Abstract
Structural characterization of the HIV envelope protein gp120 is very important to provide an understanding of the protein's immunogenicity and it's binding to cell receptors. So far, crystallographic structure determination of gp120 with an intact V3 loop (in the absence of CD4 co-receptor or antibody) has not been achieved. The third variable region (V3) of the gp120 is immunodominant and contains glycosylation signatures that are essential for co-receptor binding and viral entry to T-cells. In this study, we characterized the structure of the outer domain of gp120 with an intact V3 loop (gp120-OD8) purified from Drosophila S2 cells utilizing mass spectrometry-based approaches. We mapped the glycosylation sites and calculated glycosylation occupancy of gp120-OD8; eleven sites from fifteen glycosylation motifs were determined as having high mannose or hybrid glycosylation structures. The specific glycan moieties of nine glycosylation sites from eight unique glycopeptides were determined by a combination of ECD and CID MS approaches. Hydroxyl radical-mediated protein footprinting coupled with mass spectrometry analysis was employed to provide detailed information on protein structure of gp120-OD8 by directly identifying accessible and hydroxyl radical-reactive side chain residues. Comparison of gp120-OD8 experimental footprinting data with a homology model derived from the ligated CD4/ gp120-OD8 crystal structure revealed a flexible V3 loop structure where the V3 tip may provide contacts with the rest of the protein while residues in the V3 base remain solvent accessible. In addition, the data illustrate interactions between specific sugar moieties and amino acid side chains potentially important to the gp120-OD8 structure.
HIV-1 (Human immunodeficiency Virus) envelope protein gp120 can recognize the CD4 cell receptor and initiate viral entry into cells. Epitopes from gp120 can generate neutralizing antibodies such that antigens derived from gp120 have been examined as potential candidates for development of an HIV-1 vaccine. Structural characterization of gp120 is very important to understanding its immunogenicity and antigenicity. However, crystallographic structures for free glycosylated gp120 that include all its variable loops have been extremely difficult to achieve. Removal and/or restriction of flexible parts of proteins (including envelope glycoproteins) can enhance the overall probability of crystallization 1, 2. Therefore, several strategies have been used to facilitate crystallization of human and simian gp120; including binding to CD4 or gp120 specific antibodies 2-5, deglycosylation of gp120 2, 3, or truncation of gp120 (v1-v2 or v3) loops 4-6. However, the extensive glycosylation (~50%) and the variable polypeptide chain-loops of gp120 are critical factors for defining molecular recognition in the immune response 7, thus it is very important to obtain structural information relevant to glycosylated HIV-1 gp120 protein forms than include variable loops of interest.
Glycosylation is a very common post translational modification in higher eukaryotes and glycoproteins usually include multiple glycoforms carrying between one and dozens of different glycans with varying degrees of site occupancy. Glycosylation linked to asparagine residues in the consensus motif Asn-X-S/T (X can be any amino acid except proline) via an N-acetylglucosamine (GlcNac) linkage are called N-linked glycans. N-linked glycans are distributed among three subtypes based on their nature and location. High mannose (oligomannose) structures are composed of mannose residues attached to the core structure with composition of Man5-9GlcNac28. A second type contains N-acetyllacosamine (Galb1–3/4GlcNac) in their antenna region while a third type is represented by hybrid structures that are composed of mannose, fucose and N-acetyllacosamine attached to a trimannosyl chitobiose core. The first two categories are the most common structure for N-glycans. Glycosylation in envelope glycoproteins are mainly composed of N-linked high mannose type oligosaccharides, which control proper folding and conformational stability of the protein 9. Precise characterization of sugar occupancy via glycoproteomics analysis is challenging due to the complex structures, labile nature, and microheterogeneity inherent in glycoproteins. A comprehensive analysis typically involves three key tasks: identification of peptides/proteins, glycosylation site mapping, and evaluation of glycosylation types. Mass spectrometry is a powerful and efficient technique for completing all three tasks. Glycoprotein identification and site localization are simplified by enzymatic removal of glycans, such as peptide-N-glycosidase F (PNGase F) deglycosylation, followed by mass spectrometry 10-16 while stable isotopic labeling can be combined with enzymatic treatment to quantitate the glycosylation occupancy 14. Endo-beta-n-acetylGlu-Cosaminidase H (Endo H) treatment before mass spectrometry analysis can be used to evaluate the glycan types 17, 18 and quantitate high mannose glycosylation occupancy 15 as high mannose and hybrid structures are sensitive to Endo H treatment. Because of the labile nature of glycans, peptide identification can be challenging using traditional methods of fragmentation, such as collision-induced dissociation (CID). Therefore, alternative dissociation techniques, Electron Capture Dissociation (ECD) 19, 20 or Electron Transfer Dissociation in conjunction with CID 21 have been used to provide complementary information both on peptide sequence and glycan structure.
To develop approaches for structural analysis of gp120 constructs intended as antigens for evaluating immune responses, we characterized the glycosylation of a recombinant construct composed of the outer domain of gp120 including amino acid residues 260 to 485 (gp120-OD8) that contains the structurally important V3 loop using mass spectrometry-based approaches. We used PNGase F and Endo H to deglycosylate the protein. CID and ECD MS were combined to identify peptide sequences generated by digestion using specific proteases as well as glycan structures of gp120-OD8. A relative quantitation method combining Endo H, PNGase F treatment and LC-MS/MS was employed to estimate the degree of occupancy at each glycosylation site for high mannose and / or hybrid glycans of gp120-OD8 15.
In addition, we employed synchrotron protein footprinting, a mass spectrometry-based protein structure analysis technique, to provide detailed information on the solvent accessibility of the side chains in the gp120-OD8 structure. Hydroxyl radicals generated from radiolysis of water using millisecond pulses of a synchrotron x-ray beam directly attack accessible and reactive side chain residues of proteins in solution 22-27. After synchrotron radiolysis, oxidized side chain products can be quantified and detected by LC-MS/MS, providing measures of surface accessibility for specific side chains in the protein structure. This technique has been successfully applied to structural elucidation of proteins and protein interactions 22-30. Apparent differences in surface accessibility between this construct and surface accessibility predicted from a homology model generated from a gp120 crystal structure bound to CD4 were used to define ligand dependent variations in structure in the V3 loop region.
EXPERIMENTAL
Construction, expression and purification of gp120-OD8
A gene segment encoding the outer domain of gp120 (a.a. 260 to 485) was PCR-amplified from codon-optimized M-group consensus envelope sequence (MCON6)31. The amplified DNA fragment was cloned into pMT/Bip/V5-his vector (Invitrogen, Carlsbad, CA) between BglII and PmeI sites to yield pMT/Bip/gp120-OD8. The recombinant protein contains Bip signal sequence at the N-terminus to allow secretion and 6xHis tag at the c-terminus to facilitate purification.
S2 cell line stably expressing gp120-OD8 protein was generated according to manufacturer's protocol. Briefly, exponentially growing S2 cells were co-transfected with the pMT/Bip/gp120-OD8 and pCoBlast plasmids using the calcium phosphate method. Transfected cells were maintained at 27 °C in Schneider's Drosophila medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal Bovine serum (Hyclone, Logan, UT), penicillin/streptomycin (50 units/ml and 50 μg/ml, respectively), and blasticidin (25 μg/ml) for selection. Protein expression was induced using 10 mM Cadmium chloride (CdCl2).
Expressed gp120-OD8 secreted into cell culture medium, was purified using tandem affinity chromatography. First, gp120-OD8 was enriched from culture supernatant using Con A sepharose (GE health, Inc). gp120-OD8 was eluted from the Con A sepharose column using 500 mM Methyl α-D-mannopyranoside, Tris-Hcl pH 7.4, 500 mM NaCl. gp120-OD8 was further purified using Ni-NTA column (Qiagen, Inc, Valencia, CA). After washing the column with 20 mM Tris-HCl (pH 8.0), 500 mM NaCl and 5 mM imidazole, gp120-OD8 was eluted using the same buffer containing 250 mM imidazole. The eluted protein was dialyzed in Tris-HCl (pH 8.0), 50 mM NaCl. Finally, the protein was loaded into Q-Sepharose column. The protein, which was in the flow-through fraction, was concentrated by Amicon Ultra concentrator (Millipore, Billerica, MA), and was stored frozen at -80°C.
Evaluation of interactions between gp120-OD8 and IgG1 b12 by surface plasmon resonance (SPR)
SPR analyses were performed with Biacore 3000 (Biacore, Columbia, MD) at room temperature in HBS-P running buffer (0.01M HEPES pH 7.4, 0.15 M NaCl, 0.005% v/v surfactant P20). IgG1 b12 was covalently bound to the sensor CM5 Chip (Biacore, Columbia, MD) via carboxyl moieties on the dextran by the standard primary amine coupling method, and the resonance signal reached about 1300 resonance units (RUs). IgG1 b12 was diluted in 10 mM glycine-HCl buffer (pH 3.0) to a final concentration of10 μg/ml. A reference surface was prepared by activating and blocking a flow cell in the absence of IgG1 b12. For kinetic measurements of gp120-OD8 binding to immobilized IgG1 b12, sensorgrams were obtained by passing various concentrations of gp120-OD8 (1 nM-100 nM) over sensor surface at a flow rate of 10 μl/min using 2-min association phase and 2 min dissociation phase. The specific binding profiles of gp120-OD8 to the immobilized IgG1 b12 were obtained after subtracting the response signal from the reference flow cell. Binding kinetics of each protein was evaluated using BiaEvaluation software (Biacore, Columbia, MD) based on 1:1 Langmuir binding model.
Deglycosylation
Complete deglycosylation of gp120-OD8 was achieved using PNGase F (New England Biolabs, Ipswich, MA) treatment while Endo H (New England Biolabs, Ipswich, MA) treatment yields partially deglycosylated protein. Briefly, 1-2 μl of 1 μg/μl gp120-OD8, 1 μl glycoprotein denaturing buffer (5% SDS, 0.4 M DTT) and HPLC water were mixed to make a total of 10 μl reaction buffer. The gp120-OD8 reaction buffer was heated at 95 °C for 10 min followed by cooling on ice. After the addition of 2 μl of 0.5 M sodium phosphate (pH 7.5), 2 μl 10% NP-40, 1 μl PNGase F and 5 μl HPLC water, the gp120-OD8 reaction buffer was then subjected to deglycosylation reaction at 37 °C for 3 hours. Similarly, gp120-OD8 was denatured by the glycoprotein denaturing buffer and heated at 95 °C for partially deglycosylation reaction by use of Endo H. After the addition of 2 μl of 0.5 M sodium citrate (pH 5.5), 1 μl Endo H and 7 μl HPLC water, the gp120-OD8 reaction buffer was then subjected to partial deglycosylation at 37 °C for 3 hours. Prior to proteolysis, deglycosylated gp120-OD8 was purified by TCA precipitation to remove detergent, sugars and small molecules. 12.5 μl of TCA solution was added to 20 μl gp120-OD8 reaction buffer, and incubated on ice for 30 min followed by two washes of 200 μl ice-cold acetones. Deglycosylated gp120-OD8 was then vacuum dried and reconstituted in 10 μl of 100 mM ammonium biocarbonate buffer (pH 7.5).
Determination of deglycosylation efficiency by electrophoresis
After deglycosylation by Endo H and PNGase F, gel electrophoresis was used to monitor overall deglycosylation efficiency. 100 ng each of gp120-OD8, Endo H treated gp120-OD8 and PNGase F treated gp120-OD8 were loaded onto a 4%-20% SDS-PAGE gradient gel (Invitrogen, Carlsbad, CA). After electrophoresis, gel was fixed with 50% methanol/10% acetic acid for 10 min, washed with distilled water for 1 hour, rocked in dithiothreitol (DTT: 32.5 μl of 0. 1 M DTT in 100 ml water) for 10 min, stained with silver nitrate for 15 min, and distained in developer (6 g sodium carbonate and 100 μl formaldehyde in 200 ml water) for 5-10 min, which was quenched by addition of 10 ml of 2.3 M citric acid.
Proteolysis of glycoproteins
gp120-OD8 and deglycosylated gp120-OD8 were subjected to proteolysis using sequencing-grade modified Trypsin (Promega Biosciences, Madison, WI), Asp-N and GluC (Roche Diagnostics, Penzberg, Germany). Endo H deglycosylated gp120-OD8 was digested by three different enzymes: Trypsin, GluC and ChymoTrypsin (Roche Diagnostics, Penzberg, Germany). Before digestion, 1~2 μg gp120-OD8 was denatured by adding 10 μl of 50% ACN and heating at 95 °C for 10 min. Samples were reduced in 2.5 mM DTT at 56°C for 15 min and subsequently alkylated in 10 mM iodoacetamide in the dark at 37 °C for 30 min. DTT concentration was adjusted to 11 mM to reduce excessive iodoacetamide at 37 °C for 30 min. gp120-OD8 or deglycosylated gp120-OD8 samples were then diluted 10 times by 100 mM ammonium biocarbonate and subjected to proteolysis at a protease-to-protein ratio of 1:20 (w:w) at 37 °C for 12 hours. Digestions where GluC and Trypsin were used in conjunction, the protein was initially digested with GluC for 2 hours at room temperature with gentle shaking. Next, GluC was deactivated by heating the sample to 95 °C for 10 min and cooled on ice. Finally, Trypsin was added to the sample for overnight digestion at 37 °C. Digestion was terminated by addition of 0.1% (v: v) trifloroacetic acid (TFA). The digests were vacuum dried and desalted by C18 Ziptip before mass spectrometry analysis.
Synchrotron X-ray radiolysis
Synchrotron X-ray radiolysis was performed at beamline X28C of the National Synchrotron Light Source at the Brookhaven National Laboratory (Upton, NY). Prior to radiolysis, protein samples were dialyzed against 10 mM sodium cacodylate buffer (Electron Microscopy Science, Hatfield, PA), pH 7.4 at 4 °C. Exposure conditions were optimized by following the dose-dependent degradation of the fluorescent compound Alexa 488 (Invitrogen, Carlsbad, CA) in the presence of the sample buffer 26. An initial set of gp120-OD8 samples (3.5 μM, 5 μl volume) was exposed to mirror focused X-rays 32 by use of a multiple sample holder for 0 to 30 ms (0, 10, 20, and 30) at a beam current of 276 mA. In order to access shorter exposure times, all subsequent samples (1.7 μM, 50 μl) injection volume) were exposed using the KinTek (Austin, TX) stopped flow apparatus. Two additional sets of gp120-OD8 were then exposed to X-ray for 0-20 ms (0, 5, 10, 15, 20) at beam currents of 281 and 258 mA, respectively. After radiolysis, Met-NH2 (pH 7.0) was immediately added to a final concentration of 10 mM to avoid secondary oxidation 24. The samples were then flash frozen in liquid nitrogen and stored at -80 °C until digestion.
Liquid chromatography - Tandem mass spectrometry
MS Experiments were carried using: an LTQ-FT mass spectrometer (Thermo Finnigan, San Jose, CA). Nano-Reverse Phase Liquid Chromatography (RPLC) separations were performed on a Dionex Ultimate U3000 HPLC (Dionex, Sunnyvale, CA) with a 5 cm × 75 μm Pico Frit C18 column (New Objective, Woburn MA) directly connected to a New Objectives nanospray emitter (10 μm, New Objectives, Woburn, MA). Chromatography was performed using mobile phase A (0.1% formic acid in water) and B (80% acetonitrile, 0.04% formic acid in water) with a linear gradient of 1% per min, starting with 100% of solution A at a flow rate of 0.3 μl/min. All data were acquired in positive ion mode. CID (collision induced dissociation) and/or ECD (electron capture dissociation) were used to fragment peptides in FT-MS. In FT-MS. In these experiments, full MS scans (m/z 300-2000) were followed by eight subsequent MS2 scans on the top eight most abundant peptide ions using a normalized collision energy of 35%. In CID or ECD mode, full MS spectra were acquired from the ICR cell at a resolution of 100,000. In ECD and CID mode, full MS spectra were acquired from the ICR cell at a resolution of 25,000 to reduce the time required per scan. For each precursor ion, MS/MS data spectra were acquired by both CID and ECD scans. MS/MS datadependent CID scans were obtained from the LTQ, while the ECD spectra were produced in ICR cell at an electron energy of 1.7-2.7 eV.
Mass spectrometry data search
Tandem MS data were analyzed by use of MassMatrix 33, 34 and Mascot 35 software. gp120-OD8 tandem MS data were searched against a protein database containing the consensus sequence, MCON6 for gp120-OD8, sequences of human keratins, proteolytic enzymes used such as Trypsin or GluC and decoy sequences comprised of a reversed sequences of all the proteins listed in database. Carbamidomethylation of cysteine was included as a fixed modification. For glycosylation site identification, acetylation on the N-terminus and lysine, deamindation on asparagine and glutamine (after PNGase F treatment) or glycosylation that has a 203 Da mass shift (after Endo H treatment) were included as a variable modifications. For gp120-OD8 footprinting, acetylation on the N-terminus and lysine, deamindation on asparagine and glutamine and all possible oxidation modifications 25 were included as variable modifications. The enzyme was defined to cleave the protein after lysine, arginine, asparatic acid, glutamic acid and asparagine when Trypsin and GluC were used for digestion. When Asp-N was used for proteolysis, asparagine was included as one of the cleavage sites. Four missed cleavages were allowed for searching the data. The mass tolerance was set to 10 ppm for the precursor ion search and 1 Da for the product ion.
Calculation of glycosylation fraction and oxidation rates
The fraction of glycosylated peptides was calculated from the ratio of the chromatography area under the ion signals for the modified peptides to the sum of the glycosylated peptides and unmodified peptides. The fraction of unoxidized peptides was calculated from the ratio of the chromatography area under the ion signals for the unoxidized peptides to the sum of the unoxidized peptides and their radiolytic products 25, 30, 36. The fraction of unmodified peptide at each time point was normalized as the fraction unmodified at 0 ms is 1.0. The fraction unmodified peptide was fit to the equation Y=Y0.e-kt. Y and Y0 are the fraction of unmodified peptides at t and 0 ms, k is a first order rate constant. Dose response curves are presented as unmodified fraction versus exposure time points. The oxidation constant rate k and fitted dose response curves were calculated by ProtMapMS 36 and confirmed by manual calculation.
Structure simulation of gp120-OD8 from homology
The homology model of gp120-OD8 was built using Swiss-Model server 37-39, an automated comparative protein modeling server using residues 260 to 485 translated from the MCON6 sequence. The homology model was generated using a template of HIV-1 JR-FL gp120 core protein containing the V3 loop in complex with human CD4 and the X5 antibody (PDB ID: 2B4C) 3, which shares 76.4% sequence identity with the MCON6 gp120-OD8. The ligands were removed in the modeling. The homology model generated from this template included all but the first residue.
Side chain solvent accessibility calculation
The MSMS V 2.5 software developed by Michel F. Sanner et al was used to calculate the solvent accessible surface areas of all side chains (Å2) 40. The homology model of gp120-OD8 3 was used for this analysis.
RESULTS AND DISCUSSION
Evaluation of binding efficiency of gp120-OD8 to antibody b12
In order to examine gp120-OD8 interactions with IgG1 b12, which is an antibody specific for the CD4 binding site of gp-120, we analyzed kinetics of b12 binding to gp120-OD8 using SPR. IgG1 b12 was immobilized on a CM5 chip and gp120-OD8 solutions at various concentrations were allowed to flow over the surface. The kinetic curves (Figure 1) of gp120-OD8 were relatively homogeneous indicating a single type of binding site. The equilibrium dissociation constant (KD) of gp120-OD8 inferred from these data was 113 nM, which is about 3.9 × lower than that (438 nM) of an outer domain protein of gp120 (OD1, aa: 252-482) 41. The binding data for gp120-OD8 to the b12 antibody indicates an intact and native-like tertiary structure, at least with respect to b12 binding. The observation that OD1 was “unable” to bind to CD4 but can bind to dodecameric CD4 while gp120 core can bind to CD4 suggested that CD4 binding to gp120 need either the avidity of the cell wall or the inner domain to enhance the stability of outer-domain gp120-CD4 complex 41. Therefore, CD4 binding was not used to test the native structure of gp120-OD8 here.
Figure 1.
SPR kinetic analysis of binding of mAb IgG1b12 to gp120-OD8. mAb IgG1b12 was immobilized on a CM-5 chip, and gp120-OD8 at various concentrations was passed over the chip surface.
Identification of N-linked glycans of gp120-OD8
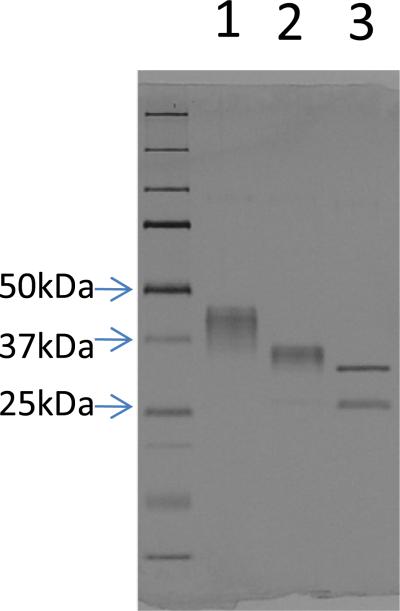
gp120-OD8 was treated with PNGase F and Endo H to examine the glycosylation of the purified protein. Deglycosylation efficiency was examined using gel electrophoresis (Figure 2). The apparent molecular mass of gp120-OD8 ranges from 48 kDa to 35 kDa (lane 1), which results from the heterogeneous glycoforms of gp120-OD8, while the apparent molecular mass of gp120-OD8 decreased to 30 kDa (lane 2) after partial deglycosylation by Endo H treatment. After complete deglycosylation by PNGase F treatment, the apparent molecular mass of gp120-OD8 decreased to 25 kDa, reflected by the bottom band in lane 3 (the upper band in Lane 3 is PNGase F, Mw: 36 kDa).
Figure 2.
4%-20% SDS-PAGE gel of gp120-OD8 (Lane 1), Endo H treated gp120-OD8 (Lane 2) and PNGase F treated gp120-OD8 (bottom band in Lane 3). Upper band in Lane 3 is PNGase F (Mw: 36 kDa)
Trypsin, GluC or Asp-N were used to digest the treated and untreated gp120-OD8. Protein digests were separated by nano-LC and detected by a high mass accuracy FT ICR/ion-trap hybrid mass spectrometer. The MS dataset was subjected to MASCOT and MassMatrix MS database searches against consensus gp120-OD8 sequence MCON6. Thirty-two unique peptides were detected from digestion with Trypsin resulting in 78% sequence coverage while forty unique peptides were detected from digestion with Asp-N resulting in 87% sequence coverage. Trypsin + GluC revealed 35 unique peptides and 85% sequence coverage, with the additional identification of peptides 102-114, 115-125, 112-130, 126-136, 137-151 due to digestion by GluC. The total sequence coverage using all the digestion methods is 100%.
After PNGase F treatment, the glycosylasparagine residues in the protein sequence lose their sugar moieties and the side chain is converted to aspartate 42, 43. Therefore, the mass difference between asparagine and aspartate (Δm = 0.9847 Da) was used to identify modification of asparagine in the database search and to localize the glycosylation sites. Twenty unique Asp-N digests from gp120-OD8 were identified as possible glycosylated peptides after PNGase F treatment. It was determined that all fifteen asparagines with a glycosylation motif converted to aspartate. However, deamindation of asparagine in vivo and in vitro results in loss of the amino ammonia group and formation of aspartate, thus deamindation can result in false positive identification of a glycosylation site 44-46. Five asparagines with a glycosylation motif were determined to be deamindated before PNGase F treatment based on analysis of untreated gp120-OD8. The non-enzymatic deamindation rate of Asn and Gln is dependent on the peptide sequence, buffer type, pH, temperature and ionic strength 46. Peptide sequences containing an “AsnGly” were found to be especially unstable 47. In this study, peptide 1-15 “RSTQLLLNGSLAEEE” containing “NG” sequence in the untreated sample exhibited a deamindation ratio greater than 70%. Therefore, identification of the glycosylation sites and use of PNGase F treatment is only used as one point of reference in the analysis of MS data of glycosylated gp120-OD8.
Endo H cleaves the chitobiose core of high mannose sugars and some hybrid oligosaccharides from N-linked glycoproteins 43. Because the recombinant proteins expressed in insect cells mainly contain high mannose sugars, removal of glycosylation using Endo H is reasonable 48-50. An N-acetylglycoseamine group remains in the asparagine after Endo H treatment which results in 203.0794 Da mass shift in the mass spectrum compared to the mass of asparagine. Therefore, 203.0794 Da was used as an indicator of asparagine modification after Endo H treatment. Glycosylasparagine sites were localized by matching mass spectra of unmodified and modified peptides unambiguously as shown in Figure 3a, 3b and Supplementary Figure 1. The mass spectra of modified and unmodified peptides showed similar y ion and b ion distribution patterns with corresponding mass shift resulting from the modification in the spectra of modified peptides. Eleven glycosylated sites were identified with a mass shift of 203.0794 Da as listed in Table 2. Neutral loss peaks of 203 Da from precursor ions in the MS/MS were observed (Figure 3a). The intensity of a neutral loss peak is usually greater than product ion that contains glycans because the labile glycosidic bonds are more easily fragmented than the peptide backbone in mass spectrometry (Figure 3a). Due to inefficient cleavage caused by the glycan groups remaining after Endo H treatment, some long peptides were observed with 2 missed cleavages. In order to obtain better sequence coverage and more accurate identification, four different digestion methods (Trypsin, Chymotrypsin, GluC-Trypsin, GluC) were used. Multiple charge states of mass spectra were detected from GluC protelytic digests with several lysine residues in the sequence. The occupancies of high-mannose or hybrid glycosylation were assessed based on the chromatographic area under the MS signal of modified species and unmodified species. The quantitative glycosylation mapping was generated by calculating the fraction of the chromatographic peak areas of modified peptides versus total peptides 15 as shown in Figure 4. In summary, eleven glycosylation sites out of fifteen predicted (possible) sites were identified. Replicates or triplicates were analyzed by mass spectrometry. The data were highly reproducible. Glycosylation occupancy of different sites in gp120-OD8 varies from 0.8% to 99.2% as listed in Table 1 and Figure 5. In some cases, we were unable to deconvolute the occupancy of the individual sites from Endo H treated samples. For example, the total glycosylation occupancies of two asparagine sites: Asn 77 and Asn 84 on peptide 67-96 were 77.2%. Therefore, PNGase F treated gp120-OD8 data were used to isolate the occupancy of the individual sites. As discussed in previous paragraph, in vivo or in vitro deamindation were detected in fully glycosylated gp120-OD8 before PNGase F treatment. Therefore, data of fully glycosylated gp120 was used to verify the accuracy of PNGase F treated data. Because no deamindation was observed for Asn 77 in fully glycosylated samples, glycosylation occupancy of Asn 77 on peptide 73-80 was determined to be 48.8% based on PNGase F treated samples. Asn 84 was deamindated in fully glycosylated gp120-OD8. Therefore, glycosylation occupancy on Asn 84 was calculated to be 28.4% by subtracting the glycosylation occupancy of Asn 77 (48.8%-PNGase F treated data) from the total glycosylation occupancies of Asn 77 and Asn 84 (77.2%-Endo H treated data) (Table 1).
Figure 3.
Identification of glycosylation sites using Endo H treatment. 3a is MS/MS spectrum of the glycopeptide precursor ion at m/z of 931.9750 Da. 3b is MS/MS spectrum of unmodified peptide of the same sequence (RSTQLLLNGSLAEEE [1-15]).
Table 2.
List of 8 unique glycopeptides containing 9 glycosylation sites identified by the CID MS and ECD MS
| Peptide sequence [residue range] | Glycosylation Sites | Glycan moieties | Theoretic Mw (Da) | Observed Mw (Da) |
|---|---|---|---|---|
| IIIRSENITNNAK[16-28] | N22 | (GlcNac)2(Man)3(Fuc)1 | 2524.2132 | 2524.2077 |
| TIIVQLNE[29-36] | N35 | (GlcNac)2(Man)3 | 1821.8474 | 1821.8445 |
| INCTRPNNNTR[40-50] | N41 and N47 | (GlcNac)2(Man)x + (GlcNac)2(Man)9-x | 3766.5055 | 3766.4984 |
| KSIHIGPGQAFYATGEIIGDIRQAHCNISR[51-80]+alkylation of C76 | N77 | (GlcNac)2(Man)9 | 5174.3534 | 5174.3134 |
| TKWNKTLQQVAKK[81-93] | N84 | (GlcNac)2(Man)7 | 3113.4504 | 3113.4368 |
| AceKLREHFNNKTIIFKPSSGGDLE[93-114] | N99 | (GlcNac)2(Man)3(Fuc)1 | 3610.7333 | 3610.7422 |
| SNITGLLLTR[190-199] | N191 | (GlcNac)2(Man)3(Fuc)1 | 2126.0221 | 2126.0184 |
| DGGNNSNKNKTETFRPGGGDMRDNWR[200-225] | N208 | (GlcNac)2(Man)3(Fuc)1 | 3961.6993 | 3961.6908 |
Figure 4.
Quantiation of glycosylation occupany of Asn 8 in peptide 1-15 (RSTQLLLNGSLAEEE) by use of Endo H treatment. Top: Chromtography peak of unmodified peptide 1-15. Peak area was calculated m/z ranging from 830.4260-830.4360. Bottom: Chromtography peak of glycopeptide 1-15. Peak area was calculated as m/z ranging from 931.9700-931.9800.
Table 1.
Overview of glycopeptides containing high mannose or hybrid structure identified by Endo H and PNGase F treatment and mass spectrometry
| Peptide sequence [residue range] | Glycosylation sites | Occupancy |
|---|---|---|
| Endo H deglycosylation | ||
| RSTQLLLNGSLAEEE [1-15] | N8 | 0.8%, 0.7%, 1.3% |
| IIIRSENITNNAKTIIVQLNE [16-36] | N22 | 98.3% , 100% |
| IIIRSENITNNAKTIIVQLNE [16-36] | N22 and N35 | 18.8%, 23.1% |
| TIIVQLNE [29-36] | N35 | 23.5%, 21.0% |
| SVEINCTRPNNNTR [37-50] | N41 | 97.8% |
| SVEINCTRPNNNTR [37-50] | N41 and N47 | 10.5% |
| SVEINCTRPNNNTRKSIHIGPGQAFYATGE [37-66] | N41 or N47 | ~100% |
| SVEINCTRPNNNTRKSIHIGPGQAFYATGE [37-66] | N41 and N47 | 10.8% |
| IIGDIRQAHCNISRTKWNKTLQQVAKKLRE [67-96] | N77 or N84 | 77.2%, 74.4% |
| HFNNKTIIFKPSSGGDLE [97-114] | N100 | 15.3%, 12.7% |
| FYCNTSGLF [127-135] | N130 | 94%, 100% |
| SNITGLLLTR [190-199] | N191 | 5.2%, 5.6%, 6.8% |
| DGGNNSNKNKTETFRPGGGDMR [200:221] | N208 | 67.7% |
| GKITCKSNITGLLLTRDGGNNSNKNKTE [184-211] | N208 | 75%, 61.2% |
| PNGase F deglycosylation | ||
| SVEINCTRPNNNTR [37-50] | N41 and N47 | N41 and N47 10.1%, 17.2% |
| QAHCNISR [73-80] | N77 | 42.1%, 55.5% |
| GTYMFNGTR [143-151] | N148 | 92%, 100% |
| STWMFNGTYMFNGTR [137-151] | N142 and N148 | 82.7% |
| SNITGLLLTR [190-199] | N191 | 39.0%, 43.3% |
Figure 5.
Quantitative glycosylation occupancy of gp120-OD8. Glycosylation of 11 sites vary from 0.8% to 99.2%. N191 might contain two types of glycosylation, N136, N142 and N148 were potential glycosylation sites but not identified in Endo H treatment. No glycosylation on N 203 was detected.
Peptide 37-50 in trypsin-digested Endo H treated sample and peptide 37-66 in GluC-digested Endo H treated sample were detected. Glycosylation occupancies of both peptide 37-50 and peptide 37-66 containing one glycosylation site were determined to be greater than 97.8% (97.8% and ~100%). Glycosylation occupancies of both peptide 37-50 and peptide 37-66 containing glycosyl Asn 41 and glycosyl Asn 47 were about 11% (10.5% and 10.8%). Asn 41 was localized as the glycosylated site in the glycosylated peptide 37-50 containing one glycosylation site. Therefore Asn 41 was determined to be highly glycosylated (97.8%) and another site Asn 47 is partially glycosylated (~11%). Additionally, these data are consistent with those of PNGase F treated samples. Peptide 190-199 was determined to be occupied with 5.9% high mannose or hybrid glycosylation in Endo H treated samples and 41.2% in PNGase F treated samples. Only 0.7% deamindation percent were observed in fully glycosylated gp120-OD8 samples. Therefore, this site might be occupied with two different types of glycosylation: 35.3% (=41.2%-5.9%) complex glycosylation and 5.9% high mannose/or hybrid glycosylation. Peptides containing Asn 136, Asn 142 and Asn148 were not detected in the Endo H treated sample and fully glycosylated samples. Heavy glycosylation and different glycosylation types on the three Asn sites may result in the undetectability of the peptides containing these Asn sites in fully glycosylated samples and Endo H treated samples. Among them Asn 142 and Asn 148 containing “NG” motif might undergo a high degree of deamindation in the sample preparation process. The sequence (aa. 126 to 156) was not detected in fully glycosylated gp120-OD8 which makes the determination of occupancy for these three potential glycosylation sites difficult although they were identified to be deamindated in PNGase F treated samples. No glycosylation of Asn 203 having NxS motif was identified in Endo H treated samples. A quantitative glycosylation map is shown in Figure 5 based on above data and analysis.
Composition analysis of glycosylation by ECD and CID MS
Glycopeptides fragment in a characteristic manner when subjected to CID. The labile glycosidic bonds of the carbohydrate moiety are much more easily fragmented than the peptide backbone (Figure 6a). Thus it is hard to obtain backbone sequence information of the glycopeptide CID MS/MS dominated by glycan neutral loss signals. Fragmentation using ECD is lower in energy and better preserves labile post-translational modifications. Therefore, ECD MS and CID MS data were used to complement each other for identifying the glycopeptides as well as interpreting the glycan moieties accurately.
Figure 6.
Identification of glycopeptides by combination of CID and ECD MS. The peptide sequence was identified as 51KSIHIGPGQAFYATGEIIGDIRQAHCNISR80, Asn 77 was identified as the glycosylation site containing 2 N-acetylgucosamine and 9Mannose. 4a is a typical CID MS/MS of the high mannose glycopeptides with many abundant neutral loss peaks and 528 Da (2 Mannose + 1 GlcNac) signature peak. 4b is ECD MS/MS of the glycopeptide providing information of peptide sequence by fragmenting peptides to C ions and Z ions.
We used two software programs to analyze mass spectra data of glycosyl-peptides. One is our in-house developed software 51, which was used to locate glycosyl-peptide mass spectra. Glycopeptide spectra generated using CID typically contain abundant neutral loss peaks; such as loss of 146 Da for fucose, 162 Da for hexose (Hex) or mannose (Man), and 203 Da for N-acetylglucosamine (GlcNac). Sugar fragment masses such as 366 Da [(GlcNac)1(Hex)1], 528 Da [(GlcNac)2(Hex)1] and 690 Da [(GlcNac)3(Hex)1] are also typical signatures of glycopeptide CID MS. We also used ProteinProspector v5.3.2 52, 53, a proteomics tool for mining sequence databases in conjunction with mass spectrometry experiments. It was used to calculate theoretical gp120-OD8 proteolytic digests based on MCON6 consensus sequence as well as to calculate theoretical CID and ECD MS product ions based on glycopeptide sequences. The type and number of the glycans were determined by calculating the mass and number of the neutral losses of product ions in the CID mass spectra of glycosyl-peptides as shown in Figure 6a and Supplementary Figure 2. The c ions and z ions in the ECD mass spectrum were used to confirm the peptide sequence as shown in Figure 6b and Supplementary Figure 2. For instance, glycopeptides with penta-charged (+5) precursor ions (m/z = 1035.6686) were detected as shown in Figure 6a. Sugar moieties were identified by multiple neutral losses of glycan signals. For example, a pattern of nine continuous “32” or “40” (m/z) differences between product ions indicated that the glycopeptides lost nine hexose moieties (m/z = 162 Da /+5 = 32.4 ± 1 or 162 Da/+4 = 40.5 ± 1). Losses of N-acetylglucosamines resulted in difference of “51” of product ions when the product ion were tetra-charged (m/z = 203 Da/+4= 50.8 ±1). It can be determined that Asp 77 firstly connected with two N-acetylglucosamines since they were fragmented (two differences of “51” in MS) following the fragmentation of nine hexoses (nine differences of “40” in MS). But from these data we are not able to unambiguously determine the connections between all the glycans. The variance is 2 ppm comparing the experimental mass with the theoretical mass of the glycopeptide calculated by addition of masses of nine hexoses and two N-acetylglucosamines to the mass of peptide 51-80, so the assignment is relatively good. This was confirmed by ECD MS (Figure 6b as the sequence “KSIHIGPQAFYA.....NISR” (peptide 51-80) through detection of Z ions and C ions from the same precursor ion. In sum, 8 unique glycosyl-peptides containing 9 glycosylation sites were identified as listed in Table 2. The glycosylation sites listed in Table 2 were confirmed to contain high mannose or hybrid glycosylation structure. These results are also consistent with the fact that the recombinant glycoproteins expressed in insect cells usually contain simple glycan types-high mannose or tri-mannose, lack of complex glycosylation 48-50. This is one feature of using insect cells to produce glycoproteins, although the insect cell system is efficient in expression of recombinant proteins with glycosylation, the occupancy may not be native.
Structural analysis of gp120-OD8
Two regions have been considered as important for gp120 co-receptor binding: i) the V3 tip, and ii) the gp120 core around the bridging sheet, the V3 base, and neighboring residues 54-56. The entire V3 loop of gp120, which includes peptides 40-50, 51-66, 52-72, 67-72 and 73-80, is an important neutralizing antibody (Nab) target for virus 57-59. The binding site for CD4 on the ligated gp120 structure is formed by the interface between the inner domain, bridging sheet and outer domain 4. The bridging sheet of gp120 includes peptide 163-183. Another loop structure V4 region includes peptides 137-151, 143-151 and 152-162. The peptides that include these three regions are shown in Figure 7.
Figure 7.
Homology Model of the gp120-OD8 including V3, V4 loops and the bridging sheet. Glycosylation sites are identified experimentally by mass spectrometry. Red: high mannose glycosylation sites. Green: oxidized residues in synchrotron radiolysis. Yellow: oxidized residues in N-terminal of V3 loop in synchrotron radiolysis which should be more exposed than other residues including those on V3 tip according to footprinting data. Blue: disulfide bond in V3 loop.
Purified and glycosylated gp120-OD8 was exposed to X-rays for intervals at 0, 10, 20, 30 milliseconds or 0, 5, 10, 15, 20 milliseconds, then deglycosylated by PNGase F, and then digested by Asp-N or GluC+Trypsin. Deglycosylation of gp120-OD8 by PNGase F treatment was performed to make the samples easier to analyze. The peptide digests were separated by LC-MS and analyzed by FT-ICR mass spectrometry. The tandem mass spectra of oxidized peptides were showed in Supplementary Figure 3 and 4. Dose-response curves were calculated by plotting the fraction of unmodified peptides (on a log scale) as a function of exposure time as shown in Supplementary Figure 5. First-order rate constants for all oxidized peptides were derived from this analysis and are shown in Table 3 and Table 4.
Table 3.
Oxidation rate constant of gp120-OD8 Asp digests
| Peptide sequence [residue range] | Modified residues and its mass shift (Da) | SASA (Å2) in homology | Rate s-1 |
|---|---|---|---|
| RSTQLLLNGSLAEEEIIIRSE [1-21] | L6 (+16/14) L11(+16)R19 (+16) | R19(104.64) | 8.5 ± 1.2 |
| NNTRKSIHIGPGQAFYATGEIIG[47-69] (V3loop) | K51 I55 F61 I67 H54 (+16) | K51(134.9) I55(129.9) F61(85.8) H54(81.7) I67(137.8) | 7.4± 1.7 |
| NISRTKW [77:83] | W83 (+32) | W83(5.1) | 7.0 ± 1.4 |
| NKTIIFKPSSGG [100:111] | K101 F104 K106 (+16) | K101(56.7) F104(33.3) K106(87.0) | 1.1 ± 0.06 |
| NSTWMF [136:141] (+1) | W139 (+32 / 4) M140 F141 (+16) | W139(28.3) M140(76.2) F141(135.9) | 29 ± 8.4 |
| NTSGLFNSTWMF [130:141] | W139(+32) M140(+16) F141(+16) | W139 (28.29) M140(76.2) F141 (135.9) | 37± 7.3 |
| NGTYMF[142:147] (+1) | F141 M146 (+ 16) | F141(135.9) M146(198.0) | 23± 3.4 |
| NSTWMFNGTYMF [136:147] (+2) | W 139 (+32) M140(+32) | W139(28.29) M140(76.2) | 36 ± 4.0 |
| NGTYMFNGTR [142:151] (+2) | M146 F147 R151(+16) | M146(198.0) F147(144.0) R151(190.6) | 31 ± 1.7 |
| NITGLLLTR [191:199] | L196 or L197 (+16) | L196(0) L197(0) | 0.36 ± 0.13 |
| DGGNNSNKNKTETFRPGGGD [200:218] | K207/K209 (+16) | K207(136.6) K209(100.2) | 1.5 ± 0.05 |
| NKTETFRPGGGDMR [208:221] | P215 M220 R221(+16) F213(-30) | P215(0) M220(24.5) R221(156.0) F213(0) | 87± 11 |
Table 4.
Oxidation rate constant of gp120-OD8 GluC and Tryptic digests
| Peptide sequence [residue range] | Modified residues and its mass shift (Da) | SASA (Å2) in homology | Rate s-1 |
|---|---|---|---|
| RSTQLLLNGSLAEEE[1-15] | R1(+16/-43) L5/L6(+14/+16) L11(+14/16) E13(-30) and E14(-30) | L5 (0) L6(12.8) L11(49.7) E13(127.3) E14(132.4) | 7.7 ± 1.4 |
| IIIRSENITNNAK[16-28] | I16(+14/16) I18(+14/+16) R19(+16/-43) I23(+16) | I16(7.6) I18(21.6) R19(104.6) I23(24.5) | 3.7 ± 0.15 |
| TIIVQLNE [29:36] | I31(+14/16) | I31(19.8) | 3.9 ± 0.37 |
| INCTRPNNNTR[40-50] | I40(+16) C42(+32/16) R50 (+16) | I40(1.2) C42(4.0) R150(145.4) | 4.8 ± 1.6 |
| KSIHIGPGQAFYATGE[51:66](V3) | K51(+16) I53(+16) I55(+16) P57(+16) A60(+16) F61(+16) Y62(+16) H54(-23/-22/+16) | K51(134.9) I53(91.8) I55(129.9) P57(107.2) A60(43.6) F61(85.8) Y62(64.7) H54(81.7) | 14 ± 1.3 |
| SIHIGPGQAFYATGE[52:66] | I53(+14) I55(+16) A60(+16) F61(+16) P57(+16) Y62(+16) H54(-23/-22/+16) | I53(91.8) I55(129.9) P57(107.2) A60(43.6) F61(85.8) Y62(64.7) H54(81.7) | 7.8 ± 0.50 |
| SIHIGPGQAFYATGEIIGDIR [52:72] (V3) | I67or I68 (+16) I71 (+16/+14) R72(+16) | I67(137.8)or I68 (143.9) I71(98.9) R72(61.2) | 59 ± 8.6 |
| TKWNKTLQQVAK[81-92] | W83(+16/+32) K85(+16) | W83(5.1) K85(61.6) | 8.9 ± 0.61 |
| LREHFNNK[94-101] | H97(+16/-22) | H97(65.1) | 14± 1.9 |
| TIIFKPSSGGDLE[102-114] | I103(+16) F 105 (+16) K106(+16) P107(+16) L113(+16) | I103(4.4) F 105 (0) K106(87.0) P 107 (19.6) L113(91.4) | 2.2 ± 0.13 |
| ITTHSFNCGGE[115:125] | I115(+16) F120 (+16) C122 (+32) | I115(55.2) F120(16.8) C122 (21.3) | 2.0 ± 0.19 |
| GTYMFNGTR[143:151] | M146(+16) Y145(+16) F147(+16) R151(+16) | M146(198.0) Y145(152.7) F147(144.0) R151(190.6) | 28 ± 5.7 |
| TITLPCR[156:162] | I157(+16) L159 (+16) P160(+16)(or R162(+16)) | I157(37.9) L 159 (2.4) P160(73.3) R162(102.1) | 1.8± 0.39 |
| ITCKSNITGLLLTR[186:199] | C188(+48) K189 (+16) | C188(17.1) K189 (111.8) | 41± 2.5 |
| SNITGLLLTR[190:199] | I192(+16) L196(+16) L197(+16) | I192(0) L196(0) L197(0) | 0.20 ± 0.16 |
| QIINMWQGVGQAMYAPPIEGK[165:185] | M169 W170 V173 M177 Y178 P180 (+16) | M169(56.79) W170(127.27) V173(127.23) M177(116.98) Y178(89.4) P180(76.26) | 41± 6.4 |
| NKTETFRPGGGDMR [208-221] | P215(+16 ) M220(+16) R221(+16) | P215(0) M220(24.5) R221(156.0) | 41 ± 15 |
Forty unique peptides were detected from digestion of Asp-N (resulting in sequence coverage greater than 81%). The enzyme will cleave at Asp residues plus Asn residues that were converted to Asp after PNGase F treatment. Twelve of the forty unique peptides were seen to be oxidized (Table 3). Oxidative modification of these peptides mainly resulted in product peptides with 16 Da or 14 Da mass increases. Changes in retention time of oxidized peptides relative to unmodified peptides was observed in the chromatography due to the change of hydrophobicity after oxidation 23.
The Asp-N peptides examined had rates of oxidation ranging from 0.36 ± 0.13 s-1 for peptide 191-199 to 87 ± 11 s-1 for peptide 208-221. Asp-N can cleave at the N terminus of aspartate generated from deamindation of asparagine. In these twelve oxidized Asp-N peptides, one was N-terminal peptide (1-21) which contains partial glycosylated asparagines; one peptide's (200-218) N terminal amino acid was aspartate; five peptides’ (47-69, 77-83, 100-111, 130-141, 208-221) N termini were deamindated asparagine generated by deglycosylation; one peptide's N terminus (191-199) was deamindated asparagine which contains both in-vivo deamindation (0.7%) and deglycosylation deamindation (40.5%); four peptides’ (136-141, 142-147, 136-147, 142-151) N-termini were deamindated asparagine, but it was not determined whether these deamindations were from deglycosylation. Thus, oxidation data of Asp-N peptides (47-69, 77-83, 100-111, 130-141, 208-221) reflect solely the oxidation behavior of glycosylated peptides, while for the other peptides the population of peptides is mixed.
Trypsin can cleave proteins after lysine and arginine, which can generate unglycosylated peptides as well as glycosylated peptides of the same overall sequence if glycosylation is incomplete. Thirty-five unique peptides were detected from digestion of Trypsin and GluC (resulting in sequence coverage greater than 85%). Eighteen of the thirty-five GluC-Tryptic peptides were detected to be oxidized. Oxidation of His 54 included formation of the +16 Da and, -22 Da and -23 Da species. The -23 Da oxidation product of His has not been reported in previous protein radiolysis experiments 22-25, 30, 34, 60-65. All the oxidized GluC-Tryptic peptide mass spectra are shown in Supplementary Figure 4. The oxidation rates of the 18 peptides are listed in Table 4 and representative dose response curves are shown in Supplementary Figure 5. The overall oxidation rates for different peptides varies significantly from 0.20 ± 0.16 s-1 for peptide 190-199 to 59 ± 8.6 s-1 for peptide 52-72. Comparison of the oxidation rates of the two sets of data from Table 3 and Table 4 revealed that the same sites of oxidation detected in different peptide sequences displayed similar oxidation rates except peptide 208-221. Secondary oxidation of methionine 25 may cause the oxidation rate of peptide 208-221 in Asp-N digests (87 ± 11 s-1 to be greater than in GluC -Tryptic digests (39 ± 6.1 s-1) as higher oxidation rates (164 ± 53 s-1) were observed when samples were deglycosylated, digested and detected by mass spectrometry in experiments at different times. Although 10 mM Met-NH2 was added to suppress the secondary oxidation typically observed in sulfur-containing residues, Met-NH2 was removed from the solution after TCA precipitation, which may have contributed to the irreproducibility of the oxidation rate for this sequence.
Correlation of Footprinting and Crystallographic Data for gp120-OD8
The relative reactivity of amino acid side chains under aerobic conditions in mass spectrometry-based footprinting experiments is known to be : Cys > Met > Trp > Tyr > Phe > Cystine > His > Leu ~ Ile > Arg ~ Lys~ Val > Ser ~ Thr ~ Pro > Gln ~ Glu > Asp ~ Asn > Ala > Gly 24. Lower reactivity residues such as Asp, Asn, Ala, Gly and residues whose oxidation products are difficult to detect such as Ser and Thr often do not provide much information for protein structure analysis. There are 6 Cys, 13 Arg, 4 Trp, 4 Tyr, 10 Phe, 4 His (neglecting the 6-His tag), 13 Leu, 23 Ile, 13 Lys and 4 Val, 7 Pro residues in gp120-OD8. Additionally, we detected 2 oxidized Ala (of 8 Ala in total) by mass spectrometry after synchrotron radiolysis. In order to provide a framework for interpreting the observed sites of oxidation, the solvent accessibility of a model of our construct, based on the structure of the core protein of gp120 with V3 loop from clade B isolate (JR-FL) was employed 3. The sequence identity of the consensus sequence of recombinant gp120-OD8 (MCON6) to the counterpart in the complex of CD4/gp120 from clade B isolate (JP-FL) is 76.4% thus providing an excellent template for modeling from this perspective, however the effects of CD4 and antibody binding on the template relative to the target must be kept in mind in the analysis.
The observed oxidations were consistent overall with the surface accessibility calculations (SASA) (Table 4). For example, the solvent accessibility and likely flexibility of the V3 and V4 loop structures was observed consistent with the oxidation of peptides 40-50, 51-66, 51-72 and 67-72 (V3 loop) and peptides 137-151, 143-151 and 152-162 (V4 loop), including multiple oxidized probe sites (Tables 3 and 4 However, variations in the oxidation rates for different V3 loop peptides argued for variations in the structure across the V3 loop. For example, V3 loop peptide 52-72 (59± 8.6 s-1) had a much higher oxidation rate than V3 tip peptides 51-66 (13.7 ± 1.3 s-1), 52-66 (7.8 ± 0.5 s-1). The V3 loop peptide 47-69 resulting from Asp-N digestion also had oxidation rate of 7.4 ± 1.7 s-1. As the oxidation behavior of this peptide was influenced by the presence of sugar residues (N47), we mainly use GluC-Tryptic peptide data for the sake of comparison in this case. The predicted SASA values of residues close to C-terminus of the V3 loop (Ile 67: 137.8 Å2, Ile 68: 143.9 Å2, Ile71: 98.88 Å2 and Arg72: 61.2 Å2) are very close to those of residues in V3 tip (Lys 51: 134.9 Å2, Ile 53: 91.8 Å2, His 54: 81.7 Å2, Ile 55: 129.9 Å2, Pro 57:107.2 Å2, Phe 61: 85.8 Å2, Tyr 62: 64.7 Å2,) in the homology model 3. Since the observed reactivitites are very different, while types of residues (in terms of absolute reactivity are similar) it suggests that residues close to the C-terminal of the V3 loop, such as Ile 67, Ile 68, Ile 71, and Arg 72 (Yellow area in Figure 7), exhibit much higher accessibility than those at the tip of the V3 loop.
Thus, the conformation of V3 loop in free gp120-OD8 must be quite different from than that derived from the homology model. Part of the V3 loop of gp120, including the V3 tip might fold back in the unliganded state of gp120. Antibody or CD4 ligation to the gp120-OD8 might cause distortion of the protein tertiary structure, resulting in the V3 tip being pushed out. There is a di-sulfide bond (42-76, Blue area in Figure 7) in the V3 loop, but it is located inside the sheet, about 8 Å (2 pairs of residues) away from other oxidized residues on V3 base such as Arg 72. So the disulfide bond should not restrict the conformation of Arg 72, Ile 71, Ile 68, and Ile 67.
We performed solvent accessibility calculations for other V3 loop models including one V3 loop model based on experimental NMR data 66. In this model Ile 67, Ile 68, Ile 71 and Arg 72 from the V3 loop show relatively higher solvent accessibility values (Ile 67: 7.673 Å2, Ile 68: 58.82 Å2, Ile 71: 25.21 Å2, Arg 72: 75.86 Å2) than many residues on the V3 tip, (Lys 51: 0.131 Å2, Ile 53: 1.083 Å2, Pro 57: 31.62 Å2, Phe 61: 0.269 Å2), consistent with our overall footprinting results (Supplementary Figure 6 and Supplementary Table 1). However, there are also some discrepancies between this model and our results. The indicated SASA value of some residues differ from the observed experiment results. For example, some of the reactive residues in V3 tip such as His 54, Ile 55, Ala 60, and Tyr 62 was detected to be oxidized in radiolysis, but SASA values of these residues in this structure are 0 Å2. This may be because this V3 loop construct is an independent folding unit lacking potential interactions with other parts of the gp120-OD8.
The V3 loop is known to be important in HIV-1 immunopathogenesis 67 and the loop protrudes when CD4 and other antibodies are bound. Our footprinting data indicates a quite different structure for the V3 loop in gp120-OD8, and is consistent with observations that addition of thrombin to virion-associated gp120 results in cleavage at the V3 loop when recombinant soluble CD4 (rsCD4) is added 68-70. Moreover, without addition of an exogenous protease, proteolytic cleavage of gp120 was induced upon binding to rsCD4. This cleavage was observed most likely at the V3 loop, depending on the rsCD4 concentration and the incubation time 71.
In our construct peptides containing CD4 binding regions had varying oxidation rates consistent with their solvent accessibility calculations, these included peptide 16-28 (3.3 ± 0.21 s-1), peptide 102-114 (2.1 ± 0.13 s-1), peptide 165-189 (41 ± 6.4 s-1), and peptide 200-218 at (1.5 ± 0.05 s-1). The bridging sheet showed a high oxidation rate (41 ± 6.4 s-1) with many reactive and accessible sites, which suggested that removal of the inner domain protein sequence leaves a large surface area of solvent accessible sites. However, there are some minor exceptions to this trend. For example, the SASA values of I192, L196 and L197 in peptide 190-199 are 0 Å2 while this peptide exhibited detectible (but very low) reactivity with a rate of 0.1 s-1. Since the oxidation rate is low, it may suggest a very modest solvent accessibility or dynamic flexibility in these residues.
Influence of glycosylation on solvent accessibility
We examined the rates of oxidation for peptides containing a glycosylation motif for potential inconsistencies with results obtained from solvent accessibility calculations to explore potential attenuation of reactivity due to sugar structures. In most cases the predicted solvent accessibility did not appear to be influenced by glycosylation. However, the specific oxidation products observed for the glycopeptide in some cases reflected a conformation influenced by glycosylation. In PNGase F deamindated Asp-N peptides 77-83 and the GluC -Tryptic peptide 83-92 only +32 Da oxidation products of Trp 83 were detected. +16 Da oxidation products of tryptic peptide 83-92 were detected in unglycosylated GluC-Trypsin peptides, but it was hard to determine the exact oxidation sites (e.g. Trp 83 or Lys 85). Similarly, analysis of PNGase F deamindated Asp-N peptides 137-141 and 130-141 showed that Trp 139 was oxidized to +32 Da and +4 Da products only (Supplementary Figure 3) while the oxidized products of Trp 139 in tryptic digests were not detected. Hydroxyl radicals attack the benzene ring in tryptophan side chains to generate +16 Da products, while when the pyrrole ring is oxidized, +16 Da, +32 Da and/or +4 Da products are generated 24. These signatures of oxidation of specific structures within a side chain have been previously reported to reflect the specific conformation of the Trp side chain in protein-protein interactions 22. Thus, the sugar on residues Asn 84 and 136 adjacent to Trp 83 and Trp 139 (respectively) may preferentially protect the Trp benzene resulting in oxidation primarily of the pyrrole ring. Because Asn 84 is partially occupied by glycosylation, it is possible to detect +16 Da oxidation products in nonglycosylated peptides on Trp 83 (undeamindated peptides after enzyme deglycosylation).
CONCLUSIONS
In this study, the structure of gp120-OD8 was characterized by identifying the glycosylation sites and structure as well as monitoring the accessible residues on the protein surface. We determined glycosylation sites of gp120-OD8 and estimated the occupancy of glycosylation by enzymatically removing the glycans before mass spectrometry analysis. Eleven out of fifteen glycosylation motifs were determined as having high mannose structures or hybrid structures of glycosylation. The oligosaccharide occupancy at each site varied from 0.7% to 99%. Glycan structure for nine glycosylation sites in eight unique glycopeptides were determined by combination of ECD and CID FT-MS data. Comparison of gp120-OD8 experimental footprinting data with structural data from a homology model derived from the ligated CD4/gp120-OD8 crystal structure revealed that the V3 loop of free gp120-OD8 is solvent accessible, like the ligated gp120, but that the V3 tip may be bound to the surface of the protein leaving the base exposed. In addition, we detected specific associations between specific sugar moieties and adjacent Trp residues in the structure.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Janna Kiselar and Parminder Kaur for their discussion in this study.
Abbreviations
- HIV
human immunodeficiency virus
- V3
the third variable region
- OD
outer domain protein
- GlcNac
N-acetylglucosamine
- PNGase F
peptide-n-glycosidase F
- Endo H
Endo-beta-n-acetylGlucosaminidase H
Footnotes
The study was funded by the National Institutes of Health (P01-AI-074286). Use of the X28C beamline at the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the NIBIB (P30-EB-09998) and the U.S. department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
SUPPORTING INFORMATION AVAILABLE
Supplementary Figures and Tables are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ostermeier C, Iwata S, Ludwig B, Michel H. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat Struct Biol. 1995;2(10):842–846. doi: 10.1038/nsb1095-842. [DOI] [PubMed] [Google Scholar]
- 2.Kwong PD, Wyatt R, Desjardins E, Robinson J, Culp JS, Hellmig BD, Sweet RW, Sodroski J, Hendrickson WA. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J Biol Chem. 1999;274(7):4115–4123. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- 3.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8(12):1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 8.Morelle W, Canis K, Chirat F, Faid V, Michalski JC. The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics. 2006;6(14):3993–4015. doi: 10.1002/pmic.200600129. [DOI] [PubMed] [Google Scholar]
- 9.Fenouillet E, Gluckman JC, Jones IM. Functions of HIV envelope glycans. Trends Biochem Sci. 1994;19(2):65–70. doi: 10.1016/0968-0004(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 10.Albach C, Damoc E, Denzinger T, Schachner M, Przybylski M, Schmitz B. Identification of N-glycosylation sites of the murine neural cell adhesion molecule NCAM by MALDI-TOF and MALDI-FTICR mass spectrometry. Anal Bioanal Chem. 2004;378(4):1129–1135. doi: 10.1007/s00216-003-2383-2. [DOI] [PubMed] [Google Scholar]
- 11.Morelle W, Faid V, Chirat F, Michalski JC. Analysis of N- and O-linked glycans from glycoproteins using MALDI-TOF mass spectrometry. Methods Mol Biol. 2009;534:5–21. doi: 10.1007/978-1-59745-022-5_1. [DOI] [PubMed] [Google Scholar]
- 12.Picariello G, Ferranti P, Mamone G, Roepstorff P, Addeo F. Identification of N-linked glycoproteins in human milk by hydrophilic interaction liquid chromatography and mass spectrometry. Proteomics. 2008;8(18):3833–3847. doi: 10.1002/pmic.200701057. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5(6):1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 15.Zeng C, Biemann K. Determination of N-linked glycosylation of yeast external invertase by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 1999;34(4):311–329. doi: 10.1002/(SICI)1096-9888(199904)34:4<311::AID-JMS773>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Khoshnoodi J, Hill S, Tryggvason K, Hudson B, Friedman DB. Identification of N-linked glycosylation sites in human nephrin using mass spectrometry. J Mass Spectrom. 2007;42(3):370–379. doi: 10.1002/jms.1170. [DOI] [PubMed] [Google Scholar]
- 17.Geyer R, Geyer H, Egge H, Schott HH, Stirm S. Structure of the oligosaccharides sensitive to endo-beta-n-acetylglucosaminidase H in the glycoprotein of Friend murine leukemia virus. Eur J Biochem. 1984;143(3):531–539. doi: 10.1111/j.1432-1033.1984.tb08402.x. [DOI] [PubMed] [Google Scholar]
- 18.Danielle LP, Paul K, Theresa M, Alain B. Characterization of Minor N-linked Glycans on Antibodies Using Endo H Release and MALDI-Mass Spectrometry. Analytical letters. 2009;42(11):1711–1724. [Google Scholar]
- 19.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptic to yield complementary sequence information. Anal Chem. 2001;73(18):4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 20.Hakansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Combined electron capture and infrared multiphoton dissociation for multistage MS/MS in a Fourier transform ion cyclotron resonance mass spectrometer. Anal Chem. 2003;75(13):3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 21.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J Proteome Res. 2005;4(2):628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiselar JG, Mahaffy R, Pollard TD, Almo SC, Chance MR. Visualizing Arp2/3 complex activation mediated by binding of ATP and WASp using structural mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(5):1552–1557. doi: 10.1073/pnas.0605380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 24.Xu G, Chance MR. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal Chem. 2005;77(14):4549–4955. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 25.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107(8):3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 26.Gupta S, Sullivan M, Toomey J, Kiselar J, Chance MR. The Beamline X28C of the Center for Synchrotron Biosciences: a national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J Synchrotron Radiat. 2007;14(Pt 3):233–243. doi: 10.1107/S0909049507013118. [DOI] [PubMed] [Google Scholar]
- 27.Goldsmith SC, Guan JQ, Almo S, Chance M. Synchrotron protein footprinting: a technique to investigate protein-protein interactions. J Biomol Struct Dyn. 2001;19(3):405–418. doi: 10.1080/07391102.2001.10506750. [DOI] [PubMed] [Google Scholar]
- 28.Zheng X, Wintrode PL, Chance MR. Complementary structural mass spectrometry techniques reveal local dynamics in functionally important regions of a metastable serpin. Structure. 2008;16(1):38–51. doi: 10.1016/j.str.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int J Radiat Biol. 2002;78(2):101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- 30.Kiselar JG, Janmey PA, Almo SC, Chance MR. Structural analysis of gelsolin using synchrotron protein footprinting. Mol Cell Proteomics. 2003;2(10):1120–1132. doi: 10.1074/mcp.M300068-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Gao F, Weaver EA, Lu Z, Li Y, Liao HX, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu JS, Decker JM, Shaw GM, Montefiori DC, Korber BT, Hahn BH, Haynes BF. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79(2):1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan MR, Rekhi S, Bohon J, Gupta S, Abel D, Toomey J, Chance MR. Installation and testing of a focusing mirror at beamline X28C for high flux x-ray radiolysis of biological macromolecules. Rev Sci Instrum. 2008;79(2 Pt 1):025101. doi: 10.1063/1.2839027. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7(1):138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Kaur P, Kiselar JG, Chance MR. Integrated algorithms for high-throughput examination of covalently labeled biomolecules by structural mass spectrometry. Anal Chem. 2009;81(19):8141–8149. doi: 10.1021/ac9013644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 40.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38(3):305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 41.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plummer TH, Jr., Tarentino AL. Purification of the oligosaccharide-cleaving enzymes of Flavobacterium meningosepticum. Glycobiology. 1991;1(3):257–263. doi: 10.1093/glycob/1.3.257. [DOI] [PubMed] [Google Scholar]
- 43.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180(2):195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 44.Wold F. In vivo chemical modification of proteins (post-translational modification). Annu Rev Biochem. 1981;50:783–814. doi: 10.1146/annurev.bi.50.070181.004031. [DOI] [PubMed] [Google Scholar]
- 45.Robinson NE. Protein deamidation. Proc Natl Acad Sci U S A. 2002;99(8):5283–5288. doi: 10.1073/pnas.082102799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson NE, Robinson AB. Mocecular Clocks Deamidation of Asparaginyl and Glutaminyl Residues in peptides and Proteins. Althouse press; 2004. [Google Scholar]
- 47.Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987;262(2):785–794. [PubMed] [Google Scholar]
- 48.Kubelka V, Altmann F, Kornfeld G, Marz L. Structures of the N-linked oligosaccharides of the membrane glycoproteins from three lepidopteran cell lines (Sf-21, IZD-Mb-0503, Bm-N). Arch Biochem Biophys. 1994;308(1):148–157. doi: 10.1006/abbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 49.Altmann F, Staudacher E, Wilson IB, Marz L. Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj J. 1999;16(2):109–123. doi: 10.1023/a:1026488408951. [DOI] [PubMed] [Google Scholar]
- 50.hang GD, Chen CJ, Lin CY, Chen HC, Chen H. Improvement of glycosylation in insect cells with mammalian glycosyltransferases. J Biotechnol. 2003;102(1):61–71. doi: 10.1016/s0168-1656(02)00364-4. [DOI] [PubMed] [Google Scholar]
- 51.Ilchenko SA, Chance MR, Whittaker LJ, Whittaker2 J. Determination of the Glycopeptide Structure of Insulin and IGF-I Receptors. 56th ASMS Conference on Mass Spectrometry and Allied Topics. 2008 [Google Scholar]
- 52.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol Cell Proteomics. 2008;7(12):2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalkley RJ, Baker PR, Hansen KC, Medzihradszky KF, Allen NP, Rexach M, Burlingame AL. Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: I. How much of the data is theoretically interpretable by search engines? Mol Cell Proteomics. 2005;4(8):1189–1193. doi: 10.1074/mcp.D500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280(5371):1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 55.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16(8):741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 56.Cormier EG, Tran DN, Yukhayeva L, Olson WC, Dragic T. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol. 2001;75(12):5541–5549. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 58.Javaherian K, Langlois AJ, McDanal C, Ross KL, Eckler LI, Jellis CL, Profy AT, Rusche JR, Bolognesi DP, Putney SD, et al. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci U S A. 1989;86(17):6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putney SD, Matthews TJ, Robey WG, Lynn DL, Robert-Guroff M, Mueller WT, Langlois AJ, Ghrayeb J, Petteway SR, Jr., Weinhold KJ, et al. HTLV-III/LAV-neutralizing antibodies to an E. coli-produced fragment of the virus envelope. Science. 1986;234(4782):1392–1395. doi: 10.1126/science.2431482. [DOI] [PubMed] [Google Scholar]
- 60.Guan JQ, Chance MR. Structural proteomics of macromolecular assemblies using oxidative footprinting and mass spectrometry. Trends Biochem Sci. 2005;30(10):583–592. doi: 10.1016/j.tibs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 61.Guan JQ, Takamoto K, Almo SC, Reisler E, Chance MR. Structure and dynamics of the actin filament. Biochemistry. 2005;44(9):3166–3175. doi: 10.1021/bi048021j. [DOI] [PubMed] [Google Scholar]
- 62.Kiselar JG, Janmey PA, Almo SC, Chance MR. Visualizing the Ca2+-dependent activation of gelsolin by using synchrotron footprinting. Proc Natl Acad Sci U S A. 2003;100(7):3942–3947. doi: 10.1073/pnas.0736004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu R, Guan JQ, Zak O, Aisen P, Chance MR. Structural reorganization of the transferrin C-lobe and transferrin receptor upon complex formation: the C-lobe binds to the receptor helical domain. Biochemistry. 2003;42(43):12447–124454. doi: 10.1021/bi0352973. [DOI] [PubMed] [Google Scholar]
- 64.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, Chance MR. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol. 2004;343(5):1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 65.Xu G, Liu R, Zak O, Aisen P, Chance MR. Structural allostery and binding of the transferrin*receptor complex. Mol Cell Proteomics. 2005;4(12):1959–1967. doi: 10.1074/mcp.M500095-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Vranken WF, Fant F, Budesinsky M, Borremans FA. Conformational model for the consensus V3 loop of the envelope protein gp120 of HIV-1 in a 20% trifluoroethanol/water solution. Eur J Biochem. 2001;268(9):2620–2628. doi: 10.1046/j.1432-1327.2001.02146.x. [DOI] [PubMed] [Google Scholar]
- 67.Moore JP, Nara PL. The role of the V3 loop of gp120 in HIV infection. Aids. 1991;5(Suppl 2):S21–33. doi: 10.1097/00002030-199101001-00004. [DOI] [PubMed] [Google Scholar]
- 68.Clements GJ, Price-Jones MJ, Stephens PE, Sutton C, Schulz TF, Clapham PR, McKeating JA, McClure MO, Thomson S, Marsh M, et al. The V3 loops of the HIV-1 and HIV-2 surface glycoproteins contain proteolytic cleavage sites: a possible function in viral fusion? AIDS Res Hum Retroviruses. 1991;7(1):3–16. doi: 10.1089/aid.1991.7.3. [DOI] [PubMed] [Google Scholar]
- 69.Stephens PE, Clements G, Yarranton GT, Moore J. A chink in HIV's armour? Nature. 1990;343(6255):219. doi: 10.1038/343219b0. [DOI] [PubMed] [Google Scholar]
- 70.Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174(2):407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Werner A, Levy JA. Human immunodeficiency virus type 1 envelope gp120 is cleaved after incubation with recombinant soluble CD4. J Virol. 1993;67(5):2566–2574. doi: 10.1128/jvi.67.5.2566-2574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.