Abstract
Biological pathways are structured in complex networks of interacting genes. Solving the architecture of such networks may provide valuable information, such as how microorganisms cause disease. Here we present a method (Tn-seq) for accurately determining quantitative genetic interactions on a genome-wide scale in microorganisms. Tn-seq is based on the assembly of a saturated Mariner transposon insertion library. After library selection, changes in frequency of each insertion mutant are determined by sequencing of the flanking regions en masse. These changes are used to calculate each mutant’s fitness. Fitness was determined for each gene of the gram-positive bacterium Streptococcus pneumoniae, a causative agent of pneumonia and meningitis. A genome-wide screen for genetic interactions identified both alleviating and aggravating interactions that could be further divided into seven distinct categories. Due to the wide activity of the Mariner transposon, Tn-seq has the potential to contribute to the exploration of complex pathways across many different species.
Introduction
Genes (and their protein products) are organized in complex networks. Genetic interaction analysis on a genome-wide scale has the potential to connect genes and to increase understanding of such networks. Interactions between genes have been explored in Saccharomyces cerevisiae by systematically knocking out all possible pairwise combinations of genes, leading to identification of networks in important pathways such as DNA integrity1 and phosphorylation2. Such comprehensive genetic interaction screens are rare in microorganisms due to lack of genome-wide tools with wide applicability. Thus the development of a high-throughput tool for gene disruption and interaction discovery in microorganisms has great potential for improving the lagging functional annotation of microbial genomes and for resolving complex biological pathways.
Three approaches have been developed to identify genetic interactions in S. cerevisiae3,4,5,6. Each makes use of an arrayed collection of single-gene deletion strains and robotic automation to construct double mutants by mating. Meiotic assortment is not applicable to bacteria; however, two analogous methods have been developed for Escherichia coli; GIANT coli and eSGA7,8. These methods use conjugation of a genome-wide arrayed collection of single gene mutants to construct all double mutants. However, the applicability of these methods to a wide range of microorganisms is problematic because there are only arrayed collections of single gene knockouts for a handful of species.
A more generally applicable method to identify genetic interactions in bacteria is TraSH9, as well as related transposon-based genetic footprinting methods10. TraSH is based on the construction of a transposon library, the generation of probes from outward-oriented T7 promoters located in the transposon ends and the subsequent detection of the probes by microarray. However, the accuracy of this and the related methods is somewhat limited because the precise location of an insertion cannot be directly determined, fitness cannot be determined for each insertion and can only be depicted as a relative fluorescence intensity ratio, and the need for a microarray restricts its use to strains for which microarrays have been developed.
Here we present Tn-seq, a robust and sensitive method for the discovery of quantitative genetic interactions in microorganisms through massively parallel sequencing. The approach does not depend on a pre-existing array of mutants but is instead based on the assembly of a saturated transposon insertion library. After growth of the library under a test condition, the change in frequency of each insertion mutant is determined by sequencing the transposon flanking regions en masse. The change in frequency reflects the effect of the insertion on fitness. Fitness of every insertion in a genome can be determined in this way and is a quantitative measure of the growth rate. We applied Tn-seq to S. pneumoniae, determining fitness for each gene and identifying those likely to be essential for basal growth. Finally, a genome-wide screen for genetic interactions of five query genes involved in transcriptional regulation and carbohydrate transport identified 97 high confidence interactions.
Results
Library selection, sequencing and fitness calculations
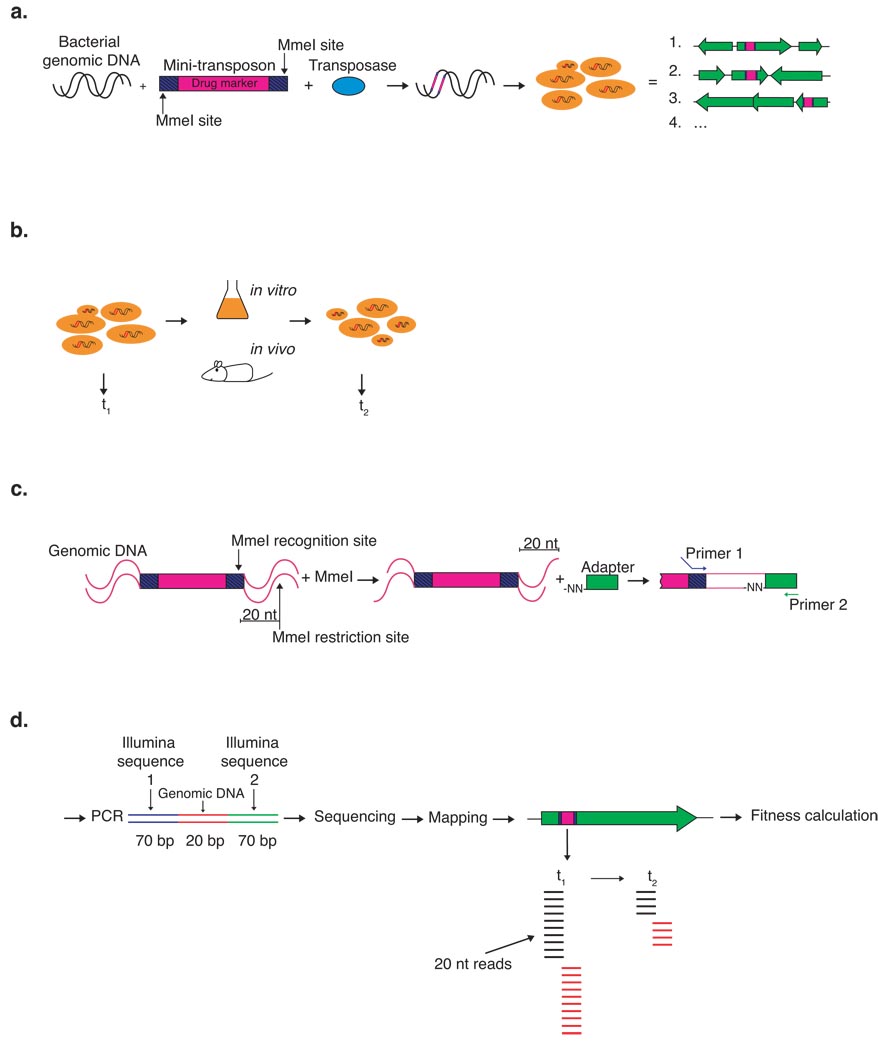
A genome-wide disruption library was established in S. pneumoniae with the Mariner Himar1 mini-transposon derivative magellan6. Magellan6 inserts randomly in the genome, requiring only a TA dinucleotide at the insertion site11. Following transposition in vitro, naturally competent S. pneumoniae were transformed with the transposed DNA resulting in a library in which each bacterium carries a single transposon insertion (Fig. 1). The library was grown in standard broth medium during which detrimental insertions decrease in frequency while advantageous ones increase. The change in frequency over time for each mutant was determined by en masse sequencing of the regions flanking the transposon insertions, such that a change in fitness translates into a change in the number of reads. This change is then used to calculate fitness for each insertion mutant (see Methods Online for details).
Figure 1.
Schematic depiction of Tn-seq. (a) A gene disruption library is constructed by first transposing the mini-transposon magellan6 into bacterial genomic DNA in vitro and subsequently transforming a bacterial population with the transposed DNA. The result is a bacterial pool where each bacterium contains a single transposon insertion in its genome. (b) DNA is isolated from a portion of the bacterial pool (t1), another portion is used to seed a culture on which selection is performed, then DNA is isolated again from recovered bacteria (t2). (c) DNA from both time points is digested with Mme I; the Mme I restriction site was introduced into the transposon’s inverted repeats. (d) A PCR amplification is performed to obtain a 160 bp sequence with 20 bp of bacterial specific DNA flanked by Illumina specific sequences, which enable sequencing. After sequencing, different samples are identified based on barcode sequence, the 20 bp reads are mapped to the genome and are counted for each insertion, thus allowing fitness to be calculated.
Single gene fitness
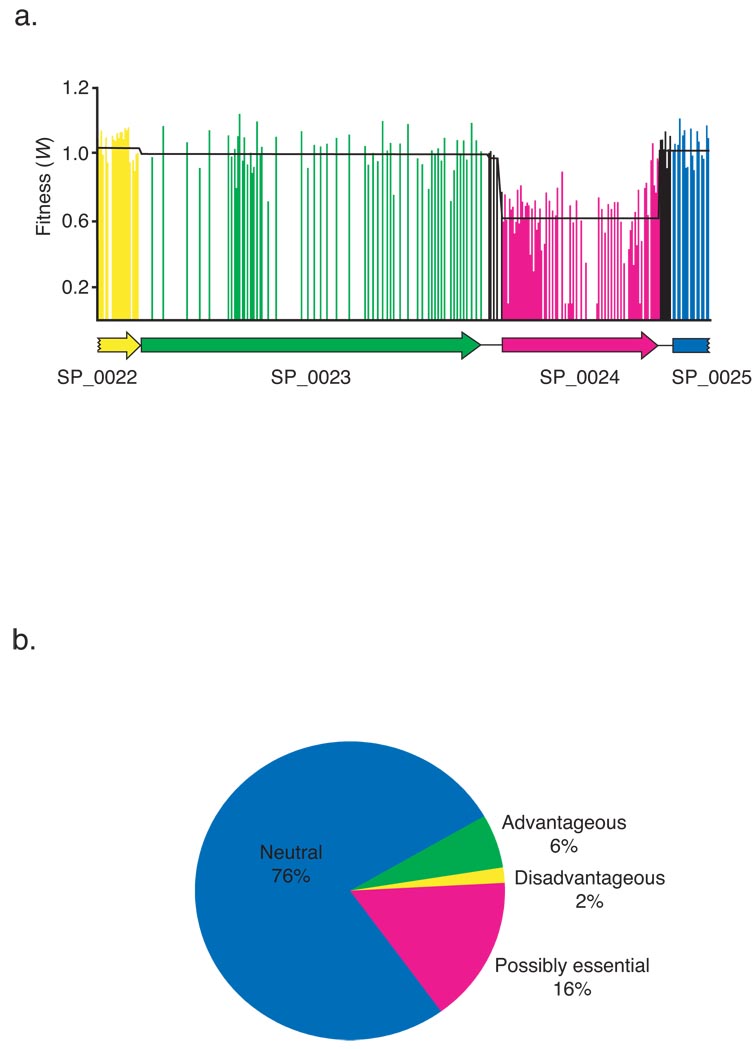
To screen for genetic interactions it is necessary to measure the fitness effects of double gene mutations. First we need to measure fitness for each single gene disruption. For this, six libraries of ~25,000 insertions each were constructed in the wild type S. pneumoniae TIGR4. The libraries were grown in standard broth for ~7 generations after which DNA was isolated and the transposon junctions were sequenced. There was no insertional bias between coding and non-coding genomic regions, and there were 36 regions that had a ≥2-fold higher or lower number of insertions (Supplementary Table 1). Figure 2A illustrates how different transposon insertions distributed over a specific genomic region affect fitness. By averaging over all insertions in a specific gene we obtain a single fitness value for each gene (Supplementary Table 2). Genes were divided into four categories; neutral (fitness = 0.96–1.04), advantageous (fitness > 1.04) disadvantageous (fitness < 0.96) and possibly essential (fitness = 0). Genes in the latter category correspond to those in which transposon insertions were absent in the sequenced library. Mutations in these genes either prevented or substantially slowed bacterial replication during outgrowth of the transposon insertion strains (also see below). Categorization was based on a one sample t-test with Bonferroni correction for multiple testing (also see Methods Online) (Figure 2B). We confirmed 68% of genes that were previously suggested to be essential in another serotype strain, R6 (Supplementary Table 2). The 32% of unconfirmed genes may be due to strain differences.
Figure 2.
Fitness determination and classification. (a) Independent insertions and their fitness effects in the genomic region from gene SP_0022 to SP_0025 are plotted. The black jagged line depicts the average fitness for each gene. (b) A pie chart categorizing every gene’s fitness in the S. pneumoniae genome into four categories.
The program GSEA12 was used to test for the enrichment of genes with a common biological function and a similar fitness effect. Eleven gene sets were negatively enriched, suggesting that they are advantageous for growth in this environment, and two were positively enriched (FDR<25%, nominal P value < 0.01), suggesting that they are deleterious for growth in this environment (Supplementary Table 3).
Validation of the method
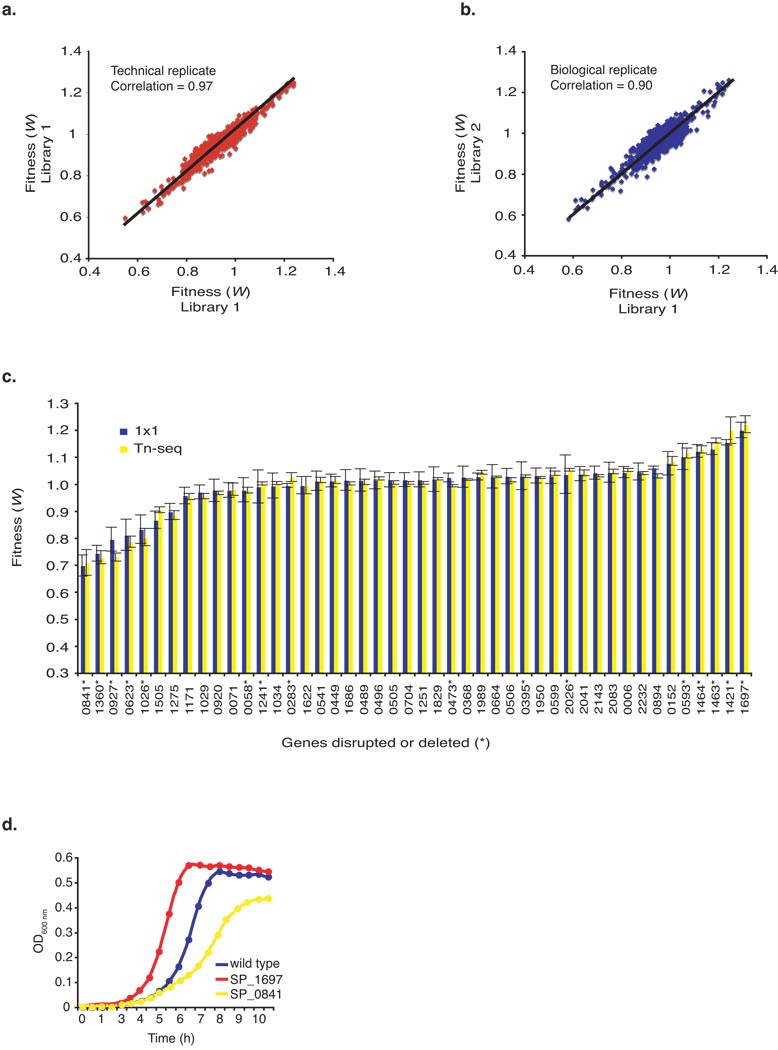
To validate the reproducibility of Tn-seq, technical and biological replicates were performed. Technical replicates compared the same libraries that were split after DNA isolation. Reproducibility was very high with a Pearson correlation coefficient of 0.97 (Fig. 3a). Biological replicates, which were done for all (six) experiments in this study, compared two or more independently established and selected-on libraries. Reproducibility between biological replicates was always high with a Pearson correlation coefficient between 0.70–0.90 (Fig. 3b), and increasing correlations were found with increased experience. These data show that Tn-seq is highly reproducible both within and between independent experiments.
Figure 3.
Validation of Tn-seq fitness measurements. (a) A technical replicate of the same library comparing the effect of sample preparation and sequencing. Pearson correlation coefficient = 0.97. (b) A biological replicate comparing two independently generated, selected upon and sequenced libraries. Pearson correlation coefficients between biological replicates ranged between 0.70 – 0.90. A representative example is shown. (c) Comparison of fitness (± s.e.m.) obtained with Tn-seq (yellow) and the classical 1×1 competition method (blue) for 30 random transposon insertions and 16 marked gene deletions (*). Numbers underneath the graph refer to Streptococcus pneumoniae TIGR4 gene numbers (SP_number). (d) Three representative growth curves; the wild type strain, a strain with an advantageous deletion (SP_1697, W=1.22) and a strain with a disadvantageous deletion (SP_0841, W=0.71).
We failed to identify genes with fitness <0.53 but >0 (Fig. 3b and Supplementary Table 2). Since the generation time of such mutants is at least twice that of wild type (shown below), it is likely that such strains are rapidly outcompeted during construction and initial expansion of the library to the point that they fall below the threshold of detection by sequencing. Hence, the “possibly essential” genes listed in Supplementary Table 2 includes both essential genes and genes that are highly deleterious when disrupted.
It is possible that fitness determined using Tn-seq differs from fitness determined using the classical 1×1 competition. For instance, frequency dependent selection, an evolutionary process where fitness of a genotype is dependent on its frequency relative to other genotypes in the population, may play a role. To address this possibility we measured fitness using 1×1 competition for 30 mutants that were randomly picked from the library. No significant differences in fitness between the two methods were observed in a student’s t-test (all P values » 0.05, n > 5) (Fig. 3c).
Depending on the site of insertion within a gene, a transposon can affect fitness in a position-dependent manner13. For example, insertions close to the 5’ or 3’ end may not disrupt the gene due to alternate start sites or truncated but still functional gene products, respectively, and could give a false indication of fitness. However, in Tn-seq, fitness is determined by averaging over multiple independent insertions within the same gene (Fig. 2a), minimizing the influence that positional effects have on fitness determination. To test for positional effects we re-analyzed the complete dataset by removing all insertions in the first or last 10% of each gene. Removing insertions in the first 10% of each gene did not change fitness, while removing insertions in the last 10% changed fitness marginally for only four genes (Supplementary Table 2). This confirms that positional effects are rare.
Transposon insertions within an operon may exert a polar effect on downstream genes leading to incorrect fitness assignments. Typically, such polar effects result from decreased expression of downstream gene(s). However, magellan6 lacks transcriptional terminators therefore allowing for read-through transcription and thus such polar effects should be minimized. Although it was not possible to screen for the presence of polar effects, we could screen for their absence. The absence of polarity is most apparent when a downstream gene in an operon has a severe fitness effect or is essential while the upstream gene has no fitness effect. We screened the genome for operons with an essential downstream gene and identified 37 such instances. Of those 37 operons, none contained an insertion in an upstream gene with a (severe) fitness defect, suggesting that polar effects are not a major confounder in our data [AU: OK as edited?] (Supplementary Table 2).
To confirm that Tn-seq accurately predicts fitness and also that a complete removal of a gene gives the same effect as averaging over multiple independent transposon insertions into the gene, we replaced the entire coding sequence of 16 genes with a drug marker. Of these, six had a Tn-seq-determined fitness between 0.95–1.05, five had a disadvantageous fitness of ≤0.8 and five had an advantageous fitness of ≥1.11. Fitness was compared between Tn-seq and the conventional 1×1 competition method and no significant differences between the two methods were found in a student’s t-test (all P values » 0.05, n > 5) (Fig. 3c).
Because the expansion factor of the population over time was incorporated into the calculation of fitness, the resulting fitness value represents the change in frequency of a mutant within a population over a single generation. Thus fitness should correlate with the growth rate of the mutant strain. For instance, a 50% decrease in fitness (W = 0.5) should result in a 2-fold increase in doubling time. Confirming this, the doubling times for ten deletion mutants with reduced fitness were determined and were found to directly correlate with fitness (Table 1 and Fig. 3d). For instance, given the wild-type doubling time of 38 minutes, mutant SP_1697 should have a doubling time of ~31 minutes ([1/1.22]*38), we measured 31 minutes, and mutant SP_0841 should have a doubling time of ~54 minutes ([1/0.71]*38), we measured 57 minutes.
Table 1.
Doubling time and fitness for wild type and ten marked deletions.
| Strain | Tn-seq |
doubling time |
|---|---|---|
| fitness ± s.e.m. | minutes ± s.e.m. | |
| wild type | 1 | 38.3 ± 1.0 |
| SP_1697 | 1.22 ± 0.031 | 31.2 ± 1.6 |
| SP_1464 | 1.13 ± 0.013 | 33.0 ± 0.9 |
| SP_1463 | 1.16 ± 0.009 | 34.1 ± 0.8 |
| SP_1421 | 1.20 ± 0.049 | 31.6 ± 0.8 |
| SP_0593 | 1.11 ± 0.019 | 33.6 ± 0.8 |
| SP_1360 | 0.73 ± 0.024 | 58.8 ± 3.0 |
| SP_1026 | 0.80 ± 0.032 | 49.3 ± 1.4 |
| SP_0927 | 0.73 ± 0.015 | 55.6 ± 3.5 |
| SP_0841 | 0.71 ± 0.048 | 57.0 ± 2.8 |
| SP_0623 | 0.79 ± 0.020 | 48.2 ± 1.6 |
Screening for genetic interactions
To assess the utility of Tn-seq in mapping genetic interactions we performed a genome-wide screen on five ‘query’ genes. Three of these are transcriptional regulators (ccpA, regR and malR) and two are ABC transporters involved in carbohydrate uptake (SP_1683 and malX). The former are all members of the LacI/GalR family of metabolic regulators.
In Bacillus subtilis, CcpA (catabolite control protein A) is a master regulator of genes involved in complex carbohydrate metabolism and controls gene expression by binding to catabolite response DNA elements (cre). Depending on where a cre-site is located, either upstream or downstream of the transcription start site, CcpA can respectively activate or repress transcription14. In S. pneumoniae inactivation of ccpA results in decreased in vitro fitness (0.84±0.04 s.e.m.) and decreased virulence in mouse models of colonization and infection15,16.
Loss of regR has a less dramatic phenotype than ccpA: although it has no in vitro growth defect it is involved in competence induction and virulence17. The third regulator MalR has so far only been implicated in the regulation of maltose uptake and metabolism18. The hierarchy among these three regulators led us to predict that we would find the most genetic interactions for ccpA, followed by regR and then malR. We also hypothesized that the dedicated ABC transporters (SP_1683 and malX) would have still fewer interactions. In addition, since malR regulates malX we expected to find overlap in the interactions of both genes.
Each of the five query genes was replaced with a drug marker and the fitness of each of the mutant strains was measured in 1×1 competition experiments and confirmed by Tn-seq. Three independent transposon libraries consisting of 10,000–25,000 insertions were then established in the background of each query gene knockout. Alleviating and aggravating genetic interactions were scored as a deviation from the multiplicative model and were divided into seven categories based on a student’s t-test with Bonferroni correction for multiple testing (Fig. 4, Supplementary Table 4 and see Methods). Relationships among genes were ordered in a single network (Fig. 4) and were supplemented with 41 additional interactions from the STRING19 database.
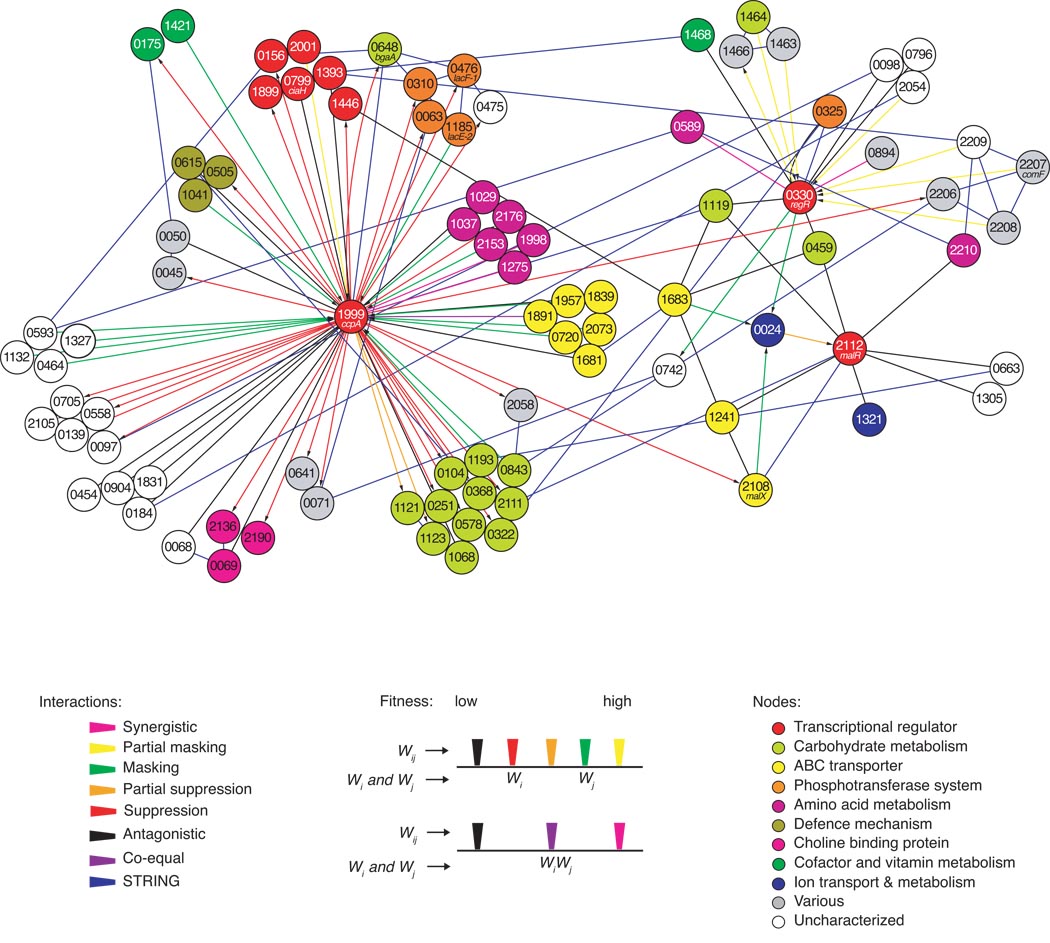
Figure 4.
Genetic interaction network of five query genes. Interactions between two genes (i and j) are depicted as a line (edge) between two genes (nodes) and were determined by Tn-seq and supplemented with interactions from the STRING19 database (confidence >0.5; only edges were added and no nodes). Genetic interactions are divided into seven categories depicted underneath the network. Wi, fitness of mutant in gene i; Wj, fitness of mutant in gene j; Wij, fitness of double mutant. In the top section, genes i and j confer different fitness; in the bottom section, genes i and j have an equal fitness effect. This scheme results in seven color coded genetic interaction categories: synergistic (Wi=Wj<Wij), partial masking (Wi<Wj<Wij), masking (Wi<Wj=Wij), partial suppression (Wi<Wij<Wj), suppression (Wi=Wij<Wj), antagonistic (Wij<Wi≤Wj) and co-equal (Wi=Wj=Wij) (see Methods for further details). Arrows depict the hypothesized direction of the interaction. Nodes are color coded according to their involvement in a biological process or molecular function. Numbers in nodes refer to Streptococcus pneumoniae TIGR4 gene numbers (SP_number).
Most interactions were scored for ccpA (64) followed by regR (17) and malR (8), while the two transporters, SP_1683 and malX, have five and three interactions respectively (Fig. 4 and Supplementary Tables 4 and 5), the latter sharing two of its three interactions with malR. Considerable overlap in interactions can also be observed between SP_1683 and malX (2 of 3), between SP_1683 and malR (4 of 5) and between SP_1683 and regR (3 of 5). The overlapping profiles suggest that all four query genes are involved in overlapping metabolic pathways, which was expected for malR and malX and possibly SP_1683 (since like malX it is also involved in carbohydrate uptake) but not for regR.
The genetic interaction screen confirms two expected interactions for regR; the STRING database interaction with the mannose phosphotransferase system (SP_0325) and, by means of comF (SP_2207) and SP_2208, the connection between regR and competence. Other regR-interacting genes suggest its involvement in various metabolic pathways as well as in other processes.
Of the five query genes, ccpA had the most genetic interactions detected, consistent with its role as a regulator of many different processes in S. pneumoniae. To determine whether there is a link between genes interacting with ccpA and the presence of a cre sequence we screened the S. pneumoniae TIGR4 sequence for matches to the B. subtilis consensus cre sequence ‘WTGNAANCGNWWNCA’14. We found 275 out of 2087 genes (13%) with putative cre sequences (Supplementary Table 2). Out of the 90 genes present in the network, cre sequences were predicted for 27, of which 24 interact with ccpA (Supplementary Table 4). Thus ccpA, with 38% (24/64) of its interaction partners carrying a cre sequence, interacts with three times more cre sequence-containing genes than would be expected from chance alone.
Half of the genes that interact with ccpA are involved in carbohydrate uptake and metabolism (32). Other large categories include uncharacterized genes (13) and transcriptional regulators (6). Three of the latter are part of the two component systems (TCS) TCS07 (SP_0156), TCS11 (SP_2001) and CiaR/H (TCS05; SP_0799). TCSs mediate communication between the environment and the cell interior. There is little known about TCS07 and TCS11 but CiaR/H has been found to regulate transcription of maltose utilization genes20. CiaR/H is also needed for S. pneumoniae to colonize the nasopharynx and is important for lung and systemic infections in mice21,22. We found an antagonistic interaction between ciaH and ccpA (fitness 0.46±0.04 s.e.m.) and suppressive interactions between ccpA and the maltose utilization genes malX/SP_2108 (fitness 0.78±0.02 s.e.m.) and malA/SP_2111 (fitness 0.71±0.03 s.e.m.). We interpret these interactions to indicate that ciaR/H and ccpA have overlapping profiles. Indeed, ciaR has a predicted cre sequence, suggesting it is subject to CcpA regulation.
Validation of genetic interactions
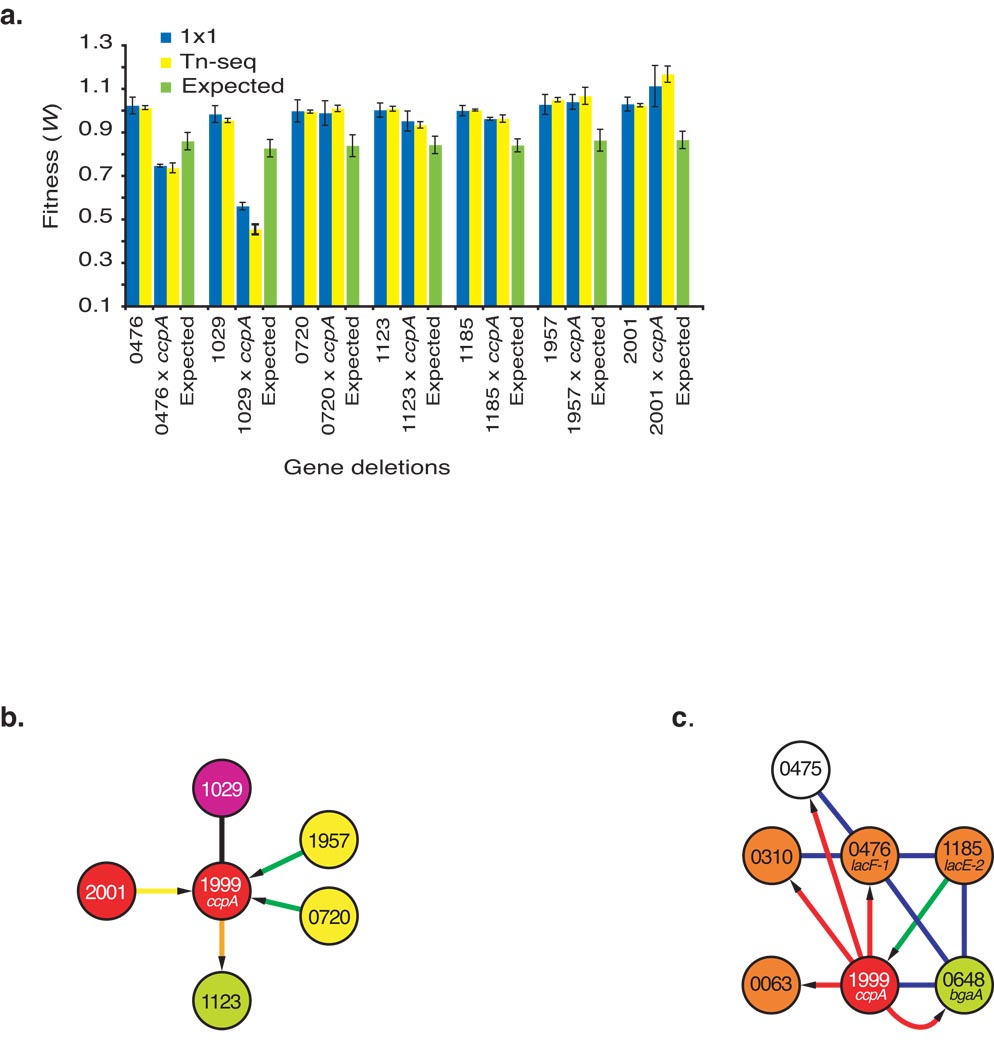
To validate the genetic interactions identified by Tn-Seq we deleted seven genes by replacement with a drug marker both in the wild type and ccpA backgrounds. Fitness values for all 14 mutant strains were confirmed by 1×1 competitions based on a student’s t-test with Bonferroni correction for multiple testing (Fig. 5a). Specifically, alleviating interactions were confirmed for SP_2001 (TCS05), ABC transporters SP_1957 and SP_0720 and metabolism gene SP_1123, and an antagonistic interaction was confirmed for amino acid metabolism gene SP_1029 (Fig. 5b). In addition, two interactions were confirmed between ccpA and the PTS genes SP_0476 (lacF-1) and SP_1185 (lacE-2), which are part of a small sub-network including four PTS genes, the hydrolase bgaA (SP_0648) and an uncharacterized gene SP_0475 (Fig. 5c).
Figure 5.
Validation of genetic interactions. (a) Fitness (± s.e.m.) of seven genes either knocked out singly or in combination with ccpA are compared between the 1×1 method (blue) and Tn-seq (yellow). The expected multiplicative fitness for each double gene knockout is depicted in green (expected fitness was determined by multiplying ccpA fitness [0.84±0.04 s.e.m.] with fitness measured for the other gene). Numbers underneath the graph refer to Streptococcus pneumoniae TIGR4 gene numbers (SP_number). (b) A detail of the genetic interaction network showing five of the validated genetic interactions and their specific interactions. (c) A sub-network from the larger genetic interaction network showing four PTS genes including two of the validated genetic interactions (SP_0476 and SP_1185), β-galactosidase (SP_0648) and a gene with unknown function (SP_0475). Interaction colors, node colors and numbers are as in Figure 4.
Discussion
Tn-seq can be used to determine the fitness conferred by single genes and to map genetic interactions in microorganisms. Unlike existing methods, Tn-seq does not depend on a preexisting genomic microarray or an array of gene knockouts, and its use of massively parallel sequencing makes it highly reproducible, sensitive and robust. Due to our method of library construction we were most successful in identifying nonessential genes with fitness above 0.53 and possibly essential genes with fitness equal to zero. Some of the genes listed as possibly essential may have a fitness between 0.53 and 0. Adjustments in library assembly should increase the dynamic range of the method (see Methods Online).
We recorded 97 high-confidence genetic interactions for five query genes in S. pneumoniae, with ccpA emerging as a so-called ‘hub’ gene. Similar to its role in B. subtilis, CcpA thus appears to be a master regulator in S. pneumoniae. Because of the sensitivity of Tn-seq we were able to determine different types of both aggravating and alleviating interactions. Specific genetic interactions can reveal direction of information flow in molecular pathways23,24. Here it is tempting to hypothesize that genes with an alleviating ccpA interaction represent deleterious genes that are repressed by CcpA in the growth condition we tested. In this scenario, knocking out ccpA results in derepression of the deleterious gene and a reduction in fitness. Subsequently knocking out the derepressed gene restores fitness.
We identified a sub-network containing interactions between ccpA, four PTS genes, the β-galactosidase bgaA and the uncharacterized gene SP_0475. In other bacterial species, cytoplasmic β-galactosidases play an important role in lactose metabolism. However, in S. pneumoniae bgaA has a cell surface localization, and has so far only been associated with virulence25. From both the Tn-seq data and its interaction with ccpA we can determine that bgaA is important in basal growth, and through its association in the sub-network with two lactose PTS genes, that it may be involved in lactose metabolism. In addition, the occurrence of the uncharacterized gene SP_0475 in the same sub-network suggests that it too may be involved in PTS-regulated carbohydrate uptake and metabolism. We expect that expanding the number of query genes and environmental conditions will contribute substantially to the understanding of the general architecture of S. pneumoniae biological networks and will be very helpful in annotating the 30% of unknown genes.
The Tn-seq data presented here illustrates the potential of the developed method; each gene’s fitness can be rapidly determined in a specific environment without the need of first constructing a full genome knockout array. Since S. pneumoniae consists of >90 different serotypes and a gene’s specific role may be strain-dependent this is an important advantage. Moreover, Tn-seq can be easily applied to different environments (e.g. different growth media, chemical or physical stresses, or infection models). Finally, the Himar1 Mariner transposon can be used in many other microorganisms including E. coli26, Staphylococcus aureus27, Haemophillus influenza28 and Mycobacterium tuberculosis29. We thus expect that our method will open up the possibility to screen for genetic interactions across many different environments, strains and species.
Supplementary Material
Acknowledgments
We thank Linc Sonenshein, Ralph Isberg, Ramkumar Iyer and Nadine Vastenhouw for both discussions and comments on the manuscript, the Tufts University core facility for Illumina sequencing and the Camilli lab, particularly David Lazinski, and André Boorsma for helpful discussions. This work was supported by a grant from the Netherlands Organization for Scientific Research (NWO - Rubicon) to T.v.O. A.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
T.v.O and A.C. designed research, analyzed the data and wrote the paper. T.v.O performed research and K.L.B. contributed to data analysis.
References
- 1.Pan X, Ye P, Yuan DS, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 2.Fiedler D, Braberg H, Mehta M, et al. Functional Organization of the S. cerevisiae Phosphorylation Network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 4.Pan X, Yuan DS, Xiang D, et al. A Robust Toolkit for Functional Profiling of the Yeast Genome. Molecular Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 5.Schuldiner M, Collins S, Thompson N, et al. Exploration of the Function and Organization of the Yeast Early Secretory Pathway through an Epistatic Miniarray Profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Roguev A, Bandyopadhyay S, Zofall M, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Typas A, Nichols RJ, Siegele DA, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Meth. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butland G, Babu M, Díaz-Mejía JJ, et al. eSGA: E. coli synthetic genetic array analysis. Nat Meth. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 9.Joshi SM, Pandey AK, Capite N, et al. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci USA. 2006;103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girgis H, Liu Y, Ryu W, et al. A Comprehensive Genetic Characterization of Bacterial Motility. PLoS Genet. 2007;3:e154. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lampe D, Churchill M, Robertson H. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs MA, Alwood A, Thaipisuttikul I, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita Y. Carbon Catabolite Control of the Metabolic Network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 15.Giammarinaro P, Paton J. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infection and Immunity. 2002;70:5454–5461. doi: 10.1128/IAI.70.10.5454-5461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapuy-Regaud S, Ogunniyi A, Diallo N, et al. RegR, a Global LacI/GalR Family Regulator, Modulates Virulence and Competence in Streptococcus pneumoniae Editor: AD O'Brien. Infection and Immunity. 2003;71:2615–2625. doi: 10.1128/IAI.71.5.2615-2625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nieto C, Espinosa M, Puyet A. The maltose/maltodextrin regulon of Streptococcus pneumoniae Differential promoter regulation by the transcriptional repressor MalR. Journal of Biological Chemistry. 1997;272:30860–30865. doi: 10.1074/jbc.272.49.30860. [DOI] [PubMed] [Google Scholar]
- 19.von Mering C, Jensen LJ, Kuhn M, et al. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Research. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascher T, Zahner D, Merai M, et al. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J Bacteriol. 2003;185:60–70. doi: 10.1128/JB.185.1.60-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Throup J, Koretke K, Bryant A, et al. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol. 2000;35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 22.Sebert ME, Palmer LM, Rosenberg MP, et al. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infection and Immunity. 2002;70:4059–4067. doi: 10.1128/IAI.70.8.4059-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drees BL, Thorsson V, Carter GW, et al. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 2005;6:R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St Onge RP, Mani R, Oh J, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King SJ, Hippe KR, Weiser J. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 26.Lampe DJ, Akerley BJ, Rubin EJ, et al. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae T, Glass EM, Schneewind O, et al. Generating a Collection of Insertion Mutations in the Staphylococcus aureus Genome Using bursa aurealis. Methods in Molecular Biology. 2008;416:103–116. doi: 10.1007/978-1-59745-321-9_7. [DOI] [PubMed] [Google Scholar]
- 28.Akerley BJ, Rubin EJ, Camilli A, et al. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sassetti C, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 30.Bricker AL, Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett. 1999;172:131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- 31.Hava D, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Research. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenski RE, Rose MRS, S.C. Tadler SC. Long-Term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. The American Naturalist. 1991;138:1315–1341. [Google Scholar]
- 35.van Opijnen T, Boerlijst MC, Berkhout B. Effects of Random Mutations in the Human Immunodeficiency Virus Type 1 Transcriptional Promoter on Viral Fitness in Different Host Cell Environments. Journal of Virology. 2006;80:6678–6685. doi: 10.1128/JVI.02547-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 37.Marcusson L, Frimodt-Møller N, Hughes D, et al. Interplay in the Selection of Fluoroquinolone Resistance and Bacterial Fitness. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000541. e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.