Abstract
Foxm1 is a member of the Forkhead Box (Fox) family of transcription factors. Foxm1 (previously called Foxm1b, HFH-11B, Trident, Win, or MPP2) is expressed in multiple cell types and plays important roles in cellular proliferation, differentiation and tumorigenesis. Genetic deletion of Foxm1 from mouse respiratory epithelium during initial stages of lung development inhibits lung maturation and causes respiratory failure after birth. However, the role of Foxm1 during postnatal lung morphogenesis remains unknown. In the present study, Foxm1 expression was detected in epithelial cells of conducting and peripheral airways and changing dynamically with lung maturation. To discern the biological role of Foxm1 in the prenatal and postnatal lung, a novel transgenic mouse line that expresses a constitutively active form of FoxM1 (FoxM1 N-terminal deletion mutant or FoxM1-ΔN) under the control of lung epithelial-specific SPC promoter was produced. Expression of the FoxM1-ΔN transgene during embryogenesis caused epithelial hyperplasia, inhibited lung sacculation and expression of the type II epithelial marker, pro-SPC. Expression of FoxM1-ΔN mutant during the postnatal period did not influence alveologenesis but caused focal airway hyperplasia and increased proliferation of Clara cells. Likewise, expression of FoxM1-ΔN mutant in conducting airways with Scgb1a1 promoter was sufficient to induce Clara cell hyperplasia. Furthermore, FoxM1-ΔN cooperated with activated K-Ras to induce lung tumor growth in vivo. Increased activity of Foxm1 altered lung sacculation, induced proliferation in the respiratory epithelium and accelerated lung tumor growth, indicating that precise regulation of Foxm1 is critical for normal lung morphogenesis and development of lung cancer.
INTRODUCTION
Transcription factor Forkhead Box m1 (Foxm1, previously known as HFH-11B, Trident, Win, or MPP2) plays important roles in cellular proliferation and differentiation during embryogenesis and development of cancer (Costa et al., 2003; Krupczak-Hollis et al., 2004; Laoukili et al., 2007; Ramakrishna et al., 2007; Teh et al., 2002; Ueno et al., 2008). Foxm1 is expressed in various embryonic cell types and its expression is dramatically increased in tumor cells during progression of liver, lung, colon, breast and prostate cancers (Kalin et al., 2006; Kalinichenko et al., 2004; Kim et al., 2006; Wonsey and Follettie, 2005; Yoshida et al., 2007). Activation of Ras/Raf/MAPK (ERK) signaling pathway drives cell cycle progression by increasing expression of cyclin proteins that bind to cyclin-dependent kinases (CDKs) (Massague, 2004). CDK-cyclin complexes and ERK phosphorylate the Foxm1 protein and increase its transcriptional activity by facilitating the binding of Foxm1 with CBP co-activator protein (Ma et al., 2005; Major et al., 2004). Foxm1 directly induces transcription of cell cycle regulatory genes essential for G1/S and G2/M progression (Costa, 2005; Laoukili et al., 2007; Wang et al., 2005). Hypoxia-Inducible Factor 1 (HIF1)(Xia et al., 2009) and c-Myc (Blanco-Bose et al., 2008) have been shown to induce Foxm1 expression, promoting cell transformation and entry into the cell cycle.
In our previous studies we demonstrated that Foxm1−/− mice die in utero between embryonic day 13.5 (E13.5) and E16.5 due to structural abnormalities in development of liver, lung, and heart (Krupczak-Hollis et al., 2004). Foxm1 deficiency in hepatoblasts and cardiomyocytes was associated with accumulation of polyploid phenotype, resulting from diminished DNA replication and failure to progress through mitosis (Korver et al., 1998; Krupczak-Hollis et al., 2004; Ramakrishna et al., 2007). Foxm1−/− livers failed to form intrahepatic bile ducts, indicating that Foxm1 is required for differentiation of hepatoblast precursor cells toward the biliary epithelial cell lineage (Krupczak-Hollis et al., 2004). Foxm1−/− embryos exhibited defects in differentiation of pulmonary mesenchyme into mature capillary endothelial cells during the canalicular stage of lung development (Kim et al., 2005). Conditional deletion of Foxm1 from precursors of cerebellar granule neurons delayed brain development due to decreased Shh-induced proliferation (Schuller et al., 2007). Severe abnormalities in postnatal β-cell mass expansion were observed in Foxm1-deleted pancreatic cells, causing impaired islet function and early onset of diabetes in mouse (Zhang et al., 2006). Foxm1 deficiency in smooth muscle cells was associated with perinatal lethality due to severe pulmonary hemorrhage, and structural abnormalities in arterial walls and esophagus (Ustiyan et al., 2009). These studies demonstrated that Foxm1 plays distinct roles in differentiation and proliferation in various tissues during embryogenesis.
We recently generated mice with an inducible deletion of Foxm1 from respiratory epithelium (SPC-rtTA/TetO-Cre/Foxm1fl/fl mice). Foxm1 inactivation in undifferentiated epithelial cells prior to the initiation of lung development did not affect branching lung morphogenesis but impaired lung maturation and decreased surfactant production, causing respiratory failure after birth (Kalin et al., 2008). However, this mouse model did not allow us to determine the role of Foxm1 in fully differentiated epithelial cells due to incomplete Foxm1 deletion in the late gestation and the postnatal period. Furthermore, given the importance of Foxm1 in the development of non-small cell lung cancer in mice and humans (Kim et al., 2006; Wang et al., 2009; Wang et al., 2008b; Yang et al., 2009), it is not clear whether activation of Foxm1 in epithelial cells is sufficient to induce lung tumors in the adult mice. To address these issues, we developed a new mouse model, expressing activated mutant FoxM1 protein in respiratory epithelial cells during different periods in the embryonic and postnatal lung.
The N-terminal region (1-232 amino acid residues) of human FoxM1 contains a transcription inhibitory domain as well as protein destruction box (D-box) and KEN box motifs that are required for degradation of the FoxM1 protein (Laoukili et al., 2008; Park et al., 2008a; Park et al., 2008b). Compared to full length FoxM1 protein, FoxM1 N-terminal deletion mutant protein (FoxM1-ΔN) displayed increased protein stability and transcriptional activity (Laoukili et al., 2008; Park et al., 2008a), causing an increase of anchorage-independent cellular proliferation in U2OS cells (Park et al., 2008a; Wang et al., 2008a). In this study, transgenic mice expressing FoxM1-ΔN under the control of lung epithelial-specific SPC or Scgb1a1 (Clara Cell Secretory Protein or CCSP) promoters were generated and used to induce Foxm1 function during different periods of embryonic and postnatal lung development.
MATERIAL AND METHODS
Construction of TetO-CMV-GFP-FoxM1 plasmid and generation of transgenic mice
The expression vector of GFP-fused human FoxM1b N-terminal deletion mutant (lacking amino acids 1-231) was obtained from Dr. Raychuadhuri (University of Illinois at Chicago) (Park et al., 2008b). GFP-FoxM1b N-terminal deletion mutant (GFP-FoxM1-ΔN) was subcloned into a plasmid containing minimal CMV promoter with a tetracycline operator (TetO7-CMV), and the 3′-untranslated region of the bovine growth hormone (bGH) gene polyadenylation (polyA) signal (Clark et al., 2001; Gossen et al., 1995) (Fig. 1C). The DNA fragment containing the (TetO)7-CMV-GFP-FoxM1-ΔN (amino acids 232-748) and polyA was isolated and microinjected into mouse oocytes at the Transgenic Mouse Facility of Cincinnati Children's Hospital Medical Center. Three TetO-GFP-FoxM1-ΔN transgenic mouse lines were established and showed similar phenotypes. Transgenic mice were bred with either SPC-rtTA (line 1) or CCSP-rtTA transgenic mice (line 2) (Chen et al., 2009; Perl et al., 2002) to generate double transgenic mice. Single transgenic mice and wild-type littermates were used as controls. Additional controls included double transgenic mice without Doxycycline (Dox) treatment. PCR analysis was used to screen for the TetO-GFP-FoxM1-ΔN transgene with sense primer made to GFP sequence: 5′-CGACAAGCAGAAGAACGGCATC-3′ and anti-sense primer annealed to human FoxM1b DNA binding domain: 5′-AGTAGGGAAAGTGGTCCTCAATCC-3′. Amplification of PCR product for TetO-GFP-FoxM1-ΔN transgene was performed by denaturation at 94°C for 3 min and then 35 cycles of amplification at 94°C for 30 sec, 55°C for 45 sec, and 72°C for 45 sec, with 7 extension at 72°C, followed by an agarose gel electrophoresis. PCR analysis and primers used identification of SPC-rtTA and CCSP-rtTA transgenes were described previously (Liu et al., 2002; Tichelaar et al., 2000). TetO-K-Ras transgenic mice and reporter mice (Rosa26R) were purchased from Jackson Lab.
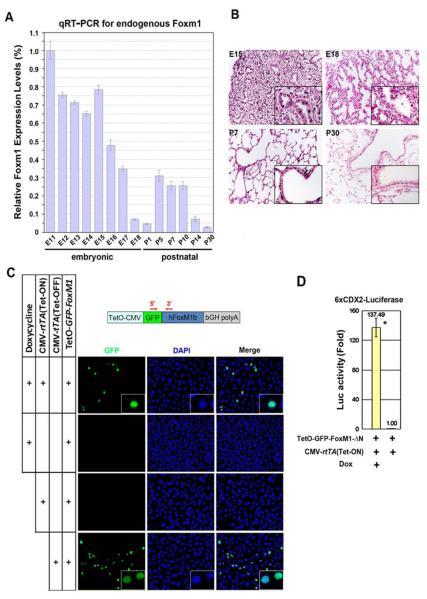
Figure 1.
Foxm1 expression in mouse lungs. (A) qRT-PCR revealed changes in endogenous Foxm1 mRNA levels throughout lung development. Lungs were isolated from wild type FVB/N mice at different stages of embryogenesis (E11-P30). The highest Foxm1 mRNA expression level (E11) was set as 1. (B) Immunohistochemistry of paraffin slides with Foxm1 antibody shows endogenous Foxm1 protein (dark brown) in alveolar and bronchial regions of E15, E18 and P7 lungs. Foxm1 protein was undetectable in P30 lungs. (C) A schematic representation of (TetO)7-GFP-FoxM1b(ΔN)-bGH polyA construct (TetO-GFP-FoxM1-ΔN). PCR primers for detection of TetO-FoxM1-ΔN transgene are shown by red arrows. U2OS cells were transfected with CMV-rtTA and TetO-GFP-FoxM1-ΔN vectors followed by Dox treatment (Tet-ON system), or co-transfected with CMV-tTA and TetO-GFP-FoxM1-ΔN vectors (Tet-OFF system). Nuclear GFP fluorescence demonstrated that GFP-FoxM1-ΔN fusion protein was specifically induced by Dox and localized in the nuclei of transfected cells. (D) U2OS cells were cotransfected with the TetO-GFP-FoxM1-ΔN and CMV-rtTA (Tet-ON) plasmids as well as 6xCDX2-Luciferase and CMV-Renilla luciferase reporters. CMV-Renilla luciferase was used as internal control. Cell extracts were assayed for luciferase activity, which is presented as fold induction ± SD (* indicates statistically significant changes, P<0.05).
Animal use and administration of Doxycycline
Mice were maintained in a pathogen-free vivarium in filtered cages at the animal facility of Cincinnati Children's Hospital Medical Center. Oral Doxycycline (Dox) was given in mouse chow. Because of the light sensitivity of Dox, Dox-containing mouse chow was replaced once a week. Sentinel mice were free of common viral and bacterial pathogens. Blood was collected from the mouse tail vein and analyzed using Hemavet 950FS hematology analyzer (Drew Scientific Inc, Dallas, TX).
Lung total RNA isolation and real-time qRT-PCR
Mouse lung tissue was homogenized in RNA Stat-60 (Tel-Test “B” Inc), and RNA was isolated following the manufacturer's specifications. RNA was treated with DNase and purified for cDNA synthesis as described previously (Kalinichenko et al., 2003; Wang et al., 2008b). cDNA samples were amplified with inventoried TaqMan Gene Expression Assays (Table 1) and analyzed by a StepOnePlus Real-Time PCR system (Applied Biosystems). Reactions were analyzed in triplicates and expression levels were normalized to β-actin mRNA levels and presented as means ± SD.
Table 1.
TaqMan gene expression assays (Applied Biosystems) used for qRT-PCR.
| Human primer | |
| FoxM1 | Hs00153543_m1 |
| Mouse primers | |
| Sox2 | Mm00488369_s1 |
| Sox4 | Mm00486320_s1 |
| Sox9 | Mm00448840_m1 |
| Sox17 | Mm00488363_m1 |
| c-Myc | Mm00487804_m1 |
| TTF1 | Mm00447558_m1 |
| CDKN1A | Mm01303209_m1 |
| PECAM1 | Mm00476702_m1 |
| α-SMA | Mm00725412_s1 |
| Foxm1 | Mm00514924_m1 |
| FoxA2 | Mm00839704_mH |
| FoxA3 | Mm00484714_m1 |
| FoxF1a | Mm00487497_m1 |
| FoxF2 | Mm00515793_m1 |
| FoxJ1 | Mm00807215_m1 |
| FoxL1 | Mm00514937_s1 |
| FoxP1 | Mm00474845_m1 |
| FoxP2 | Mm00475030_m1 |
| β-Actin | Mm00607939_s1 |
Dual luciferase assays for analysis of promoter activity
Human osteosarcoma U2OS cells were plated at 8 ×104 cells per well in a 12-well plate and used for cotransfection experiments with Lipofectamine 2000 (Invitrogen). The following plasmids were used: CMV-rtTA (Tet-ON), CMV-tTA (Tet-OFF), TetO-GFP-FoxM1-ΔN, and CDX2 promoter firefly-luciferase (6xCDX2-Luc) (Wang et al., 2008a; Ye et al., 1997). Twenty-four hours after transfection, cells were treated with 1 μg/ml of Dox (Catalog# 631311, BD Biosciences) and incubated for an additional 24 hr. Cells were harvested 48 hours after transfection and cell extracts were isolated and subjected to dual luciferase assays (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity as described (Wang et al., 2009; Wang et al., 2008b). Experiments were performed in triplicate.
Lung histology, immunohistochemistry and co-localization experiments
Embryonic and adult mouse lungs were isolated, fixed in 4% paraformaldehyde, and paraffin-embedded. Five-micrometer-thick lung sections were stained with hematoxylin and eosin (H&E) for morphological examination or used for immunohistochemistry (IHC) as described previously (Wang et al., 2009). Following antibodies were used for IHC: transgenic FoxM1 (1:2000; H-300), endogenous Foxm1 (1:1000; K-19) and CC10 (1:2000; T-18) were from Santa Cruz Biotechnology. Pro-SPC (1:500; guinea pig polyclonal), FoxJ1 (1:2000; mouse monoclonal; #3-19), TTF-1 (1:1500; mouse monoclonal; #8G7G3-1), Sox2 (1:2500; rabbit polyclonal, WRAB-SOX2), and FoxA2 (1:2000; rabbit polyclonal; #WRAB-FOXA2) were from Seven Hills Bioreagents (Cincinnati, OH) (Dave et al., 2008). Other antibodies used were: Ki-67 (1:1000; rat monoclonal; #TEC-3; DAKO); β-tubulin (1:5000; #T7451; Sigma), phospho-Histone H3 (1:2000; rabbit polyclonal; #06-570; Millipore), Sox9 (1:1000; rabbit polyclonal; #AB5535; Millipore), F4/80 (1:100; rat monoclonal; #MCA497GA; AbD Serotec), α-smooth muscle actin (1:15,000; mouse monoclonal; clone 1A4; Sigma), and PECAM-1 (1:50000; rat monoclonal; #553370; Pharmingen). Tumor areas in H&E-stained lung sections were measured using the scaling function in Axiovision Rel software. For co-localization experiments, secondary antibodies conjugated with either Texas-Red or FITC were used, and slides were counterstained with DAPI (Vector Lab). Fluorescence was detected using a Zeiss Axioplan 2 Imaging Universal Microscope with an Axiocam MRm digital camera (Axiovision Release 4.3).
Lineage-tracing experiments and X-gal staining
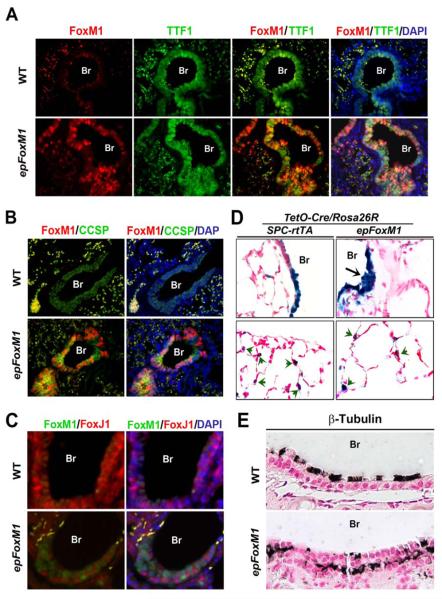
SPC-rtTA/TetO-GFP-FoxM1-ΔN double transgenic mice (epFoxM1) were bred with TetO-Cre/Rosa26-loxP-STOP-loxP-LacZ (Rosa26R) mice to generate SPC-rtTA/TetO-GFP-FoxM1-ΔN/TetO-Cre/Rosa26R quadruple transgenic mice. Dox was given from postnatal day 3 (P3) to P30, causing simultaneous FoxM1-ΔN expression and Cre-mediated excision of the LoxP-STOP-LoxP cassette from the ROSA26 locus. SPC-rtTA/TetO-Cre/Rosa26R littermates were used as controls. Lungs were fixed and stained for β-galactosidase (β-gal) activity as described (Bell et al., 2008). Paraffin sections were counterstained with nuclear fast red.
Statistical analysis
Student's t test was used to calculate P values. Statistically significant changes (P values < 0.05) were indicated with asterisks.
RESULTS
Foxm1 expression during lung development
To examine Foxm1 expression during embryonic and postnatal lung development, lungs were harvested from wild type (WT) mice at embryonic days E11-E18 or postnatal days P1-P30. Total lung RNA was isolated and then used for quantitative real-time RT-PCR (qRT-PCR). Foxm1 mRNA levels were normalized to β-actin mRNA. The highest Foxm1 mRNA levels were observed at E11, and Foxm1 mRNA gradually decreased prior to birth (Fig. 1A). Consistent with previous in situ hybridization studies (Kalin et al., 2008), Foxm1 protein was detected in bronchial and alveolar epithelial cells, and pulmonary mesenchyme as demonstrated by immunostaining with Foxm1 antibodies (Fig. 1B). Foxm1 expression was re-activated during the early postnatal period (P5-P7), being increased 4-6 fold compared to the day of birth (P1) (Fig. 1A). Foxm1 mRNA levels were further decreased to the lowest level at P30 (Fig. 1A), consistent with undetectable Foxm1 staining in adult lungs (Fig. 1B).
Conditional expression of activated FoxM1 mutant protein in respiratory epithelium
Since our results showed a significant decrease in Foxm1 expression during late gestation (Fig. 1A-B), we next performed experiments to determine whether the reduction in Foxm1 activity during this period is critical for normal lung development. We generated a transgenic construct containing an activated form of the human FoxM1 protein (FoxM1 N-terminal deletion mutant). The FoxM1 N-terminal region (1-232 amino acids) contains a transcription inhibitory domain as well as protein destruction box (D-box) and KEN box motifs that are required for FoxM1 protein degradation (Laoukili et al., 2008; Park et al., 2008a; Park et al., 2008b). FoxM1 N-terminal deletion mutant lacking D-box and KEN-box (FoxM1-ΔN) displayed increased protein stability when compared to the full length FoxM1 protein (Laoukili et al., 2008; Park et al., 2008a). To generate an inducible FoxM1-ΔN transgene, we fused the TetO7-CMV promoter region (Clark et al., 2001) to the 5′ end of GFP-Tagged human FoxM1-ΔN cDNA that was adjacent to a bovine growth hormone (bGH) gene polyadenylation (polyA) signal (Fig. 1C). To determine transcriptional activity of the TetO-GFP-FoxM1-ΔN transgenic construct, transient cotransfection experiments were performed in U2OS cells using CMV vectors expressing either the tetracycline transactivator (tTA; Tet-OFF system) or tetracycline reverse transactivator (rtTA; Tet-ON system). Twenty-four hours after transfection, cells were treated with Dox for an additional 24 hr and then assayed for GFP fluorescence. Cotransfection of TetO-GFP-FoxM1-ΔN and CMV-rtTA plasmids in the presence of Dox induced expression of the GFP-FoxM1-ΔN protein, which was located in nuclei of transfected cells (Fig. 1C). No GFP fluorescence was observed in the absence of Dox. Furthermore, expression of GFP-FoxM1-ΔN protein was induced by cotransfection of the TetO-GFP-FoxM1-ΔN and CMV-tTA plasmids (Fig. 1C). The GFP-FoxM1-ΔN protein was transcriptionally active as demonstrated by Dual Luciferase Assay using 6xCDX2-LUC plasmid (Fig. 1C-D), a known reporter for FoxM1 transcriptional activity (Ye et al., 1997).
To conditionally express FoxM1-ΔN mutant in the developing respiratory epithelium, we generated TetO-GFP-FoxM1-ΔN transgenic mice. Three distinct transgenic mouse lines were established and showed similar phenotypes. After breeding with SPC-rtTA mice, the SPC-rtTA/TetO-GFP-FoxM1-ΔN double transgenic mouse line (epFoxM1) was established and identified using PCR with primers specific to human FoxM1 cDNA and the SPC-rtTA transgene (Fig. 2B). The double transgenic mice permit expression of GFP-FoxM1-ΔN in the respiratory epithelium after treatment with Dox (Fig. 2A).
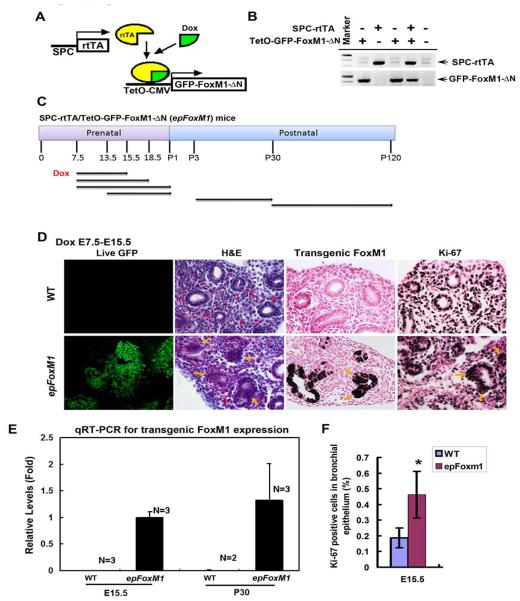
Figure 2.
Dox induced GFP-FoxM1-ΔN expression and caused epithelial hyperplasia in SPC-rtTA/TetO-GFP-FoxM1-ΔN double transgenic (epFoxM1) lungs. (A) A schematic showing Dox-mediated induction of GFP-FoxM1-ΔN transgene in epFoxM1 lung epithelium. (B) PCR analysis of transgenic embryos using primers depicted in Fig. 1C. (C) A schematic showing regiments of Dox treatment to induce GFP-FoxM1-ΔN transgene in mouse lung epithelium. Arrows indicate periods of Dox treatment and harvest times during prenatal and postnatal stages. (D) Dox treatment from E7.5 to E15.5 induced GFP fluorescence in epFoxM1 lung epithelium. GFP fluorescence was detected in nuclei of epithelial cells. Magnification is 100X. Paraffin sections of Dox-treated epFoxM1 lungs were stained with H&E or used for immunohistochemistry (IHC) with antibodies against either human (transgenic) FoxM1 or Ki-67 (dark brown nuclei). Slides were counterstained with Nuclear Fast Red (red nuclei). FoxM1-ΔN was detectable in Dox-treated epFoxM1 lungs but not in WT lungs. Epithelial hyperplasia was observed in epFoxM1 lungs (shown with arrows), while a single layer of epithelial cells was detected in WT lungs. Magnification is 400X. (E) Real time RT-PCR analysis showed transgenic GFP-FoxM1-ΔN mRNA exclusively expressed in epFoxM1 lungs treated with Dox (E7.5-E15.5 and P3-P30). (F) Increased numbers of Ki-67-positive epithelial cells in epFoxM1 lungs. Numbers of Ki-67-stained nuclei were counted in respiratory epithelium using twenty random 400x microscope fields from epFoxM1 and control mice. A statistically significant p value < 0.05 is shown with asterisk.
FoxM1-ΔN caused epithelial hyperplasia during lung development
To examine the consequences of increased FoxM1 activity on lung morphogenesis, expression of FoxM1-ΔN transgene was induced at E7.5 and lungs harvested at different stages of development (Fig. 2C). Total lung RNA was used to examine the expression levels of transgenic FoxM1 mRNA by qRT-PCR. Human FoxM1 mRNA was found in Dox-treated epFoxM1 double transgenic lungs at E15.5 and P30 (Fig. 2E), but was undetectable in lungs of Dox-treated control (WT), single transgenic littermates and age-matched epFoxM1 lungs without Dox treatment (Fig. 2E and data not shown).
When FoxM1-ΔN was induced from E7.5-E15.5, lung size and lobulation were normal. GFP fluorescence confirmed the induction of the FoxM1-ΔN transgene by Dox (Fig. 2D). The presence of FoxM1-ΔN transgenic protein in respiratory epithelial cells was also confirmed by immunohistochemistry with antibodies specific to human FoxM1 protein (Fig. 2D) (this antibody does not cross-react with endogenous mouse Foxm1 protein). FoxM1-ΔN induced epithelial hyperplasia, characterized by the presence of pseudostratified epithelium (normally a single columnar epithelium) (Fig. 2D). Increased numbers of Ki-67 positive cells were consistent with increased cellular proliferation in hyperplastic regions (Fig. 2D and 2F). Numbers of epithelial cells expressing Sox2 and Sox9 transcription factors were unchanged in epFoxM1 lungs (Supplemental. Fig. 1A).
Increased FoxM1 activity impairs lung sacculation
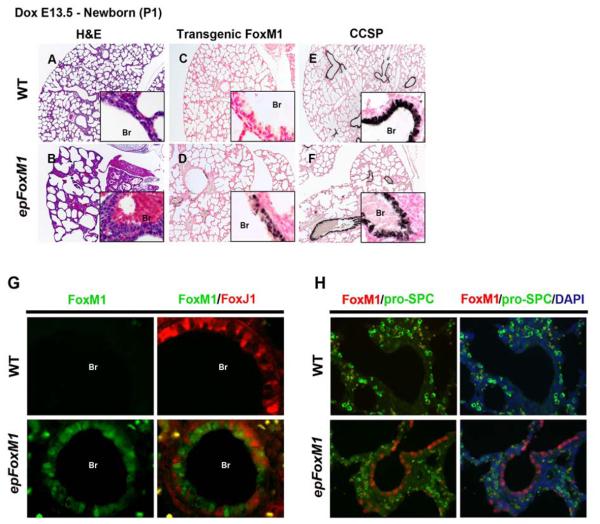
Abnormal, dilated epithelial cysts were observed throughout the peripheral lung in FoxM1-ΔN expressing mice when Dox was provided from E7.5-E18.5 or E7.5-P1 (Fig. 3B and K). FoxM1-ΔN protein was detected in the epithelial cells lining the abnormal pulmonary cysts in epFoxM1 embryos (Fig. 3D) and was absent in WT and single transgenic littermates (Fig. 3C and data not shown). Pro-SPC, a marker specific for type II epithelial cells, was absent in epithelial cells expressing FoxM1-ΔN as demonstrated by immunohistochemistry (Fig. 3F) and co-localization experiments (Fig. 3H). Interestingly, when Dox was given to mice from E13.5 until P1, expression of FoxM1-ΔN transgene was dramatically reduced (Fig. 4D), but it was still sufficient to inhibit lung sacculation (Fig. 4B). There was no significant increase in apoptosis in FoxM1-ΔN transgenic lungs as demonstrated by immunostaining with antibodies against activated (cleaved) Caspase 3 (data not shown). FoxM1-ΔN-expressing epithelial cells did not express pro-SPC (Fig. 4H). Thus, increased FoxM1 activity impaired lung sacculation and prevented the normal autogenic induction of pro-SPC in peripheral respiratory epithelial cells.
Figure 3.
Sacculation defects in epFoxM1 lungs. Embryos and newborn mice were treated with Dox from E7.5-18.5 (A-H) or E7.5-P1 (I-X), and lungs were harvested for histological and immunohistochemistry staining (IHC). (A-B) H&E staining showed sacculation defects in epFoxM1 lungs. (C-D) IHC showed FoxM1-ΔN protein in epithelium of epFoxM1 lungs. FoxM1-ΔN was not detected in control (WT) lungs (C). (E-F) pro-SPC expression was detected in epFoxM1 and control lungs. (G-H) FoxM1-ΔN (red) did not co-localize with pro-SPC (green). Erythrocytes showed autofluorescence (yellow) in alveolar regions. CCSP staining was detected in epFoxM1 and WT mouse lung airway epithelium (I-L). Immunofluorescent staining showed that FoxM1-ΔN did not co-express with FoxJ1 in bronchial epithelium (M-P). FoxM1-ΔN was found in pulmonary bronchiole (Br) of epFoxM1 mice where it was co-expressed with CCSP (Q-T). FoxM1-ΔN did not co-localize with CCSP in epithelial cells of abnormal pulmonary cysts (U-X).
Figure 4.
Expression of FoxM1-ΔN in mouse lung epithelium from E13.5 to P1 caused sacculation defects. H&E staining shows sacculation defects in epFoxM1 lungs, which were characterized by thickening of mesenchyme and increased size of peripheral saccules. Epithelial hyperplasia was observed in airway epithelium of Dox-treated epFoxM1 lungs (B) but not in WT lungs (A). While transgenic FoxM1-ΔN protein was detected in epFoxM1 lungs (D), it was undetectable in WT lungs (C). CCSP was detected in both epFoxM1 and WT airways (E-F). Immunofluorescent staining showed that FoxM1-ΔN did not co-localize with either FoxJ1 (G) or pro-SPC (H).
Since previous studies demonstrated that SPC-rtTA mice (line 1) permit expression of rtTA in a subset of Clara cells during late gestation (Perl et al., 2002; Tichelaar et al., 2000), expression of FoxM1-ΔN transgene was examined in pulmonary airways. Transgenic protein was specifically detected in epFoxM1 airway epithelium (Fig. 3O and Fig. 4G) but not in control airways (Fig. 3M and Fig. 4G). Clara cell specific protein (CCSP) was present in both epFoxM1 and control airways (Fig. 3I-L and 4E-F). FoxM1-ΔN transgene was found in CCSP-positive Clara cells of large pulmonary airways in epFoxM1 embryos (Fig. 3S-T). In contrast, FoxM1-ΔN protein did not co-localize with CCSP in distal epithelial cysts of epFoxM1 embryos (Fig. 3W-X), indicating that FoxM1-ΔN-expressing epithelial cells lining distal pulmonary cysts are not Clara cells. FoxM1-ΔN was not expressed in ciliated cells as demonstrated by co-localization experiments with FoxJ1 (Fig. 3M-P and Fig. 4G), a transcription factor selectively expressed in ciliated cells (Blatt et al., 1999).
Increased FoxM1 activity during postnatal lung development caused airway hyperplasia
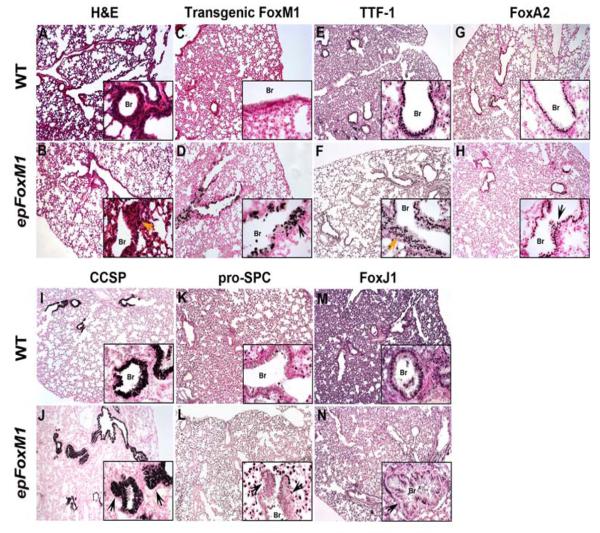
To determine consequences of increased FoxM1 activity during postnatal lung development, FoxM1-ΔN was induced from P3-P30 in epFoxM1 mice (Fig. 5D), resulting in marked hyperplasia of the airway epithelium (Fig. 5B), which was characterized by a pseudostratified epithelium at sites normally lined by a single columnar epithelium (Fig. 5A). TTF-1 and FoxA2 were expressed in the hyperplastic airway regions induced by FoxM1-ΔN (Fig. 5F and H), indicating the epithelial origin of hyperplastic cells. Moreover, the cells of hyperplastic regions expressed CCSP but not pro-SPC, indicating Clara cell hyperplasia (Fig. 5J and L). Co-localization experiments demonstrated that transgenic FoxM1-ΔN protein was co-expressed with TTF-1 (Fig. 6A) and CCSP (Fig. 6B), which is consistent with findings in normal Clara cells. Although ciliated cells were present in epFoxM1 airways as demonstrated by FoxJ1 staining (Fig. 5N), FoxM1-ΔN was not detected in ciliated cells (Fig. 6C). While β-tubulin, a protein specific to cilia (Tichelaar et al., 1999), was detected on apical surfaces of ciliated cells in control mice, β-tubulin was detected in cytoplasm of ciliated cells located near the basal lamina in epFoxM1 lungs (Fig. 6E).
Figure 5.
Expression of FoxM1-ΔN during postnatal lung development causes airway hyperplasia. epFoxM1 mice were treated with Dox from P3 to P30. Paraffin lung sections were stained with H&E (A-B) or used for immunohistochemical staining with antibodies against transgenic FoxM1 (C-D), TTF-1 (E-F), Foxa2 (G-H), CCSP (I-J), pro-SPC (K-L) or FoxJ1 (M-N). Arrows indicate hyperplastic epithelial regions in Dox-treated epFoxM1 bronchioles (Br).
Figure 6.
FoxM1-ΔN is expressed in Clara cells of epFoxM1 mice. (A) Paraffin sections were stained with antibodies against transgenic FoxM1-ΔN (red) and mouse TTF-1 (green) and then counterstained with DAPI (blue). Erythrocytes displayed yellow autofluorescence in alveolar regions. (B) Paraffin sections were immunostained with antibodies against FoxM1-ΔN (red) and CCSP (green) and then counterstained with DAPI (blue). FoxM1-ΔN co-localized with CCSP in epFoxM1 lungs. (C) FoxM1-ΔN (green) did not co-localize with FoxJ1 (red). (D) β-gal activity (blue) was detected in alveolar type II cells (green arrows) as well as hyperplastic airway regions of epFoxM1/TetO-Cre/Rosa26R lungs (black arrow) after Dox treatment from P3-P30. β-galactosidase activity was also observed in type II cells and bronchial epithelial cells of control SPC-rtTA/TetO-Cre/Rosa26R lungs. Slides were counterstained with nuclear fast red. (E) Lung sections from Dox-treated WT and epFoxM1 mice were immunostained with antibodies specific to β-tubulin (dark brown). Slides were counterstained with nuclear fast red. Br, bronchiole.
To detect epithelial progenitor cells in bronchioalveolar duct junctions, co-localization experiments with pro-SPC and CCSP antibodies were performed. Although SPC+/CCSP+ cells were found in bronchioalveolar duct junctions as well as bronchial epithelium of epFoxM1 lungs (Suppl. Fig. 1C), there were no significant differences between their numbers in epFoxM1 and control lungs (Suppl. Fig. 1D). Furthermore, to investigate the impact of FoxM1-ΔN expression on pulmonary stroma, we performed immunostaining of lung paraffin sections using antibodies specific to F4/80 (macrophage marker), α-smooth muscle actin (marker of smooth muscle cells) or Pecam-1 (endothelial marker). No differences were observed between epFoxM1 and control lungs (Suppl. Fig. 2B-C). Likewise, no differences were found in numbers of major cell types in peripheral blood (Suppl. Fig. 2A). These results showed that FoxM1-ΔN expression did not influence pulmonary stroma or numbers of immune cells in the peripheral blood.
Lineage-tracing experiments were performed to determine the origin of the hyperplastic cells induced by FoxM1-ΔN. To permanently label epithelial cells expressing FoxM1-ΔN, SPC-rtTA/TetO-GFP-FoxM1-ΔN double transgenic mice (epFoxM1) were bred with TetO-Cre/Rosa26-loxP-STOP-loxP-β-galactosidase reporter mice (Rosa26R) to generate SPC-rtTA/TetO-GFP-FoxM1-ΔN/TetO-Cre/Rosa26R quadruple transgenic mice. Mice without TetO-GFP-FoxM1-ΔN transgene (SPC-rtTA/TetO-Cre/Rosa26R) were used as controls. Dox was given from P3 until P30, simultaneously inducing FoxM1-ΔN and Cre-mediated excision of the LoxP-STOP-LoxP cassette from the ROSA26 locus. Thus, all cells expressing FoxM1-ΔN transgene during this period were permanently labeled by β-galactosidase. In control mice, β-galactosidase activity was detected in a subset of bronchial epithelial cells and type II epithelial cells of alveolar regions, which is consistent with previous studies (Fig. 6D) (Perl et al., 2002; Tichelaar et al., 2000). Hyperplastic regions induced by FoxM1-ΔN showed β-galactosidase activity, indicating that airway hyperplasia resulted from expansion of cells expressing the transgene. These results suggest a cell autonomous role for Foxm1 in the hyperplastic process.
Gene expression profile in epFoxM1 lungs
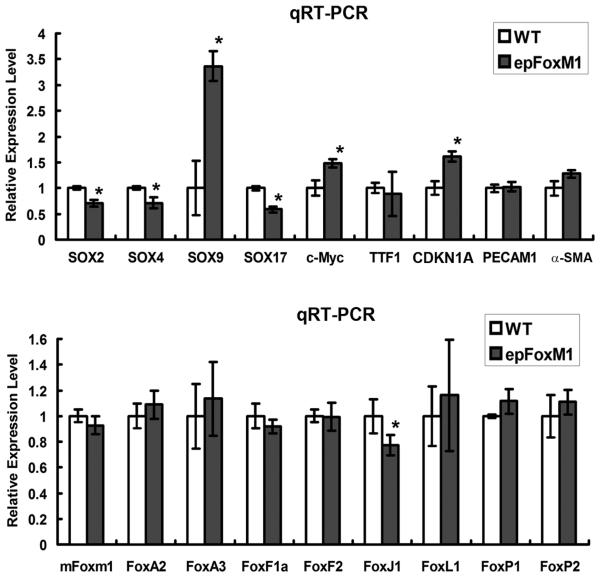
To identify potential FoxM1 target genes, expression of various transcription factors and cell cycle regulatory genes in epFoxM1 lungs was examined by qRT-PCR. Expression of cell cycle regulatory genes CDKN1A and c-Myc was increased in total lung RNA from epFoxM1 mice (Fig. 7), which is consistent with changes in cellular proliferation. Furthermore, expression of FoxM1-ΔN altered mRNA levels of various Sox transcription factors. While Sox2, Sox4 and Sox17 mRNAs were significantly decreased in epFoxM1 lungs, Sox9 mRNA was increased (Fig. 7). Since previous studies demonstrated that Sox2 and Sox17 are critical for epithelial morphogenesis in the developing lung (Park et al., 2006; Tompkins et al., 2009), reduced expression of these genes can contribute to airway defects seen in epFoxM1 mice. Interestingly, mRNA levels of Foxa2, Foxa3, Foxf1, Foxf2, Foxl1, Foxp1, Foxp2 and endogenous (mouse) Foxm1 were not changed in epFoxM1 transgenic lungs, whereas a significant reduction of Foxj1 mRNA was found (Fig. 7), a result consistent with reduced numbers of ciliated cells (Fig. 6C-E). No differences were observed in mRNA levels of TTF1 transcription factor, Pecam-1 and α-smooth muscle actin (Fig. 7). Altogether, these results indicate that FoxM1-ΔN influences expression of pulmonary genes critical for epithelial proliferation and lung morphogenesis.
Figure 7.
Expression of FoxM1-ΔN influenced mRNA levels of pulmonary genes. Lungs of mice treated with Dox from P3-P30 were used to prepare total RNA. qRT-PCR shows decreased mRNA levels of Sox2, Sox4, Sox17, and Foxj1 in epFoxM1 mice. CDKN1A, Sox9, and c-Myc mRNAs were increased. No changes were found in the expression of TTF1 and various Fox genes. β-actin mRNA was used for normalization. A p value < 0.05 is shown with asterisk (*).
Airway hyperplasia is induced by expression of FoxM1-ΔN in the adult lung
When FoxM1-ΔN was expressed in adult mice from P30-P120, focal airway hyperplasia was observed (Fig. 8B). Papillary structures were observed within airway lumens (Fig.8B, 8D and data not shown). Consistent with earlier findings, Ki-67 expression was observed in airway regions expressing FoxM1-ΔN (Fig. 8D and 8F) and CCSP protein (Fig. 8H). Single lung adenomas were observed in 2 out of 18 mice when FoxM1-ΔN was expressed until 6 months of age (Fig. 8J). The tumors were Ki-67 and phospho-Histone H3 positive (Fig. 8N and 8P), but did not express FoxM1-ΔN or CCSP (Fig. 8L and data not shown).
Figure 8.
Expression of FoxM1-ΔN transgene in the adult lung causes hyperplasia of Clara cells. Histological staining and IHC of lungs isolated from epFoxM1 and WT mice after Dox treatment for 3 months. (A-B) H&E staining displayed focal epithelial hyperplasia in epFoxM1 lungs (arrows). IHC with antibodies against FoxM1-ΔN (C-D), Ki-67 (E-F) and CCSP (G-H) showed expression of these proteins in hyperplastic airway regions of Dox-treated epFoxM1 mice (arrows). (I-P) Rare lung adenomas (Ad) were detected in epFoxM1 lungs. These tumors were positive for Ki-67 (N) and pH3 (P), but negative for FoxM1-ΔN transgene (L).
FoxM1-ΔN cooperates with activated K-Ras to increase lung tumor growth in vivo
To determine whether FoxM1-ΔN transgene increases a tumor growth in the presence of activated (oncogenic) K-Ras. TetO-GFP-FoxM1-ΔN mice were bred with SPC-rtTA/TetO-K-Ras mice (epKras), a known transgenic model of lung cancer (Fisher et al., 2001; Johnson et al., 2001). Four-week old SPC-rtTA/TetO-GFP-FoxM1-ΔN/TetO-K-Ras triple transgenic mice (epFoxM1/ epKras) were treated with Dox for 20 weeks to induce simultaneous expression of FoxM1-ΔN and activated K-Ras in respiratory epithelium. Consistent with previous studies (Fisher et al., 2001; Johnson et al., 2001), expression of activated K-Ras alone was sufficient to induce formation of lung adenocarcinomas (Fig. 9E-F), whereas expression of FoxM1-ΔN alone did not cause lung tumors but resulted in formation of epithelial hyperplasia (Fig. 9C-D). Tumor sizes in epFoxM1/ epKras mice were significantly increased compared to control epKras mice (Fig. 9I). Transgenic FoxM1-ΔN protein was specifically found in epFoxM1/ epKras tumors by immunohistochemistry (Fig. 9G-H). These results demonstrated that FoxM1-ΔN cooperates with activated K-Ras to accelerate lung tumor growth in vivo.
Figure 9.
FoxM1-ΔN cooperates with activated K-Ras to induce lung tumor growth in vivo. TetO-FoxM1-ΔN mice were bred with SPC-rtTA/ TetO-K-Ras mice to generate SPC-rtTA/ TetO-FoxM1-ΔN/ TetO-K-Ras triple transgenic mice (epFoxM1/ epKras). Single transgenic littermates were used as controls. Mice were treated with Dox for 20 weeks. (A-H) Paraffin lung sections were stained with antibodies specific for the FoxM1-ΔN transgene (brown nuclei) and counterstained with nuclear fast red (red nuclei). Expression of FoxM1-ΔN alone did not cause lung tumors but resulted in formation of epithelial hyperplasia (arrows in D). (I) Tumor sizes in epFoxM1/ epKras mice were significantly increased compared to mice expressing K-Ras alone. Tumor areas in lung sections were measured by Axiovision Rel software. Ten random lung sections were used to determine statistical significance. A p value < 0.05 is shown with asterisk (*).
Clara cell-specific expression of FoxM1-ΔN induced airway hyperplasia
To determine whether specific expression of activated FoxM1-ΔN transgene in Clara cells induced airway hyperplasia, TetO-GFP-FoxM1-ΔN transgenic mice were bred with CCSP-rtTA mice that specifically express rtTA in Clara cells in a Dox-inducible manner (Perl et al., 2002; Tichelaar et al., 2000). At 30 days of age, CCSP-rtTA/TetO-GFP-FoxM1-ΔN double transgenic mice (CCSP-FoxM1) were treated with Dox for 90 days (Fig. 10A). Expression of FoxM1-ΔN in Clara cells induced airway hyperplasia at sites expressing the transgene (Fig. 10E). Both CCSP and proliferation-specific Ki-67 were present in the lesions (Fig. 10G and 10I). Although lung tumors were not observed in the CCSP-FoxM1 transgenic mice, specific expression of FoxM1-ΔN in Clara cells was sufficient to induce airway hyperplasia.
Figure 10.
Clara cell hyperplasia in CCSP-rtTA/TetO-FoxM1-ΔN double transgenic (CCSP-FoxM1) mice. (A) A schematic showing Dox-inducible mouse model, which expresses GFP-FoxM1-ΔN transgene under the control of CCSP promoter. (B-I) H&E and IHC staining of CCSP-FoxM1 and WT lungs after 3 months of Dox treatment (P30-P120). Hyperplastic epithelial regions (arrows) were detected in CCSP-FoxM1 airways, which were positive for FoxM1-ΔN (D-E), CCSP (F-G) and Ki-67 (H-I).
DISCUSSION
Previous studies have demonstrated that Foxm1 is expressed in a variety of respiratory cell types undergoing DNA replication and mitosis (Kalin et al., 2008; Kalinichenko et al., 2003; Kim et al., 2005; Wang et al., 2009). In this study, we found that Foxm1 expression in the lung was dramatically reduced prior to birth, but was re-activated during the early postnatal period. The observed pattern of Foxm1 expression is similar to that of cell proliferation, consistent with the role of Foxm1 in the cell cycle. To address the stage-specific role of Foxm1 in respiratory epithelium, we used a constitutively active FoxM1-ΔN mutant to increase Foxm1 activity in epithelial cells under control of Dox. We found that expression of FoxM1-ΔN transgene during E7.5-E15.5 caused epithelial hyperplasia, indicating that increased Foxm1 activity is sufficient to accelerate cellular proliferation in undifferentiated respiratory epithelium. This finding is not surprising, considering that Foxm1 directly activates transcription of multiple cell cycle regulatory genes, including cyclins B1 and A2, Cdc25B phosphotase, JNK1, PLK1 and Aurora B kinase (Costa, 2005; Krupczak-Hollis et al., 2004; Laoukili et al., 2007; Wang et al., 2005; Wang et al., 2008a). We previously reported that a conditional deletion of the Foxm1 gene from developing respiratory epithelium at E7.5-E14.5 (SPC-rtTA/TetO-Cre/Foxm1fl/fl mice (Kalin et al., 2008)) caused severe abnormalities in lung maturation and respiratory failure at birth, however, no defects in epithelial proliferation were found (Kalin et al., 2008). Thus, Foxm1 appears to be dispensable for the cell cycle progression in undifferentiated respiratory epithelium, suggesting that other signaling pathways likely compensate for the loss of Foxm1 during lung development.
Previous studies demonstrated that a decrease in proliferation rates occurs prior to formation of pulmonary saccules (Perl and Whitsett, 1999). Severe sacculation defects were found in epFoxM1 lungs when the FoxM1-ΔN transgene was induced at E7.5-E18.5. Similar phenotypes were found in transgenic mice expressing activated K-Ras (Shaw et al., 2007; Tuveson et al., 2004), FGFR (Hokuto et al., 2003), or FGF-7 (Tichelaar et al., 2000), all of which are positive regulators of epithelial proliferation (Warburton et al., 2000). Therefore, sacculation defects in epFoxM1 lungs may be a direct consequence of increased proliferation of epithelial cells. Interestingly, co-localization experiments demonstrated that cells expressing the FoxM1-ΔN transgene lacked pro-SPC, a marker of mature type II epithelial cells. These results suggest that increased Foxm1 activity prevents differentiation of epithelial cells toward the type II cell lineage. Impaired differentiation of type II cells resulted in sacculation defects in transgenic mice over-expressing TTF1 (Wert et al., 2002), Sox17 (Park et al., 2006) as well as in mice deficient in Foxa2 (Wan et al., 2004), NFAT (Dave et al., 2006), or β-catenin (Mucenski et al., 2003). Therefore, abnormalities in the differentiation of type II cells may contribute to the sacculation defects seen in epFoxM1 lungs.
Expression of FoxM1-ΔN transgene during the late gestation and the postnatal period caused airway hyperplasia. We also found that the majority of cells expressing FoxM1-ΔN transgene displayed markers specific for Clara cells, demonstrating that airway hyperplasia resulted from increased Clara cells proliferation. In contrast, the transgene was not found in mature type II cells of the alveolar region. These results are surprising, especially considering previous lineage-tracing experiments demonstrating that SPC-rtTA (line 1) induced Cre-mediated recombination in a majority of type II cells and in a subset of airway Clara cells (Perl et al., 2002; Tichelaar et al., 2000). Our results showed that transgenic FoxM1-ΔN expression accelerated proliferation of Clara cells but did not influence proliferation of mature type II cells. However, the mechanism underlying transgene silencing in type II cells remains unclear. We hypothesized that overexpression of activated FoxM1 protein could be toxic to mature type II cells, nonetheless, in contrast to this hypothesis, there was no cell apoptosis found in the epFoxM1 lungs. Furthermore, a robust β-gal activity was observed in the alveolar region of epFoxM1/Rosa26R lungs, suggesting that FoxM1-ΔN transgene was expressed and then silenced in a majority of type II cells. It is possible that activated FoxM1 promotes differentiation of epithelial progenitors towards the Clara cell lineage and prevents differentiation of progenitors toward type II cell lineage.
Increased Foxm1 levels have been found in numerous types of human tumors (Laoukili et al., 2007), including non-small cell lung cancers (Gialmanidis et al., 2009; Kim et al., 2006; Yang et al., 2009). Our previous studies demonstrated that when the Foxm1fl/fl allele was deleted in either all respiratory cell types or only in pulmonary epithelial cells, the number and size of lung adenomas following urethane exposure was reduced (Kim et al., 2006; Wang et al., 2009). Over-expression of FoxM1 in all cell types using the Rosa 26 promoter accelerated formation of lung adenomas induced by the tobacco smoke carcinogen MCA (Wang et al., 2008b). Although these studies indicate an important role of Foxm1 in lung carcinogenesis, it is not clear whether an increase in Foxm1 activity alone in sufficient to cause lung tumors. In this study, we found that epFoxM1 mice developed Clara cell hyperplasia after Dox treatment. Interestingly, only 11% of epFoxM1 mice developed single lung adenomas when Dox was used for 6 months. Perhaps this result is not surprising since 13% male and 26% female mice in FVB/N background spontaneously developed lung tumors by 14 months of age (Mahler et al., 1996). Interestingly, we found that FoxM1-ΔN transgene accelerated a tumor growth in the presence of activated (oncogenic) K-Ras. These results suggest that FoxM1-ΔN cooperates with activated K-Ras to induce lung tumor growth in vivo.
In summary, expression of activated Foxm1 transgene resulted in epithelial hyperplasia and sacculation defects during lung formation in late gestation. FoxM1-ΔN expression during the postnatal period caused focal airway hyperplasia, increased proliferation of Clara cells and altered expression of Sox transcription factors. FoxM1-ΔN accelerated tumor growth induced by activated K-Ras. Precise regulation of Foxm1 expression in respiratory epithelial cells is critical for lung sacculation, proper development of airway epithelium, and lung tumorigenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Loudy for technical support and Ann Maher for excellent editorial assistance. We thank Dr. Craig Bolte (CCHMC) for critical reviewing the manuscript. We also thank Dr. Pradip Raychaudhuri (UIC) for kindly providing the GFP-FoxM1-ΔN construct. This work was supported by HL 84151-04 grant from National Institute of Health (V.V.K.), the Research Scholar Grant from American Cancer Society (V.V.K.) and the Career Development Award from National Lung Cancer Partnership (I-C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, Whitsett JA. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135:1049–58. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- Blanco-Bose WE, Murphy MJ, Ehninger A, Offner S, Dubey C, Huang W, Moore DD, Trumpp A. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008;48:1302–11. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]
- Blatt EN, Yan XH, Wuerffel MK, Hamilos DL, Brody SL. Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am J Respir Cell Mol Biol. 1999;21:168–76. doi: 10.1165/ajrcmb.21.2.3691. [DOI] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, Whitsett JA. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest. 2009;119:2914–24. doi: 10.1172/JCI39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT, Whitsett JA. FGF-10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol Lung Cell Mol Physiol. 2001;280:L705–15. doi: 10.1152/ajplung.2001.280.4.L705. [DOI] [PubMed] [Google Scholar]
- Costa RH. FoxM1 dances with mitosis. Nat Cell Biol. 2005;7:108–10. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–47. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- Dave V, Childs T, Xu Y, Ikegami M, Besnard V, Maeda Y, Wert SE, Neilson JR, Crabtree GR, Whitsett JA. Calcineurin/Nfat signaling is required for perinatal lung maturation and function. J Clin Invest. 2006;116:2597–609. doi: 10.1172/JCI27331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave V, Wert SE, Tanner T, Thitoff AR, Loudy DE, Whitsett JA. Conditional deletion of Pten causes bronchiolar hyperplasia. Am J Respir Cell Mol Biol. 2008;38:337–45. doi: 10.1165/rcmb.2007-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–62. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gialmanidis IP, Bravou V, Amanetopoulou SG, Varakis J, Kourea H, Papadaki H. Overexpression of hedgehog pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung Cancer. 2009;66:64–74. doi: 10.1016/j.lungcan.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Hokuto I, Perl AK, Whitsett JA. Prenatal, but not postnatal, inhibition of fibroblast growth factor receptor signaling causes emphysema. J Biol Chem. 2003;278:415–21. doi: 10.1074/jbc.M208328200. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, Lyubimov A, Costa RH. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin TV, Wang IC, Meliton L, Zhang Y, Wert SE, Ren X, Snyder J, Bell SM, Graf L, Jr., Whitsett JA, Kalinichenko VV. Forkhead Box m1 transcription factor is required for perinatal lung function. Proc Natl Acad Sci U S A. 2008;105:19330–5. doi: 10.1073/pnas.0806748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Tan Y, Wang IC, Major ML, Wang X, Yoder HM, Costa RH. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell types following lung injury. J Biol Chem. 2003;278:37888–94. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The Forkhead Box m1 Transcription Factor Stimulates the Proliferation of Tumor Cells during Development of Lung Cancer. Cancer Res. 2006;66:2153–61. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–86. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- Korver W, Schilham MW, Moerer P, van den Hoff MJ, Dam K, Lamers WH, Medema RH, Clevers H. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr Biol. 1998;8:1327–30. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- Krupczak-Hollis K, Wang X, Kalinichenko VV, Gusarova GA, Wang IC, Dennewitz MB, Yoder HM, Kiyokawa H, Kaestner KH, Costa RH. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, Heck AJ, Medema RH. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol. 2008;28:3076–87. doi: 10.1128/MCB.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Liu C, Morrisey EE, Whitsett JA. GATA-6 is required for maturation of the lung in late gestation. Am J Physiol Lung Cell Mol Physiol. 2002;283:L468–75. doi: 10.1152/ajplung.00044.2002. [DOI] [PubMed] [Google Scholar]
- Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118:795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- Mahler JF, Stokes W, Mann PC, Takaoka M, Maronpot RR. Spontaneous lesions in aging FVB/N mice. Toxicol Pathol. 1996;24:710–6. doi: 10.1177/019262339602400606. [DOI] [PubMed] [Google Scholar]
- Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–61. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeier W, Morrisey EE, Whitsett JA. beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J Biol Chem. 2003;278:40231–8. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. APC/C-Cdh1 Mediated Proteolysis of the Forkhead Box M1 Transcription Factor is Critical for Regulated Entry into S phase. Mol Cell Biol. 2008a doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Wang Z, Costa RH, Tyner A, Lau LF, Raychaudhuri P. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene. 2008b;27:1696–704. doi: 10.1038/sj.onc.1210814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Wells JM, Zorn AM, Wert SE, Whitsett JA. Sox17 influences the differentiation of respiratory epithelial cells. Dev Biol. 2006;294:192–202. doi: 10.1016/j.ydbio.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Perl AK, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res. 2002;11:21–9. doi: 10.1023/a:1013986627504. [DOI] [PubMed] [Google Scholar]
- Perl AK, Whitsett JA. Molecular mechanisms controlling lung morphogenesis. Clin Genet. 1999;56:14–27. doi: 10.1034/j.1399-0004.1999.560103.x. [DOI] [PubMed] [Google Scholar]
- Ramakrishna S, Kim IM, Petrovic V, Malin D, Wang IC, Kalin TV, Meliton L, Zhao YY, Ackerson T, Qin Y, Malik AB, Costa RH, Kalinichenko VV. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev Dyn. 2007;236:1000–13. doi: 10.1002/dvdy.21113. [DOI] [PubMed] [Google Scholar]
- Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, Rowitch DH. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27:8259–70. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Meissner A, Dowdle JA, Crowley D, Magendantz M, Ouyang C, Parisi T, Rajagopal J, Blank LJ, Bronson RT, Stone JR, Tuveson DA, Jaenisch R, Jacks T. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–80. [PubMed] [Google Scholar]
- Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem. 2000;275:11858–64. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- Tichelaar JW, Wert SE, Costa RH, Kimura S, Whitsett JA. HNF-3/forkhead homologue-4 (HFH-4) is expressed in ciliated epithelial cells in the developing mouse lung. J Histochem Cytochem. 1999;47:823–32. doi: 10.1177/002215549904700612. [DOI] [PubMed] [Google Scholar]
- Tompkins DH, Besnard V, Lange AW, Wert SE, Keiser AR, Smith AN, Lang R, Whitsett JA. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS One. 2009;4:e8248. doi: 10.1371/journal.pone.0008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, Hingorani SR, Zaks T, King C, Jacobetz MA, Wang L, Bronson RT, Orkin SH, DePinho RA, Jacks T. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–87. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Ueno H, Nakajo N, Watanabe M, Isoda M, Sagata N. FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development. 2008;135:2023–30. doi: 10.1242/dev.019893. [DOI] [PubMed] [Google Scholar]
- Ustiyan V, Wang IC, Ren X, Zhang Y, Snyder J, Xu Y, Wert SE, Lessard JL, Kalin TV, Kalinichenko VV. Forkhead box M1 transcriptional factor is required for smooth muscle cells during embryonic development of blood vessels and esophagus. Dev Biol. 2009;336:266–79. doi: 10.1016/j.ydbio.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Kaestner KH, Ang SL, Ikegami M, Finkelman FD, Stahlman MT, Fulkerson PC, Rothenberg ME, Whitsett JA. Foxa2 regulates alveolarization and goblet cell hyperplasia. Development. 2004;131:953–64. doi: 10.1242/dev.00966. [DOI] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Forkhead Box M1 Regulates the Transcriptional Network of Genes Essential for Mitotic Progression and Genes Encoding the SCF (Skp2-Cks1) Ubiquitin Ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes DE, Ackerson T, Major ML, Kalinichenko VV, Costa RH, Raychaudhuri P, Tyner AL, Lau LF. FoxM1 Regulates Transcription of JNK1 to Promote the G1/S Transition and Tumor Cell Invasiveness. J Biol Chem. 2008a;283:20770–8. doi: 10.1074/jbc.M709892200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Meliton L, Ren X, Zhang Y, Balli D, Snyder J, Whitsett JA, Kalinichenko VV, Kalin TV. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One. 2009;4:e6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, Kalin TV. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008b;27:4137–49. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- Wert SE, Dey CR, Blair PA, Kimura S, Whitsett JA. Increased expression of thyroid transcription factor-1 (TTF-1) in respiratory epithelial cells inhibits alveolarization and causes pulmonary inflammation. Dev Biol. 2002;242:75–87. doi: 10.1006/dbio.2001.0540. [DOI] [PubMed] [Google Scholar]
- Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- Xia LM, Huang WJ, Wang B, Liu M, Zhang Q, Yan W, Zhu Q, Luo M, Zhou ZZ, Tian DA. Transcriptional up-regulation of FoxM1 in response to hypoxia is mediated by HIF-1. J Cell Biochem. 2009;106:247–56. doi: 10.1002/jcb.21996. [DOI] [PubMed] [Google Scholar]
- Yang DK, Son CH, Lee SK, Choi PJ, Lee KE, Roh MS. Forkhead box M1 expression in pulmonary squamous cell carcinoma: correlation with clinicopathologic features and its prognostic significance. Hum Pathol. 2009;40:464–70. doi: 10.1016/j.humpath.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, Roebuck KA, Costa RH. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–41. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–31. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, Costa RH, Gannon M. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20:1853–66. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.