Abstract
To study the role of cell–ECM interactions, microscale approaches provide the potential to perform high throughput assessment of ECM microenvironments on cellular function and phenotype. Using a microscale direct writing (MDW) technique, we characterized the generation of multicomponent ECM microarrays for cellular micropatterning, localization, and stem cell fate determination. ECMs and other biomolecules of various geometries and sizes were printed onto epoxide-modified glass substrates for evaluation of cell attachment by human endothelial cells. The endothelial cells displayed strong preferential attachment to the ECM-patterned regions and aligned their cytoskeleton along the direction of the micropatterns. We next generated ECM microarrays that contained one or more ECM compositions (namely gelatin, collagen IV, and fibronectin) and then cultured murine embryonic stem cell (ESCs) on the microarrays. The ESCs selectively attached to the micropatterned features and expressed markers associated with a pluripotent phenotype, such as E-cadherin and alkaline phosphatase, when maintained in growth media containing leukemia inhibitory factor. In the presence of soluble factors retinoic acid and bone morphogenetic protein-4, the ESCs differentiated towards ectodermal lineage on the ECM microarray with differential ECM effects. The ESCs cultured on gelatin showed significantly higher levels of pan cytokeratin expression, when compared cells cultured on collagen IV or fibronectin, suggesting that gelatin preferentially promotes ectodermal differentiation. In summary, our results demonstrate that MDW is a versatile approach to print ECMs of diverse geometries and compositions onto surfaces, and it is amenable to the generation of multicomponent ECM microarrays for stem cell fate determination.
Keywords: Extracellular matrix, microscale, embryonic stem cell, differentiation, micropatterning
INTRODUCTION
The cellular microenvironment plays an important role in modulating cell reorganization, migration, proliferation, and differentiation [1, 2]. The microenvironment factors include chemical signaling factors, physical factors such as the extracellular matrix (ECM), and mechanical factors such as shear stress and strain. In particular, the ECM is a biological scaffolding material that consists of structural and functional molecules [3, 4]. Besides providing structural support to the cells, the ECM is a dynamic factor that plays a role in modulating cellular function and phenotype [5, 6]. Conventional methods to study cell–ECM interactions involve depositing the ECM of interest onto cell culture dishes followed by the culturing of cells on the surface or within the ECM. However, this approach requires large quantities of reagents and limits the ability to screen a large number of ECM compositions. As an alternative, ECM microarrays simplify the task of analyzing large numbers of ECMs simultaneously while minimizing the quantity of reagents used. Thus, microscale combinatorial techniques for screening large numbers of ECMs can provide high throughput assessment of the ECM microenvironment on cellular function and phenotype.
However, current microscale technologies for depositing ECMs are generally restricted to only one or several materials due to technical limitations. Among the commonly used microscale technologies, soft lithography utilizes elastomeric stamps containing patterns generated by silicon wafers to print or mold proteins or other materials [7–9]. The main limitation of soft lithography is the technical challenge of depositing large numbers of ECMs on the same substrate [10]. Other approaches for generating ECM microarrays include inkjet printing, dip pen nanolithography (DPN), DNA microarray robotic spotting, and microscale direct writing (MDW) [11–16]. These methods allow the direct printing of multiple proteins or other materials onto the same substrate without the use of silicon wafers. Inkjet printing typically produces protein features greater than 100 µm in size [13, 14]. DPN generates high resolution features at both the nanoscale and microscale, but it is generally limited by small printed areas and difficulty when rapidly transitioning from microscale to nanoscale printing dimensions in a single experiment [17, 18]. ECM spotters based on DNA microarray printing technology can deposit multicomponent circular features on large printed areas, but this technology is not easily amenable to non-circular features or complex nonrepeating feature arrangements [16]. Recently, Mei et al. described the development of a MDW approach for fabricating complex ECM microarrays at the subcellular feature (~5 µm) resolution using multicomponent proteins [14]. We further characterize the MDW approach by adapting this technology for generation of multicomponent ECM microarrays for cellular micropatterning, localization, and stem cell fate determination.
In this study, we investigated the printing of multicomponent ECMs and other biomolecules in diverse geometric shapes and sizes, and then applied the technology to cellular micropatterning and embryonic stem cell (ESC) fate specification. We show that MDW is a feasible computer-automated approach to print ECMs of diverse geometries and compositions onto surfaces, and it enables selective cell attachment to the micropatterned regions while maintaining cell viability and phenotype. As a demonstration of its utility, we generated multicomponent ECM microarrays and report the differential ECM effects on ectodermal differentiation of murine ESCs.
MATERIALS AND METHODS
ECM Micropatterning
Biomolecular printing of ECMs and dyes were generated with the NanoeNabler instrument (Bioforce Nanoscience, Ames, IA) (Supplementary Fig. 1A), equipped with a computer-controlled digital stage and optical microscopy system [14, 19]. The ECMs used in this study consisted of Oregon Green-488-conjugated porcine gelatin (Invitrogen, Carlsbad, CA), Oregon Green-488-conjugated human collagen IV (Invitrogen), and bovine fibronectin (Sigma, St. Louis, MO). Additionally, Hoechst 33342 and propidium iodide fluorescent nuclear dyes (all from Invitrogen) were micropatterned in selected experiments. Fluorescent bovine serum albumin (BSA) was printed as a negative control for cell attachment studies. The proteins or fluorescent dyes (1 mg/ml) were mixed at a 1:1 ratio with protein spotting buffer (Bioforce Nanosciences) and loaded into the reservoir of the cantilever-based surface-patterning tool (SPT, C60) (Supplementary Fig. 1B). The SPT has a microfluidic channel that enables deposition of the ECM by contact printing and capillary action (Supplementary Fig. 1C). The ECMs were micropatterned onto epoxide-modified glass slides (Arrayit Corp, Sunnyvale, CA) to enable covalent conjugation of ECMs onto the substrate. Computer controlled movements allowed for defined geometric shapes and complex patterns to be printed.
For ESC differentiation studies, we fabricated ECM microarrays consisting of circular features that were formed by individual circular spots in close proximity. Fluorescent gelatin, fluorescent collagen IV, non-fluorescent fibronectin, and fluorescent BSA features were printed onto epoxide-coated glass slides, in which each circular feature was 300 µm in diameter and 500 µm apart from neighboring features. Each ECM was µprinted in replicates of 5 on each microarray. After fabrication, the samples were kept in a humidified environment at 4 °C overnight. Prior to cell seeding, the substrates were pretreated in blocking buffer (Arrayit Corp), sterilized by UV for 15 minutes, and then blocked in 1% non-fluorescent BSA to prevent non-specific binding.
Morphology and Function of Primary Cells on Micropatterned ECMs
Human microvascular endothelial cells (HMEC-1, Center for Disease Control, Bethesda, MD) and primary isolated mouse embryonic fibroblasts (MEFs) were maintained in growth media consisting of Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 0.1 mM non-essential amino acids (all from Invitrogen).
For neonatal rat cardiomyocyte pacing studies, primary cardiomyocytes (Lonza, Walkersville, MD) that were approximately 80% pure were cultured in rat cardiomyocyte media (Lonza) on fibronectin in a curvilinear shape (~400µm in width). After overnight culture, the samples were paced in Tyrode’s buffer with a stimulus generator (SIU-102, Warner Instruments, Hamden, CT). The electrical stimulation consisted of a biphasic signal with peak-to-peak amplitude of 10V, pulse width of 10 ms. A frequency of 1 or 2 Hz was delivered to platinum electrodes spaced 1 cm apart. Movies were captured at 30 frames per second for 10 s duration with a Retiga 2000R color cooled camera and QCapture Pro 5.0 software (QImaging, Canada).
Embryonic Stem Cell Maintenance and Differentiation on ECM Microarrays
Mouse ESCs (D3, ATCC, Manassus, VA) were routinely cultured on top of mitotically inactivated MEF in growth media consisting of DMEM with 15% FBS (Hyclone, Logan, UT), 1% penicillin/streptomycin, 0.1 mM non-essential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), and 1000 U/ml leukemia inhibitory factor (LIF, Millipore, Billerica, MA). For differentiation experiments, ESCs were grown in a differentiation media that consisted of Alpha Minimum Eagle’s Media, 10% FBS, 1% penicillin/streptomycin, and 0.05 mM β-mercaptoethanol (Sigma, St Louis, MO), 10−5 M retinoic acid (Sigma), and 10 ng/ml recombinant human bone morphogenetic protein-4 (BMP4) (Peprotech, Rocky Hill, NJ). For all experiments, the cell were seeded onto the substrates in growth media for 1 hour at 37 °C before removing unbound cells with washes of phosphate-buffered saline (PBS) and replacing with growth or differentiation media, as indicated. For coculture experiments on 10 × 10 microarrays of fibronectin features, ESCs were seeded for 1 hour, followed by removal of unbound cells, and then replacement of growth media. After overnight incubation, the substrates were pre-treated with ESC growth media containing 0.4% gelatin for 1 hour to render the non-patterned regions favorable for cell attachment. Afterwards, MEFs were seeded onto the substrates to favor cell attachment in the non-patterned areas.
Live/Dead Viability Assay
Cell viability was assessed using the Live/Dead Viability kit (Invitrogen) in which cells were incubated with media containing 2 µM calcein-AM and 4 µM ethidium homodimer-1 for 1 hour before imaging by fluorescence microscopy. Live cells stained green, whereas dead cells were labeled red.
Immunofluorescence Staining and Alkaline Phosphatase Staining
Immunofluorescence staining was carried using methods based on our previous studies [20]. At indicated time points, the cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked in 1% BSA. For visualization of F-actin stress fibers, the cells were incubated with phalloidin-594 (Invitrogen) and then counterstained with Hoechst 33342 nuclear dye. E-cadherin (Millipore) or pan cytokeratin (Sigma) primary antibodies were applied to samples to selectively visualize undifferentiated ESCs or cells of ectodermal lineage, respectively. Incubation of primary antibodies was followed by Alexafluor-594-conjugated secondary antibodies (Invitrogen). Afterwards, the samples were counterstained with Hoechst 33342 nuclear dye before visualizing with a fluorescent microscope (Nikon, Burlingame, CA) and imaging with a SPOT RT color camera (Diagnostic Instruments, Sterling Heights, MI). For ESC differentiation studies, high-throughput fluorescent images of the samples were also captured using an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA) equipped with 532 nm and 635 nm lasers. Alkaline phosphatase expression, a marker of undifferentiated ESC phenotype, was assessed by a staining kit according to the manufacturer’s instructions (Millipore).
Data Analysis
Data are shown as mean ± standard deviation. ECM spot size was assessed by the mean perimeter and area using Image J software (NIH, Bethesda, MD) (n=100 spots). For measurement of ectodermal differentiation of ESCs on ECMs, the fluorescent images obtained from the Axon 4000B laser scanner were analyzed by GenePix Pro 6.1 software (Molecular Devices). For each ECM feature, a region of interest was drawn and the mean intensity of pan cytokeratin staining was normalized by subtracting the mean background signal value and then expressing the intensity as fold change, relative to the BSA negative control samples. Pan cytokeratin data were averaged out of 3 biological samples, in which each sample contained at least 5 replicates. Statistical analysis between multiple groups was carried out by one way analysis of variance (ANOVA) with Holm’s adjustment, and statistical significance was accepted at P<0.05.
RESULTS
Micropatterning of Spatial Geometries
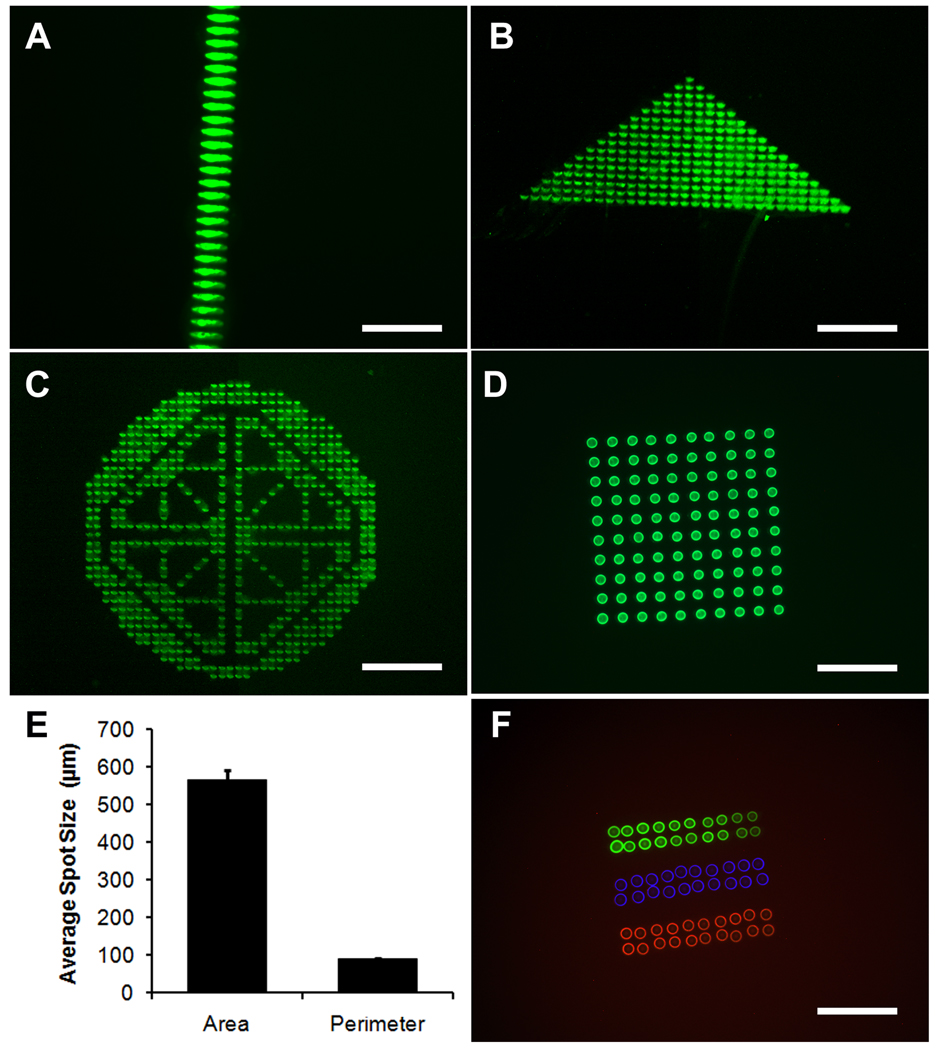
Initial studies were performed to determine the capacity of the MDW technology to print varying spatial biomolecular geometries and sizes. Since the SPT that contacts the printed surface is limited by a maximum spot diameter of 60 µm, shapes or patterns greater than 60-µm sizes were formed by the collection of distinctive spots to form larger or more complex structures, such as fluorescently conjugated gelatin lines (Fig. 1A), triangles (Fig. 1B), or other complex patterns (Fig. 1C). This method of micropatterning resembled the artistic technique of Pointillism. Individual spots of gelatin could also be fabricated in the form of a 10 × 10 microarray with high reproducibility (Fig. 1D). The average spot size within the fluorescent gelatin microarray was 565±26 µm2 in area and 88±2 µm in perimeter, which represented only 5% or 2% standard deviations, respectively, between individual spots (Fig. 1E).
Figure 1. Biomolecular micropatterning of spatial geometries.
A. Fluorescently-conjugated gelatin spots were micropatterned to form larger features with geometries of lines (A), triangles (B), or complex patterns (C). D. In addition, microarrays of fluorescently-conjugated gelatin spots could be printed. E. Quantification of mean perimeter and area of fluorescently conjugated gelatin spots (n=100). F. Multi-component printing of fluorescent gelatin (green), Hoechst 33342 (blue), and propidium iodide (red) could be printed onto the same substrate within close proximity. Scale bar: 50 µm (A); 500 µm (B–C); 200 µm (D, F).
In addition to single component printing, multicomponent printing was achieved on the same substrate in proximity to one another. As shown in Fig. 1F, multicomponent printing of fluorescent gelatin, Hoechst 33342, and propidium iodide could be deposited onto the same substrate at microscale distances apart. These data demonstrated the ability to print diverse patterns, sizes, and multiple components on the same substrate with high reproducibility.
Attachment and Phenotype of Primary Cells on Micropatterned ECMs
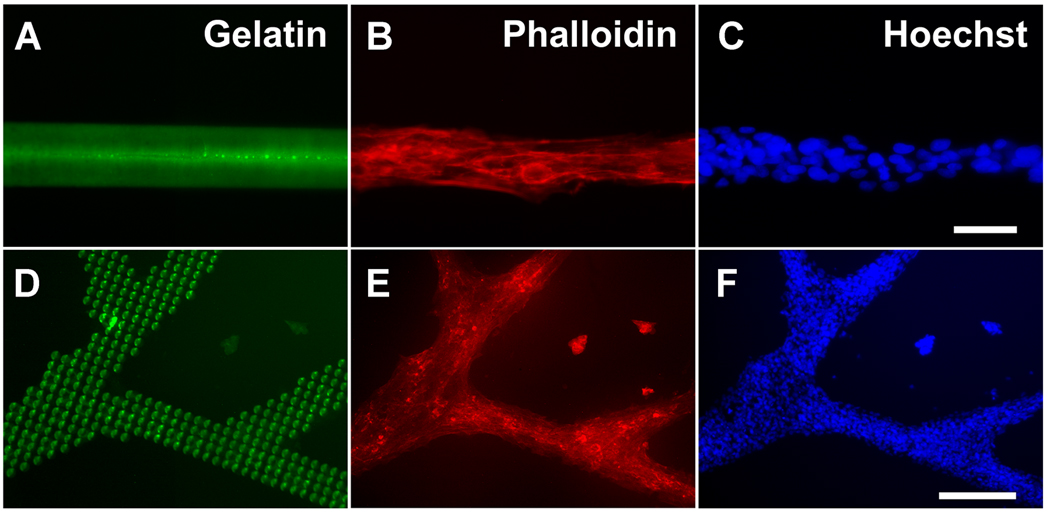
After establishing the versatility of micropatterning a variety of geometries and patterns onto the substrates, we next tested the biocompatibility and selective attachment of cells on the ECM micropatterned substrates. To test the attachment of cells to the micropatterned regions, we created lines of fluorescent gelatin that were 50 µm in width by displacing the SPT in the horizontal direction while maintaining constant contact with the glass surface. Using human endothelial cells as a model cell line for the testing of selective cell attachment, we then seeded the cells onto micropatterned lines of fluorescently labeled gelatin (Fig. 2A–C). The cells not only attached preferentially to the micropatterned regions, but they also aligned within close proximity to the direction of the gelatin pattern, based on phalloidin staining showing F-actin polarization along the direction of the gelatin lines. Furthermore, the cells selectively attached within the pattern of more complex geometries, including those that resemble vascular branching structures (Fig. 2D–F). These data demonstrated the ability of primary cells to selectively attach on the micropatterned regions and align their cytoskeleton in the direction of the micropatterns.
Figure 2. Micropatterning of endothelial cells for regulating cellular organization.
Lines of fluorescent gelatin were prepared by displacing the SPT in the horizontal direction while maintaining contact with the glass surface. Endothelial cells seeded onto the lines for 1 day preferentially attached and aligned their cytoskeleton in the direction of the protein pattern, based on phalloidin staining for F-actin (B–C). For large and complex patterns, such as gelatin spots that form branching structures (D), the endothelial cells remained preferentially adherent within the micropatterned regions (E–F). After one day of culture, the samples were processed for immunofluorescence staining of phalloidin and Hoechst. (A,D) depicts gelatin patterns; (B,E) represent phalloidin staining of endothelial cells; and (C,F) Hoechst stains for total nuclei. Scale bar: 50 µm (A–C); 250 µm (D–F).
To further investigate the maintenance of cellular function and phenotype after seeding onto micropatterned ECM geometries, we used the cardiomyocyte culture system based on the ease of pacing these cells and their spontaneously contractile phenotype. Primary isolated neonatal rat cardiomyocytes were seeded onto curvilinear fibronectin-patterned surfaces that were approximately 400 µm in width. Although the cardiomyocytes spontaneously contracted at low frequency (Supplemental Movie 1), we also used electrical pacing to contract the cardiomyocytes synchronously at both 1 Hz (Supplemental Movie 2) as well as 2 Hz (Supplemental Movie 3). These data validated the capability of this platform for supporting cellular alignment and maintenance of phenotype on micropatterned ECMs.
Attachment and Phenotype of ESCs on Micropatterned ECMs
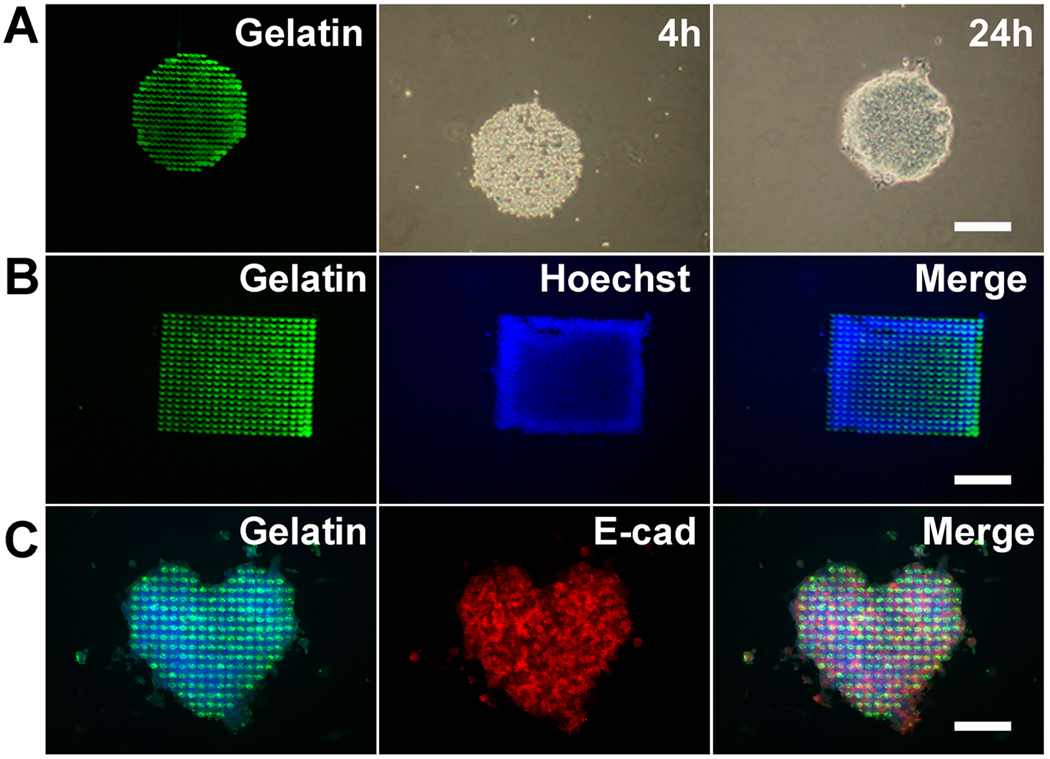
To exclude the possibility that the ECM array platform or geometric patterns could affect ESC attachment or induce differentiation, we next cultured murine ESCs on micropatterned geometries of fluorescently-labeled gelatin circles, rectangles, or curvilinear shapes in the presence of LIF (Fig. 3A–C). Regardless of the ECM geometry, the ESCs attached preferentially to the patterned regions at 4 hours after seeding and remained adherent after 24 hours (Fig. 3A). In the presence of culture media containing LIF, the majority of ESCs retained their undifferentiated phenotype on the geometric shapes of gelatin after 24 hours (Fig. 3B), as indicated by their intense expression of E-cadherin [21], a murine ESC marker of pluripotency (Fig. 3C).
Figure 3. Attachment and phenotype of ESCs on biomolecular patterns of gelatin in the presence of LIF.
A. ESCs selectively attached on biomolecular patterns of fluorescently labeled gelatin. The cells attached within 4 hours and then spread onto gelatin patterns after 24 hours. Similar trends of cell attachment were found on rectangular patterns after 24 hours (B). C. ESCs cultured on gelatin patterns in the presence of growth media containing LIF demonstrated positive expression of E-cadherin (E-cad), a commonly used surface marker of pluripotency. Scale bar: 500µm (A–B); 200µm (C).
ESCs could also maintain their phenotype on ECM microarrays. Using 10 × 10 microarrays of fibronectin features that were each 80 µm in diameter and 300 µm apart, we cultured ESCs on the fibronectin arrays in growth media containing LIF (Fig. 4A). After 2 days, the colonies grew in size without evidence of cytotoxicity, as shown by the Live/Dead viability assay in which almost all cells were viable (green) cells with few dead (red) cells (Fig. 4B). To support the undifferentiated phenotype of ESCs on the fibronectin microarray culture environment, we coated the ESC-seeded microarrays with non-fluorescent gelatin to render the regions without fibronectin favorable for cell attachment. Then we incubated MEFs onto the fibronectin arrays containing pre-seeded ESCs. Twenty four hours after the addition of MEFs, the MEFs attached to the regions containing non-fluorescent gelatin and surrounded the ESC colonies (Fig. 4C). In this coculture system, the ESC colonies remained in an undifferentiated phenotype, based on the formation of colonies with defined borders that were surrounded by MEFs. In contrast, colonies with diminished colony borders or reduced nuclear-to-cytoplasm ratio were not observed. The ESC colonies also maintained strong expression of alkaline phosphatase (Fig. 4D), which is abundant in undifferentiated ESCs, whereas the surrounding MEFs did not express alkaline phosphatase. These data suggest that the ECM microarray platform did not intrinsically promote ESC differentiation, but rather supported ESC pluripotency in the presence of media containing LIF.
Figure 4. Maintenance of ESC pluripotency on fibronectin microarrays in coculture with MEFs.
A. ESCs attached to the fibronectin microarray after 1 day. B. ESCs remained viable on the microarray after 2 days, based on robust calcein uptake (green). C. Phase-contrast images depict coculture of ESCs and MEFs on ECM microarray after 1 day. D. Alkaline phosphatase staining of ESCs in coculture with MEFs after 2 days indicates pluripotent phenotype. Scale bar: 500µm (A,C); 200µm (B); 300µm (D).
ECM Microarrays for Stem Cell Fate Specification
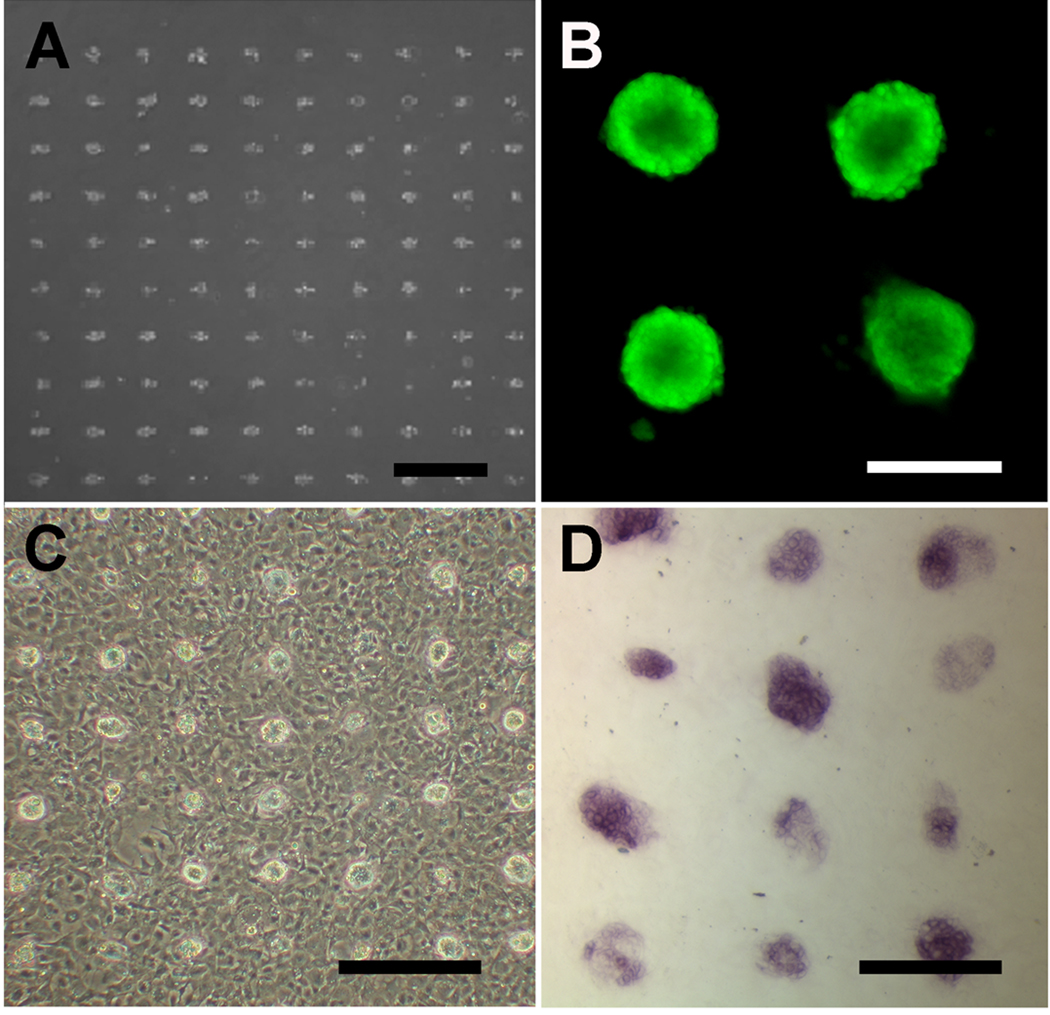
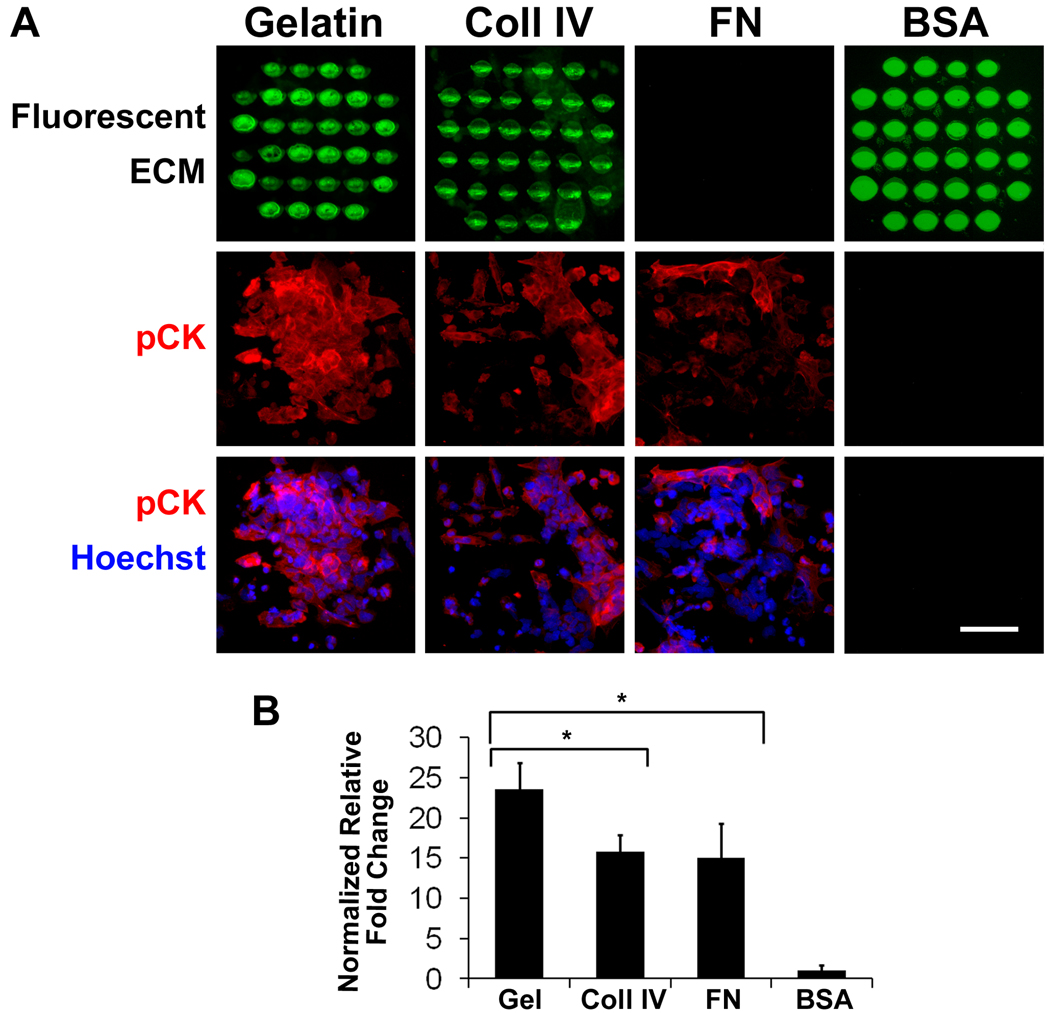
After demonstrating that the ECM microarray platform could support the attachment and viability of ESCs without inducing differentiation, we fabricated multicomponent microarrays to assess ECM-mediated differential induction of ectodermal differentiation in ESCs. To show proof-of-concept, we generated ECM microarrays consisting of fluorescently labelled gelatin, fluorescently labelled collagen IV, non-fluorescent fibronectin, and fluorescently labelled BSA. Each circular feature was 300 µm in diameter and 500 µm apart. After 2 days of culture in a differentiation media that supports ectodermal differentiation, we analyzed the differential effect of ECM composition on the differentiation of ESCs. We immunofluorescently stained the samples for an ectodermal marker, pan cytokeratin, and captured immunofluorescent images of each feature by fluorescence microscopy (Fig. 5A) as well as by laser scanning cytometry for image analysis. Our analysis showed that the normalized relative intensity of pan cytokeratin staining was significantly higher in differentiating ESCs on the gelatin substrate, compared to the cells on collagen IV and fibronectin (P<0.05), suggesting that gelatin significantly enhanced the differentiation of ESCs towards ectodermal lineages (Fig. 5B). As expected, the BSA negative control substrate did not support any cell attachment. These data provided evidence that ECM microarrays could be fabricated using MDW technology for the assessment of matrix-mediated induction of ESC lineage specification.
Figure 5. Ectodermal differentiation of ESCs on ECM microarray.
ESCs were differentiated on fluorescent gelatin, fluorescent collagen IV (coll IV), non-fluorescent fibronectin (FN), and fluorescent BSA for 2 days in the presence of RA and BMP4 (A). A. Immunofluorescence images depict fluorescently labelled ECMs, pan cytokeratin staining (pCK) and Hoechst 33342 nuclear dye. Since fibronectin was not fluorescently conjugated, no fluorescent image of the ECM could be captured. B. The mean fluorescence intensity of pCK was quantified and expressed as normalized fold change, relative to that of the BSA negative control. * indicates statistically significant relationship (n=3, P<0.05). Scale bar: 100µm.
DISCUSSION
We demonstrated that MDW is a practical and versatile system for ECM micropatterning and for the generation of ECM microarrays. The salient features of this technology include the printing of multicomponent ECM compositions in nonrepeating geometries that can span the area of a standard microscope slide; the micropatterning of ECMs for regulating cell attachment, phenotype, and function; and the fabrication of ECM microarrays for assessment of matrix-mediated induction of ESC differentiation. To date, there are limited numbers of groups that have utilized this type of MDW technology. Mei et al. developed multicomponent ECM microarrays with 6–9 µm feature sizes to assess the attachment and spreading of skeletal myoblasts on varying gap distances and protein compositions [14]. In contrast, we demonstrated the features of ECM micropatterning at the supracellular scale (~30–300 µm features).
The role of ECMs on the modulation of ESC self-renewal or differentiation has only recently been studied. Using traditional DNA microarray technology, several investigators have assessed the role of ECM composition on ESC differentiation towards hepatic, cardiac, and ectodermal differentiation [15, 16, 22]. More recently, ECM microarrays have been adapted in a multiwell format for combined assessment of matrix and soluble inducing factors on cardiac differentiation of murine ESCs [22]. Besides assessing stem cell fate specification, the ECM microarray platform has also been shown to define the ECM and soluble factor microenvironment that promotes human ESC pluripotency and proliferation [23].
In comparison to DNA robotic spotting technology, we provide another method to generate ECM microarrays for stem cell fate determination using MDW, which can be further adapted to generate ECM microarrays with varying geometries. Although a simplified ECM microarray containing 4 compositions was demonstrated, this technology can be extended to larger quantities of ECM as well as compositions consisting of multiple ECM components. Furthermore, this system can incorporate repeated time point measurements using fluorescent reporter genes for non-invasive temporal assessment of cell fate specification. Despite the limitations of this simplified ECM microarray, our data provides evidence of ECM-mediated effects on ectodermal differentiation of ESCs.
In addition to the generation of ECM microarrays for stem cell fate specification, the MDW technology can be utilized for a number of other biological applications. For example, to improve cell survival in ischemic tissues in vivo, the ECM microarray can serve as an in vitro platform for identifying the ECM requirements that support cell viability and proliferation under conditions that mimic ischemia, such as hypoxia and reduced serum media. In addition, the MDW technology can be used for tissue engineering constructs with preferential ECM geometries and cellular alignments that mimic physiological tissues. Scaffold designs incorporating MDW can promote efficient homing of cells in predefined patterns. Other applications address questions of basic biology, such as the role of cell–cell, cell–matrix, and cell–soluble factors on cell phenotype and function. These applications are interesting and warrant future research.
Cell–ECM interactions are involved in a wide range of biological processes, from the formation of embryonic organs to pathological remodeling in disease states. In chick embryos, for example, fibronectin is critical for embryonic development and is expressed in the dorsal aorta of embryonic day 6 (E6) [24], and laminin is prevalent in the aortic vascular wall at E10 [24]. In the adult vasculature, for example, collagen IV is highly abundant in the basement membrane of blood vessels [25]. Owing to the importance of cell–ECM interactions during embryonic development and physiological maintenance, ECMs likely play an important role in specifying stem cell fate.
The mechanism by which ECMs mediate the induction of stem cell differentiation is not well characterized. ECM-mediated regulation of cell behavior is largely due to integrins, which are heterodimeric transmembrane adhesion receptors. The extracellular domain of integrins binds to ECMs, whereas the intracellular domain connects to the actin cytoskeleton through the focal adhesion complex. Emerging evidence implicates integrins as mechanotransducers that relay external cues from the ECM into the intracellular space and activate downstream signaling pathways that regulate stem cell renewal and differentiation [26, 27]. Other mediators include focal adhesion kinase, which have been shown to modulate cardiogenesis in ESCs [28]. As further research in cell–ECM interactions continue, we anticipate these mechanisms will reveal interesting insights regarding the process by which cells sense and respond to matrix stimuli.
Although MDW is a powerful and versatile platform for studying the role of ECM spatial patterning and chemical composition in directing cellular organization and stem cell fate specification, it is limited by several technical issues. The use of MDW with two-dimensional ECM printing does not mimic the three-dimensional ECM microenvironment, which can be assessed using microwell technology [29, 30]. Future application of MDW with a microwell platform may enable the generation of ECM spatial geometries and chemical compositions in a three-dimensional microenvironment. Another limitation of MDW is the open cell culture system that enables paracrine factors from neighboring cells of an ECM microarray to influence each other. To overcome this limitation, individual ECM compositions can be isolated from one another using gaskets to create an ECM microarray with multiwell platform [22]. Despite these limitations, our results highlight the utility of MDW as a reproducible platform for studying cell-ECM interactions
In summary, we have demonstrated a MDW approach for generating micropatterned ECMs and ECM microarrays. This method is amenable to the deposition of varying ECM compositions, geometries, and sizes. MDW shows tremendous potential for micropatterning applications related to cellular patterning and cell-matrix interactions that influence stem cell renewal or differentiation. We anticipate that this technology will have increasing value for the study of cellular function and behaviour.
Supplementary Material
A. NanoeNabler instrument. B. Schematic of surface patterning tool (SPT) shows protein reservoir and deposition onto substrate. C. Bright-field image shows deposition process with SPT in view.
Spontaneous contraction of neonatal cardiomyocytes on geometric micropatterns of fibronectin at baseline conditions in the absence of electrical stimulation.
Pacing of neonatal cardiomyocytes on geometric micropatterns of fibronectin at 1 Hz.
Pacing of neonatal cardiomyocytes on geometric micropatterns of fibronectin matrix at 2 Hz.
ACKNOWLEDGEMENTS
This work was supported by research grants from the California Institute for Regenerative Medicine (RC1-00151-1), the National Institutes of Health (U01HL100397, U01HL099775, RC2HL103400, 1R21HL089027), and the National Science Foundation (EFRI 0735551). N.F.H. was supported by postdoctoral fellowships from the American Heart Association and by the National Institutes of Health (HL953552). O. A. was supported by a fellowship from the Advanced Residency Program at Stanford program. Use of the Nanoenabler was kindly provided by Bioforce Nanoscience, Ames, IA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR DISCLOSURE STATEMENT
No conflicts of interest reported.
REFERENCES
- 1.Ingber DE. Tensegrity: the architectural basis of cellular mechanotransduction. Annu Rev Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- 2.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 3.Bissell MJ, Aggeler J. Dynamic reciprocity: how do extracellular matrix and hormones direct gene expression? Prog Clin Biol Res. 1987;249:251–262. [PubMed] [Google Scholar]
- 4.Rosso F, Giordano A, Barbarisi M, Barbarisi A. From cell-ECM interactions to tissue engineering. J Cell Physiol. 2004;199:174–180. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]
- 5.Daley WP, Peters SB, Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 6.Ingber DE. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ Res. 2002;91:877–887. doi: 10.1161/01.res.0000039537.73816.e5. [DOI] [PubMed] [Google Scholar]
- 7.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 8.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 9.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tien J, Nelson CM, Chen CS. Fabrication of aligned microstructures with a single elastomeric stamp. Proc Natl Acad Sci U S A. 2002;99:1758–1762. doi: 10.1073/pnas.042493399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piner RD, Zhu J, Xu F, Hong S, Mirkin CA. "Dip-Pen" nanolithography. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 12.Lee KB, Lim JH, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J Am Chem Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 13.Roth EA, Xu T, Das M, Gregory C, Hickman JJ, Boland T. Inkjet printing for high-throughput cell patterning. Biomaterials. 2004;25:3707–3715. doi: 10.1016/j.biomaterials.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 14.Mei Y, Cannizzaro C, Park H, Xu Q, Bogatyrev SR, Yi K, Goldman N, Langer R, Anderson DG. Cell-compatible, multicomponent protein arrays with subcellular feature resolution. Small. 2008;4:1600–1604. doi: 10.1002/smll. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 16.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 17.Huo F, Zheng Z, Zheng G, Giam LR, Zhang H, Mirkin CA. Polymer pen lithography. Science. 2008;321:1658–1660. doi: 10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salaita K, Lee SW, Wang X, Huang L, Dellinger TM, Liu C, Mirkin CA. Sub-100 nm, centimeter-scale, parallel dip-pen nanolithography. Small. 2005;1:940–945. doi: 10.1002/smll.200500202. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Lynch M, Huff JL, Mosher C, Vengasandra S, Ding G, Henderson E. Microfabricated quill-type surface patterning tools for the creation of biological micro/nano arrays. Biomed Microdevices. 2004;6:117–123. doi: 10.1023/b:bmmd.0000031748.13353.10. [DOI] [PubMed] [Google Scholar]
- 20.Huang NF, Patel S, Thakar RG, Wu J, Hsiao BS, Chu B, Lee RJ, Li S. Myotube Assembly on Nanofibrous and Micropatterned Polymers. Nano Lett. 2006;6:537–542. doi: 10.1021/nl060060o. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita J, Itoh H, Hirashima M, Ogawa M, Nishikawa S, Yurugi T, Naito M, Nakao K. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 22.Flaim CJ, Teng D, Chien S, Bhatia SN. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 2008;17:29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 23.Brafman DA, Shah KD, Fellner T, Chien S, Willert K. Defining Long-Term Maintenance Conditions of Human Embryonic Stem Cells With Arrayed Cellular Microenvironment Technology. Stem Cells Dev. 2009;18:1141–1154. doi: 10.1089/scd.2008.0410. [DOI] [PubMed] [Google Scholar]
- 24.Risau W, Lemmon V. Changes in the vascular extracellular matrix during embryonic vasculogenesis and angiogenesis. Dev Biol. 1988;125:441–450. doi: 10.1016/0012-1606(88)90225-4. [DOI] [PubMed] [Google Scholar]
- 25.Francis ME, Uriel S, Brey EM. Endothelial cell-matrix interactions in neovascularization. Tissue Eng Part B Rev. 2008;14:19–32. doi: 10.1089/teb.2007.0115. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi Y, Furue MK, Okamoto T, Ohnuma K, Myoishi Y, Fukuhara Y, Abe T, Sato JD, Hata R, Asashima M. Integrins regulate mouse embryonic stem cell self-renewal. Stem Cells. 2007;25:3005–3015. doi: 10.1634/stemcells.2007-0103. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, He X, Corbett SA, Lowry SF, Graham AM, Fassler R, Li S. Integrins are required for the differentiation of visceral endoderm. J Cell Sci. 2009;122:233–242. doi: 10.1242/jcs.037663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakuno D, Takahashi T, Lammerding J, Lee RT. Focal adhesion kinase signaling regulates cardiogenesis of embryonic stem cells. J Biol Chem. 2005;280:39534–39544. doi: 10.1074/jbc.M505575200. [DOI] [PubMed] [Google Scholar]
- 29.Lutolf MP, Doyonnas R, Havenstrite K, Koleckar K, Blau HM. Perturbation of single hematopoietic stem cell fates in artificial niches. Integr Biol (Camb) 2009;1:59–69. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochsner M, Dusseiller MR, Grandin HM, Luna-Morris S, Textor M, Vogel V, Smith ML. Micro-well arrays for 3D shape control and high resolution analysis of single cells. Lab Chip. 2007;7:1074–1077. doi: 10.1039/b704449f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. NanoeNabler instrument. B. Schematic of surface patterning tool (SPT) shows protein reservoir and deposition onto substrate. C. Bright-field image shows deposition process with SPT in view.
Spontaneous contraction of neonatal cardiomyocytes on geometric micropatterns of fibronectin at baseline conditions in the absence of electrical stimulation.
Pacing of neonatal cardiomyocytes on geometric micropatterns of fibronectin at 1 Hz.
Pacing of neonatal cardiomyocytes on geometric micropatterns of fibronectin matrix at 2 Hz.