Abstract
Toll-like receptor (TLR)-7 agonists show prominent immunostimulatory activities. The synthesis of a TLR7-active N1-(4-aminomethyl)benzyl substituted imidazoquinoline 5d served as a convenient precursor for the covalent attachment of fluorophores without significant loss of activity. Fluorescence microscopy experiments show that the fluorescent analogues are internalized and distributed in the endosomal compartment. Flow cytometry experiments using whole human blood show differential partitioning into B, T, and natural killer (NK) lymphocytic subsets, which correlate with the degree of activation in these subsets. These fluorescently-labeled imidazoquinolines will likely be useful in examining the trafficking of TLR7 in immunological synapses.
Keywords: Imidazoquinoline, Toll-like receptor, TLR7, Fluorescent probe, Flow cytometry
Toll-like receptors (TLRs) are pattern recognition receptors that recognize specific molecular patterns present in molecules that are broadly shared by pathogens, but are structurally distinct from host molecules.1 There are 10 TLRs in the human genome.2 The ligands for these receptors are highly conserved microbial molecules such as lipopolysaccharides (LPS) (recognized by TLR4), lipopeptides (TLR2 in combination with TLR1 or TLR6), flagellin (TLR5), single stranded RNA (TLR7 and TLR8), double stranded RNA (TLR3), CpG motif-containing DNA (recognized by TLR9), and profilin present on uropathogenic bacteria (TLR 11).3 The activation of TLRs by their cognate ligands leads to activation of innate immune effector mechanisms, including the production of pro-inflammatory cytokines, up-regulation of MHC molecules and co-stimulatory signals in antigen-presenting cells, resulting in amplification of specific adaptive immune responses involving both T- and B-cell effector functions.4-6 Thus, TLR stimuli serve to link innate and adaptive immunity4 and can therefore be exploited as powerful adjuvants in eliciting both primary and anamnestic immune responses.
Our point of departure in the systematic evaluation of TLR agonists as vaccine adjuvants7-9 focuses on identifying chemotypes which are strongly immunostimulatory, and yet devoid of dominant proinflammatory cytokine-inducing activities;7 the TLR7-agonistic imidazoquinolines have thus far seemed ideal in meeting these requirements.7 Continuing work on characterizing the immunostimulatory activities of TLR7 agonists show, as expected, clear involvement of plasmacytoid dendritic cells,10 but we have also observed a set of accessory cell-independent direct responses in CD4+ and CD8+ T and CD3-CD56+ natural killer (NK) lymphocytes (to be published elsewhere). We are specifically desirous of examining the uptake, intracellular distribution, and trafficking of the imidazoquinoline in immunological synapses,11;12 and it became necessary to develop probes of TLR7 that are fluorescently labeled.
Our earlier SAR study on the TLR7-agonistic activities had exhaustively explored C2 and C4 substituents on the imidazoquinoline scaffold (Fig. 1),13 but proved unsuccessful in identifying potential positions that would tolerate the introduction of bulky aryl groups without compromising activity.
Figure 1.
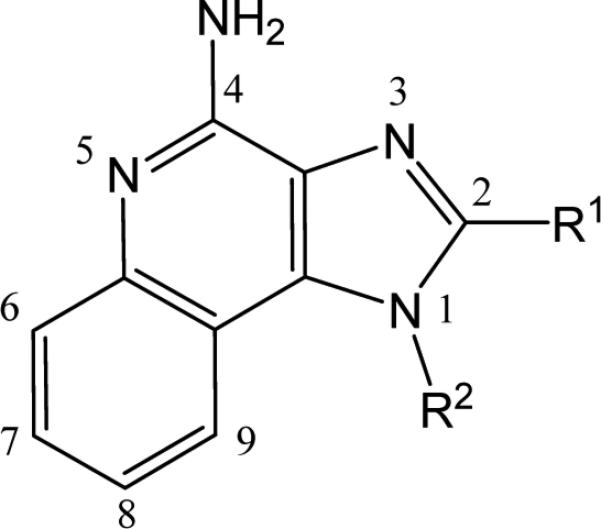
General structure of 1H-imidazo[4,5-c]quinolin-4-amine. The C2 and N1 substituents that were found to correspond to optimal TLR7-agonistic activity were n-butyl and benzyl, respectively (Ref. 13).
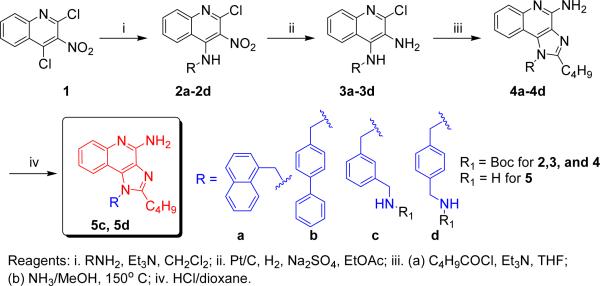
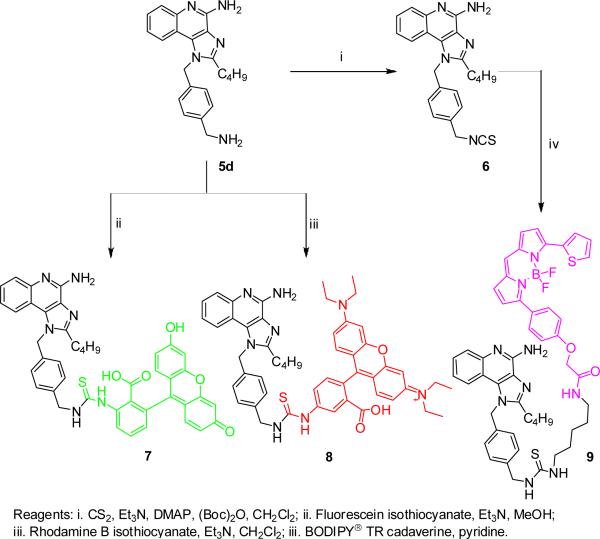
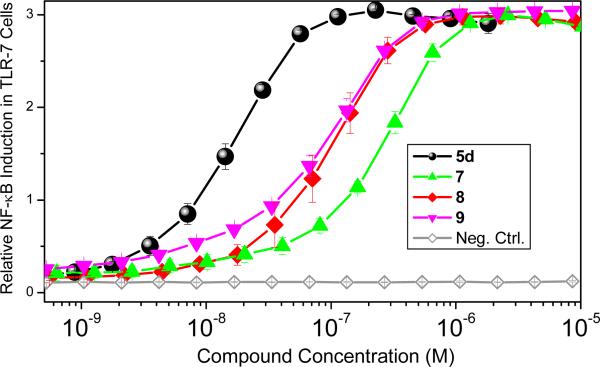
Our attention subsequently turned to exploring the effect of varying substituents at N1, while holding the C2-n-butyl and C4-NH2 groups constant since these have been shown to correspond to maximal activity.13 The N1-naphthylenemethyl-substituted compound 4a was inactive, and the N1-biphenyl-4-methyl compound 4b was weakly active (EC50: 396 nM); the N1-(4-aminomethyl)benzyl substituted analogue 5d was considerably more active (EC50: 20 nM; Scheme 1) than its N1-(3-aminomethyl)benzyl regioisomer 5c (EC50: 110 nM). The free primary amine on the N1 substituent of 5d was covalently coupled directly to commercially-available fluorescein isothiocyanate and rhodamine B isothiocyanate (Scheme 2). Conversely, the amine on 5d was converted first to the isothiocyanate 6, allowing the subsequent coupling of amine-bearing fluorophores, such as the bora-diazaindacene dye, BODIPY-TR-cadaverine (Scheme 2). All three fluorescent conjugates retain TLR7-agonistic activity, although their potencies are slightly attenuated relative to the parent compound, 5d (Fig. 2); the EC50 values of 7, 8, and 9 are, respectively, 247 nM, 115 nM, and 108 nM.
Scheme 1.
Syntheses of N1-substituted imidazoquinolines.
Scheme 2.
Syntheses of fluorescent analogues of 5d.
Figure 2.
Activities of 5d, 7, 8, and 9 in reporter gene assays using human TLR7.
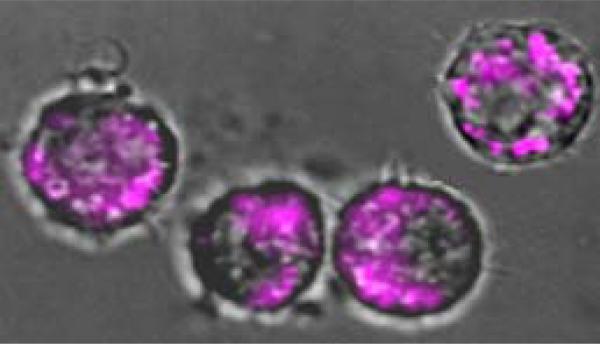
Incubation of murine macrophage J774.A1 cells with 8 or 9, followed by intravital epi- and confocal fluorescence microcopy showed prominent perinuclear localization, which is consistent with the expected endosomal distribution of TLR7.14 Shown in Fig. 3 is a representative epifluorescence micrograph of J774 cells treated with 9 at 100 nM concentration.
Figure 3.
Murine J774 cells treated with 100 nM of 9. An overlay of phase-contrast and epifluorescence images is depicted. An excitation filter at 562 nm and a long-pass emission filter (601-800 nm) were used.
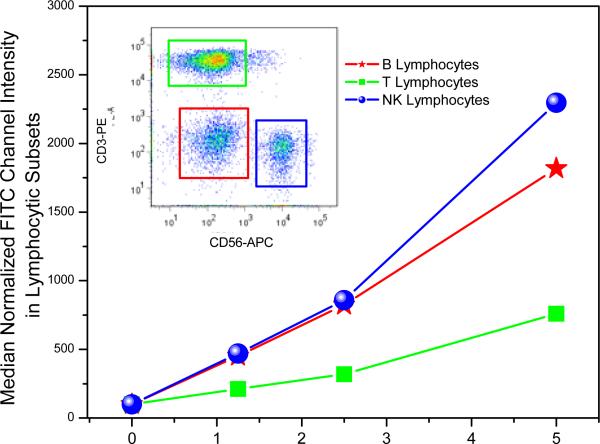
Our earlier immunoprofiling of the TLR7-agonistic imidazoquinolines had shown a very prominent activation of B- and NK-cells, but minimal activation of T cells,7 and we asked if a possible reason could be differential uptake of the TLR7 agonist in lymphocytic subsets. Flow cytometric analysis of the FITC-labeled 7 in experiments employing whole human blood indeed show a prominent uptake of 7 in CD3-CD56+ NK and CD3-CD56- B lymphocytes as compared to CD3+CD56- T lymphocytes (Fig. 4).
Figure 4.
Uptake of 7 in lymphocytic subsets as examined by flow cytometry. Whole human blood was incubated with graded concentrations of 7 for 30 min, lymphocytes stained with cell surface markers (anti-CD3-phycoerythrin [PE], and anti-CD56-PE-allophycocyanin). Erythrocytes were lysed, and 105 total events were acquired per sample.
The syntheses of fluorescent imidazoquinoline analogues that retain TLR7-agonistic activity are expected to be useful probes in examining their potential immunostimulatory and adjuvantic properties.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIAID contract HHSN272200900033C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Experimental methods and characterization of compounds (1H, 13C, and mass spectra).
References
- 1.Kawai T, Akira S. Semin.Immunol. 2007 Jan 31;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai Y, Takeuchi O, Akira S. J.Infect.Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- 3.Akira S. Immunol.Rev. 2009;227:5–8. doi: 10.1111/j.1600-065X.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K, Kaisho T. Nature Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 5.Cottalorda A, Verschelde C, Marcais A, Tomkowiak M, Musette P, Uematsu S, Akira S, Marvel J, Bonnefoy-Berard N. Eur.J.Immunol. 2006;36:1684–1693. doi: 10.1002/eji.200636181. [DOI] [PubMed] [Google Scholar]
- 6.Kaisho T, Akira S. Biochim.Biophys.Acta. 2002 Feb 13;1589:1–13. doi: 10.1016/s0167-4889(01)00182-3. [DOI] [PubMed] [Google Scholar]
- 7.Hood JD, Warshakoon HJ, Kimbrell MR, Shukla NM, Malladi S, Wang X, David SA. Hum.Vaccin. 2010;6:1–14. doi: 10.4161/hv.6.4.10866. [DOI] [PubMed] [Google Scholar]
- 8.Shukla NM, Kimbrell MR, Malladi SS, David SA. Bioorg.Med.Chem.Lett. 2009 Apr 15;19:2211–2214. doi: 10.1016/j.bmcl.2009.02.100. [DOI] [PubMed] [Google Scholar]
- 9.Warshakoon HJ, Hood JD, Kimbrell MR, Malladi S, Wu WY, Shukla NM, Agnihotri G, Sil D, David SA. Hum.Vaccin. 2009 Jun 16;5:381–394. doi: 10.4161/hv.5.6.8175. [DOI] [PubMed] [Google Scholar]
- 10.Liu YJ. Annu.Rev.Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 11.Padhan K, Varma R. Immunology. 2010;129:322–328. doi: 10.1111/j.1365-2567.2009.03241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manz BN, Groves JT. Nat.Rev.Mol.Cell Biol. 2010;11:342–352. doi: 10.1038/nrm2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla NM, Malladi SS, Mutz CA, Balakrishna R, David SA. J.Med.Chem. 2010 Jun 10;53:4450–4465. doi: 10.1021/jm100358c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai T, Akira S. J.Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.