Abstract
PDCD2 is a conserved eukaryotic protein implicated in cell cycle regulation by virtue of its interactions with HCFC1 and the NCOR1/SIN3A corepressor complex. Pdcd2 transcripts are enriched in ES cells and other somatic stem cells, and its ortholog is essential for hematopoietic stem cell maintenance in Drosophila. To characterize the physiological role(s) of mammalian PDCD2, we created a disruption allele in mice. Pdcd2−/− embryos underwent implantation but did not undergo further development. Inner cell masses (ICMs) from Pdcd2−/− blastocysts failed to outgrow in vitro. Furthermore, embryonic stem cells (ESCs) require PDCD2 as demonstrated by the inability to generate Pdcd2−/− ESCs in the absence of an ectopic transgene. Upon differentiation of ESCs by retinoic acid treatment or LIF deprivation, PDCD2 levels declined. In conjunction with prior studies, these results indicate that in vivo, PDCD2 is critical for blastomere and ESC maintenance by contributing to the regulation of genes in a manner essential to the undifferentiated state of these cells.
Keywords: mouse development, embryonic stem cells, cell differentiation, preimplantation
Introduction
Pdcd2 (Programmed cell death 2; originally called Rp8 in rats) was identified in a screen for mRNAs upregulated upon apoptosis of rat thymocytes (Owens et al., 1991). However, subsequent experiments failed to support a correlation between Pdcd2 expression and apoptosis (Fan et al., 2004; Kawakami et al., 1995; Minakhina et al., 2007; Steinemann et al., 2003). While there is little published information on the role of Pdcd2, studies have revealed that it is directly repressed by BCL6, which is associated with translocation breakpoints in B and T cell lymphomas (Baron et al., 2002; Baron et al., 2007). PDCD2 appears to be a transcriptional regulator involved in cell cycle control during cell proliferation and differentiation. It physically interacts with host cell factor 1 (HCFC1, also called HCF-1) (Scarr and Sharp, 2002), which functions as a coactivator or corepressor in the regulation of multiple phases of the cell cycle (Tyagi et al., 2007).
PDCD2 plays critical roles during embryonic development in Drosophila. Zfrp8 (the Drosophila ortholog of Pdcd2) mutant exhibits developmental delay, larval and pupal lethality (Minakhina et al., 2007). Our previous studies also suggested the potential importance of Pdcd2 in mouse embryonic development. Pdcd2 is a likely candidate for the t haplolethal 1 (Thl1) locus (Howell et al., 2005), identified by deletions on proximal Chr 17 (Browning et al., 2002). Thl1 hemizygosity causes strain background-dependent embryonic lethality. Microarray profiling of mouse tissues and developmental stages showed that the site of highest Pdcd2 transcription is blastocyst stage embryos (Su et al., 2004), implicating its participation in preimplantation development.
Although the identification of certain key factors such as OCT3/4, SOX2, and NANOG (Boyer et al., 2005; Loh et al., 2006) has given insight into the maintenance of an undifferentiated state in mouse and human ESCs, numerous auxiliary transcription factors have been implicated to play a role in ESC biology (Lunyak and Rosenfeld, 2008). PDCD2 was found to be enriched in three types of mouse stem cells (embryonic, neural, and hematopoietic) (Ramalho-Santos et al., 2002) and human ESCs compared to their differentiated derivatives (Skottman et al., 2005). Functional evidence for a key role for PDCD2 in stem cells was recently reported in Drosophila, whereby the ortholog Zfrp8 was found to be essential for maintenance of hamatopoietic stem cells but not differentiated daughters (Minakhina and Steward, 2010). PDCD2 interacts physically with HCFC1 (HCF-1) and NCOR1 (Scarr and Sharp, 2002), which are components of histone modification complexes and are required for embryonic and adult stem cell self-renewal (Dejosez et al., 2008; Hermanson et al., 2002; Jepsen et al., 2007). Taken together, these data implicate PDCD2 as a potential auxiliary factor in stem cell maintenance and differentiation.
Here, we disrupted Pdcd2 in mice via gene targeting to elucidate the physiological roles of PDCD2 in embryonic development, and to determine if Pdcd2 functions in a dose-dependent manner during embryogenesis as implied by deletion analysis of its genomic region. We found that whereas hemizygosity for Pdcd2 did not disrupt embryogenesis, complete absence of PDCD2 led to the failure of ICM outgrowth and peri-implantation lethality. Furthermore, we found that PDCD2 is essential for ESC viability, but that its levels decrease upon differentiation. These and other data suggest that PDCD2 has a role in stem cell self-renewal.
Materials and Methods
Targeting vector construction
Two homologous arm fragments of 2.5-kb and 4.3-kb were amplified by PCR from a mouse 129 strain BAC clone, bMQ245J21 (The Sanger Institute) containing the entire Pdcd2 gene. The primers contained restriction sites as follows. For the 2.5-kb fragment: 5’-CCCCGCGGCAATCCCATCTCACCTCACC-3’ (Sac II site) and 5’-CTAGCTAGCCAGGATGAAAGGGACACGAT-3’ (Nhe I site); for the 4.3 kb fragment: 5’-ACGCGTCGACTACCCACCCACTCCTGATTC-3’ (Sal I site) and 5’-ATTTGCGGCCGCTCTGTTCTGGCATGTTGA GC-3’ (Not I site). The PCR products were digested with those restriction enzymes and subcloned upstream and downstream of a PGK-neo expression cassette to generate the targeting vector. Homologous recombination between the WT allele and targeting vector resulted in the disruption of Pdcd2 by replacing exon 2 with PGK-neo (Fig. 1a).
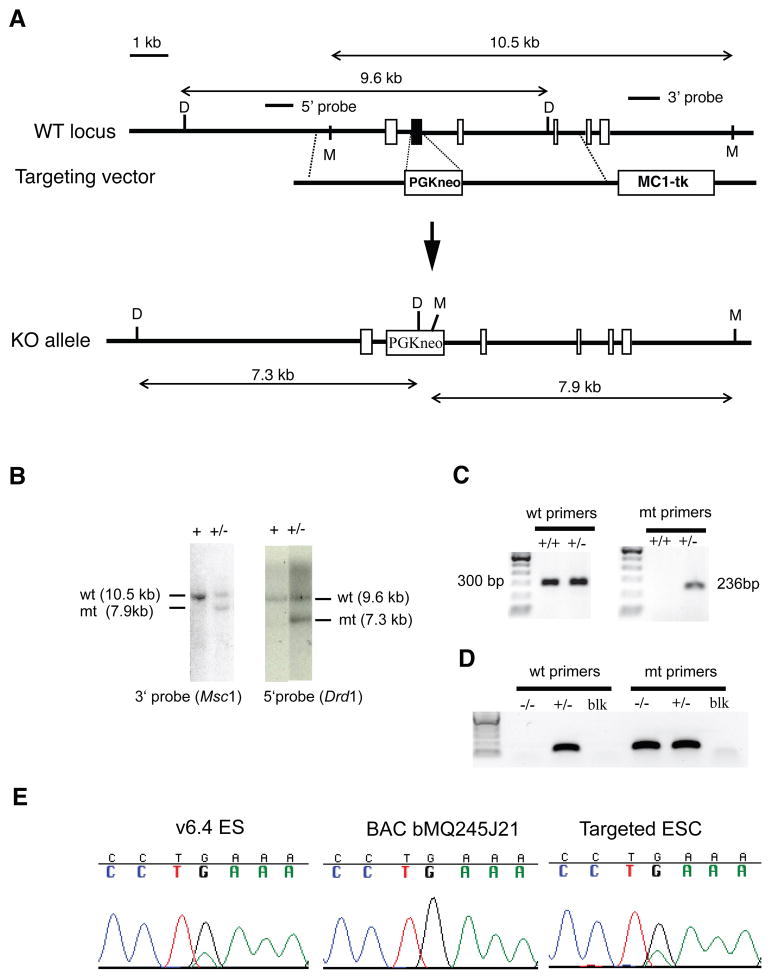
Figure 1. Targeted disruption of mouse Pdcd2.
(A) A map of the genomic locus surrounding the second exon (solid black) is shown. Other exons are depicted as empty boxes. The targeting construct contains 2.5 kb of 5’ flanking sequence, a PGK-Neo cassette, 4.3 kb of 3’ flanking sequence, and the herpes simplex thymidine kinase (MC1-TK) cassette for negative selection. Correct homologous recombination will yield the indicated “KO allele.” M = MscI. D = DrdI. (B) Southern blot analysis of ESC genomic DNA digested with the indicated enzymes and hybridized with the indicated probes depicted in “A.” (C) PCR genotype analysis with primer pairs for the mutant (mt) or wild-type (wt) alleles. (D) Nested PCR genotyping of blastocysts. “blk” = no embryo control. (E) DNA sequencing traces of a region containing a 129 vs B6 SNP in the Pdcd2 targeted region (the SNP is included in the 3’ targeting arm, in intron 3). The v6.4 ES cells used for gene targeting are (B6X129) F1. The targeting DNA was from strain 129 (amplified from BAC bMQ245J21). Targeting of the 129 allele of Pdcd2 with the 129 vector leaves heterozygosity of the SNP, as shown by sequencing (E).
Gene targeting of ES cells and generation of mutant mice
(129XC57BL/6J) F1 v6.4 ES cells (107 cells in 0.8 ml PBS) were electroporated with 20μg of AscI-linearized targeting vector DNA. Transformants were selected with G418 (Geneticin, 250 μg/ml; Gibco-BRL, Rockville, Md.) and 0.2 μM FIAU (2'-fluoro 2'-deoxy-5-iodouracil-ß-D-arabinofuranoside; Moravek Biochemicals, Brea, Calif.). Correctly targeted clones were identified by Southern blot analysis with 5' and 3' probes as indicated in Figs. 1a, b). 129 allele-targeted ESC clones were verified by SNP analysis (Fig. 1e) and microinjected into C57BL/6J (“B6”) blastocysts to generate chimeric mice. These chimeras were subsequently crossed with B6 females, and heterozygous offspring bearing the targeted allele were intercrossed to produce progeny for phenotype analyses.
Animals were genotyped by PCR. The sizes of PCR products for WT and knockout alleles are 300-bp and 236-bp, respectively (Fig. 1c). Common forward primer: 5’-GAATCGTGT CCCTTTCATCC-3’; WT allele reverse primer: 5’-ATGCGTTGTTCCTTGGTAGC-3’; Targeted allele reverse primer: 5’-ATTGCATCGCATTGTCTGAG-3’. E3.5 embryos were genotyped by nested PCR as follows. The first round of amplification (25 cycles at 55° annealing) was performed with primer pairs Pdcd2WtnestL (5’-CTGAACCCTGGACGTAGGAC-3’) plus Pdcd2WtnestR (5’-TTTTCGGTGCTTCACCTCAT-3’) for WT and Pdcd2WtnestL plus Pdcd2KonestR (5’-GGGAGGATTGGGAAGACAAT-3’) for mutant. Two microliters of first round PCR product was used as template for the second round PCR (35 cycles) using primer pairs Pdcd2WTnest2L (5’-CGCGAGTGGTTGTATTCAGG-3’) plus Pdcd2WTnest2R (5’-GCTTTTAAACCCGGGAAGAG-3’) for WT and Pdcd2WTnest2L plus Pdcd2KOnest2R (5’-GTGGGCTCTATGGCTTCTGA-3’) for mutant. The amplified products are 119- and 128-bp for the wild type and mutant, respectively (Fig. 1d).
RNA in situ hybridization
In situ hybridizations were performed as described (Welsh and O'Brien, 2000). Pdcd2 probes were synthesized using the DIG RNA Labeling Kit (Roche). The primers used to amplify the probe were 5’-ATGACCCAGCAGTGGAGATT-3’ and 5’-GCGACCTCATTTGGTTTCAG-3’.
Immunolabelling and genotyping of preimplantation embryos
The indirect immunofluorescence procedure was performed essentially as described (Vitale et al., 2005). The embryos were incubated with a mouse monoclonal anti-CDX2 antibody (BioGenex AM392-5M) overnight at 4°, then with incubated with secondary antibody Alexa Fluor 594 goat anti-mouse immunoglobulin G (Invitrogen) for 1 hour. After washing, they were subsequently DAPI stained, washed, then imaged using confocal microscopy. Cells that were DAPI-positive but CDX2-negative were considered to be ICM cells. After microscopy, DNA was isolated as described (Ralston et al., 2010) and genotyped by nested PCR as outlined above.
Isolation of nuclear and cytoplasmic fractions
ESCs were lysed in 10 mM Tris pH 7.4, 10 mM NaCl, 10 mM MgCl2, 0.05% Nonidet P-40, and protease inhibitor for cytosol preparations (Gondran et al., 1999). Then nuclear pellets were resuspended in CSK buffer (He et al., 1990)(10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mm EGTA, 0.5% Triton X-100, 1 mM DTT and protease inhibitors) on ice for 10 minutes. After 2 minute centrifugation at 5000×g, the supernatant, or nucleoplasm, was removed and the pellets were incubated with 7 μl Benzonase® Nuclease (Qiagen) in 13μl distilled water for 15 min at room temperature. Extraction buffers CE3 and CE4 in Qproteome cell compartment kit (Qiagen) were employed for chromatin-binding protein and cytoskeleton fraction, respectively.
Cell culture and ES cell differentiation
Mouse embryonic fibroblasts (MEFs), ESCs, and blastocysts were cultured as described previously (Mu et al., 2008). Blastocysts were collected after 5 days of culture for DNA isolation and genotyping. For the induction of ESC differentiation, 2 × 105 cells were plated on 60-mm dishes in ESC with LIF supplementation for 24 hours, then LIF was removed and replaced with retinoic acid (RA) at final concentrations indicated in the text. For measurement of alkaline phosphatase activity, the StemTAG Alkaline Phosphatase Activity Assay Kit (Cell Biolabs, Inc. San Diego, CA) was used according to the manufacturer’s instructions.
RT-PCR analysis
Total RNA was extracted from ESCs using RNeasy mini kits (Qiagen) and quantified using a NanoDrop apparatus. cDNA synthesis was primed with oligo(dT) on 1 μg of total RNA with SuperScript III (Invitrogen), according to the manufacturer's instructions. The mRNA levels of Oct4, Nanog, and Sox2 were semi-quantified by RT-PCR using β-actin as an internal control. The primers were those used by Gu et al. (Gu et al., 2005)
RNA interference
An siRNA pool, consisting of four individual siRNAs against Pdcd2 mRNA, was purchased from Dharmacon (Lafayette, CO). Immortalized MEFs were seeded on 6-well plates at 5 × 105 cells/well and grown to ~50% confluence. The cells were then transfected with siRNA against Pdcd2 or a negative control primer set at a final concentration of 100 nM using DharmaFECTTM 3 transfection reagent (Dharmacon) . ESCs were transfected with siRNA using lipofectamine 2000 (Invitrogen) according to manufacture’s protocol. The cells were harvested for assessing knockdown efficiency, cell proliferation, or differentiation 72 hours post transfection.
Western blot analysis
Protein was extracted from MEFs, ESCs, and tissues using radioimmunoprecipitation assay buffer, and the concentration was quantified with a BCA kit (Pierce). Amounts of 15 μg of total protein were separated by SDS-polyacrylamide gel electrophoresis, electrotransferred onto a pure nitrocellulose membrane (Bio-Rad), and probed with the relevant antibodies. Binding was detected by using a Pierce ECL kit. The band intensities were quantified by using NIH Image J software. The employed antibodies were rabbit anti-PDCD2 (ProteinTech Group, Inc.), rabbit anti-CREB (Cell signaling), mouse anti-β-actin (Sigma), and rabbit anti-GAPDH (Sigma).
Creation of homozygous mutant Pdcd2 ESCs
The remaining WT Pdcd2 allele was disrupted in Pdcd2+/− ESCs by homologous recombination or selection for spontaneous loss-of-heterozygosity (LOH). For homologous recombination, we used the same targeting vector as that described earlier, except that the neo resistance marker was replaced with a puromycin resistance gene. ESCs were selected in the medium containing 250 μg/ml G418 and 5 μg/ml puromycin (Sigma). Disruption of Pdcd2 by spontaneous LOH was performed as described (Mortensen et al., 1992) using 1mg/ml G418. For rescue experiments, Pdcd2+/− ESCs were electroporated with plasmids containing Pdcd2 cDNA and a blasticidin resistance gene. Cells were treated with 5μg/ml blasticidin (Invitrogen), and surviving colonies were screened by Western blot analysis of PDCD2 to identify Pdcd2-overexpressing cell lines. Those ESC lines were used for homozygous disruption of Pdcd2 as mentioned above.
Results
Pdcd2 deficiency is not sufficient to cause t complex haplolethality
To elucidate the function of Pdcd2 in mammalian development, we generated a disruption allele in ESCs by homologous recombination (Fig. 1a-c). The targeting strategy deletes exon 2, containing the MYND zinc-finger domain of PDCD2, and causes a frameshift that likely results in a null mutation. Targeted ES cells were readily obtained by homologous recombination (12.5% of G418-positive clones; Table 2), and used to generate mice transmitting the mutant locus (Pdcd2tm1Jcs, abbreviated Pdcd2−).
Table 2.
Disruption of remaining WT Pdcd2 allele in Pdcd2+/− ESCs by homologous recombination or selection for LOH in elevated G418.
| ESC line | Method | Colonies | Targeted/ LOH | Rate |
|---|---|---|---|---|
| Pdcd2+/− | HR | 48 | 6 | 12.5% |
| Pdcd2+/− | HR | 192 | 0 | 0 |
| Pdcd2+/− | G418 inc. | 96 | 0 | 0 |
| Flag-Pdcd2; Pdcd2+/− #5 | G418 inc. | 36 | 7 | 19.4% |
| Flag-Pdcd2; Pdcd2+/− #8 | G418 inc. | 24 | 19 | 79.2% |
| Flag-Pdcd2; Pdcd2+/− #8 | HR | 42 | 4 | 9.5% |
HR = homologous recombination. “inc.” = increase.
Pdcd2 resides in a 140 kb region containing a locus called t complex haplolethal 1 (Thl1) (Howell et al., 2005). Deletion of the 129 allele of Thl1 from (129XB6)F1 ESCs resulted in late gestation lethality of deletion-bearing embryos produced from matings of chimeras to B6 females, but not females of other strains (Browning et al., 2002). Pdcd2 was identified as a likely Thl1 candidate based on analysis of genes in the 140 kb critical region (Howell et al., 2005). To determine if Pdcd2 is indeed Thl1, we identified an ESC clone in which the 129 allele was targeted (Fig. 1e), generated chimeric males with 100% agouti coats (indicative of ESC contribution), mated them to B6 females, and determined inheritance of the mutant allele. Nearly equal numbers of Pdcd2 +/+ and Pdcd2 +/− pups were born (28 and 26, respectively; Table 1), all with visibly normal phenotypes. These results suggest that hemizygosity for Pdcd2 alone does not cause the Thl1 phenotype.
Table 1.
Offspring genotypes from Pdcd2 heterozygote intercrosses
| Stage | Genotype |
Total | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Weanling | 24 | 55 | 0 | 79 |
| 12.5 dpc | 9 | 14 | 0 | 23 |
| 7.5 dpc | 12 | 22 | 0 | 39 |
| 3.5 dpc | 13 | 26 | 7* | 46 |
Weanlings are ~ 21 days of age.
Among the 7 embryos, 2 were blastocysts, and 5 were at cleavage or morula stages as described in the text. “dpc” = days post coitus.
PDCD2 deficiency causes early embryonic lethality associated with defective ICM proliferation
Intercrosses of Pdcd2+/ mice failed to produce homozygous mutant weanlings (Table 1), and premature death of newborns (pre-genotyping) was not noted. To characterize the nature of the probable embryonic lethality, we then examined E7.5 and E12.5 embryos from intercrosses. Again, no homozygotes were seen, and there were no indications of abnormal, resorbing embryos at either stage (Table 1). We noted that in three of the E7.5 timed matings, the aggregate number of ovarian corpora lutea (27) equaled the number of uterine decidua, but 10 lacked any embryos. These observations indicated that mutant embryos implanted, but there was little or no development of the embryo proper. This phenotype is suggestive of a defect in the embryonic, but not extraembryonic (trophectoderm) lineage.
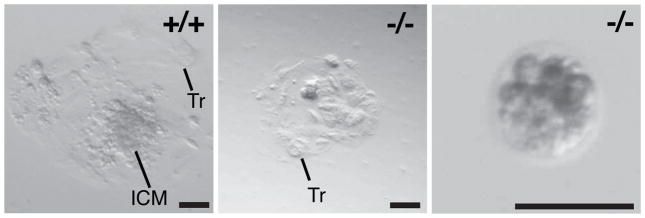
To test this idea, we collected E3.5 embryos (which should be at the blastocyst stage) and placed them into culture to perform inner cell mass (ICM) outgrowth assays. Of the 46 embryos collected, 7 were Pdcd2 −/− (not a statistically significant shortfall). Five of the homozygous mutant embryos were delayed at the cleavage or morula stages, and the remaining 2 were at the blastocyst stage (Table 1). All the Pdcd2+/+ and Pdcd2+/− embryos (39) attached to the culture dishes, hatched, and underwent ICM outgrowth. Whereas the two Pdcd2−/− blastocysts attached and hatched to produce a monolayer of viable trophoblast cells, they failed to exhibit ICM outgrowth (Fig. 2a). The five cleavage/morula stage mutant embryos did not attach or hatch, and their ICMs/blastomeres displayed decompaction (Fig. 2).
Figure 2. PDCD2 is essential for mouse preimplantation development.
E3.5 embryos were plated and placed in culture for 5 days to assess inner cell mass (ICM) outgrowth. The genotypes of three representative blastocysts are as indicated. The black bars indicate 100μm. Tr = trophoblast cell. Note that the mutant embryo in the right panel failed to attach to the culture dish and hatch. Also, the blastomeres appear to be de-compacting.
To determine if the failure of ICM outgrowth was due to a specific depletion of ICM cells or a preferential differentiation into trophectoderm, morula and blastocyst stage embryos were collected, stained with a trophectoderm specific marker (CDX2), and then genotyped (Supplemental Table 1). Three mutant embryos were identified of 25 that were successfully genotyped. Two morula stage embryos were isolated, containing an average of 22 total cells, 3.5 of which were CDX2-negative (putative ICM precursors). Four similar stage WT embryos had a similar overall number of cells (average = 23), with slightly more (4.75) being CDX2 negative. Only one mutant blastocyst was identified with 15/27 ICM cells, compared to an average of 11/33.6 for WT. These results suggest that when compared to similarly staged WT embryos, there is no dramatic skewing of either the embryonic/extra-embryonic cell ratio or the total number of cells. However, as indicated by the embryo culture experiments, the mutant embryos are more predisposed to arrest before progression to the blastocyst stage.
Downregulation of Pdcd2 upon ES cell differentiation
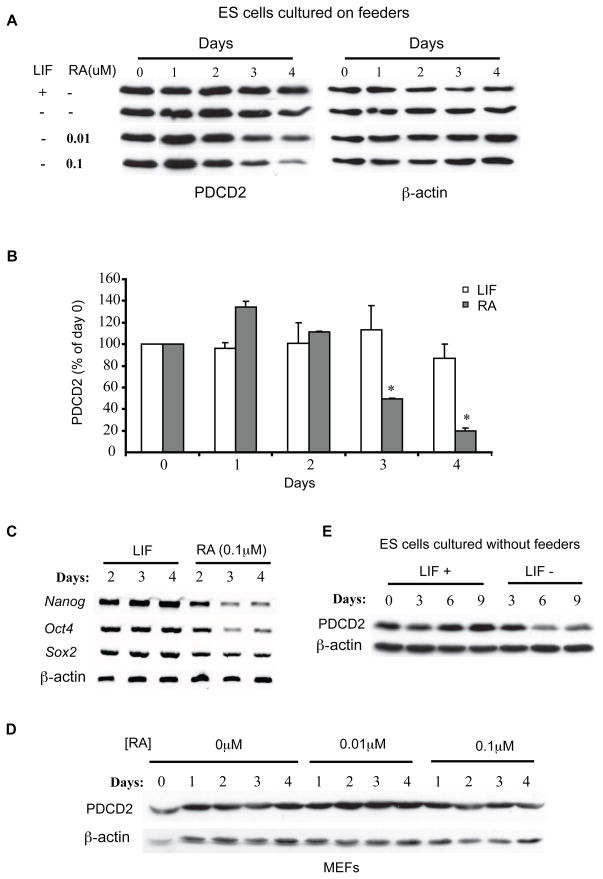
The failure of ICM outgrowth in Pdcd2−/− embryos could mean that PDCD2 is essential for cellular growth or metabolism. Alternatively, gene expression data suggests a potential role in stem cell maintenance. Microarray studies have shown that in mice, Pdcd2 is most highly transcribed in blastocyst stage embryos (Su et al., 2004). Additionally, it is more highly transcribed in stem cells (embryonic, neural, and hematopoietic) than their corresponding differentiated derivatives (Ramalho-Santos et al., 2002; Skottman et al., 2005). To explore the possibility that PDCD2 may have a particular function in stem cell maintenance, and to corroborate the microarray studies, we measured PDCD2 levels in ESCs before and after inducing them to differentiate with retinoic acid (RA). After RA treatment (0.1μM) for three and four days in the absence of leukemia inhibitory factor (LIF), PDCD2 protein levels in ES cells were decreased by 50% and 80%, respectively (Figs. 3a, b). The induction of ES cell differentiation by RA was confirmed by the decreased expression of the “stemness” markers Oct4, Nanog, and Sox2 (Fig. 3c). To exclude the possibility that the decrease in Pdcd2 expression was caused by direct transcriptional repression by RA rather than by differentiation of the ESCs, we assessed PDCD2 protein levels in RA-treated MEFs by Western blot analysis. PDCD2 levels were unaffected by RA treatment of MEFs (Fig. 3d). LIF deprivation of ESCs grown without feeders, a condition resulting in differentiation, caused reduction of PDCD2 beginning 4-6 days after withdrawal (Fig. 3a, e). These observations demonstrate a correlation between Pdcd2 expression and ESC pluripotency.
Figure 3. Downregulation of Pdcd2 upon ES cell differentiation.
(A) Western blot analysis of PDCD2 in mouse ES cells induced to differentiate by addition of retinoic acid (RA) for the number of days indicated. Blots were first probed with PDCD2 antibody and reprobed with antiβ-actin. The ESCs were cultured on mitotically-inactivated MEFs, which prevent differentiation even in the absence of exogenous LIF. (B) Quantification of band intensities in “A” for the 0.1μM [RA] treatment level. The values are shown as mean ± SEM (% of day 0); *P<0.05, N=3. (C) RT-PCR analysis of “stemness” markers Nanog, Oct4, and Sox2 in the RA-treated or LIF-supplemented ES cells from “A”. (D) Western blot analysis of PDCD2 in immortalized MEFs supplemented with RA for the indicated number of days. (E) Western blot analysis of PDCD2 in ES cells grown without LIF supplementation for the number of days indicated. Blots were reprobed with anti-β-actin.
Small amounts of PDCD2 are adequate for ESC self renewal and pluripotency
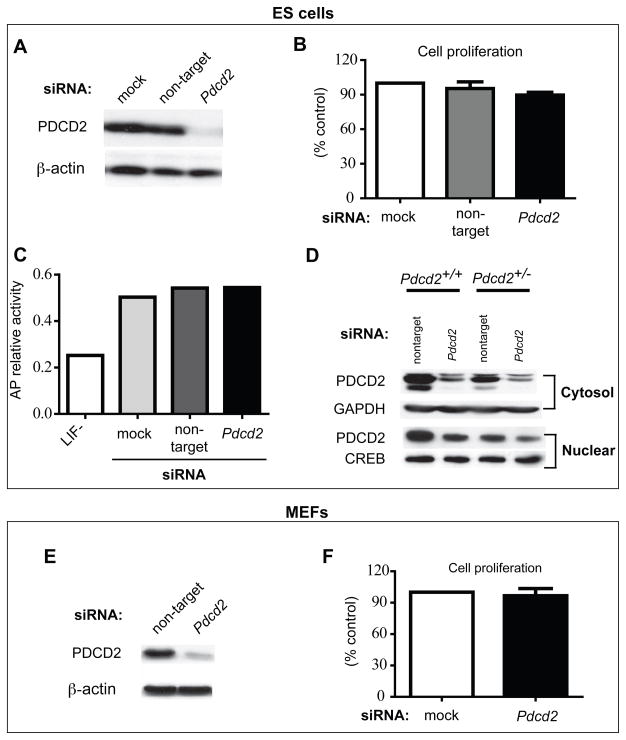
The requirement of PDCD2 for ICM outgrowth suggests that it would also be required for ESC viability. To test this, we performed siRNA knockdown of Pdcd2 in ESCs and observed the effect on cell proliferation and differentiation. Western blot analysis showed that total cellular PDCD2 protein was dramatically reduced by Pdcd2 siRNA treatment (Fig. 4a). However, the decreased PDCD2 did not impair cell proliferation (Fig. 4b). Additionally, there was no marked loss in alkaline phosphatase (AP) activity, which would indicate differentiation had occurred as in the case of LIF removal (Fig. 4c).
Figure 4. Small amounts of PDCD2 are sufficient for cell survival.
siRNA knockdown of Pdcd2 in ESCs and immortalized MEFs was performed and monitored by various metrics. The knockdown efficiency was assessed in ESCs and MEFs by Western blot analysis (A, E). 72 hours after siRNA transfection, the cells were harvested and counted for proliferation assays (B, F). The cell number was quantified from three independent experiments and shown as the mean ± SEM (% of control). (C) Effect of decreased PDCD2 on ESC pluoropotency was assessed by alkaline phosphatase (AP) activity. The LIF withdrawal group is a positive control for cell differentiation. (D) Western blot analysis of cytosolic and nuclear PDCD2 in Pdcd2 knockdown ESCs. Blots were probed with antibodies indicated. GAPDH and CREB are loading controls for cytosolic and nuclear proteins, respectively. “Non-target” refers to transfection with siGENOME® Non-Targeting siRNA Pools.
The lack of effect from PDCD2 knockdown was surprising, given the requirement for this protein by ICM cells (from which ESCs are derived). Since PDCD2 contains a MYND domain, a motif that mediates interactions with nuclear proteins involved in transcriptional regulation (namely repression) (Gelmetti et al., 1998; Gottlieb et al., 2002; Ladendorff et al., 2001; Sims et al., 2002; Wang et al., 1998), we explored whether nuclear and cytosolic PDCD2 levels were equally altered in siRNA-treated cells. The ratio of nuclear to cytosolic PDCD2 was much higher in knockdown cells compared to controls, such that the levels of nuclear PDCD2 were only marginally reduced in knockdown cells (Fig. 4d). These Western analyses (compare Fig. 4a and d) indicate that the majority of PDCD2 in normal cells is cytosolic, but that there is a mechanism to maintain a determined level of nuclear PDCD2 using the cytosolic stores. These experiments suggest that only a small fraction of normal PDCD2 levels are sufficient to provide physiological function in ESCs.
PDCD2 is indispensible for ESCs
Since RNAi knockdown was unable to completely eliminate PDCD2 in ESCs, we attempted to address the requirement for PDCD2 in ESCs by making homozygous mutant ESCs. Two strategies were attempted: 1) disrupting the second allele of Pdcd2 in Pdcd2+/− ESCs by gene targeting; and 2) selecting for spontaneous LOH in the presence of elevated G418 concentration.
As described earlier, Pdcd2 was targeted efficiently in wild type ES cells, with 12.5% of transformants having undergone homologous recombination with the vector (Table 2). However, attempted targeting of the remaining WT locus in Pdcd2+/− ESCs, using a vector containing identical homology arms, was unsuccessful. None of the 196 transformed colonies underwent homologous recombination with the vector (Table 2). Selection for LOH also failed; none of the 96 colonies that grew in elevated G418 were homozygous for the mutant allele (Table 2). To verify that the failure to isolate homozygous mutants was due to cell lethality from PDCD2-deficiency rather than a technical issue with mutating the remaining Pdcd2 allele in the Pdcd2+/− ESCs, we repeated the experiments in cells containing a stably integrated FLAG-tagged Pdcd2 cDNA expression construct. In two independent lines, selection for LOH (i.e. homozygosity for the targeted allele) at the Pdcd2 locus was efficiently achieved (Table 2) as detected by PCR analysis of the Pdcd2 locus itself (Fig. 5a top two panels, samples 1–3 in both ESC lines) and LOH of flanking polymorphic microsatellite markers in the F1 ESCs (Fig. 5a, bottom two panels). Targeting of the remaining WT allele by homologous recombination was also attained with high efficiency (9.5%) in Pdcd2+/− cells containing the rescuing Pdcd2 expression construct (Table 2; Fig. 5b). Western blot analysis confirmed a lack of endogenous PDCD2 protein in homozygous disrupted ESCs of Pdcd2 via gene targeting (Fig. 5c). Approximately normal levels of the epitope tagged PDCD2 were present in these lines, presumably providing normal protein activity. From these data, we conclude that PDCD2 is essential for both ICM and ESC viability and proliferation.
Figure 5. Screening and verification of PDCD2 homozygous mutant ESC lines.
(A) Shown are analyses of ESC clones isolated following selection in elevated G418. These were derived from two subclones of the original targeted (Pdcd2+/−) ESCs (indicated at top of panel), that were stably transfected with a FLAG-Pdcd2 expression construct. Samples 1–5: ESC colonies; 6: B6 control; 7: 129 control. To detect LOH, the selected colonies were genotyped with probes specific for the WT and KO alleles of Pdcd2. LOH was confirmed with microsatellite markers D17Jcs27 and D17Mit213, which are located upstream (~378 kb) and downsteam (~1.1 Mb) of Pdcd2, respectively, showing loss of the B6 allele. Such events are likely to have occurred by either chromosome loss/reduplication or mitotic recombination. (B) Derivation of Pdcd2−/− ESCs by homologous recombination in cells containing a FLAG-Pdcd2 expression construct. “ctrl” is Pdcd2+/− ESC DNA. (C) Homozygous knockout of Pdcd2 was verified by Western blot analysis of PDCD2 on ESC protein extracts. Colonies 2, 6, 19, and 24 lack endogenous PDCD2.
Embryonic expression pattern, tissue distribution, and cellular localization of PDCD2 in ES cells
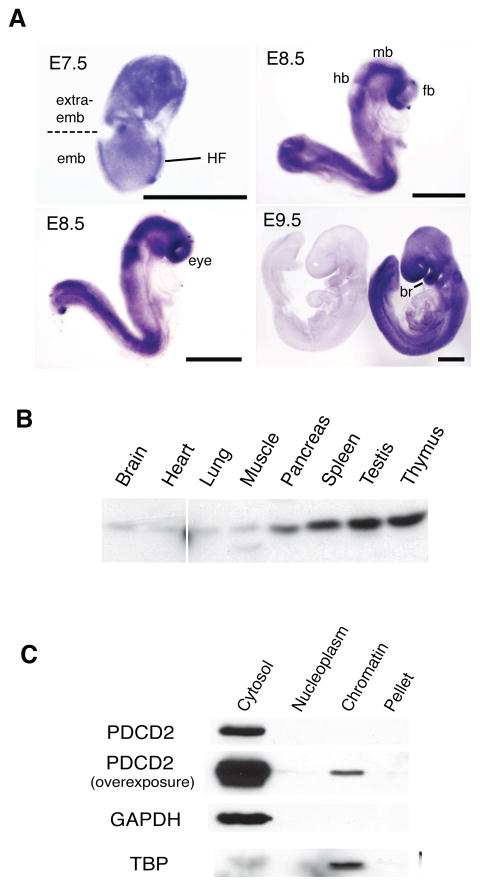
To gain insight into the possible roles of PDCD2 in pre- and postimplantation development, we examined the expression and localization of Pdcd2 mRNA in various tissues and stages of development. In E7.5 embryos, in situ hybridization indicated that Pdcd2 transcription was highest in extraembryonic tissue and the embryonic head fold region (Fig. 6a). At E8.5, Pdcd2 exhibited a specific expression pattern in the neural fold region, which gives rise to the fore-, mid-, and hindbrain (Fig. 6a). Pdcd2 mRNA was also abundant in the eyes, posterior trunk, and first branchial arch (precursor to the middle ear; Fig. 6a). These data indicate a role for PDCD2 in organogenesis.
Figure 6. Embryonic expression pattern, tissue distribution and cellular localization of PDCD2.
(A) Pdcd2 expression pattern in whole mount embryos. Pdcd2 mRNA was detected by in situ hybridization of an antisense RNA probe to WT embryos. The left E9.5 embryo in the bottom right panel was hybridized to a sense RNA control probe. HF = head fold; hb=hindbrain; mb=midbrain; fb=forebrain; br=1st branchial arch. Black bars=500 μm. (B) Western blot analysis of PDCD2 in tissues dissected from one-month old mice. 20μg of protein was loaded in each lane. Liver and kidney have intense nonspecific bands that obscure the PDCD2 species, and are not shown. (C) Western blot analysis of PDCD2 in ES cell fractions. The loaded protein was extracted from equal numbers of ES cells. Blots were probed with antibodies indicated. GAPDH and TBP are loading controls for cytosolic and nuclear proteins, respectively.
PDCD2 protein levels were detected by Western blot analysis in various tissues (Fig. 6b). PDCD2 appeared to be most highly expressed in testis and thymus, but protein was also evident in spleen, pancreas, brain, lung, heart, and muscle. In mouse ESCs, we found that the majority of PDCD2 exists in the cytosol, with only trace amounts of nuclear PDCD2 being chromatin-bound (Fig. 6c). This is consistent with the Western analyses presented in Fig. 4d, and the observations in Drosophila indicating predominantly cytoplasmic localization of Zfrp8 (Minakhina et al., 2007).
DISCUSSION
This report shows that ablation of mouse Pdcd2 disrupted embryonic development beginning at the cleavage and blastocyst stages. The basis for this lethality was a failure of blastomere and ICM proliferation. Additionally, PDCD2 is essential for growth of ESCs, which are ICM-derived. Data presented here and by others raise the possibility that PDCD2 is required particularly for stem cell growth and self-renewal, as opposed to growth of all cell types. The Drosophila melanogaster ortholog Zfrp8 is essential for maintenance of hematopoietic stem cells, but not differentiated daughters (Minakhina and Steward, 2010). Although Pdcd2 is expressed in many mouse tissues, it is most highly transcribed in pre-implantation embryos. Furthermore, we showed that Pdcd2 expression in ES cells decreased upon induction of differentiation by RA treatment or LIF deprivation. This is consistent with a report that mouse embryonic, neural, and hematopoietic stem cells have higher levels of Pdcd2 mRNA compared to their differentiated derivatives (Ramalho-Santos et al., 2002). Severe knockdown of PDCD2 in MEFs did not affect viability and proliferation, although similar levels of depletion didn’t affect ESCs either. Therefore, a potential requirement for PDCD2 in differentiated cells is unclear.
Regarding the biochemical function of PDCD2 in early embryos, the body of evidence indicates it is involved in regulating gene expression. PDCD2 interacts physically with both HCFC1 and NCOR1 (also known as N-Cor), the latter being a component of the NCOR1/SIN3A corepressor complex that recruits histone deacetylases (HDACs) to chromatin. Consistent with a gene regulatory function, we found that the nuclear fraction of PDCD2 was linked to ESC growth and survival in our siRNA knockdown experiments. Scarr et al reported that PDCD2 overexpression negatively regulated transfected HCFC1 in a cell line containing a temperature-sensitive HCFC1 allele (Scarr and Sharp, 2002). The HCFC1N subunit (produced by processing of a larger precursor protein) promotes G1 phase progression (Goto et al., 1997) via the E2F pathway (Tyagi et al., 2007) in certain cell culture models. E2F transcriptional regulators control human cell proliferation by repressing and activating the genes required for cell cycle progression through S phase in particular. The association and binding of HCFC1/E2F1 to E2F-responsive promoters are cell cycle selective. During the G1-to-S transition, HCFC1 recruits MLL and the SET1 H3K4 methyltransferase to E2F-responsive promoters in place of E2F1-bound RB1 (a.k.a. pRB, or retinoblastoma)-mediated repressive complexes, inducing H3K4 methylation and transcriptional activation (Tyagi et al., 2007).
Given that PDCD2 reportedly inhibits HCFC1, how would loss of PDCD2 prevent ESC and ICM cell proliferation? Cell cycle progression is exceptionally rapid in 4–16 cell stage embryos, with a short G1 phase (1–2 hours) causing rapid transition into S phase. Successful G1/S transition and S phase progression requires inactivation of the Rb1 (Retinoblastoma) gene product (RB1) (Knudsen et al., 1998; Sherr, 1996), a negative growth regulator that inhibits the G1/S transition. The association of RB1 with cell cycle regulation of preimplantation embryos has been demonstrated in two reports showing that RB1 expression is downregulated between the four-cell and morula stages (Iwamori et al., 2002; Xie et al., 2005). Overexpression of Rb1 in mouse embryos at these stages inhibited their development (Iwamori et al., 2002). In light of these data, we can offer two models to explain PDCD2’s role in growth of preimplantation embryos. One is that PDCD2 negatively regulates RB1. This could occur conceivably via inhibition of HCFC1, which has been shown to stimulate RB1 transcription in myoblasts, thereby causing cell cycle exit and myogenic differentiation (Delehouzee et al., 2005). The second model takes into account the observations that HCFC1 can cause transcriptional repression by interacting with E2F4 at certain promoters, and that the SIN3A HDAC is present when HCFC1 is bound to E2F4 (Tyagi et al., 2007). PDCD2, which interacts with both HCFC1 and the NCOR/SIN3A complexes, may mediate the E2F4/HCFC1/SIN3A relationship at repressed genes. In embryos, these repressed genes could include inhibitors of the cell cycle (such as RB1) or pro-differentiation genes.
Embryonic stem cells, like preimplantation embryos, have a short G1 phase of roughly 1.5h during which hypophosphorylated RB1 is virtually undetectable (Savatier et al., 1994). PDCD2 might regulate the G1/S transition in ES cells the same way as in early embryogenesis. Our experiments showed that ESC viability and proliferation was retained following RNAi – mediated knockdown, apparently by shifting the ratio of remaining PDCD2 from the cytoplasm to the nucleus.
Because of the early lethality in Pdcd2−/− embryos, we were unable to characterize the potential in vivo role(s) for PDCD2 during later stages of development or in adult tissues. Given that Pdcd2 is transcribed in many cell types, it might function in cell proliferation, differentiation, and the determination of cell fate during organogenesis or adult cell homeostasis as in Drosophila hematopoiesis (Minakhina et al., 2007; Minakhina and Steward, 2010). Analyses of a conditional allele will be useful in addressing this issue. Disruptions of the PDCD2 interactors NCOR, or NCOR- and HCFC1-associated proteins such as SIN3A and SIN3B, also lead to developmental defects. Ncor−/− embryos exhibit defects in developmental progression of erythrocyte, thymocyte and neural events (Jepsen et al., 2000). Sin3A deletion results in lethality at E6.5 and impaired T cell development (Cowley et al., 2005; Dannenberg et al., 2005). David et al revealed that Sin3b plays a critical role in the control of cell cycle exit, terminal differentiation, and specific lineages in mammals (David et al., 2008).
Our studies of PDCD2 function were originally motivated to test its possible identity to Thl1, a strain-dependent haplolethal locus in the mouse t complex that was revealed by studies of a nested series of chromosomal deletions induced in ESCs (Browning et al., 2002). Thl1 embryonic lethality was partially rescued by BAC RP24-346I22 containing 4 Refseq genes, Pmsb1, Pdcd2, Tbp1, and Prdm9 (Howell et al., 2005). Since Prdm9 nulls have meiotic defects, and Tbp1 has no haploinsufficient phenotype, one remaining candidate is Pmsb1. This gene encodes a core β subunit of the 20S proteasome, which is an essential component of the ATP-dependent proteolytic pathway in eukaryotic cells and is responsible for the degradation of cellular proteins (Coux et al., 1996). Alternatively, it is possible that Thl1 lethality might be a synthetic effect of hemizygous deletion of two or more than two genes, including Pdcd2.
Conclusions
Collectively, we have demonstrated an essential role for PDCD2 in preimplantation development of the inner cell mass and in embryonic stem cell viability and growth. Although it is most prevalent in the cytoplasm, it is chromatin-bound PDCD2 that appears to be critical for ESC growth. Our results combined with those of others suggest that PDCD2 promotes cell proliferation/self-renewal of pluripotent embryonic cells and ESCs cells via transcriptional control of the cell cycle. Future studies will address if these activities are required for other stem cell types, and if PDCD2 has critical functions in differentiated cells as well.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant HD24374 from the National Institute of Child Health and Human Development.
Abbreviations
- ESC
embryonic stem cell
- ICM
inner cell mass
- RA
retinoic acid
- MEF
mouse embryonic fibroblast
- LIF
leukemia inhibitory factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears S, Kim DC, Simone F, Birkenbach M, Montag A, Sadhu A, Zeleznik-Le N, McKeithan TW. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci U S A. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron BW, Zeleznik-Le N, Baron MJ, Theisler C, Huo D, Krasowski MD, Thirman MJ, Baron RM, Baron JM. Repression of the PDCD2 gene by BCL6 and the implications for the pathogenesis of human B and T cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:7449–7454. doi: 10.1073/pnas.0701770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning VL, Bergstrom RA, Daigle S, Schimenti JC. A haplolethal locus uncovered by deletions in the mouse T complex. Genetics. 2002;160:675–682. doi: 10.1093/genetics/160.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci U S A. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delehouzee S, Yoshikawa T, Sawa C, Sawada J, Ito T, Omori M, Wada T, Yamaguchi Y, Kabe Y, Handa H. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells. 2005;10:717–731. doi: 10.1111/j.1365-2443.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- Fan CW, Chan CC, Chao CC, Fan HA, Sheu DL, Chan EC. Expression patterns of cell cycle and apoptosis-related genes in a multidrug-resistant human colon carcinoma cell line. Scand J Gastroenterol. 2004;39:464–469. doi: 10.1080/00365520310008809. [DOI] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondran P, Amiot F, Weil D, Dautry F. Accumulation of mature mRNA in the nuclear fraction of mammalian cells. FEBS Lett. 1999;458:324–328. doi: 10.1016/s0014-5793(99)01175-8. [DOI] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol Cell Biol. 2005;25:8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Howell GR, Munroe RJ, Schimenti JC. Transgenic rescue of the mouse t complex haplolethal locus Thl1. Mamm Genome. 2005;16:838–846. doi: 10.1007/s00335-005-0045-8. [DOI] [PubMed] [Google Scholar]
- Iwamori N, Naito K, Sugiura K, Tojo H. Preimplantation-embryo-specific cell cycle regulation is attributed to the low expression level of retinoblastoma protein. FEBS Lett. 2002;526:119–123. doi: 10.1016/s0014-5793(02)03121-6. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick SM, Mandel G, Glass CK, Rose DW, Rosenfeld MG. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Furukawa Y, Sudo K, Saito H, Takami S, Takahashi E, Nakamura Y. Isolation and mapping of a human gene (PDCD2) that is highly homologous to Rp8, a rat gene associated with programmed cell death. Cytogenet Cell Genet. 1995;71:41–43. doi: 10.1159/000134058. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Buckmaster C, Chen TT, Feramisco JR, Wang JY. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladendorff NE, Wu S, Lipsick JS. BS69, an adenovirus E1A-associated protein, inhibits the transcriptional activity of c-Myb. Oncogene. 2001;20:125–132. doi: 10.1038/sj.onc.1204048. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Hum Mol Genet. 2008;17:R28–36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- Minakhina S, Druzhinina M, Steward R. Zfrp8, the Drosophila ortholog of PDCD2, functions in lymph gland development and controls cell proliferation. Development. 2007;134:2387–2396. doi: 10.1242/dev.003616. [DOI] [PubMed] [Google Scholar]
- Minakhina S, Steward R. Hematopoietic stem cells in Drosophila. Development. 2010;137:27–31. doi: 10.1242/dev.043943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen RM, Conner DA, Chao S, Geisterfer Lowrance AA, Seidman JG. Production of homozygous mutant ES cells with a single targeting construct. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Wang W, Schimenti JC. An allelic series uncovers novel roles of the BRCT domain-containing protein PTIP in mouse embryonic vascular development. Mol Cell Biol. 2008;28:6439–6451. doi: 10.1128/MCB.00727-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GP, Hahn WE, Cohen JJ. Identification of mRNAs associated with programmed cell death in immature thymocytes. Mol Cell Biol. 1991;11:4177–4188. doi: 10.1128/mcb.11.8.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- Scarr RB, Sharp PA. PDCD2 is a negative regulator of HCF-1 (C1) Oncogene. 2002;21:5245–5254. doi: 10.1038/sj.onc.1205647. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Weihe EK, Zhu L, O'Malley S, Harriss JV, Gottlieb PD. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem. 2002;277:26524–26529. doi: 10.1074/jbc.M204121200. [DOI] [PubMed] [Google Scholar]
- Skottman H, Mikkola M, Lundin K, Olsson C, Stromberg AM, Tuuri T, Otonkoski T, Hovatta O, Lahesmaa R. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- Steinemann D, Gesk S, Zhang Y, Harder L, Pilarsky C, Hinzmann B, Martin-Subero JI, Calasanz MJ, Mungall A, Rosenthal A, Siebert R, Schlegelberger B. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer. 2003;37:421–426. doi: 10.1002/gcc.10231. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Vitale A, Perlin J, Leonelli L, Herr J, Wright P, Digilio L, Coonrod S. Mouse cPLA2gamma, a novel oocyte and early embryo-abundant phospholipase A2 gamma-like protein, is targeted to the nuclear envelope during germinal vesicle breakdown. Dev Biol. 2005;282:374–384. doi: 10.1016/j.ydbio.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh IC, O'Brien TP. Loss of late primitive streak mesoderm and interruption of left-right morphogenesis in the Ednrb(s-1Acrg) mutant mouse. Dev Biol. 2000;225:151–168. doi: 10.1006/dbio.2000.9814. [DOI] [PubMed] [Google Scholar]
- Xie Y, Sun T, Wang QT, Wang Y, Wang F, Puscheck E, Rappolee DA. Acquisition of essential somatic cell cycle regulatory protein expression and implied activity occurs at the second to third cell division in mouse preimplantation embryos. FEBS Lett. 2005;579:398–408. doi: 10.1016/j.febslet.2004.10.109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.