Abstract
Prephenate is the direct precursor of phenylpyruvate and 4-hydroxyphenylpyruvate in the biogenesis of phenylalanine and tyrosine by action of the decarboxylative, aromatizing enzymes prephenate dehydratase and dehydrogenase, respectively. The recent characterization of BacA in bacilysin biosynthesis as a non-aromatizing decarboxylase reveals a new route from prephenate in the biosynthesis of nonproteinogenic amino acids. This study describes two additional enzymes, AerD from Planktothrix agardhii and SalX from Salinispora tropica, that utilize the central building block prephenate for flux down distinct pathways to amino acid products, representing a new metabolic fate for prephenate and establishing a new family of non-aromatizing prephenate decarboxylases.
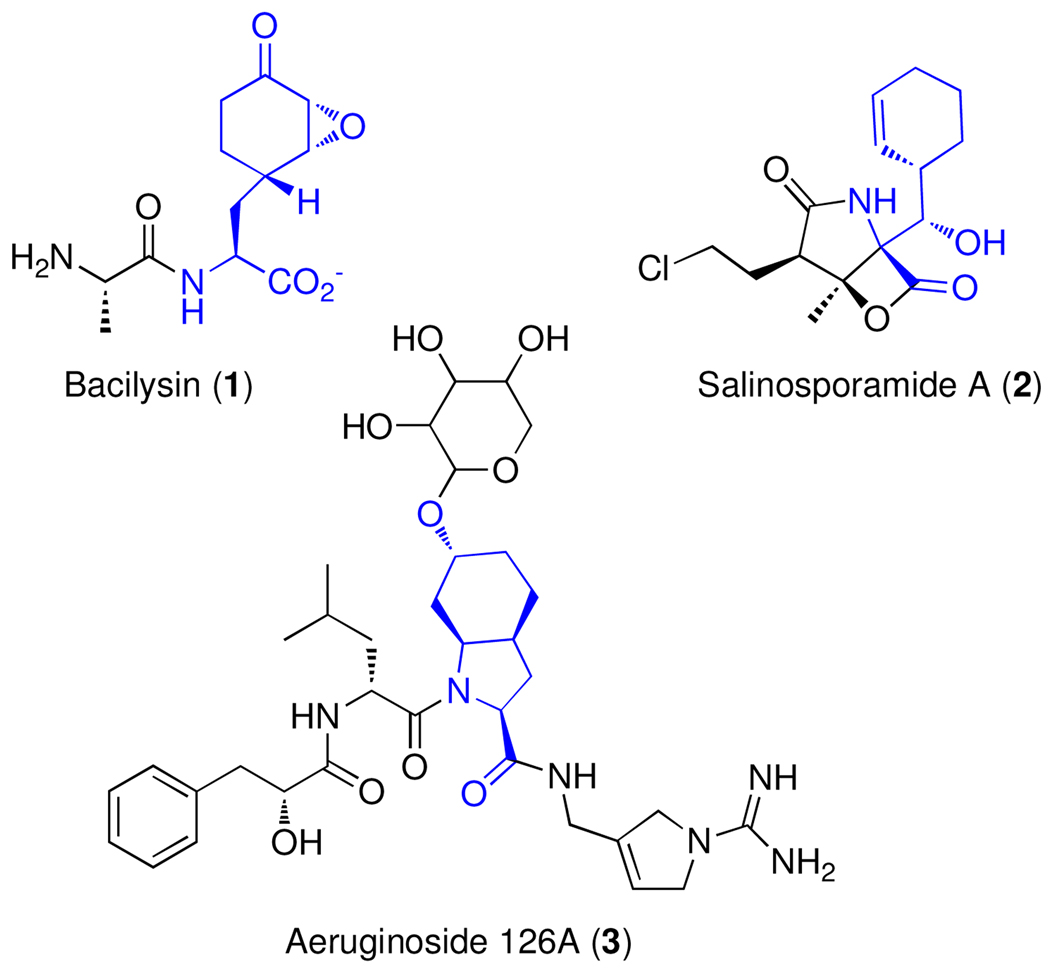
Prephenate has been known for decades as the product of the celebrated 3,3-sigmatropic rearrangement of chorismate, a reaction that is classically followed by formation of the aromatic amino acids phenylalanine and tyrosine (1). Prephenate dehydratase and prephenate dehydrogenase are the enzymes that direct prephenate to Phe and Tyr by decarboxylation and aromatization of the cyclohexenyl ring with concomitant loss of the C-7 hydroxide or hydride, respectively. Recently we characterized the Bacillus subtilis prephenate dehydratase homolog BacA, which is involved in the biosynthesis of bacilysin (1, Figure 1), and catalyzes the first step of a 4-enzyme pathway that transforms prephenate to a distinct non-aromatic amino acid product (2). Like prephenate dehydratase, BacA decarboxylates prephenate at C-4 yet uniquely protonates C-6 or C-6’ to yield the endocyclic diene dihydro-4-hydroxyphenylpyruvate (H2HPP, 5, Figure 2A) (2). This diene is then subject to nonenzymatic isomerization to the exocyclic diene in a reaction that can be dramatically accelerated by BacB to yield the indicated H2HPP regioisomer 6. Here we identify two homologs to BacA from distinct microbial secondary metabolite pathways that catalyze the same non-aromatizing decarboxylation of prephenate, thus establishing a new family of enzymes. The first of these is AerD from the cyanobacterium Planktothrix agardhii, which is involved in the production of the nonribosomal glycopeptides aeruginoside 126A and B (3, Figure 1) (3). These natural products contain the unusual amino acid residue 2-carboxy-6-hydroxy-octahydroindole (Choi) (7) (4, 5). The second enzyme is SalX in the marine actinomycete Salinispora tropica biosynthetic pathway to the proteasome inhibitor salinosporamide A (2) (6–8). Specifically, SalX is relevant to the production of the nonproteinogenic amino acid building block cyclohexenylalanine (9; H4Phe) (9).
Figure 1.
Natural products containing amino acids (blue) derived from prephenate.
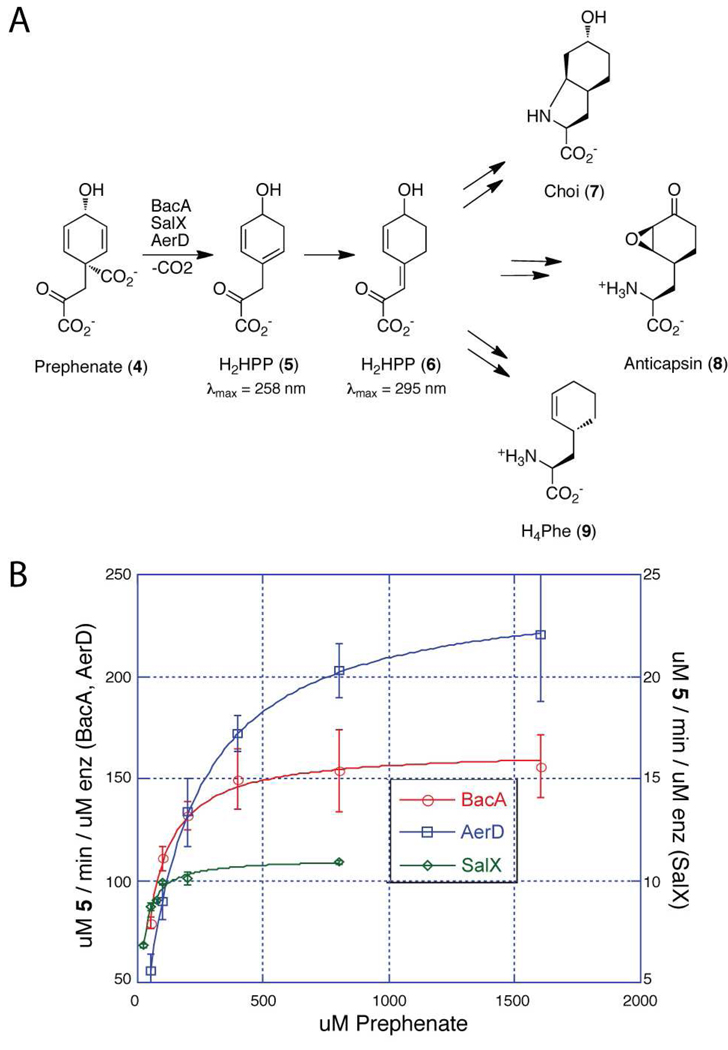
Figure 2.
A) Transformation of prephenate first to H2HPP 6, which is eventually incorporated into various amino acids. B) Kinetics of BacA, AerD and SalX.
In a study of nonribosomal peptide synthetase gene clusters in the cyanobacterium P. agardhii CYA126/8, gene insertions led to the discovery and identification of novel aeruginoside glycopeptides (3). Aeruginosin natural products, which are potent inhibitors of serine proteases, typically contain the unusual amino acid residue Choi (4). Three of the genes in the aeruginosin cluster, aerDEF, were found to be relevant to Choi production (3). Ishida et al. noted the similarity of aerDEF to the bacABC genes in the bacilysin gene cluster in B. subtilis. However, at the time of the aeruginosin gene cluster discovery, the specific roles of the BacABC proteins were unknown, so it was unclear what catalysts the congeneric aer genes were encoding. For example, it was hypothesized that arogenate, not prephenate, might be the entry point onto the Choi pathway (3). As noted by Ishida et al., AerD is 33.8% identical to BacA, so we sought to determine whether AerD was in fact a catalytic homolog of BacA. Because the aeruginoside producer P. agardhii CYA126/8 and its DNA were not readily available, we chose to use a synthetic gene for AerD (DNA 2.0), which was optimized for expression in Escherichia coli (Figure S2). This gene was cloned into a pET-28b vector equipped with an N-terminal His6 tag. As shown in Figure S3, excellent overproduction of soluble protein ensued, allowing purification of 14 mg/liter of culture. We assayed AerD with prephenate as a substrate utilizing UV scanning kinetics and readily observed the conversion to an A258 nm species, indicative of endocyclic H2HPP diene 5 (Figure 2, S4a). As we had found in studies with BacA from B. subtilis, H2HPP 5 non-enzymatically isomerized to the thermodynamically favored conjugated exocyclic diene 6 (2). 1H NMR analysis validated that AerD ultimately generates H2HPP 6 (Figure S4b, Table S1). The kcat (245 ± 1 min−1) and Km (170 ± 3 µM) values for prephenate conversion to initial endocyclic diene 5 for AerD compare favorably with those for BacA. Thus it is very likely prephenate is the physiological substrate for AerD in the initial step in construction of the bicyclic amino acid Choi.
Salinosporamide A (2) from S. tropica CNB-440 has a high density of functionality built into its scaffold. The bicyclic β-lactone-γ-lactam framework is a latent warhead and the C-2 chloroethyl side chain acts as a covalent trap for irreversible capture of the proteasome (10). The third element of the salinosporamide scaffold is the nonproteinogenic 3-hydroxyl-L-cyclohexenylalanyl residue (referred to here as a β-hydroxylated H4Phe (9)), which governs the binding specificity of the natural product to the proteasome (11). Examination of the salinosporamide biosynthetic gene cluster has indicated a set of sal genes that could be involved in H4Phe formation, among them SalX (8). Although our initial proposal suggested SalX might function similar to a prephenate dehydratase for a putative dihydroprephenate substrate (12), we decided to test it for conversion of prephenate to 5 and then 6, given 35% sequence identity to BacA. Toward this end, the salX gene was amplified from S. tropica genomic DNA and ligated into an N-His8 vector for expression in E. coli. As shown in Figure S3, moderate overproduction of soluble enzyme was obtained at 5 mg/liter of culture (Figure S3). UV assays revealed that SalX also converts prephenate to endocyclic diene H2HPP 5 as the initial product before spontaneous isomerization to exocyclic diene 6 (Figure 2, S4a), which was further confirmed by 1H NMR (Figure S4b, Table S2). While SalX had a 16–22 fold lower kcat than AerD and BacA, it also had a lower Km for prephenate. The SalX kcat/Km catalytic efficiency ratio of 0.73 min−1 µM−1 is only 2–4 fold lower than BacA and AerD, making it likely that prephenate is a physiologic substrate for SalX. To further probe whether prephenate is indeed the natural substrate for SalX and if H2HPP 6 is an intermediate in H4Phe production, feeding studies were preformed with a previously described S. tropica salX− disruption mutant that lacks salinosporamide production (9). H2HPP 6 was generated and purified from an in vitro transformation of prephenate by SalX and then added aseptically to a 50 mL culture of the ΔsalX S. tropica mutant. After 5 days the culture was extracted and HPLC analysis indicated restoration of salinosporamide production (Figure S5), which was confirmed by LCMS analysis with authentic standards. This chemical complementation confirmed that 6 is a pathway intermediate of 2 biosynthesis in S. tropica.
We establish here a new family of prephenate decarboxylases that produces a non-aromatized H2HPP product. The three new members of this family, BacA, AerD and SalX, are capable of initiating divergent pathways that transform the primary metabolite prephenate into distinct non-proteinogenic amino acids used in natural product production. The intriguing aspect of these enzymes is that while decarboxylation occurs similar to prephenate dehydratases and dehydrogenases, these enzymes avoid aromatization by delivery of a proton to C-6 or C-6’ to yield a diene product. The net result is conversion of the 5,8-cyclohexadiene-7-ol in prephenate to a 4,8-cyclohexadiene-7-ol in the H2HPP product 5. Isotopic labeling studies in S. tropica demonstrated that both halves of the cyclohexenyl ring of prephenate are distinguished during the biosynthetic transformation, suggesting a regioselectivity on the part of SalX (12). A similar biosynthetic observation was established in studies of the conversion of prephenate to the antibiotic 2,5-dihydrophenylalanine (13). However, additional detailed stereochemical analyses will be necessary to determine if all three enzymes share the same regio- and stereoselectivity in regards to protonation. Two possible mechanistic scenarios arise, one in which the reaction could involve a concerted pericyclic reaction where the C-4 carboxylate becomes protonated and in turn donates a proton to C-6 (or C-6’) as CO2 is evolved, or a second mechanism in which a BH+ conjugate acid in the enzymatic active site acts as the proton source. We are presently pursuing crystallization trials with BacA, AerD, and SalX to shed light on the mechanism and determine how these enzymes avoid the aromatization of prephenate that is typical of prephenate dehydratase and prephenate dehydrogenase-catalyzed transformations.
An initial BLAST search finds many other examples of proteins homologous to these enzymes, several of which are located near potential biosynthetic enzymes in host genomes such as Nodularia spumigena, Clostridium celluloytican, Photorhabdus luminescens, and Erwinia amylovora, the latter of which is known to produce 2,5-dihydrophenylalanine (DHPA, (14). DHPA is also produced in a variety of Streptomyces species, with recent data indicating production in P. luminescens (15) as well. Isotopic feeding studies in S. arenae demonstrated that DHPA is derived from prephenate (13), which suggests this enzyme family may be diverting prephenate into unique small molecule products across a broad array of microorganisms.
Supplementary Material
ACKNOWLEDGMENT
We thank Michael Acker for helpful advice and discussions and an anonymous reviewer for suggestions in regards to a pericyclic mechanism. This work was generously supported by the NIH (CA127622 to B.S.M. and GM49338 to C.T.W.), and by the NSF Graduate Research Fellowship Program (S.A.M.).
Footnotes
SUPPORTING INFORMATION PARAGRAPH
Materials and Methods, gene clusters for Aeruginoside 126 and Salinosporamide A, gene sequence for aerD, SDS-PAGE of protein expression, representative scanning kinetics for AerD and SalX, 1H NMR data for the SalX and AerD products, feeding studies with S. tropica, alignment of BacA, AerD and SalX with homologs from other organisms, and alignment of BacA, AerD and SalX. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Knaggs AR. The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 1999;16:525–560. doi: 10.1039/a707501d. [DOI] [PubMed] [Google Scholar]
- 2.Mahlstedt SA, Walsh CT. Investigation of anticapsin biosynthesis reveals a four-enzyme pathway to tetrahydrotyrosine in Bacillus subtilis. Biochemistry. 2010;49:912–923. doi: 10.1021/bi9021186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishida K, Christiansen G, Yoshida WY, Kurmayer R, Welker M, Valls N, Bonjoch J, Hertweck C, Borner T, Hemscheidt T, Dittmann E. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 2007;14:565–576. doi: 10.1016/j.chembiol.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ersmark K, Del Valle JR, Hanessian S. Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew. Chem. Int. Ed. Engl. 2008;47:1202–1223. doi: 10.1002/anie.200605219. [DOI] [PubMed] [Google Scholar]
- 5.Ishida K, Welker M, Christiansen G, Cadel-Six S, Bouchier C, Dittmann E, Hertweck C, Tandeau de Marsac N. Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Appl. Environ. Microbiol. 2009;75:2017–2026. doi: 10.1128/AEM.02258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. U S A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 8.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U S A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGlinchey RP, Nett M, Eustaquio AS, Asolkar RN, Fenical W, Moore BS. Engineered biosynthesis of antiprotealide and other unnatural salinosporamide proteasome inhibitors. J. Am. Chem. Soc. 2008;130:7822–7823. doi: 10.1021/ja8029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groll M, Huber R, Potts BC. Crystal structures of Salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of beta-lactone ring opening and a mechanism for irreversible binding. J. Am. Chem. Soc. 2006;128:5136–5141. doi: 10.1021/ja058320b. [DOI] [PubMed] [Google Scholar]
- 11.Nett M, Gulder TA, Kale AJ, Hughes CC, Moore BS. Function-oriented biosynthesis of beta-lactone proteasome inhibitors in Salinispora tropica. J. Med. Chem. 2009;52:6163–6167. doi: 10.1021/jm901098m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beer LL, Moore BS. Biosynthetic convergence of salinosporamides A and B in the marine actinomycete Salinispora tropica. Org. Lett. 2007;9:845–848. doi: 10.1021/ol063102o. [DOI] [PubMed] [Google Scholar]
- 13.Shimada K, Hook DJ, Warner GF, Floss HG. Biosynthesis of the antibiotic 2,5-dihydrophenylalanine by Streptomyces arenae. Biochemistry. 1978;17:3054–3058. doi: 10.1021/bi00608a017. [DOI] [PubMed] [Google Scholar]
- 14.Feistner GJ. (l)-2,5-dihydrophenylalanine from the fireblight pathogen Erwinia amylovora. Phytochemistry. 1988;27:3417–3422. [Google Scholar]
- 15.Kontnik R, Crawford JM, Clardy J. Exploiting a Global Regulator for Small Molecule Discovery in Photorhabdus luminescens. ACS Chemical Biology. 2010;5:659–665. doi: 10.1021/cb100117k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.