD (A) or first day of DD (B). The relative levels of dper RNA were measured. Results from at least two independent experiments were pooled, and the error bars indicate SEM.
Delayed nuclear entry of dPER(S661A) in key clock neurons of the brain
Recent evidence indicates the presence of anatomically and functionally distinct pacemaker neurons in the brain that interact to regulate different aspects of the daily activity rhythm in D. melanogaster. These dper-expressing clock cells include the small and large ventral lateral neurons (LNvs), the lateral dorsal neurons, and three groups of dorsal neurons (DN1–DN3) (for review, see Nitabach and Taghert, 2008). The small LNvs are central for the maintenance of activity rhythms in constant-dark conditions and produce PDF, a circadian-relevant neuropeptide that can serve as a convenient cytoplasmic marker of these cells (Renn et al., 1999). Wild-type dPER is first observed accumulating in the cytoplasm of small LNvs during the early night and displays a mixed cytoplasmic–nuclear localization at approximately ZT19–Z20, which is followed a little later, such as by ZT22, by a more punctate pattern that is essentially exclusively located in the nucleus (Curtin et al., 1995; Shafer et al., 2002) (Fig. 6). In sharp contrast, the nuclear accumulation of dPER(S661A) in the LNvs is delayed, e.g., at ZT22, the S661A mutant is mainly found in the cytoplasm or exhibits a mixed cytoplasmic–nuclear pattern (Fig. 6A–C) that eventually translocates to the nucleus by ZT24/ZT0 (Fig. 6A). Thus, the S661A mutation does not block nuclear entry/accumulation but delays its by several hours. The delayed nuclear entry time of dPER(S661A) can readily explain the period lengthening effects of this mutation on the daily activity rhythm (Table 2). Indeed, delayed nuclear accumulation of dPER in the LNvs during LD is strongly associated with longer behavioral periods in DD (Curtin et al., 1995; Martinek et al., 2001; Lin et al., 2002; Akten et al., 2003). Thus, the lengthening of behavioral periods in the per0;p{dper/S661A} flies (Table 2) is most intimately linked to primary effects on the timing of dPER nuclear entry in brain pacemaker neurons critical for driving behavioral rhythms.
Figure 6.
The timing of when dPER(S661A) enters the nucleus of key pacemaker neurons is delayed. Transgenic flies expressing either a wild-type (WT; wper0;p{dper}) or S661 mutant (S661A; wper0;p{dper/S661A}) version of dper were collected at the indicated times in LD (A) or at ZT22 (B, C) and processed for whole-mount immunohistochemistry. Shown are representative staining patterns obtained for the small LNvs. A, Two independent examples are shown for the S661A mutant. B, C, For each genotype, the subcellular localization of dPER at ZT22 was determined for 24 small LNvs from at least five flies; three representative examples are shown for each genotype (B), and the staining distributions for all 24 small LNvs were tabulated (C). dPER was visualized with anti-HA antibodies (shown in green). PDF was visualized with an anti-PDF antibody (shown in red) and serves as a convenient cytoplasmic marker for the small LNvs. Note that, whereas punctate nuclear staining of control dPER is clearly observed by ZT22, the subcellular localization of dPER(S661A) is much more dispersed at this time with primarily cytoplasmic or mixed cytoplasmic–nuclear staining, eventually exhibiting primarily nuclear staining by ZT24.
Discussion
In animal circadian pacemakers, daily changes in the phosphorylated state of PER proteins drive time-of-day-specific changes in a number of its attributes, such as levels and subcellular localization, molecular oscillations that are inextricably linked to normal clock progression (Bae and Edery, 2006; Gallego and Virshup, 2007). Time-delayed autoinhibitory feedback loops, whereby the negative arm inhibits the positive arm after some interval, are central to the oscillatory mechanisms underlying circadian clocks. The timing of when PER proteins translocate from the cytoplasm to the nucleus is thought to be a key aspect of the time-delay circuit in animal clocks, an event that regulates the pace of these clocks and initiates repression of CLOCK:BMAL1/CYC transactivation, critical for generating appropriately phased cyclical gene expression. In addition, the subcellular localization of dPER/TIM defines how the clock in Drosophila responds to light. Exposure to light during the early night delays the phase of the clock by retarding the timing of dPER/TIM translocation to the nucleus, whereas similar treatments in the late night accelerate the nuclear clearance of dPER/TIM, which advances the phase of the clock (Lee et al., 1996; Myers et al., 1996). A complex and integrated network involving TIM and several kinases (i.e., DBT, CK2, and SGG) regulate the nuclear accumulation of dPER (see Introduction).
We identify Ser661 as a critical phosphorylation signal that controls the timing of dPER nuclear entry. S661 and its flanking Pro residue are conserved in dPER from all Drosophilids analyzed to date (Fig. 1A), suggesting that this phosphorylation event is widely used to regulate dPER subcellular distribution. Because of structural constraints imposed by the flanking proline residue, all Pro-directed kinases are in the CMGC group of kinases, which include MAPKs, DYRKs, and CDKs (Kannan and Neuwald, 2004). Indeed, in vitro kinase assays indicate that S661 can be phosphorylated by the classic MAPK ERK1/2 (Fig. 3). Although we have yet to identify the physiologically relevant Pro-directed kinase(s) phosphorylating S661 on dPER, studies in cultured cells and in vitro indicate that phosphorylation of S661 can catalyze the subsequent phosphorylation of S657 by SGG/GSK-3β (Figs. 2, 3). Phosphorylation at S661 generates a good consensus for a GSK-3β/SGG site at S657, because this kinase usually requires a priming phosphate located at n + 4 (where n is the site of phosphorylation by GSK-3β/SGG) (Cohen and Frame, 2001). Abolishing phosphorylation at S661 delays the nuclear entry of dPER in key pacemaker neurons (Fig. 6), yielding clocks with lengthened endogenous periods (Table 2) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Phosphorylation of S661 appears to be constitutive in flies because its relative occupancy highly correlates with overall levels of dPER (Fig. 4D). The phosphorylated status of S657/S661 has very little to no effect on dPER stability (Figs. 1, 2, 4), indicating that this phosphorylation “module” regulates the timing of dPER nuclear translocation in a manner unrelated to changes in dPER stability. Results based on the attenuated hyperphosphorylation of the dPER(S661A) mutant (Figs. 1, 2, 4) further support our recent findings indicating that global hyperphosphorylation of dPER is not closely linked to stability (Chiu et al., 2008). In addition, as assayed in cultured cells, SGG has no noticeable effect of dPER stability (Fig. 2), in contrast to DBT and CK2 (Ko et al., 2002; Nawathean and Rosbash, 2004). Collectively, our findings support a model whereby phosphorylation at S661 primes subsequent phosphorylation by SGG at S657 and likely other sites, modifications that stimulate the nuclear entry/retention of dPER.
The function of GSK-3β/SGG in the Drosophila circadian system is multifaceted, involving effects on the core oscillator mechanism and photic entrainment pathways. A role in the clockworks was initially demonstrated by showing that ectopic expression of sgg in clock cells leads to shortened free-running behavioral rhythms, whereas hypomorphic alleles lengthen these rhythms, phenotypes associated with either advanced or delayed nuclear translocation of dPER/TIM in central pacemaker neurons, respectively (Martinek et al., 2001). This study also identified TIM but not dPER as a direct target of SGG, inferred from the observation that SGG stimulates the hyperphosphorylation of TIM, an event thought to enhance its light-mediated degradation (Martinek et al., 2001). However, a link between TIM phosphorylation and the timing of dPER/TIM nuclear translocation was not shown. Moreover, recent findings suggest that CRY but not TIM is a direct target of SGG (Stoleru et al., 2007). The possibility that SGG phosphorylates CRY might also underlie its role in photic entrainment (Martinek et al., 2001; Yuan et al., 2005; Stoleru et al., 2007). Our results suggest a more direct role for SGG in stimulating the nuclear entry/retention of dPER. That dPER might be a direct target of SGG was also recently suggested by pharmacological studies in S2 cells (Fang et al., 2007).
Although our findings strongly suggest a hierarchical phosphorylation cascade, S657 is not highly conserved (Fig. 1A), suggesting the possibility for species-specific phosphorylation programs. Nonetheless, GSK-3β can also exhibit “discontinuous” phosphorylation at sites far removed from its conventional n + 4 consensus (Gwack et al., 2006). Thus, although some Drosophila species lack a typical GSK-3β site near to their highly conserved “661” site (Fig. 1A), it is possible that, after priming, SGG phosphorylates at more distant sites. This could also explain why the S661A mutation is associated with a reduction in global hyperphosphorylation of dPER, whereas this is not the case for the S657A mutation (Figs. 1, 2, 4 and data not shown). It is presently unclear what other sites besides S657 are phosphorylated by SGG in an S661-dependent manner or whether the attenuated global phosphorylation of dPER(S661A) is solely attributable to SGG-specific sites. That the period lengthening effects of the S661A mutation are more severe compared with S657A (Table 2) is in line with the idea that, after priming at S661, multiple SGG-dependent sites collectively contribute to regulating the timing of dPER nuclear entry. Thus, the absence of a classic GSK-3β/SGG site nearby the conserved S661 phospho-acceptor does not rule out the possibility that S661 is a key phospho-switch universally operating in Drosophila that regulates the timing of dPER nuclear entry by catalyzing sequential phosphorylation by GSK-3β/SGG at multiple sites.
How might phosphorylation at the “S661 priming-dependent cluster” regulate the timing of dPER nuclear translocation? Although this is not clear, recent work using cultured cells suggested that the physical interaction of dPER and TIM appears to initiate a “maturation” process in the cytoplasm that lasts several hours and somehow makes TIM and dPER competent to enter the nucleus independent of each other (Meyer et al., 2006). It is possible that phosphorylation of dPER at S657, S661, or both is part of the maturation process that stimulates the nuclear entry/retention of dPER. CK2 also appears to promote dPER nuclear translocation by direct phosphorylation of dPER, which almost certainly includes a “CK2 cluster” on dPER (amino acids 149–153) (Lin et al., 2005). It will be of interest to determine whether mutations in the CK2 and S661 phospho-clusters have synergistic effects on the timing of dPER nuclear translocation. Although the identity of the S661 kinase and its subcellular localization will be important to determine, SGG is localized in the cytoplasm of pacemaker neurons (Yuan et al., 2005). A cytoplasmic function for SGG in the Drosophila clock is further supported by the observation that overexpressing SGG in pacemaker cells mainly affects the delay portion of light-induced phase shifts, which corresponds to times in the day when dPER/TIM are located in the cytoplasm (Martinek et al., 2001).
Intriguingly, a hierarchical phosphorylation program involving a Pro-directed priming site followed by GSK-3β phosphorylation is also observed for the mammalian clock proteins CRY1/2 (Harada et al., 2005). Thus, phosphorylation at a Ser/Thr–Pro site that subsequently primes phosphorylation by GSK-3β might be a common phospho-module used in regulating the metabolism/activity of clock proteins. Understanding how GSK-3β regulates animal clocks is of medical importance because lithium ion therapy is a powerful treatment for bipolar disorders, which are closely linked to malfunctions in the human circadian system (McClung, 2007). In Drosophila, rodents, and humans, lithium treatment, which inhibits GSK-3β activity, lengthens circadian rhythms (Padiath et al., 2004; Dokucu et al., 2005; McClung, 2007). This is consistent with our findings showing that preventing phosphorylation of the “S657–661 phospho-module” delays behavioral rhythms in Drosophila. GSK-3β has been shown to phosphorylate several components of the mammalian clock, such as the orphan receptor REV–ERBα (Yin et al., 2006), mCRYs (Harada et al., 2005), mPER2 (Iitaka et al., 2005), and mammalian CLOCK (Spengler et al., 2009). Of note, GSK-3β promoted the nuclear translocation of mPER2 in cultured mammalian cells (Iitaka et al., 2005), similar to what is observed in Drosophila (Martinek et al., 2001). Together with our results, this implies that GSK-3β has a conserved role in regulating the timing of when PER proteins translocate to the nucleus. To the best of our knowledge, our results are the first to identify and evaluate the in vivo significance of a GSK-3β phospho-site on a clock protein and raise the intriguing possibility that the period-altering effects of lithium therapy are primarily mediated by modulating the timing of when PER proteins translocate to the nucleus, an event central to setting the pace and phase of animal clocks (Bae and Edery, 2006). The identification of SGG sites on human PER proteins might facilitate the development of more efficient therapies to combat mood disorders.
Footnotes
This work was supported by National Institutes of Health Grants NS049862 and NS061952 (J.C.) and 034958 (I.E.). Work in the laboratory of A.K. was supported by the Deutsche Forschungsgemeinschaft and the sixth European Union framework program EUCLOCK. Work in the laboratory of E.Y.K. was supported by Basic Science Research Program through the National Research Foundation of Korea funded by Ministry of Education, Science, and Technology Grants 2008-0058506, 2009-0067106, and NRF-2008-521-C00220. We thank Justin Blau for kindly providing monoclonal anti-PDF antibody and Steven Reppert for pAct–dper deletion constructs.
References
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Bae K, Edery I. Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem. 2006;140:609–617. doi: 10.1093/jb/mvj198. [DOI] [PubMed] [Google Scholar]
- Bao S, Rihel J, Bjes E, Fan JY, Price JL. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci. 2001;21:7117–7126. doi: 10.1523/JNEUROSCI.21-18-07117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DC, Reppert SM. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol. 2003;13:758–762. doi: 10.1016/s0960-9822(03)00286-0. [DOI] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Yu L, Taghert PH. Lithium- and valproate-induced alterations in circadian locomotor behavior in Drosophila. Neuropsychopharmacology. 2005;30:2216–2224. doi: 10.1038/sj.npp.1300764. [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, Rao A. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- Hall JC. Genetics and molecular biology of rhythms in Drosophila and other insects. Adv Genet. 2003;48:1–280. doi: 10.1016/s0065-2660(03)48000-0. [DOI] [PubMed] [Google Scholar]
- Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- Kannan N, Neuwald AF. Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2alpha. Protein Sci. 2004;13:2059–2077. doi: 10.1110/ps.04637904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Kimmel AR. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Genet Dev. 2000;10:508–514. doi: 10.1016/s0959-437x(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Edery I. Analyzing the degradation of PERIOD protein by the ubiquitin-proteasome pathway in cultured Drosophila cells. Methods Enzymol. 2005;393:394–408. doi: 10.1016/S0076-6879(05)93018-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Ko HW, DiMassa S, Kim EY, Bae K, Edery I. Cis-combination of the classic per(S) and per(L) mutations results in arrhythmic Drosophila with ectopic accumulation of hyperphosphorylated PERIOD protein. J Biol Rhythms. 2007;22:488–501. doi: 10.1177/0748730407306929. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin. Mol Cell Biol. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lin JM, Schroeder A, Allada R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci. 2005;25:11175–11183. doi: 10.1523/JNEUROSCI.2159-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24:3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Padiath QS, Paranjpe D, Jain S, Sharma VK. Glycogen synthase kinase 3beta as a likely target for the action of lithium on circadian clocks. Chronobiol Int. 2004;21:43–55. doi: 10.1081/cbi-120027981. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. On temperature independence in the Clock system controlling emergence time in Drosophila. Proc Natl Acad Sci U S A. 1954;40:1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Schlosser A, Vanselow JT, Kramer A. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal Chem. 2005;77:5243–5250. doi: 10.1021/ac050232m. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler ML, Kuropatwinski KK, Schumer M, Antoch MP. A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle. 2009;8:4138–4146. doi: 10.4161/cc.8.24.10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernández MP, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Hung HC, Maurer C, Kay SA. Second messenger and Ras/MAPK signalling pathways regulate CLOCK/CYCLE-dependent transcription. J Neurochem. 2006;98:248–257. doi: 10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Lin F, Zheng X, Sehgal A. Serotonin modulates circadian entrainment in Drosophila. Neuron. 2005;47:115–127. doi: 10.1016/j.neuron.2005.05.027. [DOI] [PubMed] [Google Scholar]