Abstract
Following introduction of Haemophilus influenzae type b (Hib) conjugate vaccines, meningitis caused by serotypes other than Hib has gained importance. We conducted active hospital-based surveillance for meningitis over an 11-year period in Salvador, Brazil. H. influenzae isolates were serotyped and analyzed by PCR, pulsed-field gel electrophoresis and DNA sequencing to identify strains with a specific deletion (IS1016) in the bexA gene (IS1016-bexA). We identified 43 meningitis cases caused by non-type b H. influenzae: 28 (65%) were caused by type a (Hia), 9 (21%) by non-capsulated strains and 3 (7%) each by types e and f. Hia isolates clustered in two clonal groups; clonal group A strains (n=9) had the IS1016-bexA deletion. Among children <5 years, meningitis caused by Hia from clonal group A had higher case-fatality than clonal group B. Despite small numbers, these results indicate that the presence of IS1016-bexA deletion is associated with enhanced virulence in non-type b H. influenzae.
Keywords: Haemophilus influenzae, non-type b H. influenzae, meningitis, Hib conjugate vaccine, virulence, IS1016-bexA deletion, molecular epidemiology
INTRODUCTION
Introduction of Haemophilus influenzae type b (Hib) conjugate vaccines into childhood immunization programs has dramatically reduced the incidence of Hib meningitis in countries using Hib vaccines [1–3]. Hib conjugate vaccines are highly efficacious against invasive Hib disease [4], decrease Hib carriage among vaccinated children and reduce transmission and invasive disease among non-immunized children [5].
Hib conjugate vaccines do not prevent H. influenzae disease due to other serotypes, raising the potential for the emergence of H. influenzae disease due to virulent organisms with non-type b capsules [6–11]. Detection of meningitis due to non-type b H. influenzae has increased following widespread use of Hib conjugate vaccines as a result of improved surveillance and use of molecular techniques, which have reduced the serotyping errors associated with slide agglutination [12–14]. Molecular methods have also been used to identify genetic elements in invasive non-type b H. influenzae isolates, including presence of a partial deletion of IS1016 in the bexA gene commonly found in Hib isolates. The IS1016-bexA deletion is a putative virulence factor that has been identified in invasive Hia isolates from patients with severe disease in The Gambia and United States [9, 10, 15], but not in all areas where Hia strains have been isolated [11, 16]. Acquisition of virulence factors from Hib strains could possible lead to the emergence of non-type b H. influenzae disease.
Previously, we reported a transient increase in meningitis due to H. influenzae serotype a after introduction of Hib conjugate vaccine in Salvador, the third largest urban center in Brazil [17, 18]. Clinical outcomes of meningitis cases due to non-type b H. influenzae were similar to those of Hib cases [17, 18]. To investigate the role of the IS1016-bexA deletion in clinical outcomes of meningitis cases due to non-type b H. influenzae, we analyzed data from 11 years of active, hospital-based meningitis surveillance in Salvador, Brazil.
METHODS
Surveillance
Meningitis is a nationally notifiable disease in Brazil, with mandatory reporting of all suspect meningitis cases to public health authorities. We conducted active surveillance for meningitis among patients admitted to Couto Maia Hospital in Salvador, Brazil. According to state health guidelines, all suspected cases of meningitis in the region are referred to Couto Maia Hospital for diagnostic procedures, including lumbar puncture and examination of cerebrospinal fluid. Couto Maia Hospital accounted for 98% of reported meningitis cases among persons from the metropolitan area of Salvador during the study period [19].
We analyzed data for H. influenzae meningitis cases identified between March 9, 1996 and September 8, 2007. A case of H. influenzae meningitis was defined as a patient who had: (1) clinical presentation of meningitis, characterized by fever, meningismus, and altered mental status; (2) abnormal cerebrospinal fluid examination; and (3) cerebrospinal fluid or blood culture positive for H. influenzae. The study team reviewed laboratory records 5 days a week to identify new culture isolations of H. influenzae. Patients were enrolled in the study according to informed consent procedures approved by the Institutional Review Boards of the Oswaldo Cruz Foundation, Brazilian Ministry of Health and the New York-Presbyterian Hospital, United States. We used a standardized data entry form to collect information on demographics, clinical presentation, laboratory results and outcome from the patient’s medical records. Number of doses of Hib conjugate vaccine received prior to hospitalization and dates of vaccination were obtained from patient immunization records.
Strains identification and serotyping
H. influenzae was identified by Gram stain morphology and growth requirement for hemin and nicotinamide adenine dinucleotide. Commercial antiserum (Becton, Dickinson and Company, Franklin Lakes, NJ) was used to determine capsular serotype. Each isolate was tested for slide agglutination with the complete panel of type a to type f–specific antisera (Becton, Dickinson and Company) and a saline control. A semi-nested polymerase chain reaction (PCR) method was used to amplify serotype-specific and nonspecific DNA sequences from the H. influenzae capsular loci [20]. Isolates were defined as non-capsulated if agglutination was not observed with the six type–specific antisera and if PCR capsular loci sequences conserved among serotypes were not detectable by PCR [20].
Pulsed-field gel electrophoresis characterization
H. influenzae non-type b clinical isolates were examined by pulsed-field gel electrophoresis (PFGE) after digestion of bacterial DNA with Sma I (New England Biolabs), as previously described [21, 22]. The Sma I fingerprints were analyzed using GelCompar II software (Applied Maths, Kortrijk, Belgium). A 1.5% band position tolerance was used for gel comparisons. Cluster analysis was performed using the unweighted-pair-group (UPGMA) method and the relatedness between isolates was interpreted according to the criteria of Tenover [23].
Identification of the IS1016-bexA partial deletion and sequencing
H. influenzae non-type b isolates and a random sample of 20 Hib isolates were evaluated by PCR for identification of a partial deletion of the bexA gene, using the IS1016 and bexA primers as previously described [15]. For DNA sequencing, PCR products were purified with the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA, USA) and subjected to sequence analysis. The DNA sequences from both strands were edited, assembled, and aligned using MEGA4 and BioEdit software. The sequences were compared to those of the Hib strains AF549213 [24], S62752 [25] and the type a strain DQ086152 [10], available in the NCBI Gene bank.
Multilocus sequence typing
Multilocus sequence typing (MLST) was performed for two H. influenzae type a isolates that were randomly-selected among the isolates which had and did not have the IS1016-bexA deletion. Chromosomal DNA was extracted using a Qiagen genomic Kit (Qiagen Inc.). PCR was used to amplify 450-bp internal fragments of seven housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA), according to previously described methods [26]. Sequences were submitted to the online MLST database (http://www.mlst.net), which in turn assigned alleles at each locus and a sequence type.
Statistical analysis
Data were entered and analyzed using Epi-Info (Version 3.3.2; Center for Disease Control and Prevention, Atlanta, US). Fisher’s exact test and Wilcoxon rank-sum test were used for comparison of proportions and continuous data, respectively. A significant difference was defined by a two-tailed P-value less than 0.05.
Mean annual incidence of H. influenzae meningitis was compared for the period prior to introduction of Hib vaccination (March 1996 to July 1999) and after Hib vaccine introduction (August 1999 to September 2007). Incidence was calculated for the metropolitan area of Salvador by dividing the number of cases among residents of metropolitan Salvador by the estimated population from the 2000 national census [27].
RESULTS
During the study period, we identified 615 cases of H. influenzae meningitis. Among the 573 (93%) cases for which an isolate was serotyped, 43 (8%) episodes were caused by H. influenzae non-type b strains (Table 1). The majority of H. influenzae non-type b isolates were type a (28 isolates, 65%), followed by non-capsulated (9 isolates, 21%), type e (3 isolates, 7%) and type f (3 isolates, 7%). The proportion of H. influenzae meningitis cases due to a non-type b isolate increased from 2% (8 of 424) to 23% (35 of 149) after the introduction of routine Hib immunization (P<0.001). This increase was largely explained by the 91% reduction in the incidence of Hib meningitis between the pre and post vaccine periods (from 2.45 to 0.24 cases per 100,000 population, P<0.001). The incidence of meningitis due to non-type b H. influenzae increased after the introduction of the Hib conjugate vaccine, mainly because of an increase in disease due to Hia. Meningitis cases due to Hia did not cluster spatially with respect to the neighborhood of residence during pre and post vaccine periods.
Table 1.
Cases and incidences of Haemophilus influenzae meningitis in Salvador, Brazil, according to period of identification and serotype.
| Isolate type | Pre-vaccine period (n=424)a | Post-vaccine period (n=149)b | Total period (n=573) | |||
|---|---|---|---|---|---|---|
| No. cases (%)c | Incidenced | No. cases (%) | Incidenced | No. cases (%)c | Incidenced | |
| Type b | 416 (98) | 2.45 | 114 (77) | 0.24 | 530 (92) | 0.90 |
| Non-type b | 8 (2) | 0.04 | 35 (23) | 0.07 | 43 (8) | 0.06 |
| Type a | 5 (1) | 0.02 | 23 (15) | 0.05 | 28 (5) | 0.04 |
| Type e | 0 (0) | 0.00 | 3 (2) | 0.01 | 3 (1) | 0.00 |
| Type f | 1 (0) | 0.00 | 2 (1) | 0.01 | 3 (1) | 0.00 |
| Non-capsulated | 2 (0) | 0.02 | 7 (5) | 0.01 | 9 (2) | 0.01 |
NOTE. The Hib conjugate vaccine was introduced in the childhood immunization program in August 9, 1999.
From March 9, 1996 to August 8, 1999.
From August 9, 1999 to September 9, 2007.
Sum of percents are not equal to 100% due to rounding.
Mean annual cumulative incidence (per 100,000 population) was calculated for the cases of H. influenzae type b (365), type a (16), type e (2), type f (2) and non-capsulated (4) which resided within Metropolitan Salvador.
Hia and Hib meningitis occurred mainly among children <5 years of age while meningitis due to H. influenzae types e, f and non-capsulated strains occurred in older age groups (Table 2). Case-fatality of Hia and Hib meningitis cases was also higher than for meningitis cases due to other serotypes. (Table 2). The age group distribution and case fatality rate for H. influenzae type a cases did not differ between the pre and post-vaccine period.
Table 2.
Characteristics of H. influenzae meningitis cases from Salvador, Brazil, according to serotype.
| Characteristics | Type b (n=530) | Type a (n=28) | Other types (n=15)a | |||
|---|---|---|---|---|---|---|
| No. Responsesb | N (%) or median (IQR) | No. Responsesb | N (%) or median (IQR) | No. Responsesb | N (%) or median (IQR) | |
| Male sex | 527 | 301 (57) | 28 | 18 (64) | 15 | 8 (53) |
| Age <5 years | 520 | 473 (91) | 28 | 24 (86) | 15 | 4 (27)c |
| <2 years | 520 | 327 (63) | 28 | 14 (50) | 15 | 3 (20)c |
| 2–4 years | 520 | 146 (28) | 28 | 10 (36) | 15 | 1 (7) |
| CSF examinationd | ||||||
| Cells (×103/mm3) | 525 | 5.8 (2.2–10.0) | 28 | 8.4 (1.5–10.0) | 13 | 6.6 (2.8–8.6) |
| Glucose (mg/dL) | 529 | 20 (20–35) | 28 | 20 (20–30) | 15 | 20 (20–38) |
| Protein (mg/dL) | 529 | 300 (200–400) | 28 | 290 (185–500) | 15 | 300 (180–400) |
| Outcome | ||||||
| Neurological deficite | 481 | 70 (15) | 28 | 4 (14) | 15 | 1 (7) |
| ICU admission | 504 | 108 (21) | 28 | 4 (14) | 14 | 4 (29) |
| Death | 518 | 88 (17) | 28 | 4 (14) | 14 | 0 (0) |
NOTE: IQR, interquartile range; CSF, cerebrospinal fluid.
Includes 15 meningitis cases due to H. influenzae non-capsulated (9) type e (3) and type f (3) strains.
Number for which information was obtained.
Significant difference (P value <0.01) when compared with H. influenzae type b meningitis cases.
Initial examination performed during hospital admission.
Neurological deficit on discharge among survivors included ataxia (34 cases, all serotype b), motor deficit (16 cases, serotype b [15] and other type [1]), auditory deficit (9 cases, serotype b [6] and a [3];), hydrocephalus (7 cases, serotype b [6] and a [1]), and others (9 cases, all serotype b).
We were able to obtain information on immunization status for 26 (74%) of the 35 meningitis cases due to non-type b H. influenzae identified in the post-vaccine period. While 75% (13 of 17) of the cases due to H. influenzae type a isolate had received two or three Hib vaccine doses, only 11% (1 of 9) of the cases due to H. influenzae type e, f and non-capsulated isolates received the same number of Hib vaccine doses (P<0.01).
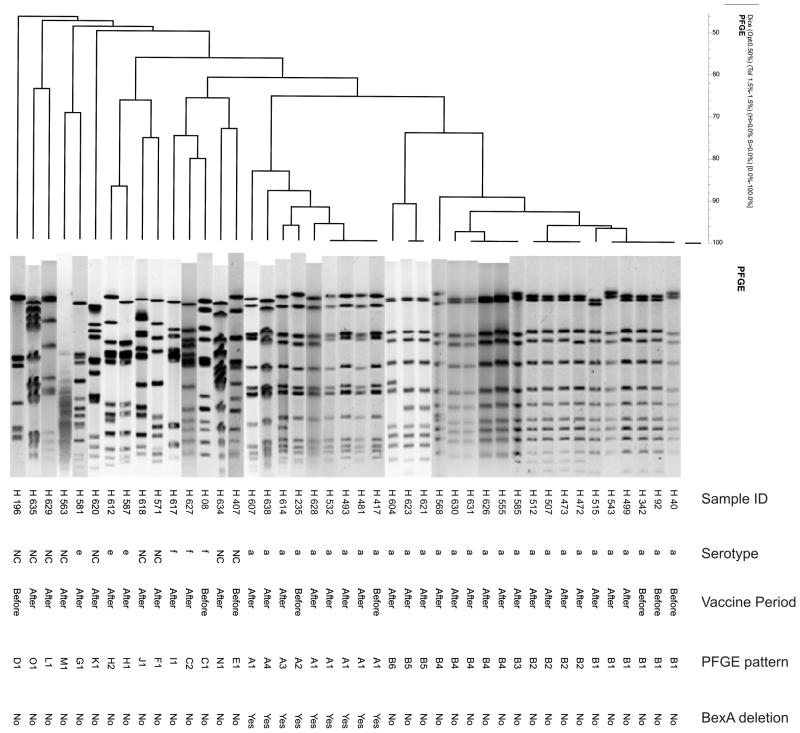
PFGE analysis for the 43 H. influenzae non-type b isolates discriminated fifteen distinct patterns (Figure 1). The 28 H. influenzae type a isolates had two different patterns, cluster A (9 isolates) and cluster B (19 isolates), while H. influenzae types e, f and non-capsulated strains were heterogeneous. (Figure 1). MLST analysis determined that PFGE clusters A and B corresponded to sequence type (ST) 4 and 23, respectively. PCR analysis identified the 339bp IS1016-bexA partial deletion product in nine of the 43 H. influenzae non-type b isolates. All of the nine H. influenzae isolates containing the IS1016-bexA deletion were serotype a and belonged to PFGE cluster A (ST4). Among the 28 Hia isolates, 5 and 23 were isolated during the pre- and post-vaccine periods, respectively. The proportion of Hia isolates with the IS1016-bexA deletion was 40% (2 of 5) and 30% (7 of 23) in the pre and post-vaccine periods, respectively, and this difference was not significantly different (P=0.65).
FIGURE 1.
Dendrogram showing the genetic relationships among the 43 non-type b H. influenzae isolates obtained from meningitis cases in Salvador, Brazil, as determined by pulsed-field gel electrophoresis. The columns, from left to right, show the isolate identification number, serotype, period of isolation in relation to introduction of Hib conjugate vaccine, the PFGE pattern designation, and presence of the IS1016bexA partial deletion. Note: NC, non-capsulated.
Meningitis cases caused by Hia isolates belonging to cluster A or B were similar with respect to gender, age and characteristics of cerebrospinal fluid (Table 3). However, case-fatality for meningitis cases caused by Hia isolates that had the IS1016-bexA deletion was 33% (3 of 9), versus 5% (1 of 19) for cases caused by Hia strains with complete IS1016-bexA (P=0.06) (Table 3). Among children <5 years of age with H. influenzae type a meningitis, 38% (3 of 8) of cases from which the isolate contained the IS1016-bexA deletion died whereas none of the 16 cases from which the isolate did not contain the IS1016-bexA deletion died (P=0.03) (Table 3).
Table 3.
Characteristics for the H. influenzae type a meningitis cases identified through active surveillance in Salvador, Brazil, according to the presence on the isolate of the IS1016-bexA partial gene deletion.
| Characteristics | Presence of IS1016-bexA | deletion in H. influenzae type a isolates | P value |
|---|---|---|---|
| With (n=9) | Without (n=19) | ||
| N (%) or median (IQR) | |||
| Male sex | 5 (56) | 13 (68) | 0.68 |
| Age <5 years | 8 (89) | 16 (84) | 1.00 |
| CSF exam | |||
| Cells (×103/mm3) | 10.0 (4.5–10.0) | 7.8 (1.2–10.0) | 0.27 |
| Glucose level (mg/dL) | 20 (20–28) | 20 (20–30) | 0.55 |
| Protein level (mg/dL) | 280 (100–500) | 300 (200–500) | 0.94 |
| Neurological deficit | 1 (17) | 7 (39) | 0.62 |
| Death | 3 (33) | 1 (5) | 0.08 |
| <5 years of agea | 3 (38) | 0 (0) | 0.03 |
NOTE:
Among the cases of H. influenzae type a with and without the IS1016-bexA deletion, 8 and 16 had <5 years of age, respectively.
Sequencing of the PCR products confirmed the presence of an IS1016-bexA deletion in the nine H. influenzae type a ST4 isolates. The size and location of the deletion, as well as the flanking region sequences, was identical to that previously-reported for an invasive serotype a strain that was isolated from Georgia, USA in 2005 (Genbank accession number DQ086152) (Figure 2). However, the sequence of the regions flanking the IS1016-bexA deletion for the ST4 isolates differed at four nucleotide sites from corresponding sequences for two previously-reported Hib strains (Genbank accession numbers AF549213 [HI 1007 – Georgia, USA] and S62752 [RM 7004 – Gambia]) and 3 of 4 Hib stains isolated during surveillance in Salvador. One Hib strain isolated from Salvador had a flanking region sequence that differed at only one nucleotide from the corresponding sequence in serotype a ST4 isolates (Figure 2).
FIGURE 2.
Nucleotide sequence of the IS1016-bexA deletion region of H. influenzae type b and type a strains. The arrowhead identifies the site of the deletion (bp 66) with the sequence of the bexA gene and IS1016 denoted in capital and lower case letter, respectively. The consensus sequence was obtained from the H. influenzae type b isolates (Genbank AF549213) from the US and the Gambia (Genbank S62752) and compared with a H. influenzae type a isolate from the US (Genbank DQ086152) and four H. influenzae type b (H503, H636, H637 and H492) and nine H. influenzae type a (H493, H532, H235, H417, H481, H607, H614, H628 and H638) isolates from Salvador, Brazil. Dots indicate identical nucleotides to the consensus sequence. The box denotes nucleotides specific to H. influenzae type a.
DISCUSSION
Widespread use of Hib conjugate vaccines have substantially reduced the incidence of Hib meningitis [1–3, 28, 29], resulting in increased awareness of meningitis due to other H. influenzae serotypes [7, 8, 11]. As Hib conjugate vaccines are effective in reducing Hib nasopharyngeal carriage [30, 31], it was hypothesized that non-type b strains could potentially occupy the niche left by Hib and consequently increase the risk of invasive disease by non-type b strains. To date, however, there has been little evidence of a substantial replacement of Hib disease by disease caused by other serotypes, a phenomenon known as serotype replacement [17, 32].
Among the capsulated H. influenzae strains that are not type b, type a has the capsular polysaccharides most closely related to that of type b. In animal challenge studies, reports have found that Hia is the most virulent capsulated H. influenzae after Hib [33]. The H. influenzae type a meningitis cases from this study affected similar age groups and had similar case-fatality rates to the Hib cases. In contrast, cases due to serotypes e, f and non-capsulated occurred at older ages and tend to have a better prognosis. These findings are consistent with prior clinical and epidemiological characterizations of invasive disease due to H. influenzae non-type b and support the hypothesis that type a isolates are the most virulent capsulated H. influenzae serotype after type b [33]. While H. influenzae type a invasive infections typically occur in healthy children [9, 10, 12, 16, 34], type e, f and non-capsulated types mostly occur in adults with underlying conditions such as cancer [8, 35, 36].
In this study, we found that patients with H. influenzae type a meningitis have an increased risk of death when the IS1016-bexA partial deletion was present in the clinical isolate. The association did not appear to be confounded by other prognostic factor such as patients’ age and disease duration prior to hospitalization. This finding is both plausible and analogous with what is known about virulence factors for Hib, for which the IS1016-bexA deletion stabilizes duplicated loci and leads to increased production of capsular polysaccharide [25, 33]. Hib capsular loci amplification has been found to inhibit complement-mediated bacteriolysis and opsonization [37]. Capsular amplification and the IS1016-bexA deletion have been identified in Hia invasive isolates [9, 10, 12, 15]. However, this study provides the first evidence for the significant association between the IS1016-bexA deletion and poor clinical outcome from Hia invasive disease.
However, the IS1016-bexA partial deletion was present in the minority of the H. influenzae type a isolates (9 of 28). Other investigations have also identified isolates of H. influenzae type a causing invasive infections resembling Hib invasive disease in the absence of the IS1016-bexA partial deletion [11, 16]. Additional studies in other geographical settings and with larger sample sizes are warranted to confirm the role of the IS1016-bexA deletion as a virulence factor in H. influenzae type a invasive disease. Furthermore, we did not evaluate whether the presence of the IS1016-bexA deletion was associated with neurological sequelae, hearing impairment or other markers of disease severity. Finally, further studies are needed to determine whether clinical isolates with the IS1016-bexA deletion exhibit enhanced virulence in animal models for Hia infection.
Results of this study suggest that Hia strains causing meningitis in Salvador have been stable over time. Sequence type 23 (ST 23) has been isolated in Malaysia, Canada and New Guinea [11, 26], suggesting worldwide spread of these clones. Interestingly, sequence type 4 (ST4), previously isolated in Kenya and The Gambia [26], was the first non-type b strain identified as having the IS1016-bexA partial deletion [15]. In addition, Sill et al described in Canada a case of H. influenzae type a invasive disease due to a ST4 strain containing the IS1016-bexA partial deletion [38]. This isolate was closely related on the basis of PFGE analysis to two Hia strains possessing the IS10116-bexA deletion that were isolated from cases of invasive disease in Georgia, USA [10]. Future studies are needed to investigate whether the ST4 clone is entirely responsible for the global spread of H. influenzae type a strains containing the IS1016-bexA partial deletion. These findings highlight the need to continue surveillance for H. influenzae invasive disease to monitor for the potential emergence of non-type b H. influenzae virulent clones.
Acknowledgments
We thank overall the study patients and their families; the clinical, laboratory and administrative staff of Hospital Couto Maia, especially Ana Maria Maia and Neide Oliveira Silva. Ricardo Martinez and Tatiana Lobo for their participation in data collection and processing; Neci Ivo Ramos, Nilda Lúcia Nunes Ivo, Maria Auxiliadora Macedo de Lima Machado and Helena Macedo in providing information on Hib immunization program and meningitis case notifications. We would also like to thank Hermes P. da Silva Filho, PhD for support on the sequence analysis and Brendan Flannery, PhD for manuscript review.
Funding: Grants from the Brazilian National Research Council (491345/2005-4), Research Support Foundation for the State of Bahia (FAPESB – 1431040054051) and National Institute of Health, USA (D43 TW00919 and R01 TW007303).
Footnotes
Conflict of interest statement: The authors do not have any commercial or other association that might pose a conflict of interest.
References
- 1.Adegbola RA, Secka O, Lahai G, et al. Elimination of Haemophilus influenzae type b (Hib) disease from The Gambia after the introduction of routine immunisation with a Hib conjugate vaccine: a prospective study. Lancet. 2005;366:144–50. doi: 10.1016/S0140-6736(05)66788-8. [DOI] [PubMed] [Google Scholar]
- 2.Farhoudi D, Lofdahl M, Giesecke J. Invasive Haemophilus influenzae type b disease in Sweden 1997–2003: epidemiological trends and patterns in the post-vaccine era. Scand J Infect Dis. 2005;37:717–22. doi: 10.1080/00365540510012800. [DOI] [PubMed] [Google Scholar]
- 3.CDC. Progress toward elimination of Haemophilus influenzae type b invasive disease among infants and children--United States, 1998–2000. MMWR Morb Mortal Wkly Rep. 2002;51:234–7. [PubMed] [Google Scholar]
- 4.Eskola J, Kayhty H, Takala AK, et al. A randomized, prospective field trial of a conjugate vaccine in the protection of infants and young children against invasive Haemophilus influenzae type b disease. N Engl J Med. 1990;323:1381–7. doi: 10.1056/NEJM199011153232004. [DOI] [PubMed] [Google Scholar]
- 5.Hviid A, Melbye M. Impact of routine vaccination with a conjugate Haemophilus influenzae type b vaccine. Vaccine. 2004;22:378–82. doi: 10.1016/j.vaccine.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Perdue DG, Bulkow LR, Gellin BG, et al. Invasive Haemophilus influenzae disease in Alaskan residents aged 10 years and older before and after infant vaccination programs. Jama. 2000;283:3089–94. doi: 10.1001/jama.283.23.3089. [DOI] [PubMed] [Google Scholar]
- 7.Urwin G, Krohn JA, Deaver-Robinson K, Wenger JD, Farley MM. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiologic characteristics in the H. influenzae serotype b vaccine era. The Haemophilus influenzae Study Group. Clin Infect Dis. 1996;22:1069–76. doi: 10.1093/clinids/22.6.1069. [DOI] [PubMed] [Google Scholar]
- 8.Cerquetti M, Ciofi degli Atti ML, Cardines R, Salmaso S, Renna G, Mastrantonio P. Invasive type e Haemophilus influenzae disease in Italy. Emerg Infect Dis. 2003;9:258–61. doi: 10.3201/eid0902.020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adderson EE, Byington CL, Spencer L, et al. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: emerging pathogen in the vaccine era? Pediatrics. 2001;108:E18. doi: 10.1542/peds.108.1.e18. [DOI] [PubMed] [Google Scholar]
- 10.Kapogiannis BG, Satola S, Keyserling HL, Farley MM. Invasive infections with Haemophilus influenzae serotype a containing an IS1016-bexA partial deletion: possible association with virulence. Clin Infect Dis. 2005;41:e97–103. doi: 10.1086/498028. [DOI] [PubMed] [Google Scholar]
- 11.Tsang RS, Mubareka S, Sill ML, Wylie J, Skinner S, Law DK. Invasive Haemophilus influenzae in Manitoba, Canada, in the postvaccination era. J Clin Microbiol. 2006;44:1530–5. doi: 10.1128/JCM.44.4.1530-1535.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogilvie C, Omikunle A, Wang Y, St Geme IJ, 3rd, Rodriguez CA, Adderson EE. Capsulation loci of non-serotype b encapsulated Haemophilus influenzae. J Infect Dis. 2001;184:144–9. doi: 10.1086/322001. [DOI] [PubMed] [Google Scholar]
- 13.Satola SW, Collins JT, Napier R, Farley MM. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J Clin Microbiol. 2007;45:3230–8. doi: 10.1128/JCM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaClaire LL, Tondella ML, Beall DS, et al. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J Clin Microbiol. 2003;41:393–6. doi: 10.1128/JCM.41.1.393-396.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroll JS, Moxon ER, Loynds BM. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J Infect Dis. 1994;169:676–9. doi: 10.1093/infdis/169.3.676. [DOI] [PubMed] [Google Scholar]
- 16.Hammitt LL, Block S, Hennessy TW, et al. Outbreak of invasive Haemophilus influenzae serotype a disease. Pediatr Infect Dis J. 2005;24:453–6. doi: 10.1097/01.inf.0000160954.90881.29. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro GS, Reis JN, Cordeiro SM, et al. Prevention of Haemophilus influenzae type b (Hib) meningitis and emergence of serotype replacement with type a strains after introduction of Hib immunization in Brazil. J Infect Dis. 2003;187:109–16. doi: 10.1086/345863. [DOI] [PubMed] [Google Scholar]
- 18.Ribeiro GS, Lima JB, Reis JN, et al. Haemophilus influenzae meningitis 5 years after introduction of the Haemophilus influenzae type b conjugate vaccine in Brazil. Vaccine. 2007;25:4420–8. doi: 10.1016/j.vaccine.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Case notification records. Salvador, Brazil: Secretary of Health for the State of Bahia; 2005. [Google Scholar]
- 20.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–6. doi: 10.1128/jcm.32.10.2382-2386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curran R, Hardie KR, Towner KJ. Analysis by pulsed-field gel electrophoresis of insertion mutations in the transferrin-binding system of Haemophilus influenzae type b. J Med Microbiol. 1994;41:120–6. doi: 10.1099/00222615-41-2-120. [DOI] [PubMed] [Google Scholar]
- 22.Saito M, Umeda A, Yoshida S. Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1999;37:2142–7. doi: 10.1128/jcm.37.7.2142-2147.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satola SW, Schirmer PL, Farley MM. Complete sequence of the cap locus of Haemophilus influenzae serotype b and nonencapsulated b capsule-negative variants. Infect Immun. 2003;71:3639–44. doi: 10.1128/IAI.71.6.3639-3644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroll JS, Moxon ER, Loynds BM. An ancestral mutation enhancing the fitness and increasing the virulence of Haemophilus influenzae type b. J Infect Dis. 1993;168:172–6. doi: 10.1093/infdis/168.1.172. [DOI] [PubMed] [Google Scholar]
- 26.Meats E, Feil EJ, Stringer S, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–36. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.IBGE. Anuário Estatístico do Brasil. Vol. 56. Rio de Janeiro: Instituto Brasileiro de Geografia e Estatística; 1996. [Google Scholar]
- 28.Dagan R, Fraser D, Roitman M, et al. Effectiveness of a nationwide infant immunization program against Haemophilus influenzae b. The Israeli Pediatric Bacteremia and Meningitis Group. Vaccine. 1999;17:134–41. doi: 10.1016/s0264-410x(98)00165-0. [DOI] [PubMed] [Google Scholar]
- 29.Peltola H, Aavitsland P, Hansen KG, Jonsdottir KE, Nokleby H, Romanus V. Perspective: a five-country analysis of the impact of four different Haemophilus influenzae type b conjugates and vaccination strategies in Scandinavia. J Infect Dis. 1999;179:223–9. doi: 10.1086/314535. [DOI] [PubMed] [Google Scholar]
- 30.Forleo-Neto E, de Oliveira CF, Maluf EM, et al. Decreased point prevalence of Haemophilus influenzae type b (Hib) oropharyngeal colonization by mass immunization of Brazilian children less than 5 years old with hib polyribosylribitol phosphate polysaccharide-tetanus toxoid conjugate vaccine in combination with diphtheria-tetanus toxoids-pertussis vaccine. J Infect Dis. 1999;180:1153–8. doi: 10.1086/315018. [DOI] [PubMed] [Google Scholar]
- 31.Millar EV, O'Brien KL, Levine OS, Kvamme S, Reid R, Santosham M. Toward elimination of Haemophilus influenzae type B carriage and disease among high-risk American Indian children. Am J Public Health. 2000;90:1550–4. doi: 10.2105/ajph.90.10.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajanca P, Canica M. Emergence of nonencapsulated and encapsulated non-b-type invasive Haemophilus influenzae isolates in Portugal (1989–2001) J Clin Microbiol. 2004;42:807–10. doi: 10.1128/JCM.42.2.807-810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Z, Romero-Steiner S, Carlone GM, Robbins JB, Schneerson R. Haemophilus influenzae type a infection and its prevention. Infect Immun. 2007;75:2650–4. doi: 10.1128/IAI.01774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millar EV, O'Brien KL, Watt JP, et al. Epidemiology of invasive Haemophilus influenzae type A disease among Navajo and White Mountain Apache children, 1988–2003. Clin Infect Dis. 2005;40:823–30. doi: 10.1086/428047. [DOI] [PubMed] [Google Scholar]
- 35.Bruun B, Gahrn-Hansen B, Westh H, Kilian M. Clonal relationship of recent invasive Haemophilus influenzae serotype f isolates from Denmark and the United States. J Med Microbiol. 2004;53:1161–5. doi: 10.1099/jmm.0.45749-0. [DOI] [PubMed] [Google Scholar]
- 36.Campos J, Hernando M, Roman F, et al. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J Clin Microbiol. 2004;42:524–9. doi: 10.1128/JCM.42.2.524-529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noel GJ, Brittingham A, Granato AA, Mosser DM. Effect of amplification of the Cap b locus on complement-mediated bacteriolysis and opsonization of type b Haemophilus influenzae. Infect Immun. 1996;64:4769–75. doi: 10.1128/iai.64.11.4769-4775.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sill ML, Zhou J, Law DK, et al. Molecular characterization of four Haemophilus influenzae serotype a strains isolated from patients in Quebec, Canada. Can J Microbiol. 2007;53:1191–4. doi: 10.1139/W07-088. [DOI] [PubMed] [Google Scholar]