Abstract
Filamin, known primarily for its actin cross-linking function, is a stretch-sensitive structural and signaling scaffold that binds transmembrane receptors and a wide variety of intracellular signaling proteins. The C. elegans filamin ortholog, FLN-1, has a well conserved overall structure, including an N-terminal actin-binding domain, and a series of 20 immunoglobulin (Ig)-like repeats. FLN-1 partially colocalizes with actin filaments in spermathecal and uterine cells. Analysis of phenotypes resulting from a deletion allele and RNAi depletion indicates FLN-1 is required to maintain the actin cytoskeleton in the spermatheca and uterus, and to allow the exit of embryos from the spermatheca. FLN-1 deficient animals accumulate embryos in the spermatheca, lay damaged and unfertilized eggs, and consequently exhibit dramatically reduced brood sizes. The phospholipase PLC-1 is also required for the exit of embryos from the spermatheca, and analysis of doubly mutant animals suggests that PLC-1 and FLN-1 act in the same pathway to promote proper transit of embryos from the spermatheca to the uterus. Given the modular protein structure, subcellular localization, genetic interaction with PLC-1, and known mechanosensory functions of filamin, we postulate that FLN-1 may be required to convert mechanical information about the presence of the oocyte into a biochemical signal, thereby allowing timely exit of the embryo from the spermatheca.
Keywords: Filamin, FLN-1, Ovulation, Spermatheca, Actin, PLC-1
Introduction
Filamins are large, dimeric proteins composed of an N-terminal actin-binding domain (ABD) and multiple C-terminal immunoglobulin (Ig)-like filamin repeats (Gorlin et al., 1990). The final Ig-like domain mediates dimerization (Himmel et al., 2003; Stossel et al., 2001). The ABD consists of two tandem calponin homology domains. Human filamins contain 24 Ig-like domains segregated into two rod domains (rod 1 and 2) and connected by flexible hinges (hinge 1 and 2). Although much work has been done to understand the structure and function of filamin, surprisingly little is known about the mechanism by which filamin functions in vivo. In Drosophila melanogaster the filamin ortholog cheerio is required for proper maintenance of ovarian ring canals (Li et al., 1999; Sokol and Cooley, 2003), and the Dictyostelium discoideum filamin is required for photosensory signaling (Annesley et al., 2007). A number of cell migration (Fox et al., 1998), muscle (Ferrer and Olive, 2008), and bone (Stefanova et al., 2005) disorders have been linked to filamin mutations in humans. Similarly, filamin mutations in mice result in a wide variety of defects, including bone and microvasculature defects (Hart et al., 2006; Zhou et al., 2007).
Filamins organize the actin cytoskeleton into an elastic three-dimensional network and connect the cytoskeleton to transmembrane proteins, including integrins (Calderwood et al., 2001). Additionally, filamins interact with many intracellular signaling proteins, such as migfilin, FilGAP, Trio and small GTPases (Feng and Walsh, 2004; Popowicz et al., 2006). Experimental evidence suggests that filamins may act as mechanical stress sensors (Gehler et al., 2009; Kainulainen et al., 2002; Shifrin et al., 2009; Stossel et al., 2001). Because filamins can concurrently bind transmembrane proteins, filamentous actin, and intracellular signaling components, they are ideally placed to regulate the response of cells or tissues to changing mechanical forces. Filamin may transduce mechanical signals through stretch-sensitive changes in conformation or arrangement of its Ig-like domains that reveal cryptic binding sites for downstream effectors (Nakamura et al., 2007).
The C. elegans gonad, essentially consisting of a contractile tube, is an ideal model system for in vivo study of filamin function. The C. elegans gonad consists of two symmetrical, U-shaped arms connected by a common uterus (Hubbard and Greenstein, 2000; McCarter et al., 1997; Strome, 1986). Germ cells proliferate in the distal gonad, and mature into oocytes as they move proximally. The gonad is enveloped by an outer basal lamina, and most portions of the gonad are surrounded by a layer of contractile, myoepithelial sheath cells (Strome, 1986). The proximal gonad is connected to the spermatheca, which serves as the site of sperm storage and fertilization. The spermatheca consists of three distinct regions: the distal constriction, a central bag-like chamber, and the spermatheca-uterine (sp-ut) valve (McCarter et al., 1997; Strome, 1986).
During each ovulatory cycle, the most proximal oocyte is ovulated into the spermatheca, fertilized, and then released into the uterus (McCarter et al., 1999). Ovulation requires the coordination of signaling between the sperm, oocyte, proximal sheath cells, and the distal spermatheca. Major Sperm Protein (MSP), secreted by the sperm, promotes sheath cell contractions and oocyte meiotic maturation (Miller et al., 2001). Ovulation begins with intense sheath cell contractions and concomitant dilation of the distal spermatheca. The distal spermatheca is pulled over the proximal oocyte, and the oocyte passes through the distal spermathecal constriction to enter the spermatheca, where fertilization occurs immediately (McCarter et al., 1999). Following fertilization, the embryo exits the spermatheca via the sp-ut valve. Oocyte entry is controlled by LIN-3/EGF signaling from the oocyte to the sheath and the distal spermatheca (Clandinin et al., 1998). In the sheath and spermatheca, the LET-23/EGF receptor likely activates PLC-3/Phospholipase C-γ, which generates IP3 causing release of Ca2+ by the ITR-1/IP3 receptor (Clandinin et al., 1998; Yin et al., 2004). The IP3 signal is negatively regulated by IPP-5/inositol 5-phosphatase and LFE-2/inositol (1,4,5) trisphosphate-3-kinase (Bui and Sternberg, 2002; Clandinin et al., 1998). Ca2+ release is thought to control the contraction of the sheath cells and dilation of the distal spermatheca. With the exception of the role of PLC-1, very little is known about the molecular events that control the exit of embryos from the spermatheca (Kariya et al., 2004).
In this study, we demonstrate that FLN-1/filamin plays a critical structural and mechanosensory role in the C. elegans somatic gonad. Using RNAi depletion and analysis of a deletion allele, fln-1(tm545), we demonstrate that loss of fln-1 results in severe spermathecal exit defects and a dramatically reduced brood size. The expression pattern and phenotypic analysis show fln-1 is required in the spermatheca for normal release of embryos and for maintenance of the actin cytoskeleton in the spermatheca and uterus. Interestingly, our analysis suggests that in addition to its structural role in the gonad, filamin may play a role in phosphatidylinositol signaling.
Results
fln-1 encodes three protein isoforms
The C. elegans genome contains two clusters of filamin-like sequences (WormBase WS205) (Harris et al., 2010). One set of predicted open reading frames (ORFs) is on chromosome IV (Y66H1B.2, Y66H1B.5, and Y66H1B.3) and the other is on chromosome X (C23F12.1 and C23F12.2). RNAi knockdown of Y66H1B.2, Y66H1B.5, and Y66H1B.3 resulted in striking defects including a dramatically reduced brood size (described below). Given this strong and reproducible phenotype, we chose to focus on the Y66H1B.2, Y66H1B.5, and Y66H1B.3 filamin cluster for further analysis.
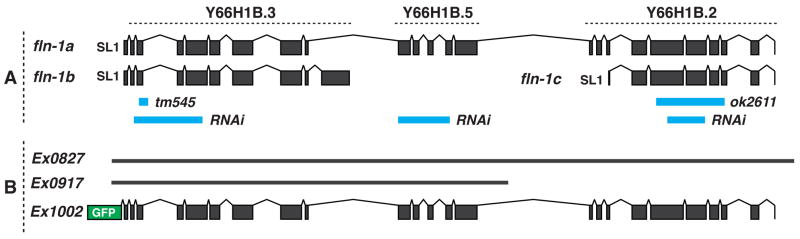
Extensive cDNA sequencing of transcripts that span Y66H1B.3, Y66H1B.5, and Y66H1B.2 suggests these ORFs represent a single gene, which we have named fln-1 (FiLamiN) (WormBase WS213) (Figure 1A). The full-length transcript includes all three computationally predicted ORFs and encodes a full-length C. elegans filamin ortholog (fln-1a) (Figure 1A). In addition to the full-length transcript, we detected two shorter transcripts (fln-1b and c) (Figure 1A). Y66H1B.3 and Y66H1B.2, but not Y66H1B.5, are trans-spliced to SL1 as determined by SL1 PCR and sequencing, which suggests the independent transcription of fln-1a/b and fln-1c (Figure 1A). The full-length filamin protein is predicted to be 2257 amino acids, while the C- and N-terminal truncations are predicted to be 1084 and 836 amino acids, respectively. The full-length filamin contains a well-conserved N-terminal ABD and 20 Ig-like repeats based on sequence alignments to human and the Drosophila filamin cheerio.
Figure 1. Gene structure of the fln-1 locus.
This figure is a diagrammatic representation of the fln-1 locus derived from cDNA sequencing data. Rectangles and lines represent exons and introns, respectively. Solid, thick lines represent the genomic sequences. A) Predicted ORFs Y66H1B.3, Y66H1B.5, and Y66H1B.2 are indicated. Deletion alleles (tm545 and ok2611) and RNAi targeting constructs are shown as blue lines. B) The rescuing constructs Ex0827 and Ex0917 are amplified genomic sequences, and Ex1002 is an N-terminal GFP fusion to the fln-1a open reading frame. Approximately 19 kb of chromosome IV is shown.
fln-1 is required for normal fertility
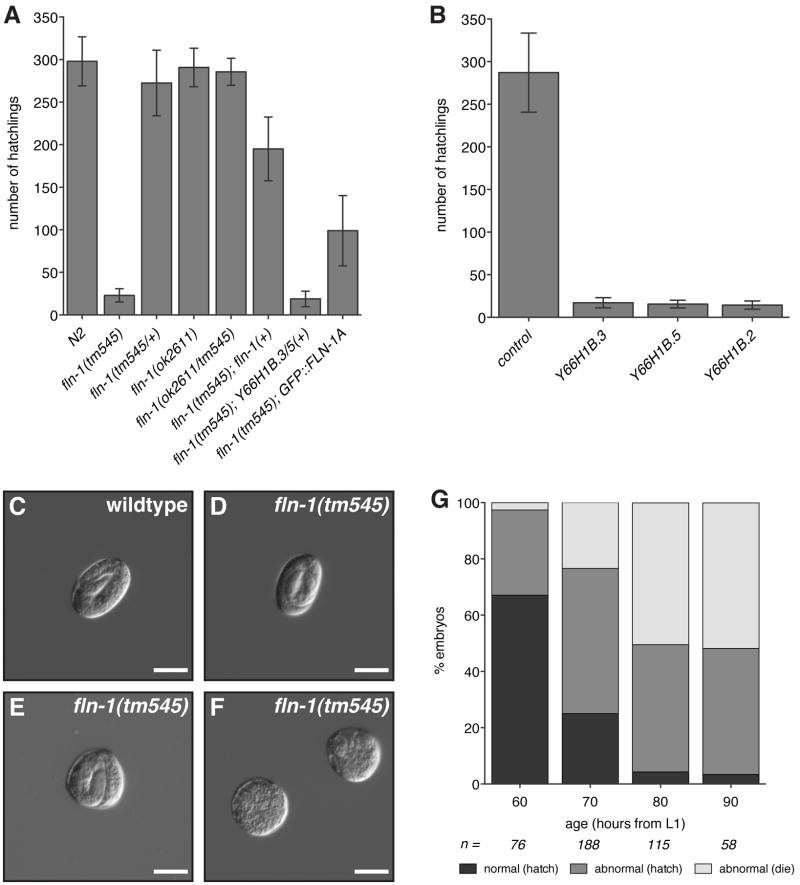
In order to genetically analyze the function of fln-1, we obtained two fln-1 deletion alleles, tm545 and ok2611 from the Japanese National Bioresource Project and the C. elegans Gene Knockout Consortium, respectively. The tm545 allele is a 263/17 bp deletion/insertion in the third exon of fln-1a, which causes a frameshift and a premature stop codon. The truncated protein is predicted to be 102 amino acids (Figure 1A). The ok2611 allele is an in-frame 1763 bp deletion at the 3′ end of fln-1a, and is predicted to remove 455 amino acids (Figure 1A). The most striking defect of tm545 hermaphrodites is a dramatic reduction in brood size (23 ± 7.9, n=26) compared to wildtype animals (298 ± 28.8, n=11) (Figure 2A). This phenotype is 100% penetrant in fln-1(tm545) animals. In contrast, ok2611 animals are superficially wildtype and do not exhibit a brood size defect (290.8 ± 22.6, n=9) (Figure 2A). The brood sizes of heterozygous tm545/+ animals (272.6 ± 38.5, n=9) and tm545/ok2611 trans-heterozygotes (285.6 ± 15.9, n=7) are not significantly different from wildtype (Figure 2A). These results strongly suggest that the region removed by ok2611 is not required for the function of filamin in fertility.
Figure 2. Filamin is required for normal brood size.
A) Brood size comparison of wildtype, fln-1 mutant and rescued animals. B) RNAi depletion of Y66H1B.3, Y66H1B.5 or Y66H1B.2 phenocopies the tm545 brood size defect. Columns indicate the average number of hatchlings, and error bars indicate the standard deviation. C–F) wildtype animals produce wildtype, oval-shaped embryos (C). tm545 animals initially produce normal embryos (D), but progressively worsen producing round (E), and eventually inviable embryos (F). Bar indicates 25 μm. G) Shape and hatching of tm545 embryos was determined at the indicated maternal ages. Older tm545 animals produce a higher proportion of misshapen and arrested embryos (G). During this time course, age-matched wildtype animals produce only normally shaped embryos, of which all hatch (n=561). Control wildtype animals are omitted from (G) for simplification. Number of tm545 embryos examined for each time point is indicated.
To confirm that the brood size defect of fln-1(tm545) is caused by disruption of fln-1 function, we constructed transgenic nematodes carrying fln-1 genomic DNA on an extrachromosomal array (xbEx0827). fln-1(tm545) animals carrying the full-length fln-1 genomic region (Figure 1B) produce an average brood size of 195.1 ± 37.4 progeny (n=10) (Figure 2A). This robust rescue indicates the brood size defect in tm545 animals is attributable to the deletion in fln-1. To investigate whether full-length fln-1 is required for rescue of the brood size defect we attempted to rescue fln-1(tm545) with a genomic fragment encompassing only Y66H1B.3 and Y66H1B.5 (xbEx0817). The fln-1(tm545) animals expressing this construct are not significantly different from non-transgenic siblings, with an average brood size of 18.8 ± 9.1 (n=18) progeny (Figure 2A). RNAi-mediated knockdown of fln-1 with three independent RNAi constructs targeting Y66H1B.3, Y66H1B.5, or Y66H1B.2 (Figure 1A) phenocopies the tm545 allele, reducing the brood size to 14.8 ± 5.2 (n=12), 15.6 ± 5 (n=14), and 15.5 ± 4.5 (n=10) progeny respectively (Figure 2B). These results indicate fln-1(tm545) is a strong hypomorphic allele, and suggest expression of full-length filamin is required for normal brood size.
Brood size defects are generally the result of somatic gonad, germ line, or embryogenesis defects. To investigate whether fln-1 is required zygotically we mated fln-1(tm545) hermaphrodites to wildtype males. The brood size of fln-1(tm545) hermaphrodites mated to wildtype males (28.6 ± 6.2, n=7) is not significantly different from unmated hermaphrodites (23 ± 7.9, n=26; Student’s t-test, p=0.09), indicating paternally supplied fln-1 is not sufficient to restore normal brood size. To determine if depletion of filamin in the germline can account for the observed brood size defect we used the rrf-1(pk1417) mutant strain, which is defective for somatic RNAi, but has an apparently normal germline RNAi response (Sijen et al., 2001). The rrf-1(pk1417) animals have been used previously to demonstrate a somatic role for fos-1 in the spermatheca (Hiatt et al., 2009). We found that rrf-1(pk1417) mutants treated with fln-1 RNAi did not show a significantly different brood size (270.3 ± 45.1, n=8) from control RNAi treated rrf-1(pk1417) animals (307.2 ± 28.1, n=6; Student’s t-test, p=0.1). Although this data does not entirely rule out a role for FLN-1 in the germ line, it does suggest that depletion of fln-1 in the germ line does not contribute substantially to the observed brood size defects.
Wildtype animals produce embryos of consistent size and shape (Figure 2C). In contrast, most embryos laid by fln-1(tm545) hermaphrodites are misshapen (Figure 2D–F). Interestingly, some of the abnormally shaped embryos hatch (Figure 2E), while others fail at variable points during embryogenesis (Figure 2F). To characterize the embryo shape and possible embryogenesis defects we synchronized wildtype and fln-1(tm545) animals and examined embryo shape and hatching at 60, 70, 80, and 90 hours following L1 diapause recovery (Figure 2G). Throughout the time course, wildtype animals produce normally shaped and viable embryos (100%, n=561). The 60-hour time-point is shortly before the onset of egg laying and corresponds to the initial ovulations. At 60 hours fln-1(tm545) animals produce 67% normal embryos, 30% abnormally shaped but viable embryos, and 3% abnormally shaped embryos that fail to hatch. The proportion of abnormally shaped embryos and arrested abnormal embryos increases with maternal age. By 90 hours, 97% of the embryos are abnormally shaped, and 51% of these fail to hatch. Importantly, none of the normally shaped embryos arrest in fln-1(tm545) animals. These results strongly suggest that misshapen embryos and embryonic lethality are the result of a maternal defect during ovulation that results in physical damage to the embryos, rather than a defect during embryonic development.
fln-1 is expressed in the somatic gonad
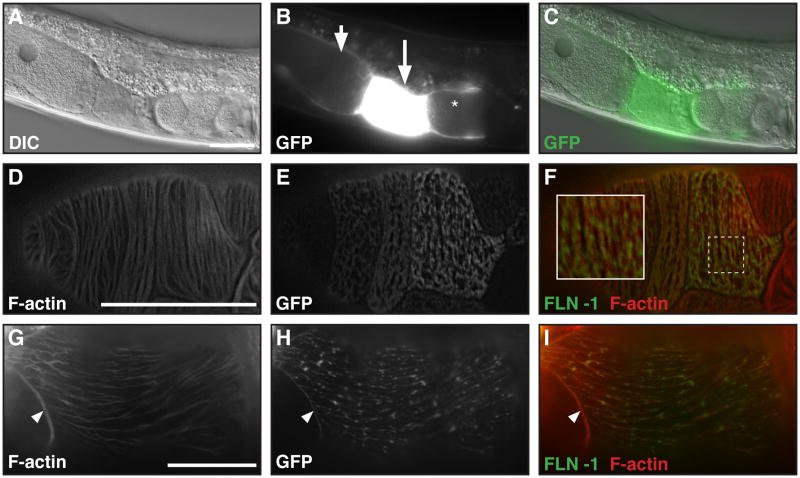
To investigate the expression pattern of fln-1 we created transgenic nematodes expressing GFP under the control of the fln-1 promoter. GFP expression begins in early L4 stage and continues through adulthood in the uterus, spermatheca, and proximal gonadal sheath (Figure 3A–C). In addition to the somatic gonad, weak and variable GFP expression is evident in the posterior intestinal cells, anal depressor muscle, unidentified neurons, and the pharynx (data not shown). Due to silencing of repetitive extrachromosomal arrays in the germline, we cannot exclude the possibility that fln-1 is also expressed in the germline. To determine the subcellular localization of FLN-1 we created transgenic nematodes carrying full-length FLN-1A fused to GFP at the N-terminus under the control of the native promoter and 3′ UTR. GFP::FLN-1A localizes to punctate filaments in the spermathecal and uterine cells, and partially co-localizes with F-actin (Figure 3D–F and 3G–I, respectively). Importantly, the GFP::FLN-1A fusion protein (xbEx1002) rescues the brood size defect of fln-1(tm545) (99 ± 41.3, n=37) (Figure 2A), suggesting that the fusion protein is functional and represents normal FLN-1A subcellular localization. Incomplete rescue is likely due to overexpression of the transgene from the extrachromosomal arrays and mosaicism.
Figure 3. FLN-1 is expressed in the somatic gonad and colocalizes with F-actin.
A) DIC image of an adult hermaphrodite carrying a transcriptional fln-1::gfp fusion, and (B) a corresponding GFP image. The fln-1::gfp transcriptional fusion is expressed strongly in the proximal sheath cells (arrowhead), spermatheca (arrow), and the uterus (asterisk) in adult animals. C) Merged GFP and DIC images. D–I) GFP::FLN-1A translational fusion expressed in the spermatheca (D) and uterus (G) and co-stained for F-actin (E and H). GFP::FLN-1A appears to colocalize with F-actin in the spmeratheca (F) and the uterus (I). Arrows in G–I indicate the uterine lariat. F and I) Merged F-actin (red) and GFP::FLN-1A (green). Bar indicates 25 μm.
A primary function of filamin in other systems is to link the actin cytoskeleton to transmembrane receptors, such as integrins (Critchley, 2000). We hypothesized that filamin may be required to anchor the F-actin to integrins in the spermatheca. Filamin has been shown to interact with β-integrin cytoplasmic tails in vitro, and localizes to F-actin and focal adhesions in cultured cells (Kiema et al., 2006). To determine whether filamin and integrin co-localize in C. elegans we used the GFP::FLN-1A fusion and the β-integrin monoclonal antibody MH25 (Figure S1). Wildtype dissected gonads stained strongly for integrins in the sheath cells (Figure S1A–C) where they are organized into large dense bodies (Figure S1A)(Hall et al., 1999; Ono et al., 2007); however, the integrin staining in the spermatheca was much weaker and very diffuse (Figure S1D–F). We observed a similar staining pattern for talin (not shown). To our knowledge, dense bodies have not been described in the spermatheca. In contrast, GFP::FLN-1A is expressed strongly in the spermatheca and appears to localize in a punctate filament pattern (Figure S1D). Given the weak and diffuse staining of PAT-3 in the spermatheca we cannot conclude that filamin co-localizes with integrin in the spermatheca (Figure S1F).
fln-1 is required for normal exit of embryos from the spermatheca
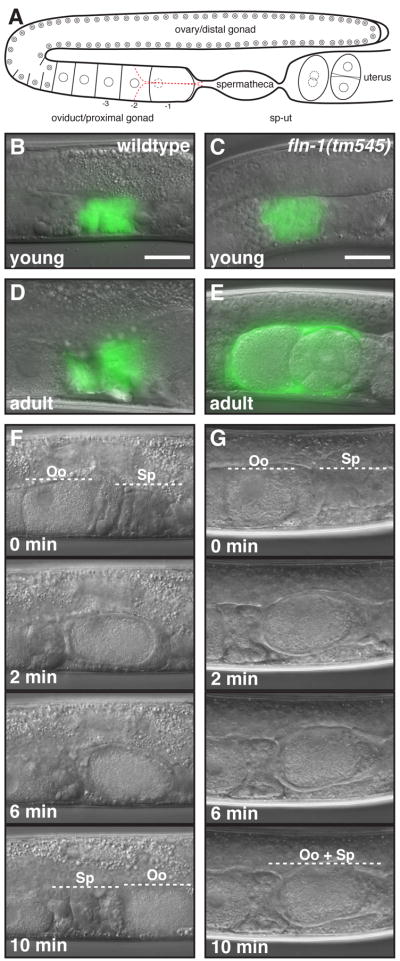
We hypothesized that the reduced fertility and abnormal embryonic morphology in fln-1(tm545) animals might be the result of abnormal exit of embryos from the spermatheca. In wildtype animals, oocytes are ovulated into the spermatheca, fertilized, and then released into the uterus via the spermatheca-uterine valve (Figure 4A). The entire ovulation and fertilization process in wildtype animals is completed in less than ten minutes (Figure 4F, Movie 1A) (McCarter et al., 1999). In contrast, fln-1(tm545) animals showed a fully penetrant (n=29) spermathecal exit defect due to failure of the sp-ut valve to dilate (Figure 4G, Movie 1B). Our ovulation recordings of fln-1(tm545) animals do not show any overt abnormalities during the entry process. Importantly, the exit defect is rescued by the fln-1(+) and the GFP::FLN-1A transgenes (Movie 2A and B, respectively).
Figure 4. Ovulation time-lapse of fln-1(tm545) animals.
A) Schematic representation of the hermaphrodite gonad. B–C) Young wildtype and fln-1(tm545) spermatheca marked with fkh-6::gfp. D–E) Adult wildtype and fln-1(tm545) spermatheca marked with fkh-6::gfp. F) Wildtype ovulation sequence captured by DIC time-lapse microscopy. G) fln-1(tm545) ovulation sequence is identical to the wildtype ovulation during the entry (0 min) and fertilization process (2 min), but the embryos fail to exit the spermatheca (10 min). Bar indicates 25 μm.
Due to the exit defect, multiple embryos accumulate in the spermatheca, which causes subsequent ovulations to fail. Embryos remain trapped in the spermatheca for at least six hours, the duration of the longest experiment. The initial three to four ovulations are normal in terms of entry, fertilization, and embryo development. Subsequent oocytes fail to enter the spermatheca due to accumulated embryos and are often fragmented by sheath cell contractions. In unanesthetized animals the spermatheca-uterine valve appears to prolapse between the second and third ovulations, presumably due to backpressure of oocytes and embryos, possibly aided by cycles of body-wall muscle contraction and relaxation. In order to assess the degree of trapping in unanesthetized animals, wildtype and fln-1(tm545) animals expressing spermatheca-specific fkh-6::gfp (Chang et al., 2004) were monitored while freely moving on plates. In fln-1(tm545) animals the spermatheca remains obviously distended and occupied by multiple embryos and oocytes (100%, n=160). Although wildtype animals also have distended spermatheca during ovulation, the spermatheca returns to normal size following oocyte exit. The short duration of ovulation results in only a small percentage of the wildtype population with distended spermathecae (5%, n=120). As previously discussed, the majority of the fln-1(tm545) brood is generated during the first several ovulation cycles, with some abnormally shaped, but surviving embryos generated afterward (Figure 2D–G). Following valve prolapse, fln-1(tm545) lay many unfertilized oocytes suggesting inefficient fertilization. The misshapen embryos often result from pinched-off oocyte cytoplasm, but inappropriate sheath cell contractions and compaction in the spermatheca also contribute.
fln-1 is not required for spermathecal development
Given the reduced fertility of fln-1(tm545) animals and the prominent expression of fln-1::gfp in the proximal gonad, we speculated filamin may be required for proper development of these tissues. To assess development of the proximal gonad in fln-1(tm545) animals, we examined young-adult animals prior to the first ovulation using DIC microscopy. The spermatheca consists of three distinct regions, a distal neck-like constriction, a large central bag, and the spermatheca-uterine (sp-ut) valve (McCarter et al., 1997). The sp-ut valve is consists of a syncytial toroidal cell (sujn) and a core syncytial cell (sujc) (Kimble and Hirsh, 1979). The core cell is displaced during the first ovulation, and its fate is unknown.
Prior to the first ovulation, the morphology of the distal and central spermatheca is indistinguishable between wildtype and fln-1(tm545) animals by DIC (Figure 4B, C). In addition, the apical junctions between spermathecal cells were visualized with jam-1::gfp and the anti-JAM-1 monoclonal antibody (MH27) (Koppen et al., 2001). Junctions were indistinguishable from wildtype in fln-1(tm545) and fln-1 RNAi animals (data not shown). We also used fkh-6::gfp (Chang et al., 2004; Gissendanner et al., 2008), a spermatheca developmental marker, to assess morphogenesis of the spermatheca. In wildtype animals fkh-6::gfp is expressed in the distal and central spermatheca, but not in the sp-ut. An identical staining pattern is observed in fln-1(tm545) animals (Figure 4B, C). In older adult wildtype animals the spermatheca appears as a compact structure (Figure 4D). In contrast, the fln-1(tm545) spermatheca is distended due to accumulated embryos (Figure 4E). The spermatheca remains distended and occupied by embryos and fragmented oocytes for the duration of adulthood. Importantly, the spermatheca does not disintegrate at any point. .
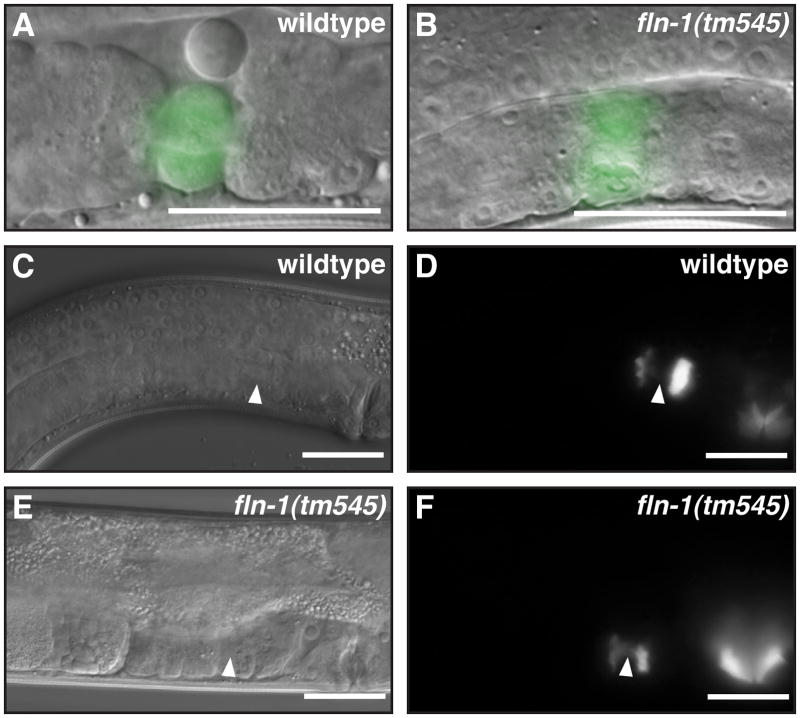
fln-1 is required for correct morphology of the spermatheca-uterine valve
We also examined the spermatheca-uterine (sp-ut) valve using DIC microscopy in wildtype and fln-1(tm545) animals (Figure 5A, B). In wildtype animals the sp-ut is a toroid produced by the donut-shaped sujn cell, with a rod-like opening in the center formed by the sujn and sujc cells (Figure 5A) (Kimble and Hirsh, 1979). In contrast, the sp-ut valve of fln-1(tm545) animals appeared creased or folded and lacked the characteristic wildtype morphology, suggesting that filamin is required to maintain the proper morphology of the sujn cell (Figure 5B). To visualize the morphology of the sujn cell in fln-1(tm545) animals we used tag-312::gfp, which is expressed in the sujn and the uterus. We thought the crumpled appearance of the sujn cell might indicate a closed valve, through which oocytes might not be able to exit. To visualize the inside of the valve, we used a cog-1::gfp (Palmer et al., 2002) fusion to label the sujc cell, which projects filopodia from the uterus into the spermatheca through the sp-ut valve (Figure 5C, D). The expression pattern of cog-1::gfp was essentially identical in wildtype and fln-1(tm545) animals, suggesting that the valve does form a channel connecting the spermatheca and the uterus (Figure 5E, F). In addition, we regularly observe sperm passing from the spermatheca into the uterus of fln-1(tm545) animals during ovulation. In motile animals, embryos do eventually exit the spermatheca through the valve into the uterus, and do not break out through the spermathecal wall.
Figure 5. FLN-1 is required for normal spermatheca-uterine valve morphology.
A) Wildtype sp-ut valve marked with tag-312::gfp. B) fln-1(tm545) sp-ut valve marked with tag-312::gfp. C–D) cog-1::gfp is expressed in the sujc cells (arrowheads) and in the vulva. E–F) cog-1::gfp is normally expressed in the sujc cells (arrowheads) of fln-1(tm545) animals and in the vulva. Bar indicates 25 μm.
Even though the valve appears to be open and of apparently normal diameter in fln-1(tm545) animals, the crumpled morphology of the apical surface of the sujn cells suggests that the valve might not be functional. A non-functional valve could account for the trapped oocyte phenotype seen in the filamin mutants. Alternatively, fln-1 function may be required in the spermatheca itself as well as in the valve for proper exit of fertilized oocytes. We used the GFP::FLN-1A transgene in the fln-1(tm545) background to determine which spermathecal cells require FLN-1 for normal ovulation. Animals which visibly expressed GFP::FLN-1A only in the sp-ut valve had a morphologically normal valve, but contained trapped embryos (100%; n=8). In contrast, animals expressing GFP::FLN-1A in the spermatheca and the sp-ut valve were essentially wildtype and contained no trapped embryos (90%; n=29). Therefore, expression of GFP::FLN-1A in the sp-ut valve apparently restores normal morphology of the valve, but is not sufficient to rescue the exit defect. These results are consistent with the hypothesis that filamin is required to maintain the structure of the valve, but is also necessary in the spermatheca. These results suggest that filamin is required to maintain the structure of the sp-ut valve, but may also be required in the spermatheca itself to transduce signals in addition to playing a structural role.
F-actin is disorganized in the spermatheca and uterus of fln-1(tm545) mutants
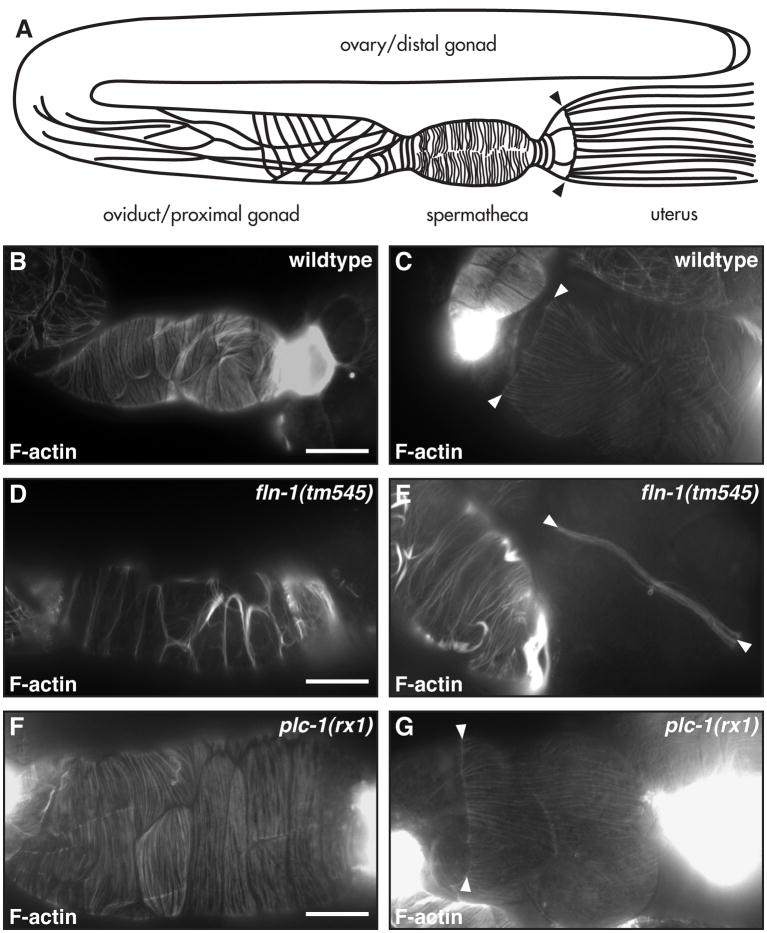
The most well characterized role of filamins is organization of the actin cytoskeleton (Stossel et al., 2001). The actin cytoskeleton is essential for proper ovulation in C. elegans, and is required for the structure and function of the myoepithelial sheath cells, spermatheca, and uterus (McCarter et al., 1999; Ono et al., 2007). To visualize actin in the gonadal sheath, spermatheca, and uterus of wildtype and fln-1(tm545) animals, we stained dissected C. elegans gonads with Texas Red-X phalloidin. Sheath cell actin was indistinguishable from wildtype in fln-1(tm545) animals, with the expected longitudinally arranged actin filaments in the distal sheath cells and more circumferential filaments in the contractile proximal sheath cells (Figure 6A). In wildtype spermathecal cells, regularly spaced circumferential actin bundles can clearly be visualized (Figure 6B). In stark contrast, in adult fln-1(tm545) animals, spermathecal F-actin was bundled into thick, cortical bundles and localized to the cell-cell junctions (Figure 6D). In the uterus of wildtype animals, F-actin is organized predominantly longitudinally and originates at the spermatheca-uterine lariat (Figure 6C) (Strome, 1986). Interestingly, the spermatheca-uterine lariat is severely disorganized in filamin mutant animals and lacks the characteristic branching filaments (Figure 6E). fln-1 is the first gene known to be required for maintenance of the spermatheca-uterine lariat, and may play a role in strengthening the uterus to withstand the pressure of accumulating embryos.
Figure 6. FLN-1 is required for maintenance of F-actin in the spermatheca and uterus.
A) Diagram of F-actin in the somatic gonad. B) Wildtype spermathecal and uterine F-actin is arranged roughly circumferentially. C) Wildtype uterine actin is arranged longitudinally and originates at the uterine lariat (arrowheads). D) fln-1(tm545) actin is bundled into thick cortical filaments at the cell-cell junctions. E) fln-1(tm545) actin in the uterus appears thicker and is collapsed to the uterine lariat. F) plc-1(rx1) F-actin in the spermatheca is normal. G) plc-1(rx1) uterine lariat is normal. Arrowheads indicate the lariat. Bar indicates 25 μm.
The first ovulation event coincides with a dramatic rearrangement of the spermathecal actin cytoskeleton. Pre-ovulation spermathecal actin is not organized into tight circumferential filaments (Figure 7A). After the first ovulation, these filaments are formed and persist through many cycles of fertilization (Figure 7B–E). This suggests that the adult spermathecal cytoskeleton is established shortly before or during the first ovulation. Spermathecal actin in fln-1(tm545) animals, before the first ovulation, resembles that of wildtype animals, but is not entirely normal (Figure 7F). Following the first ovulation of fln-1(tm545) animals the F-actin appears to reorganize as in wildtype animals, although the filaments are not as robust (Figure 7G). Young adult fln-1(tm545) spermathecae lack the cortical bundles characteristic of later stage animals (Figures 6D and 7J). Our F-actin staining at various ages suggests that spermathecal actin progressively degenerates following the first failed ovulation (Figure 7H–J). In addition, the early time points (55–65 hours) show a greater degree of variability than the later time points (72 hours). The variability is likely due to rapid changes that occur between the first and second ovulations. These results suggest that filamin is primarily required to maintain the F-actin cytoskeleton in response to stretching by the oocyte during ovulation. Our data do not rule out a role for filamin in the initial F-actin organization.
Figure 7. fln-1(tm545) spermathecal F-actin progressively worsens.
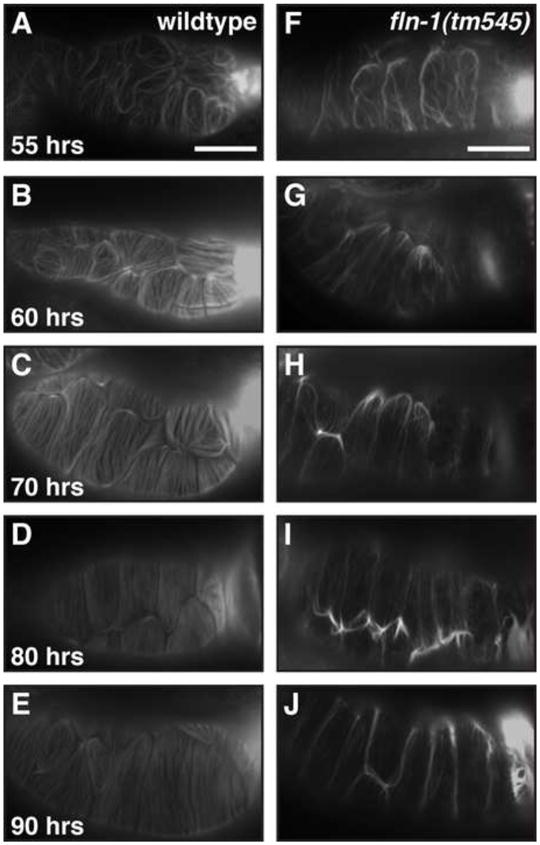
A–E) Wildtype spermatheca F-actin at 55 (A), 60 (B), 70 (C), 80 (D), and 90 (E) hours of age. F–J) fln-1(tm545) F-actin at 55 (F), 60 (G), 70 (H), 80 (I), and 90 (J) hours of age. Onset of ovulation is between 55 and 60 hours, therefore (A) and (F) are before the first ovulation, while (B) and (G) are following the first ovulation. Corresponding DIC images (not shown) were used to determine number of ovulations that had occurred. Bar indicates 25 μm.
The phospholipase PLC-1 is also required for the exit of embryos from the spermatheca (Kariya et al., 2004). The plc-1(rx1) putative null animals phenocopy the exit defect of fln-1(tm545) animals, and retain multiple embryos in the spermatheca. PLC-1 is broadly expressed, and has been shown to have a role in embryogenesis, in addition to its role during ovulation (Hiatt et al., 2009; Vazquez-Manrique et al., 2008). We hypothesized that, if stretching induced by accumulated embryos causes the actin to be redistributed to the cell boundaries, the actin cytoskeleton in plc-1(rx1) spermathecal cells might be similarly disrupted. Surprisingly, spermathecal and uterine F-actin in plc-1(rx1) animals was indistinguishable from wildtype, despite stress caused by the retained embryos (Figure 6F, G). F-actin staining of fln-1(tm545); plc-1(rx1) double mutant animals was indistinguishable from that of fln-1(tm545) single mutants (data not shown). Also, the sp-ut valve in plc-1(rx1) animals appears normal by DIC microscopy. These results suggest that PLC-1 may not play a significant role in the maintenance of the actin cytoskeleton or in sp-ut valve morphology, but instead PLC-1 may be required to produce a signal that allows exit of the fertilized embryo from the spermatheca. fln-1(tm545); plc-1(rx1) double mutant animals are viable and show a decreased brood size (16 ± 4, n=15), which is not significantly different from the plc-1(rx1) single mutant animals (12.4 ± 5.9, n=9; Student’s t-test, p=0.1). These data suggest that fln-1 and plc-1 act in the same genetic pathway to allow exit of embryos from the spermatheca, perhaps through modulation of phosphoinositol signaling.
Discussion
Filamin is a large cytoskeletal protein that functions as a physical and signaling scaffold, and has been implicated in a wide variety of human disorders. We show that the C. elegans filamin ortholog FLN-1 is required to organize actin in the spermatheca and uterus, and to control the exit of embryos from the spermatheca. Filamin mutation or RNAi-mediated depletion of fln-1 results in a highly penetrant spermathecal exit defect during ovulation. Filamin-deficient animals accumulate embryos in the spermatheca, and consequently exhibit dramatically reduced brood sizes. The ovulation exit defect manifests as abnormally shaped, variably arrested embryos, which may be a general characteristic of spermathecal exit mutants. The proportion of misshapen and inviable embryos rapidly increases after the first four to five ovulations, suggesting the disrupted exit process in fln-1(tm545) animals physically damages the embryos.
FLN-1 is not required for spermathecal or uterine development per se; however, filamin is required for normal spermatheca-uterine valve morphology. The wildtype sp-ut valve is a toroid formed by the fusion of the sujn cells, which appears as an ellipse with a dense core and smooth edges when viewed by DIC microscopy. The sp-ut valve in fln-1(tm545) animals lacks the characteristic wildtype morphology on the basal surface, and is more crumpled on the apical surface. Our analysis shows that the sujn and sujc cells are present, and that the valve connects the spermatheca to the uterus. In motile animals, embryos exit the spermatheca after the second or third ovulation weakening the sp-ut valve and causing valve prolapse. The spermatheca remains occupied by multiple embryos and oocytes following valve prolapse. Expression of GFP::FLN-1A only in the valve restores the normal morphology and prevents valve prolapse, but is not sufficient for normal exit. This is similar to the plc-1 phenotype where the valve does not prolapse, but the embryos do not exit the spermatheca in a timely manner. These results suggest that a morphologically normal valve is not sufficient for exit of embryos from the spermatheca.
Filamin-deficient animals show F-actin disorganization in the spermatheca and uterus that worsens as embryos accumulate. This phenotype suggests that filamin is necessary to maintain the actin cytoskeleton under stress. Prior to the first ovulation, spermathecal F-actin is comparatively normal. PLC-1/Phospholipase C-ε mutant animals phenocopy the filamin exit defect and accumulate embryos; however, the F-actin appears unaffected. These results suggest that physical stress is not sufficient to cause F-actin disorganization and that the effect is specific to loss of filamin. The cytoskeleton is necessary for many aspects of cell behavior, such as trafficking and signaling, making it difficult to differentiate between direct and indirect effects of F-actin disorganization. Perturbation of the actin cytoskeleton by loss of filamin may have diverse downstream effects. Based on progression of the F-actin degeneration in the spermatheca it seems unlikely that F-actin disorganization is the sole factor responsible for the exit defect. All first ovulation embryos are trapped in the spermatheca despite relatively normal F-actin organization. Although our attempts to image F-actin in the sp-ut valve have been unsuccessful due to the size of the valve, we know that rescuing the valve morphology, and presumably any F-actin defects, is not sufficient for normal exit. Our data, and the previous plc-1 data, show that normal structure is not sufficient for the exit process, and suggest that an underlying signaling pathway plays an important role.
In filamin mutant animals filamentous actin is mislocalized to cell-cell junctions, which suggests the cytoskeleton may not be properly anchored throughout the cell. Our analysis of the GFP::FLN-1A fusion protein shows FLN-1 partially co-localizes to F-actin in the spermatheca and the uterus in a punctate filamentous pattern. Surprisingly, our β-integrin immunostaining shows diffuse staining in the spermatheca and a lack of clear dense bodies, despite strong staining of the sheath cells. Given the weak and diffuse integrin staining in the spermatheca we cannot conclude that integrin and filamin co-localize. The possibility that FLN-1 may not need to associate with integrin to promote proper spermathecal function is also suggested by the fln-1(ok2611) allele, which removes the integrin-association repeat of FLN-1 but does not affect ovulation. The pattern of dense body staining in the sheath and the spermatheca is consistent with the different functions of the two tissues. The sheath contracts rapidly and strongly, while the spermatheca shows no such contractions. Integrin-based adhesions in the spermatheca are likely required for development of the spermatheca, but they may not be required for the function of the adult spermatheca (Ono et al., 2007).
The entry process is regulated by EGF signaling from the oocyte to the sheath cells and the distal spermatheca. Upon activation of LET-23/EGFR, PLC-3/PLC-γ is probably activated by LET-23 (Clandinin et al., 1998; Yin et al., 2004). In contrast, the potentiating signal for sp-ut dilation and embryo release from the spermatheca into the uterus is unknown. Upon entry of the oocyte into the spermatheca, the spermathecal cells become stretched. We predict this stretching could result in conformational changes to the filamin dimer and revelation of cryptic binding sites. Alternatively, filamin may have a role in cell response to stretch not directly sensed by the filamin molecule, such as activation of stretch-gated ion channels (Orr et al., 2006). For example, filamin interacts with flow-sensitive polycystin ion channels in epithelial and endothelial cells (Sharif-Naeini et al., 2009), suggesting that in addition to being a direct mechanosensor, filamin may modulate the cytoskeleton to sensitize or desensitize cells to physical force.
Our working model is that entry of the oocyte into the spermatheca triggers a series of events involving FLN-1 and PLC-1 that leads to relaxation of the sp-ut valve and exit of the embryo into the uterus. Based on our time-lapse recordings of ovulation, the spermatheca and the sp-ut valve do not appear to actively contract or dilate, but appear to gradually relax as the oocyte moves forward. The gradual relaxation of the spermatheca and the sp-ut is in sharp contrast to the forceful and swift gonadal sheath contractions, suggesting a different mechanism. Consistent with the idea that IP3 signaling controls sp-ut dilation, depletion of IP3 by LFE-2 over-expression under the heatshock promoter (Clandinin et al., 1998) or PLC-1 loss-of-function causes an exit defect (Kariya et al., 2004). We therefore hypothesize that filamin-depleted animals are also unable to produce or respond to IP3 signals, resulting in the exit defect. In support of this idea, filamin has been shown to interact with phosphoinositol signaling pathways in cell culture, usually as a downstream effector (Bourguignon et al., 2006; Dyson et al., 2001; Takabayashi et al.).
Taken together, our results suggest that filamin is required to maintain circumferential actin bundles in the spermatheca under stress and to act as a stretch-sensitive signaling scaffold. Filamin may potentiate or transduce the phosphoinositol and calcium signals that result in the sp-ut valve dilation and exit of oocytes from the spermatheca. This work provides evidence that FLN-1/filamin plays a structural and signaling role in the C. elegans somatic gonad. Our results directly demonstrate the importance of filamin in tissues undergoing stress in vivo. Studies are underway to more precisely determine the position of FLN-1 in the phosphoinositol signaling pathway, and to identify additional genes that are required for the exit of oocytes from the spermatheca. Given the striking conservation of FLN-1 sequence, structure, and interaction partners from worm to human, elucidation of the function of filamin in C. elegans will continue to provide important insights into the fundamental regulation of mechanosensation and tissue function.
Materials and Methods
C. elegans strains and culture
All nematode strains were cultured on NGM agar plates with OP50 E. coli at 20°C. Nematode observations and manipulations were performed at 20°C unless otherwise noted. The C. elegans filamin allele fln-1(tm545) was obtained from the Japanese National Bioresource Project and outcrossed three times to wildtype animals to create the strain UN0810 fln-1(tm545). All other strains were obtained from the Caenorhabditis Genetics Center, with the exception of PS4195, which was kindly provided by Paul Sternberg. For a list of strains used in this study, please see Table S1.
Genetic crosses
fln-1(ok2611)/fln-1(tm545) trans-heterozygous animals were generated by crossing fln-1(tm545) males to fln-1(ok2611) hermaphrodites. F1 animals of the putative genotype tm545/ok2611 were segregated for brood size assays. The first ten F2 progeny were genotyped using PCR to confirm the presence of tm545 and ok2611 alleles.
Construction of the fln-1(tm545); plc-1(rx1) double mutant was performed by crossing fln-1(tm545) males to plc-1(rx1) hermaphrodites. F1 animals of the putative genotype tm545/+ were subcloned. F2 animals were subcloned and the genotype determined by PCR. Homozygosity of the alleles was confirmed by PCR genotyping of the progeny.
RNA interference
RNA interference was performed by feeding animals dsRNA-expressing HT115 DE3 E. coli essentially as described (Cram et al., 2006). Eggs were obtained from gravid hermaphrodites using alkaline hypochlorite solution and transferred to NGM/Carbenicillin/IPTG plates seeded with RNAi bacteria. RNAi experiments were performed at 20°C. RNAi targeting constructs for Y66H1B.3, Y66H1B.5, and Y66H1B.2 were constructed by PCR amplification of wildtype cDNA using engineered restriction sites, and subsequently cloned into pPD129.36 (Fire Vector Kit). Empty pPD129.36 vector was used as a negative control in RNAi experiments. All primer sequences and cloning details available upon request.
Brood size assays
Total number of hatchlings produced by a single animal was determined by segregating L4 parental animals to individual, freshly seeded plates and aspirating larvae each day, beginning two days after transfer and continuing for at least five days.
Embryo shape and embryonic lethality assay
Animal populations were synchronized by hatching embryos in the absence of food for 20 hours in M9 buffer. The arrested L1 animals were transferred to NGM plates seeded with OP50 E. coli. Worms were collected in M9, dissected, and mounted on 1.5% agarose pads at 60, 70, 80, and 90 hours after being placed on food. Embryo shapes were scored immediately using DIC microscopy, then allowed to hatch overnight at 20°C. The slides were examined again within 24 hours to determine which embryos hatched.
Time-lapse microscopy
Animals were anesthetized with 0.01% tetramisole and 0.1% tricaine in M9 buffer (Kirby et al., 1990; McCarter et al., 1997), mounted on 1.5% agarose pads and imaged using a 60x oil-immersion objective with a Nikon Eclipse 80i microscope equipped with a SPOT RT3 CCD camera (Diagnostic Instruments; Sterling Heights, MI, USA). Images were captured at a rate of 0.5 frames per second (FPS) using SPOT Advanced version 4.6.4.6 (Diagnostic Instruments; Sterling Heights, MI, USA) software. The individual frames (TIFF with JPEG compression) were resized to 400 × 300 pixels and assembled into time-lapse movies using SPOT Advanced software at 10 FPS. The Audio Video Interleave (AVI) files generated by SPOT Advanced were imported into QuickTime Pro version 7.6.2 (Apple; Cupertino, CA, USA) and compressed using H.264, then exported as Moving Picture Experts Group, Standard 4 (MPEG-4) movies. The time-lapse movies are 20-fold faster than real-time.
Construction of fln-1::gfp transgenic animals
GoTaq Green MasterMix (Promega; Madison, WI, USA) was used to amplify a 957 bp region 5′ to Y66H1B.3 flanked by engineered restriction sites HindIII and BamHI. The HindIII-BamHI fragment was cloned into pPD95.77 (Fire Vector Kit) to create pUN70. The plasmid was isolated from E. coli and directly used in microinjection at an approximate concentration of 100 μg/mL. Transgenic strains were created by standard germline transformation technique (Mello et al., 1991) of wildtype animals to create strain UN0811 xbEx0811[fln-1::gfp].
Construction of GFP::FLN-1A transgenic animals
Full-length fln-1a transcript was amplified by PCR from wildtype cDNA using Phusion DNA Polymerase (New England Biolabs; Ipswich, MA, USA). The amplicon was then inserted into pUN88 between the fln-1 promoter and 3′ UTR to create pUN97 using the In-Fusion enzyme (Clontech; Mountain View, CA, USA). pUN97 was purified and mixed with co-injection marker rol-6(su1006) (pRF4) to a final concentration of approximately 50 μg/mL. The DNA mixture was injected into wildtype animals to establish the extrachromosomal array xbEx1002[fln-1::gfp::fln-1a, rol-6(su1006)]. The rescuing construct was introduced into the fln-1(tm545) background by genetic cross to create strain UN1003.
fln-1 genomic rescue
LongAmp (New England Biolabs, Ipswich, MA, USA) was used to amplify the 19 kb genomic region encompassing ORFs Y66H1B.3, Y66H1B.5, and Y66H1B.2, including 1.5 kb of 5′ and 3′ flanking sequences. Transgenic animals were created by standard microinjection germline transformation. The long-range amplicon was agarose gel-purified and mixed with co-injection marker sur-5::gfp (pTG96) and water to a final concentration of 100 μg/mL. The DNA mixture was injected into wildtype animals to establish the extrachromosomal array xbEx0827[fln-1(+), sur-5::gfp]. The rescuing construct was introduced into the fln-1(tm545) background by genetic cross to create strain UN0903. The same process was used to construct an extrachromosomal array with a truncated fln-1 genomic region encompassing Y66H1B.3 and Y66H1B.5, xbEx0917[fln-1(Y66H1B.3/5), sur-5::gfp].
Construction of sujn-specific marker
To construct an sujn-specific GFP marker we used WormBase expression data to identify candidate genes expressed in the sujn cell. We used Phusion DNA Polymerase to amplify the putative tag-312 promoter (1 kb upstream) and cloned it into pPD95_77. The plasmid was isolated from E. coli and mixed with co-injection marker rol-6(su1006) (pRF4) to a final concentration of approximately 50 μg/mL. Transgenic strains were created by standard germline transformation technique (Mello et al., 1991) of wildtype animals to create strain UN1019 xbEx1019[tag-312::gfp, rol-6(su1006)]. The transgene was introduced into fln-1(tm545) background by genetic cross.
Immunofluorescence
F-actin and PAT-3 immunoflourescence was performed as described (Ono et al., 2007). Briefly, partially synchronized populations were dissected using a 25-gauge hypodermic needle in PBS, and dissected gonads were fixed in 3.7% formaldehyde in PBS for 20 minutes at room temperature. For F-actin staining, following the formaldehyde fixation the animals were washed twice with PBST (PBS + 0.1% Triton X-100) for 20 minutes, and then incubated with 0.4 U/mL of Texas Red-X phalloidin in PBS (Invitrogen, Carlsbad, CA, USA) overnight at 4°C or 4 hours at room temperature, washed with PBS, and mounted on 1.5% agarose pads for observation. For PAT-3 staining, fixed animals were incubated with MH25 anti-PAT-3 antibodies (1:100 dilution) overnight at 4°C, followed by Cy3-conjugated donkey anti-mouse (Jackson ImmunoResearch Laboratories; West Grove, PA, USA) for 4 hours at room temperature, washed twice with PBS, and mounted on 1.5% agarose pads for observation. Fluorescence microscopy was performed on a Nikon Eclipse 80i microscope equipped for epifluorescence. Images were captured with a SPOT RT3 CCD camera using SPOT Advanced software (Diagnostic Instruments; Sterling Heights, MI, USA). This fixation procedure preserves GFP fluorescence.
Supplementary Material
A) Wildtype ovulation begins with intense sheath cell contractions; the sheath contractions and the simultaneous dilation of the distal spermatheca force the proximal-most oocyte into the spermatheca, where it is fertilized. Following fertilization the embryo exits the spermatheca via the spermatheca-uterine valve. B) tm545 animals exhibit no defects during the ovulation entry process, but the embryo fails to exit the spermatheca. Bar indicates 25 μm.
A) fln-1(tm545) animals carrying the entire genomic region of fln-1 show normal ovulation with no trapped embryos. B) fln-1(tm545) animals expressing GFP::FLN-1 show normal ovulation. Movies show GFP expression before ovulation (A and B) and during ovulation for (B).
A list of strain names and corresponding genotypes used in this study.
A) PAT-3 stained in the gonadal sheath cell using MH25 antibody, and (B) a corresponding GFP::FLN-1A image. C) Merged PAT-3 (red) and GFP::FLN-1A (green). D) PAT-3 stained in the spermatheca, and (E) a corresponding GFP-FLN-1A image. F) Marged PAT-3 (red) and GFP::FLN-1A (green). Bar indicates 15 μm.
Acknowledgments
Many C. elegans strains were provided by the Caenorhabditis Genetic Center, which is funded by the National Center for Research Resources, National Institutes of Health. We thank Paul Sternberg for providing multiple strains, and Anne Hart for helpful discussions. We thank Jose Orozco for help with experiments. I.K. and E.J.C. are partially supported by Northeastern University and a grant from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Annesley SJ, Bandala-Sanchez E, Ahmed AU, Fisher PR. Filamin repeat segments required for photosensory signalling in Dictyostelium discoideum. BMC Cell Biol. 2007;8:48. doi: 10.1186/1471-2121-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. 2006;281:14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- Bui YK, Sternberg PW. Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol Biol Cell. 2002;13:1641–1651. doi: 10.1091/mbc.02-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- Chang W, Tilmann C, Thoemke K, Markussen FH, Mathies LD, Kimble J, Zarkower D. A forkhead protein controls sexual identity of the C. elegans male somatic gonad. Development. 2004;131:1425–1436. doi: 10.1242/dev.01012. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–533. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Shang H, Schwarzbauer JE. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci. 2006;119:4811–4818. doi: 10.1242/jcs.03274. [DOI] [PubMed] [Google Scholar]
- Critchley DR. Focal adhesions - the cytoskeletal connection. Curr Opin Cell Biol. 2000;12:133–139. doi: 10.1016/s0955-0674(99)00067-8. [DOI] [PubMed] [Google Scholar]
- Dyson JM, O’Malley CJ, Becanovic J, Munday AD, Berndt MC, Coghill ID, Nandurkar HH, Ooms LM, Mitchell CA. The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembraneous actin. J Cell Biol. 2001;155:1065–1079. doi: 10.1083/jcb.200104005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Olive M. Molecular pathology of myofibrillar myopathies. Expert Rev Mol Med. 2008;10:e25. doi: 10.1017/S1462399408000793. [DOI] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Gehler S, Baldassarre M, Lad Y, Leight JL, Wozniak MA, Riching KM, Eliceiri KW, Weaver VM, Calderwood DA, Keely PJ. Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell. 2009;20:3224–3238. doi: 10.1091/mbc.E08-12-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner CR, Kelley K, Nguyen TQ, Hoener MC, Sluder AE, Maina CV. The Caenorhabditis elegans NR4A nuclear receptor is required for spermatheca morphogenesis. Dev Biol. 2008;313:767–786. doi: 10.1016/j.ydbio.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, Fernandes J, Han M, Kishore R, Lee R, Muller HM, Nakamura C, Ozersky P, Petcherski A, Rangarajan A, Rogers A, Schindelman G, Schwarz EM, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Yook K, Durbin R, Stein LD, Spieth J, Sternberg PW. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- Hiatt SM, Duren HM, Shyu YJ, Ellis RE, Hisamoto N, Matsumoto K, Kariya K, Kerppola TK, Hu CD. Caenorhabditis elegans FOS-1 and JUN-1 regulate plc-1 expression in the spermatheca to control ovulation. Mol Biol Cell. 2009;20:3888–3895. doi: 10.1091/mbc.E08-08-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel M, Van Der Ven PF, Stocklein W, Furst DO. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry. 2003;42:430–439. doi: 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kainulainen T, Pender A, D’Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. J Biol Chem. 2002;277:21998–22009. doi: 10.1074/jbc.M200715200. [DOI] [PubMed] [Google Scholar]
- Kariya K, Bui YK, Gao X, Sternberg PW, Kataoka T. Phospholipase Cepsilon regulates ovulation in Caenorhabditis elegans. Dev Biol. 2004;274:201–210. doi: 10.1016/j.ydbio.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol. 1990;142:203–215. doi: 10.1016/0012-1606(90)90164-e. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Li MG, Serr M, Edwards K, Ludmann S, Yamamoto D, Tilney LG, Field CM, Hays TS. Filamin is required for ring canal assembly and actin organization during Drosophila oogenesis. J Cell Biol. 1999;146:1061–1074. doi: 10.1083/jcb.146.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Ono S. Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev Dyn. 2007;236:1093–1105. doi: 10.1002/dvdy.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell. 2006;10:11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Inoue T, Sherwood DR, Jiang LI, Sternberg PW. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev Biol. 2002;252:202–213. doi: 10.1006/dbio.2002.0850. [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Sharif-Naeini R, Folgering JH, Bichet D, Duprat F, Lauritzen I, Arhatte M, Jodar M, Dedman A, Chatelain FC, Schulte U, Retailleau K, Loufrani L, Patel A, Sachs F, Delmas P, Peters DJ, Honore E. Polycystin-1 and -2 dosage regulates pressure sensing. Cell. 2009;139:587–596. doi: 10.1016/j.cell.2009.08.045. [DOI] [PubMed] [Google Scholar]
- Shifrin Y, Arora PD, Ohta Y, Calderwood DA, McCulloch CA. The role of FilGAP-filamin A interactions in mechanoprotection. Mol Biol Cell. 2009;20:1269–1279. doi: 10.1091/mbc.E08-08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Cooley L. Drosophila filamin is required for follicle cell motility during oogenesis. Dev Biol. 2003;260:260–272. doi: 10.1016/s0012-1606(03)00248-3. [DOI] [PubMed] [Google Scholar]
- Stefanova M, Meinecke P, Gal A, Bolz H. A novel 9 bp deletion in the filamin a gene causes an otopalatodigital-spectrum disorder with a variable, intermediate phenotype. Am J Med Genet A. 2005;132:386–390. doi: 10.1002/ajmg.a.30484. [DOI] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabayashi T, Xie MJ, Takeuchi S, Kawasaki M, Yagi H, Okamoto M, Tariqur RM, Malik F, Kuroda K, Kubota C, Fujieda S, Nagano T, Sato M. LL5{beta} directs the translocation of Filamin A and SHIP2 to sites of PtdIns(3,4,5)P3 accumulation and PtdIns(3,4,5)P3 localization is mutually modified by co-recruited SHIP2. J Biol Chem. 2010 doi: 10.1074/jbc.M109.081901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Manrique RP, Nagy AI, Legg JC, Bales OA, Ly S, Baylis HA. Phospholipase C-epsilon regulates epidermal morphogenesis in Caenorhabditis elegans. PLoS Genet. 2008;4:e1000043. doi: 10.1371/journal.pgen.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Gower NJ, Baylis HA, Strange K. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3938–3949. doi: 10.1091/mbc.E04-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Tian F, Sandzen J, Cao R, Flaberg E, Szekely L, Cao Y, Ohlsson C, Bergo MO, Boren J, Akyurek LM. Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc Natl Acad Sci U S A. 2007;104:3919–3924. doi: 10.1073/pnas.0608360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Wildtype ovulation begins with intense sheath cell contractions; the sheath contractions and the simultaneous dilation of the distal spermatheca force the proximal-most oocyte into the spermatheca, where it is fertilized. Following fertilization the embryo exits the spermatheca via the spermatheca-uterine valve. B) tm545 animals exhibit no defects during the ovulation entry process, but the embryo fails to exit the spermatheca. Bar indicates 25 μm.
A) fln-1(tm545) animals carrying the entire genomic region of fln-1 show normal ovulation with no trapped embryos. B) fln-1(tm545) animals expressing GFP::FLN-1 show normal ovulation. Movies show GFP expression before ovulation (A and B) and during ovulation for (B).
A list of strain names and corresponding genotypes used in this study.
A) PAT-3 stained in the gonadal sheath cell using MH25 antibody, and (B) a corresponding GFP::FLN-1A image. C) Merged PAT-3 (red) and GFP::FLN-1A (green). D) PAT-3 stained in the spermatheca, and (E) a corresponding GFP-FLN-1A image. F) Marged PAT-3 (red) and GFP::FLN-1A (green). Bar indicates 15 μm.