Abstract
Rotavirus (RV), a leading cause of severe diarrhea, primarily infects intestinal epithelial cells (IEC) causing self-limiting illness. In order to better understand innate immunity to RV, we sought to define the extent to which IEC activation of anti-viral responses required viral replication or could be recapitulated by inactivated RV or its components. Using model human intestinal epithelia, we observed that RV-induced activation of signaling events and gene expression typically associated with viral infection was largely mimicked by administration of UV-inactivated RV. Use of anti-IFN neutralizing antibodies revealed that such replication-independent anti-viral gene expression required type I interferon signaling. In contrast, RV-induction of NF-κB-mediated IL-8 expression was dependent upon viral replication. The anti-viral gene expression induced by UV-RV was not significantly recapitulated by RV RNA or RV VLP even though the latter could enter IEC. Together, these results suggest that RV proteins mediate viral entry into epithelial cells leading to intracellular detection of RV RNA that generates an anti-viral response.
Keywords: Type I interferon, dsRNA, pattern-recognition receptors, STAT-1, IRF 3/7
INTRODUCTION
Rotavirus (RV) is the most common cause of severe dehydrating diarrheal disease in young children worldwide, causing up to 100 million cases and > 600,000 deaths each year (1, 2). RV infections are also common in adults, especially those who interact with children, but generally cause only mild symptoms (3). RV, a member of the Reoviridae family of viruses, is a non-enveloped double- stranded (ds) RNA virus containing three concentric protein layers (4, 5). Its genome consists of 11 dsRNA segments that each code for different rotaviral proteins. Rotaviral proteins are divided into two groups, namely, structural proteins (VP 1–4, 6–7) that compose the viral structure and non-structural proteins (NSP1–6), which are synthesized during infection and function to facilitate viral replication or pathogenesis (4). VP4, a spike-shaped hemagglutinin protein that emanates through the outermost (VP7) protein layer, plays a role in cellular attachment and must be cleaved by proteases normally present in the host intestine for rotavirus to be infectious (4, 5). NSP proteins play a role in driving RV-induced diarrhea and suppressing host immune responses (4).
RV infection is generally localized to the gastrointestinal tract and typically resolves within 7 days (1, 6). RV infection of mice lacking functional B and T lymphocytes often results in chronic infection, highlighting the importance of adaptive immunity in host defense to this pathogen (6, 7). However, Franco and Greenberg (7) observed that 40% of SCID mice (on a C57BL/6 background) cleared RV infection. Additionally, Eiden et al. (8) observed clearance of RV in athymic Balb/c mice that was not accompanied by anti-RV antibodies. These findings suggest an important role for innate immunity in controlling RV infection. Such innate immune control of RV may be mediated by intestinal epithelial cells (IEC) since these cells are the predominant target of RV infection. The role of IEC in clearing RV infection is likely independent of RV-induced diarrhea in that the kinetics of clearance during primary infection are similar in newborn and adult mice even though only the former exhibit diarrhea (4). Numerous studies have noted that RV robustly induces NF-κB mediated epithelial expression of the chemokine IL-8, yet given that one of the hallmarks of RV infection is the absence of neutrophil infiltration (9–11), it is hard to envision IEC production of this chemokine plays a major role in clearance of RV. Rather, based on the large body of data on host responses to viral infections in general, it seems more plausible that type 1 interferon (IFN) responses contribute to innate immune-mediated clearance of RV. In accordance, gene-profiling studies of RV-infected IEC observed elevated expression of a panel of genes related to type I IFN responses (12). Pre-treatment of cultured cells with type 1 IFNs limited RV infection (13). Levels of type I IFNs increase in RV-infected children and animals and administration of exogenous type I IFN reduces disease sequellae in cattle and pigs (4, 14–16). Moreover, RV uses the non-structural protein NSP1 to suppress IFN signaling (17, 18). Mutations in NSP1 that ablate RV’s ability to interfere with IFN-related signaling attenuate RV’s spread to uninfected cells (17), further supporting the notion that IFN signaling is a potential hindrance to this pathogen. While mice deficient in type I and II IFN receptors are able to clear RV (19), loss of the transcription factor STAT1, which mediates much of the gene expression induced by type I IFN, severely impairs control of RV (20). Thus, although there is considerable redundancy in host defense mechanisms against RV, it seems likely that IEC activation of STAT1 and induction of genes regulated by type I IFN play an important role in host defense against this virus. Therefore, we sought to investigate the mechanism by which IEC activate such anti-viral signaling events in response to RV infection.
Viral infection has long been known to alter host gene expression by inducing endoplasmic reticulum (ER) stress, sometimes called “ER overflow,” in which the rapid switch in translation from host-encoded to viral-encoded proteins activates host stress-related transcription factors (21). This model predicts that a UV-inactivated virus, which cannot replicate or express its genes, would not alter host gene expression. In accordance, inactivation of the RV genome via UV-irradiation markedly reduced RV’s ability to induce NF-κB activation and IL-8 secretion (10, 11). More recently, it has become appreciated that a large portion of microbe-induced gene expression does not require a viable microbe per se but, rather, results from host pattern-recognition receptors (PRR) such as the toll-like receptors (TLR) detecting various microbial components. For example, IEC detection of motile bacteria is largely mediated by TLR5 detection of bacterial flagellin (22). However, IEC appear to be hyporesponsive to a number of other TLR agonists, highlighting the stark differences in the mechanism by which different microbes might be detected by IEC (23). IEC have been observed to respond to synthetic dsRNA, poly (I:C), resulting in expression of both IL-8 and a panel of genes associated with type I IFN responses, suggesting IEC have the potential to respond to RV via detection of viral RNA (24). However, whether such observations could apply to an actual virus-IEC interaction remained unclear. Thus, the goal of this study was to define the extent to which IEC response to RV results from viral replication or, rather is largely a consequence of IEC detection of RV components. We observed that RV-induced IEC anti-viral signaling was almost entirely driven by detection of viral components likely resulting from VP4-mediated viral entry followed by detection of RV RNA.
RESULTS
Apical infection by rotavirus induces epithelial anti-viral signaling
Epithelial cell-mediated innate immune responses play a potentially important role in protecting the host against rotavirus (RV). Yet, although some studies of RV-induced innate immune activation have focused on activation of NF-κB mediated genes such as IL-8, rotavirus-induced anti-viral signaling in intestinal epithelial cells has not been extensively studied. Thus, we infected human HT-29 intestinal epithelial cells (IEC) with RV (MOI 0.5–1) and temporally assayed signaling events typically associated with a variety of viral infections. Mock-infected control samples were exposed to low levels of trypsin, which may activate protease activated receptors (PARs), however such treatment was included because it is required for cleavage of RV capsid protein VP4 and thus allows for robust infectivity (25). RV infection induced transient phosphorylation of transcription factors IRF3 and STAT1 with a maximal response being observed between 8–16 hours post-inoculation (hpi) (Figure 1A). RV also induced epithelial secretion of IFN-β (Figure 1B) and, in accordance with other studies, IL-8 (Figure 1C). Interestingly, RV-induced IL-8 production was relatively delayed suggesting it may not reflect immediate IEC sensing of the virus. These signaling events, which were not observed in mock-infected cells, roughly correlated with levels of the viral protein VP6 and are consistent with the finding that viral replication is required for RV-induced NF-κB activation (11). We next performed these experiments in polarized IEC, which result when IEC are cultured on collagen-coated permeable supports, in the hope that doing so might provide insight into mechanisms underlying such RV-induced anti-viral signaling. For example, use of polarized IEC previously enabled us to uncouple Salmonella invasion from activation of innate immunity, ultimately allowing definition of mechanisms underlying the latter process (26). Consistent with previous findings (27), RV infection was markedly more efficient when the virus was administered to the apical surface of epithelia (Figure 1D). Such reduced infectivity of basolaterally-administered RV correlated with marked attenuation of IRF7, STAT1, and PKR activation and induced secretion of IFN-β and IL-8 (Figure 1D, E, F). Such preferential infection and induction of anti-viral signaling by apical RV argues against RV activating innate immune signaling via a basolateral pattern-recognition receptor, as occurs in response to Salmonella (28), but seems consistent with the possibilities that anti-viral signaling might be triggered by an apical receptor, an intracellular receptor, or result from viral replication causing ER stress.
Figure 1. Anti-viral protein expression in RV-infected intestinal epithelia.
Human intestinal epithelia (HT29) were grown to confluence in 6 well plates (A, B, C) or collagen-coated permeable supports (D, E, F) and infected with RV (MOI 0.5–1). Control samples were treated with trypsin diluted in SFM (Mock) or SFM alone (C). Cell lysates and supernatants were collected 0–48 hours post-inoculation (hpi). Western blot analyses were performed to assess viral protein (VP6) synthesis and protein expression of the indicated anti-viral markers in cell lysates (A, D). ELISA assays were used to measure IFN-β (B, E) and IL-8 (C, F) secretion in supernatants over time. Data in A, B, D, and E are results of a single experiment and representative of 3 separate experiments that gave similar results. Data in C and F reflects the mean +/− standard error of the mean (SEM) of 3 parallel experiments. Statistically significant differences P< 0.05 are denoted as starred values (*).
Inactivated rotavirus induces anti-viral gene expression similar to live rotavirus
To determine the extent to which RV-induced anti-viral signaling required viral replication or could be mimicked by structural components of RV, we examined the epithelial response to UV-irradiated RV (UV-RV), which is structurally intact but rendered non-replicative (29, 30). The inability of UV-RV to replicate in IEC was verified by monitoring levels of VP6 over time (Figure 2A). In accordance with previous studies, such UV-inactivation of the RV genome substantially reduced induction of IL-8 (Figure 2C). However, in contrast, signaling events typically associated with viral infection including activation of STAT1 and IRF3/7 and induction of IFN-β secretion, were elicited at least as robustly by UV-RV (Figure 2A, B). Similar activation of anti-viral signaling by RV and UV-RV was also observed in polarized epithelia in response to apical stimulation (data not shown). To confirm these events were indeed induced by UV-RV as opposed to UV cross-linked cell debris that might have been present in our virus preparation, we performed a control experiment showing that UV-irradiation of a mock viral preparation, which contained MA104 cell debris but not RV, did not elicit antiviral signaling induced by UV-RV (Figure 2D). Next, we determined if trypsinization, which is known to be required for viral entry, is also required for anti-viral signaling in response to UV-RV. Indeed, robust activation of anti-viral signaling by both RV and UV-RV required the stimulating agonist to be treated with trypsin prior to IEC stimulation (Figure 3). Lastly, we observed that, analogous to the case for RV, activation of innate immune signaling in response to UV-RV was more robust when UV-RV was applied to the apical rather than basolateral surface of polarized epithelia (Figure 4). Together, these results suggest that UV-RV induces anti-viral signaling via a mechanism similar to live virus, and further supports the notion that type I IFN is activated by IEC detection of RV structural components rather than viral replication.
Figure 2. Anti-viral protein expression exhibited in RV-infected and UV-RV stimulated epithelia.

Confluent intestinal epithelia (HT29) were grown in 6 well plates and treated with RV and UV-RV (MOI 1). Control samples were exposed to trypsin diluted in SFM (Mock), irradiated cellular debris from a mock preparation of UV-RV (Mock Irradiation), or SFM alone (C). Cell lysates and supernatants were collected at various time points (0–48 hpi). Western blot analyses were performed to assess viral protein (VP6) synthesis and protein expression of the indicated anti-viral markers in cell lysates (A, D). ELISA assays were used to measure secretion of IFN-β (B) and IL-8 (C) in supernatants at 48 hpi. Data in A, B and D are results of a single experiment and representative of 3 separate experiments that gave similar results. Data in C is the mean +/− SEM of 3 parallel experiments. Statistically significant differences P< 0.05 are denoted as starred values (*).
Figure 3. Anti-viral protein expression exhibited in epithelia treated with RV and UV-RV in the presence or absence of trypsin.
Intestinal epithelial monolayers (HT29) grown in 6 well plates were treated with RV and UV-RV (MOI 1) in the presence or absence of trypsin (24 hpi). Control samples received equivalent amounts of trypsin diluted in SFM (Mock) or SFM alone. Trypsin and trypsin-free treatments are denoted as (+) and (−) symbols, respectively. Western blot analysis was used to detect anti-viral gene expression in cell lysates (A). ELISA assays were performed to measure secretion of IFN-β (B) and IL-8 (C) in supernatants at 24 hpi. Data in A shows results of a single experiment and is representative of 3 separate experiments that gave similar results. Data in B and C is the mean +/− SEM of 3 parallel experiments. Statistically significant differences P< 0.05 are denoted as starred values (*).
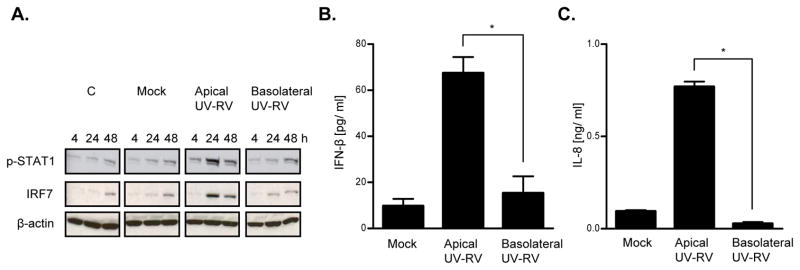
Figure 4. Anti-viral protein expression exhibited in epithelia treated apically and basolaterally with UV-RV.
Intestinal epithelial monolayers (HT29) were grown on collagen-coated permeable supports and infected either apically or basolaterally with UV-RV (MOI 1). Control samples received equivalent amounts of trypsin diluted in SFM (Mock) or SFM alone (C). Cell lysates and supernatants were collected at various time points (0–48 hpi). Western blot analyses were performed to assess protein expression of the indicated anti-viral markers in cell lysates (A). ELISA assays were used to measure IFN-β (B) and IL-8 (C) secretion in supernatants at 48 hpi. Data in A shows results of a single experiment and is representative of 3 separate experiments that gave similar results. Data in B and C reflects the mean +/− SEM of 3 parallel experiments. Statistically significant differences P< 0.05 are denoted as starred values (*).
We next sought to define the extent to which RV-induced gene expression in general can be mimicked by UV-RV and determine the role of type I IFN in RV-induced changes in IEC gene expression. IEC were mock-infected or exposed to RV, UV-RV, or RV in the presence of neutralizing antibodies to type 1 IFN (anti-IFN α/β) for 24 h, at which time gene expression was assayed by cDNA microarray. The gene chip employed for this purpose permits simultaneous examination of 12 different samples allowing us to assay 4 different experimental conditions in biological triplicates, thus permitting statistical analysis to be performed directly on the microarray data. Such microarray analyses indicated that RV upregulated 1190 genes by at least 1.3 fold relative to mock-treated uninfected cells (cut-off was arbitrarily chosen based on our previous experience with microarray-based studies of IEC (24)). The entire microarray data set is available on-line (posted on GEO) and some of the common means of examining microarray data such as unsupervised clustering analysis are shown in supplemental data (Supplemental Figure 1). In Figure 5A, we sought to display our microarray data in a manner that would most facilitate addressing our central questions. Specifically, we generated a “heat map” that displays gene expression in each of the 12 samples (4 conditions, 3 replicates) relative to the average expression of mock-treated cells. The relative uniformity of the 3 replicates in each condition indicates the high degree of similarity among our biological replicates. Genes were ordered (top to bottom) based on their relative dependence upon type I IFN (ratio of expression upon exposure to RV alone vs. RV in the presence of anti-IFN α/β). The majority of genes that were induced by RV are thus type I IFN-dependent in that their expression was reduced by the neutralizing antibody. The heat map shows that the vast majority of such IFN-dependent gene expression did not appear to require viral replication in that almost of all of these genes were similarly induced by both RV and UV-RV. Such type-I IFN dependent, replication-independent, RV-induced gene expression included a panel of classic anti-viral genes such as IRF7, IFN-β, STAT1, Mx1, OAS-2 and MHC I (Table I). Use of qRT-PCR verified the upregulated mRNA levels of some of these genes (Figure 5B, C, D, E). In contrast to the induction of such classic anti-viral genes, expression of IL-8 was partially dependent upon viral replication (Figure 2C) and independent of type I IFN (Table I). Thus, a large portion of RV-induced gene expression in IEC, particularly upregulation of genes typically associated with viral infection, is independent of viral replication and dependent upon type I IFN.
Figure 5. Transcription profiles of epithelia stimulated with RV, UV-RV, and RV in the presence of Type 1 IFN (α/β) antibodies.
Intestinal epithelial (HT29) cell monolayers were grown in 6 well plates and infected with RV and UV-RV (MOI 1), and RV (MOI 1) plus Type 1 IFN antibodies (anti-IFN α/β). Control samples received equivalent amounts of trypsin diluted in SFM (Mock). Experiments were performed in biological triplicates. At 24 hpi, cell lysates were extracted for RNA and microarray analyses were performed to assess global transcription of genes. Heat map illustration of genes induced by RV and UV-RV with > 1.3 fold change relative to mock (A). qRT-PCR results used to confirm mRNA synthesis of select anti-viral genes at 24 hpi (B, C, D, E). Data in A shows results of 3 parallel experiments. Data in B, C, D, and E reflects the mean +/− SEM of 3 parallel experiments. Statistically significant differences P< 0.05 are denoted as starred values (*).
Table I. Fold change induction of anti-viral markers relative to mock.
Anti-viral gene expression in cells treated with RV, UV-RV, and RV + anti-IFN α/β. Intestinal epithelial (HT29) cell monolayers were stimulated with RV and UV-RV (MOI 1), RV (MOI 1) plus Type 1 IFN antibodies (anti-IFN α, anti-IFN β), and mock treatments as described in Figure 5. At 24 hpi, lysates were collected in TRIzol to assess global transcription of genes (Figure 5A), including anti-viral markers of interest as indicated above. mRNA expression of select anti-viral genes was confirmed via qRT-PCR analysis (Figure 5B, C, D, E). Fold induction values greater than standard deviation calculations reflect statistically significant differences from mock (P < 0.05).
| RV | UV-RV | RV + anti-IFN α/β | |
|---|---|---|---|
| TLR3 | 1.5 ± 0.07 | 2.2 ± 0.37 | 1.13 ± 0.09 |
| Mda-5 | 8.7 ± 1.2 | 11.1 ± 0.84 | 4.24 ± 0.71 |
| RIG-I | 3.7 ± 0.33 | 6.6 ± 0.39 | 2.41 ± 0.29 |
| IRF3 | 1.3 ± 0.06 | 1.3 ± 0.06 | 1.16 ± 0.07 |
| IRF7 | 5.9 ± 0.24 | 7.7 ± 0.90 | 3.92 ± 0.20 |
| IFN-α | 1.1 ± 0.06 | 1.5 ± 0.04 | 0.95 ± 0.08 |
| IFN-β | 11 ± 0.88 | 31.6 ± 4.4 | 6.24 ± 0.84 |
| STAT1 | 10.5 ± 1.1 | 12.8 ± 0.85 | 4.42 ± 0.28 |
| Mx1 | 35.6 ± 0.44 | 45.2 ± 3.9 | 12.3 ± 0.37 |
| PKR | 3.3 ± 0.18 | 3.3 ± 0.22 | 1.72 ± 0.18 |
| OAS-2 | 43.6 ± 2.3 | 76.5 ± 4.2 | 11.6 ± 1.2 |
| MHC I | 11.6 ± 0.92 | 24.1 ± 1.7 | 4.1 ± 0.64 |
| IL-8 | 60 ± 5.9 | 24 ± 2.2 | 58 ± 5.3 |
| IFITM1 | 44.4 ± 1.9 | 112.6 ± 4.9 | 8 ± 0.92 |
| IFITM3 | 28.9 ± 2.3 | 37.3 ± 2.9 | 6.4 ± 0.83 |
| CXCL10 | 35.5 ± 3.7 | 108 ± 11.3 | 17.6 ± 2.3 |
| ISG15 | 10.4 ± 3.0 | 15.4 ± 3.3 | 5.03 ± 0.90 |
RV components fail to recapitulate RV-induced IEC anti-viral signaling
Next, we sought to better define the structural determinant of RV that played an important role in activating IEC anti-viral signaling. Specifically, we compared epithelial responses to UV-RV, RV virus-like particles (VLPs), and purified RV RNA. The VLPs used here are protein shells comprised of 4 major RV structural proteins (VP 2/4/6/7) that lack nucleic acid (31). RV VLPs were used at protein concentrations of 0.5–5μg/ml, which is equivalent to the concentration of UV-RV that corresponds to MOIs of 1–10. Purified RV RNA was used at concentrations of 0.5–5 μg/ml, which is approximately 100 times the amount of RNA in UV-RV that corresponds to MOIs of 1–10. In contrast to UV-RV, neither RV VLPs or RNA induced detectable elevations in levels of phospho-STAT1 or IRF7 (Figure 6A). RV VLPs and RNA also failed to recapitulate the induction of IFN-β or IL-8 elicited by UV-RV (Figure 6B, C). To more broadly understand the extent to which these components of UV-RV might recapitulate its ability to activate gene expression in epithelial cells, we measured changes in epithelial gene expression via microarray analysis. A modest concentration of UV-RV (MOI 0.5) induced 401 genes by > 1.3 fold relative to mock-treated control cells including key anti-viral genes such as Mda-5, IFN-β, MHC I (Tables II and III). Only a small portion of the genes upregulated by UV-RV were similarly induced by treatment with RV VLPs or RNA (Table II). In accordance, induction of anti-viral gene expression by RV viral components was also not observed to be comparable to UV-RV (Table III), and these trends were confirmed by measuring mRNA synthesis of select anti-viral genes via qRT-PCR (Figure 6D, E). Thus, neither RV RNA nor VLPs could substantially recapitulate the changes in gene expression induced by inactive but structurally intact RV.
Figure 6. Anti-viral protein expression exhibited in epithelia treated with UV-RV, RV VLPs and RV RNA.
Confluent intestinal epithelia (HT29) were grown in 6 well plates and treated with UV-RV (MOI 0.5–1), RV RNA (0.5–5 μg/ml), and VLPs (0.5–5 μg/ml). Control samples received equivalent amounts of trypsin diluted in SFM (Mock). Cell lysates and supernatants were collected at various time points (0–24 hpi). Western blot analyses were used to detect anti-viral gene expression in cell lysates (A). ELISA assays were performed to measure IFN-β (B) or IL-8 (C) secretion in supernatants at 24 hpi. qRT-PCR analyses were utilized to confirm mRNA synthesis of select anti-viral genes from microarray experiments (see Table III). Data in A are results of a single experiment and representative of 3 separate experiments that gave similar results. Data in B, C, D, and E shows the mean +/− SEM of 3 parallel experiments. Statistically significant differences P< 0.05 are denoted as starred values (*).
Table II. Genes induced by UV-RV, RV VLPs, and RV RNA relative to mock.
Cellular Gene Transcription Induced by UV-RV, RV VLP, and RV RNA. Intestinal epithelial (HT29) cell monolayers were stimulated for 24 h with UV-RV (MOI 0.5), RV VLPs (2.5 ug/ml), and RV RNA at low (0.1 μg/ml) and high (1 μg/ml) concentrations (denoted as RV RNA “lo” and “hi,” respectively). Mock-treated samples received equivalent amounts of trypsin in SFM. Experiments were performed in biological duplicates. Lysates were collected in TRIzol to assess mRNA expression via microarray as described in text. The number of genes induced by UV-RV, RV VLPs and RV RNA with > 1.3 fold change is shown.
| # genes | Avg. induction | |
|---|---|---|
| UV-RV | 401 | 1.5 |
| RV VLPs | 64 | 1.4 |
| RV RNA lo | 72 | 1.4 |
| RV RNA hi | 97 | 1.5 |
Table III. Fold change induction of anti-viral markers relative to mock.
Antiviral Gene Expression in Cells Treated with UV-RV, RV VLPs and RV RNA. Cells were stimulated with UV-RV, RV VLPs, RV RNA and mock treatments as described in Table 2. After 24 h, lysates were collected in TRIzol to assess global transcription of genes (depicted in Table 2), including anti-viral markers of interest as indicated above. mRNA expression of select anti-viral genes was confirmed via qRT-PCR analysis (Figure 6D, E). Fold induction values greater than standard deviation calculations reflect statistically significant differences relative to mock (P < 0.05).
| UV-RV | RV VLPs | RV RNA lo | RV RNA hi | |
|---|---|---|---|---|
| Mda-5 | 1.8 ± 0.18 | 1.4 ± 0.21 | 1.2 ± 0.03 | 1.4 ± 0.00 |
| IFN-β | 6.4 ± 0.12 | 1.2 ± 0.06 | 1.1 ± 0.02 | 1.3 ± 0.21 |
| MHC I | 1.7 ± 0.14 | 1.2 ± 0.02 | 1.1 ± 0.03 | 1.3 ± 0.12 |
RV and UV-RV display similar cell entry kinetics
To better understand the interaction of UV-RV and RV VLPs with polarized epithelia and activation of epithelial anti-viral signaling in response to RV, we examined the interaction of RV, UV-RV, and RV VLPs with model epithelia via confocal microscopy for up to 4 hpi (Figure 7A, B). RV and UV-RV (MOI 10), and RV VLPs roughly equivalent to the estimated protein concentration of the RV preparation, were applied to the apical surface of epithelia for 1 and 4 hpi, followed by washing off of non-adsorbed or non-adhered materials. As expected, planar images of IEC taken 3 μm below the apical surface revealed the presence of RV, UV-RV and RV VLPs in the sub-apical region within 1 hpi (Figure 10A). RV increased in abundance by 4 hpi, reflecting viral replication, while UV-RV levels decreased likely due to degradation of viral proteins and an inability to replicate (Figure 7B). Thus, consistent with models by which RV is internalized by IEC, RV’s primary structural proteins are sufficient to mediate its entry into IEC. Taken together, we interpret our results to suggest that RV activation of anti-viral signaling requires viral structural proteins to mediate entry of viral RNA into epithelial cells where it can be detected by host pattern-recognition receptor(s).
Figure 7. RV, UV-RV, and VLP cell entry during early stages of infection.
Intestinal epithelial monolayers (HT29) were grown on collagen-coated permeable supports and treated apically with RV and UV-RV (MOI 10), and an amount of VLPs roughly equivalent to the estimated protein concentration in the RV preparation. Control samples were apically treated with an equivalent amount of trypsin in SFM (Mock) for 4 hpi. At 1 and 4 hpi, cells were fixed, stained and examined via confocal fluorescence microscopy for presence of rotaviral proteins (green) and F-actin (red) in the sub-apical region of the cells (3 μm below the apical surface). Sub-apical images of cells stimulated with RV, UV-RV, and RV VLPs at 1 hpi, magnification 60X (A). Sub-apical images of cells treated with RV and UV-RV at 1 and 4 hpi, magnification 40X (B). Data in A and B are results of a single experiment and representative of 3 separate experiments. Scale reflects distance of 10 μm.
DISCUSSION
The ability of adult humans and mice to efficiently clear rotavirus without major sequellae serves as an example of effective mucosal immunity. Thus, understanding host immunity to RV may provide insights into understanding the pathogenesis of viruses that are not dispatched in such an expedient manner. Such adept handling of RV likely reflects considerable redundancy in the mechanisms that protect the host against RV. In accordance, and in contrast to the case for many viruses, mice lacking either adaptive immunity or type I IFN still exhibit substantial control of RV infection (7, 19). Although not required for clearance, type I IFNs modulate the course of RV infection in a variety of experimental systems (4, 13–15) and deletion of STAT1, a transducer of IFN-related signals, severely impairs control of RV (20). Thus, activation of STAT1/type I IFN signaling is likely one important means of host defense against RV. Therefore, the goal of this study was to investigate how the predominant cell type infected by RV, the intestinal epithelial cell (IEC), activates such anti-viral signals in response to RV infection.
We observed that, in contrast to RV activation of NF-κB, which is driven by synthesis of viral proteins that may cause ER stress (11), RV-induced anti-viral signaling was almost entirely driven by IEC detection of viral components. Indeed, while adding RV and inactivated RV (UV-RV) at the same MOI initially delivered similar levels of viral components, UV-RV antigens were degraded within a few hours while RV replication resulted in increasing levels of viral proteins over the ensuing 24 h. Yet, a greater level of anti-viral signaling was observed in response to UV-RV, largely reflecting higher induction of type I IFN. That such higher levels of viral proteins in RV-infected cells did not result in higher IFN levels than was seen in cells exposed to UV-RV likely reflects the ability of RV to suppress IFN signaling/expression via NSP1-mediated degradation of IRF 3, 5, and 7 (17, 18). Such ability of RV to suppress what might otherwise be a very robust anti-viral signal in response to the presence of even modest amounts of viral components speaks to the potential of these innate immune signals to limit RV infection.
While a large portion of RV-induced IEC gene expression was dependent upon type I IFN, including numerous genes typically associated with viral infection, RV nonetheless induced numerous genes whose induction did not require type I IFN. Such RV-induced gene expression was not substantially recapitulated by UV-RV suggesting it results from ER stress and/or requires higher levels of viral components than was achieved by treatment with UV-RV. RV replication-dependent induced genes includes several NF-κB regulated pro-inflammatory genes including the neutrophil chemoattractant IL-8, which was one of the genes most highly induced by RV. That RV infection is not typically associated with cellular inflammation makes the role of such chemokine induction somewhat enigmatic. Yet, given that purified bacterial flagellin, which primarily activates NF-κB mediated pro-inflammatory gene expression provided mice with temporary protection against RV infection (32), we speculate that ER stress-mediated NF-κB activation may also be a redundant mechanism by which IEC limit RV infection.
The potent induction of anti-viral signaling observed in response to UV-RV was not mimicked by RV VLPs or RV RNA. The failure of RV VLPs to activate anti-viral signaling could conceivably reflect that RV non-structural proteins, which are not present in VLPs, play a role in activation of innate immunity. However, it seems more likely that IEC detection of RV RNA is required to generate robust anti-viral signaling. The failure of RV-RNA to robustly activate anti-viral signaling likely resulted from lack of significant uptake by IEC. In accordance, IEC uptake and responses to synthetic dsRNA, poly(I:C), required that poly(I:C) be administered at high concentrations and correlated with its uptake (24). In vivo, and perhaps in vitro, free RNA that is not rapidly taken up by IEC would likely be quickly degraded by RNAses. In contrast, surface expression of VP4 on RV, UV-RV, and RV VLPs results in efficient internalization upon encountering the sialic acid residues abundant on the apical surface of gut epithelia (27), enabling intracellular detection of RV dsRNA. The identity of such sialylated proteins, and the question of whether they engage other proteins during RV internalization, remains elusive. Highlighting the difference in the way that free dsRNA would interact with epithelia, we note that in contrast to RV and UV-RV, which preferentially enter IEC and activate anti-viral signaling when applied to the apical surface, uptake of poly(I:C) and subsequent signaling was much greater when administered to the basolateral surface (24). Following RV entry, its RNA could be recognized by endosomal pattern-recognition receptors (PRR) such as TLR3 or by cytosolic detectors of dsRNA such as RIG-I and Mda-5. Protein kinase R (PKR) is known to play a role in RV-induced gene expression but whether PKR is a true dsRNA receptor or is purely a participant in the dsRNA-activated signaling cascade remains unclear.
In conclusion, inactivation of the RV genome does not ablate but rather augments its induction of anti-viral signaling in IEC. These findings parallel observations made by us and others that treatment of epithelial cells with heat-killed bacteria and/or purified bacterial components induce greater activation of innate immune signaling than live bacteria. Such a finding supports the general paradigm that innate immune responses are largely driven by host PRR and do not require viable microbes per se, but rather, anti-microbial responses occur despite the complex mechanisms employed by microbes to suppress them. Consequently, inactivated microbes and/or their components might not only be useful for eliciting adaptive immune responses to protect against future infection, but may also help an infected host overcome the microbial innate immune suppression that can hinder pathogen clearance.
METHODS
Reagents
Rabbit anti-RV sera to purified rhesus rotavirus was prepared as described previously (33). Mab to VP6, clone 6E7, was previously described (34). Antibodies to total and phosphorylated STAT1, IRF3 and PKR were obtained from Cell Signaling Technology (Beverly, MA). Total IRF7 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to human interferon alpha and beta (anti-IFN α/β) were obtained from the National Institute of Allergy and Infectious Diseases (NIAID) Reference Reagent Laboratory through ATCC (Manassas, VA). Anti-β-actin and psoralen AMT (4′-aminomethyl-4,5′, 8-trimethylpsoralen) were purchased from Sigma-Aldrich (St. Louis, MO). The ELISA kit used to assay IFN-β was obtained from PBL Biomedical Laboratories (Piscataway, NJ). IL-8 was assayed via R&D systems Duoset IL-8 reagents (Minneapolis, MN).
Cell culture
Human intestinal epithelial cells (HT29), herein referred to as “IEC,” were cultured as previously described on both standard tissue-culture plastics or collagen-coated permeable supports to result in polarized IEC (35).
Preparation of rotavirus and components
Rhesus RV (RRV) was propagated in MA104 cells as previously described (36). Virus was prepared in bulk, aliquoted, and stored at −80°C until use. To prepare UV-inactivated RV (UV-RV), aliquots of RRV were pre-treated with 40 μg/ml psoralen AMT and then irradiated by long-wave UV-light (365 nm) for two hours as previously described (29). RV virus-like particles (VLPs) were isolated from Sf9 cells that had been infected with baculovirus recombinants expressing cDNAs encoding RV Rf VP2, and SA11 VP4, VP6, and VP7 as previously described (37). RV RNA was extracted from purified virions as previously described (38, 39).
Cell treatment with RV and its components
Prior to infection, RV, UV-RV, RV VLPs and RV RNA were diluted in serum-free medium (SFM) and incubated with 10 μg/ml trypsin (Mediatech, Inc., Manassas, VA), except where indicated otherwise, for 30 min in a 37°C water bath. Control samples were treated with an equivalent amount of trypsin diluted in SFM (Mock) or SFM alone. Where indicated, neutralizing antibodies to human interferon alpha and beta (anti-IFN α/β) (1:100) were added to some preparations of trypsinized RV prior to infection. IEC, HT29 cells, were grown to 90–100% confluence in 6 well plates or collagen-coated permeable supports, washed several times with SFM, and inoculated with virus, viral components, or mock controls for 1 h at 37°C/5% C02 to allow for adsorption. Following adsorption, cells were washed again several times with SFM and then incubated with 2 μg/ml trypsin in SFM for 0–48 hours post-inoculation (hpi). Cells stimulated with RV in the presence of anti-IFN α/β were treated with 2 μg/ml trypsin in SFM plus the same concentration of Type 1 IFN antibodies used during viral adsorption. For experiments comparing RV VLPs and RNA, longer cell stimulation was required to allow enough time for RNA to enter cells; thus, following adsorption these cells were not washed with SFM and instead components were retained in the presence of 2 μg/ml trypsin for up to 24 h. At the indicated time points, supernatants were collected and stored at −20°C for IL-8/IFN-β ELISA. Cells were washed several times with PBS and resuspended in radioimmunoprecipitation assay II buffer (RIPA II) (20mM Tris-HCl, 2.5 mM EDTA, 1% Triton X-100, 10% glycerol, 1% deoxycholate, 0.1 % SDS, 50 mM NaF, 10 mM Na2P2O7, and 2 mMNaV04 plus protease inhibitor cocktail) for western blotting or TRIzol for RNA isolation and subsequent qRT-PCR/microarray analysis.
Immunoblotting
At various time points (0–48 hpi), cells were rinsed several times in PBS, lysed in RIPA II buffer and cleared by centrifugation (10 min at 15, 000 × g, 4°C). Total protein concentrations were estimated by BioRad Protein Assay. Cell lysates were assayed for anti-viral markers (IRF3, IRF7, STAT1 and PKR) by 12% SDS-PAGE immunoblotting and membranes were stripped and probed for β-actin as a loading control. Immunoblots were visualized with the ECL system (GE Healthcare, Piscataway, NJ).
ELISA
Supernatants were collected at different time points (0–48 hpi) and stored at −20°C until use. Human IL-8 secretion in the supernatants was measured by the Duoset kit from R&D systems (Minneapolis, MN). The human IFN-β enzyme immunoassay kit from PBL Biomedical Laboratories (Piscataway, NJ) was used for the quantification of IFN-β in the supernatants, according to the manufacturer’s instructions.
Microarray analyses
Cells were exposed to RV or the indicated component for 24 h, washed several times with PBS, resuspended in TRIzol, subjected to DNase I digestion and purified for RNA by using a commercially available RNeasy Minikit from Qiagen (Valencia, CA). RNA concentration was measured using spectrophotometry and quality was assessed by Agilent BioAnalyzer analysis. Microarray analyses were performed at the Emory Biomarker Microarray Core, where qualified RNA samples were reverse-transcribed, amplified, labeled, and used to probe human HT-12 chips purchased from Affymetrix, Inc. (Santa Clara, CA). Briefly, samples were assayed using a Molecular Devices Gene Pix (4100A) and raw fluorescence readings were processed by an algorithm designed to reduce spurious readouts of gene activation. Microarray data was quantile normalized using freely available scripts written in R (http://R-project.org). Significantly altered genes were identified using SAM (Significance of Analysis of Microarray) analyses and assessed by hierarchal clustering and principle component analysis using Spotfire Decision Site for Functional Genomics software to determine relatedness of gene expression patterns resulting from cell stimulation by RV or the indicated component.
qRT-PCR
At the indicated time points, total RNA was extracted from cells using TRIzol reagent and reverse-transcribed using a commercial kit (TaqMan Reverse Transcription kit; PerkinElmer, Boston, MA) according to the manufacturer’s directions. The RT cDNA reaction products were subjected to quantitative real-time quantitative PCR (qRT-PCR) (SYBR Green PCR Core kit; PerkinElmer, Boston, MA) with primers for IFN-β (Sense 5′-CTCTCCTGTTGTGCTTCTCC-3′, Antisense 5′-GTCAAAGTTCATCCTGTCCTTG-3′), Mda-5 (Sense 5′-TCAGCCAAATCTGGAGAAGG-3′, Antisense 5′-CTTCATCTGAATCACTTCCC-3′), STAT1 (Sense 5′-GTTAGACAAACAGAAAGAGC-3′, Antisense 5′-TCTGTTGTGCAAGGTTTTGC-3′), OAS-2 (Sense 5′-CAACAAATGCTTCCTAGAGC-3′, Antisense 5′-ACGAGATCGGCATCAGAGCC-3′) (Invitrogen; Carlsbad, CA), and 18S ribosomal RNA (Sense 5′-CGGCTACCACATCCAAGGAA-3′, Anti-sense 5′-GCTGGAATTACCGCGGCT-3′) (PerkinElmer; Boston, MA) as previously described (40). Expression level of anti-viral genes was normalized to 18S rRNA levels of the same sample. Fold difference was the ratio of the normalized value of each sample to that of uninfected control cells. All PCR samples were performed in triplicate.
Confocal microscopy
IEC were grown to confluence on collagen-coated permeable supports, apically treated with the indicated stimuli, and fixed in 10% formalin for 15 min. Cells were washed 3X in PBS, permeabilized in 0.5% Triton X-100/PBS for 10 min at RT, and blocked overnight in 3% BSA in PBS (4°C). Cells were incubated for 1 h with polyclonal rabbit anti-RRV (1:10,000) or monoclonal mouse anti-VP6 (1:100) in blocking buffer, washed with PBS 3X, and probed with anti-rabbit and anti-mouse FITC secondary antibody in PBS (1:50) (Jackson ImmunoResearch Laboratories, West Grove, PA), respectively, for 1 h at RT. Alexa-conjugated phalloidin stain (Alexa Fluor 633; Invitrogen, Carlsbad, CA) was used as a counterstain for F-actin and was included in the secondary antibody preparation at a dilution of 1:500. After staining with secondary antibody and phalloidin, cells were washed 3X in PBS and mounted on slides with fluorescent anti-fade medium (VectaShield; Burlingame, CA). Stained cell monolayers were examined using a Zeiss LSM510 laser scanning confocal microscope (Zeiss Microimaging Inc., Thornwood, NY) coupled to a Zeiss 100M axiovert and ×63 or ×100 Pan-Apochromat oil lenses. Fluorescent dyes were imaged sequentially in frame-interlace mode to eliminate cross talk between channels. Images shown are representative of at least 3 experiments, with multiple images taken per slide.
Supplementary Material
Acknowledgments
We thank Drs. Rheinallt Jones and Huxia Wu (Emory University, Atlanta, GA) for their advice and technical assistance. This work was supported by NIH grant AI083420 to AG, AI080656 to MKE, a Digestive Disease Research and Development Center (DDRDC) grant DK06439, and a Digestive Diseases Research Core Center (DDRCC) grant DK66338. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
Abbreviations
- RV
rotavirus
- UV-RV
irradiated rotavirus
- VLPs
virus like particles
- IEC
intestinal epithelial cell
- IRF
Interferon regulatory factor
- PRR
pattern-recognition receptor
- STAT
signal transducer and activator of transcription
- PKR
protein kinase R
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
DISCLOSURE
The authors declared no conflicts of interest.
References
- 1.Glass RI, Parashar UD, Bresee JS, Turcios R, Fischer TK, Widdowson MA, et al. Rotavirus vaccines: current prospects and future challenges. Lancet. 2006 Jul 22;368(9532):323–32. doi: 10.1016/S0140-6736(06)68815-6. [DOI] [PubMed] [Google Scholar]
- 2.Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis. 2003 May;9(5):565–72. doi: 10.3201/eid0905.020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson EJ, Weber SG. Rotavirus infection in adults. Lancet Infect Dis. 2004 Feb;4(2):91–9. doi: 10.1016/S1473-3099(04)00928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009 May;136(6):1939–51. doi: 10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayaram H, Estes MK, Prasad BV. Emerging themes in rotavirus cell entry, genome organization, transcription and replication. Virus Res. 2004 Apr;101(1):67–81. doi: 10.1016/j.virusres.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Riepenhoff-Talty M, Dharakul T, Kowalski E, Michalak S, Ogra PL. Persistent rotavirus infection in mice with severe combined immunodeficiency. J Virol. 1987 Oct;61(10):3345–8. doi: 10.1128/jvi.61.10.3345-3348.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco MA, Greenberg HB. Immunity to rotavirus in T cell deficient mice. Virology. 1997 Nov 24;238(2):169–79. doi: 10.1006/viro.1997.8843. [DOI] [PubMed] [Google Scholar]
- 8.Eiden J, Lederman HM, Vonderfecht S, Yolken R. T-cell-deficient mice display normal recovery from experimental rotavirus infection. J Virol. 1986 Feb;57(2):706–8. doi: 10.1128/jvi.57.2.706-708.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheth R, Anderson J, Sato T, Oh B, Hempson SJ, Rollo E, et al. Rotavirus stimulates IL-8 secretion from cultured epithelial cells. Virology. 1996 Jul 15;221(2):251–9. doi: 10.1006/viro.1996.0374. [DOI] [PubMed] [Google Scholar]
- 10.Casola A, Estes MK, Crawford SE, Ogra PL, Ernst PB, Garofalo RP, et al. Rotavirus infection of cultured intestinal epithelial cells induces secretion of CXC and CC chemokines. Gastroenterology. 1998 May;114(5):947–55. doi: 10.1016/s0016-5085(98)70314-2. [DOI] [PubMed] [Google Scholar]
- 11.Rollo EE, Kumar KP, Reich NC, Cohen J, Angel J, Greenberg HB, et al. The pithelial cell response to rotavirus infection. J Immunol. 1999 Oct 15;163(8):4442–52. [PubMed] [Google Scholar]
- 12.Cuadras MA, Feigelstock DA, An S, Greenberg HB. Gene expression pattern in Caco-2 cells following rotavirus infection. J Virol. 2002 May;76(9):4467–82. doi: 10.1128/JVI.76.9.4467-4482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bass DM. Interferon gamma and interleukin 1, but not interferon alfa, inhibit rotavirus entry into human intestinal cell lines. Gastroenterology. 1997 Jul;113(1):81–9. doi: 10.1016/S0016-5085(97)70083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecce JG, Cummins JM, Richards AB. Treatment of rotavirus infection in neonate and weanling pigs using natural human interferon alpha. Mol Biother. 1990 Dec;2(4):211–6. [PubMed] [Google Scholar]
- 15.Schwers A, Vanden Broecke C, Maenhoudt M, Beduin JM, Werenne J, Pastoret PP. Experimental rotavirus diarrhoea in colostrum-deprived newborn calves: assay of treatment by administration of bacterially produced human interferon (Hu-IFN alpha 2) Ann Rech Vet. 1985;16(3):213–8. [PubMed] [Google Scholar]
- 16.Wang Y, Dennehy PH, Keyserling HL, Tang K, Gentsch JR, Glass RI, et al. Rotavirus infection alters peripheral T-cell homeostasis in children with acute diarrhea. J Virol. 2007 Apr;81(8):3904–12. doi: 10.1128/JVI.01887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barro M, Patton JT. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):4114–9. doi: 10.1073/pnas.0408376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barro M, Patton JT. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J Virol. 2007 May;81(9):4473–81. doi: 10.1128/JVI.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng N, Kim B, Fenaux M, Nguyen H, Vo P, Omary MB, et al. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J Virol. 2008 Aug;82(15):7578–90. doi: 10.1128/JVI.00391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vancott JL, McNeal MM, Choi AH, Ward RL. The role of interferons in rotavirus infections and protection. J Interferon Cytokine Res. 2003 Mar;23(3):163–70. doi: 10.1089/107999003321532501. [DOI] [PubMed] [Google Scholar]
- 21.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006 Mar;13(3):393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 22.Zeng H, Carlson AQ, Guo Y, Yu Y, Collier-Hyams LS, Madara JL, et al. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J Immunol. 2003 Oct 1;171(7):3668–74. doi: 10.4049/jimmunol.171.7.3668. [DOI] [PubMed] [Google Scholar]
- 23.Gewirtz AT. Intestinal epithelial toll-like receptors: to protect. And serve? Curr Pharm Des. 2003;9(1):1–5. doi: 10.2174/1381612033392422. [DOI] [PubMed] [Google Scholar]
- 24.Vijay-Kumar M, Gentsch JR, Kaiser WJ, Borregaard N, Offermann MK, Neish AS, et al. Protein kinase R mediates intestinal epithelial gene remodeling in response to double-stranded RNA and live rotavirus. J Immunol. 2005 May 15;174(10):6322–31. doi: 10.4049/jimmunol.174.10.6322. [DOI] [PubMed] [Google Scholar]
- 25.Estes MK, Graham DY, Mason BB. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981 Sep;39(3):879–88. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewirtz AT, Siber AM, Madara JL, McCormick BA. Orchestration of neutrophil movement by intestinal epithelial cells in response to Salmonella typhimurium can be uncoupled from bacterial internalization. Infect Immun. 1999 Feb;67(2):608–17. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciarlet M, Crawford SE, Estes MK. Differential infection of polarized epithelial cell lines by sialic acid-dependent and sialic acid-independent rotavirus strains. J Virol. 2001 Dec;75(23):11834–50. doi: 10.1128/JVI.75.23.11834-11850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001 Aug 15;167(4):1882–5. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 29.Groene WS, Shaw RD. Psoralen preparation of antigenically intact noninfectious rotavirus particles. J Virol Methods. 1992 Jul;38(1):93–102. doi: 10.1016/0166-0934(92)90172-a. [DOI] [PubMed] [Google Scholar]
- 30.Shaw RD, Hempson SJ, Mackow ER. Rotavirus diarrhea is caused by nonreplicating viral particles. J Virol. 1995 Oct;69(10):5946–50. doi: 10.1128/jvi.69.10.5946-5950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang B, Estes MK, Barone C, Barniak V, O’Neal CM, Ottaiano A, et al. Heterotypic protection from rotavirus infection in mice vaccinated with virus-like particles. Vaccine. 1999 Feb 26;17(7–8):1005–13. doi: 10.1016/s0264-410x(98)00317-x. [DOI] [PubMed] [Google Scholar]
- 32.Vijay-Kumar M, Aitken JD, Sanders CJ, Frias A, Sloane VM, Xu J, et al. Flagellin treatment protects against chemicals, bacteria, viruses, and radiation. J Immunol. 2008 Jun 15;180(12):8280–5. doi: 10.4049/jimmunol.180.12.8280. [DOI] [PubMed] [Google Scholar]
- 33.Gerna G, Sarasini A, Parea M, Arista S, Miranda P, Brussow H, et al. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J Clin Microbiol. 1992 Jan;30(1):9–16. doi: 10.1128/jcm.30.1.9-16.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blutt SE, Matson DO, Crawford SE, Staat MA, Azimi P, Bennett BL, et al. Rotavirus antigenemia in children is associated with viremia. PLoS Med. 2007 Apr;4(4):e121. doi: 10.1371/journal.pmed.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nash S, Parkos C, Nusrat A, Delp C, Madara JL. In vitro model of intestinal crypt abscess. A novel neutrophil-derived secretagogue activity. J Clin Invest. 1991 Apr;87(4):1474–7. doi: 10.1172/JCI115156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw RD, Stoner-Ma DL, Estes MK, Greenberg HB. Specific enzyme-linked immunoassay for rotavirus serotypes 1 and 3. J Clin Microbiol. 1985 Aug;22(2):286–91. doi: 10.1128/jcm.22.2.286-291.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford SE, Labbe M, Cohen J, Burroughs MH, Zhou YJ, Estes MK. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994 Sep;68(9):5945–52. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990 Mar;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992 Jun;30(6):1365–73. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans CO, Young AN, Brown MR, Brat DJ, Parks JS, Neish AS, et al. Novel patterns of gene expression in pituitary adenomas identified by complementary deoxyribonucleic acid microarrays and quantitative reverse transcription-polymerase chain reaction. J Clin Endocrinol Metab. 2001 Jul;86(7):3097–107. doi: 10.1210/jcem.86.7.7616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.