Abstract
DNA polymerase λ (Pol λ) is a novel X-family DNA polymerase that shares 34% sequence identity with DNA polymerase β (Pol β). Pre-steady state kinetic studies have shown that the Pol λ•DNA complex binds both correct and incorrect nucleotides 130-fold tighter on average than the Pol β•DNA complex, although, the base substitution fidelity of both polymerases is 10−4 to 10−5. To better understand Pol λ’s tight nucleotide binding affinity, we created single- and double-substitution mutants of Pol λ to disrupt interactions between active site residues and an incoming nucleotide or a template base. Single-turnover kinetic assays showed that Pol λ binds to an incoming nucleotide via cooperative interactions with active site residues (R386, R420, K422, Y505, F506, A510, and R514). Disrupting protein interactions with an incoming correct or incorrect nucleotide impacted binding with each of the common structural moieties in the following order: triphosphate ≫ base > ribose. In addition, the loss of Watson-Crick hydrogen bonding between the nucleotide and template base led to a moderate increase in the Kd. The fidelity of Pol λ was maintained predominantly by a single residue, R517, which has minor groove interactions with the DNA template.
Keywords: X-family DNA polymerase, pre-steady state kinetics, nucleotide binding, non-natural nucleotide analogs, DNA polymerase fidelity
INTRODUCTION
DNA polymerases, which are organized into the A, B, C, D, X, and Y families,1; 2 have been shown to share a minimal kinetic mechanism for DNA polymerization.3 This minimal mechanism indicates that DNA polymerases discriminate between a correct or incorrect nucleotide (dNTP) substrate predominantly during two steps: dNTP binding affinity to the enzyme•DNA complex and the rate of dNTP incorporation. Interestingly, DNA polymerases exhibit a wide range of binding affinity values for correct and incorrect dNTPs, thereby suggesting that these enzymes employ different discriminatory mechanisms. Using transient state kinetic methods, the equilibrium dissociation constants (Kd) of an incoming nucleotide have been determined for replicative,4; 5; 6; 7; 8; 9 repair,10; 11; 12 and lesion bypass DNA polymerases.13; 14; 15; 16; 17; 18; 19; 20 For example, the discrimination factor for nucleotide binding, defined as the ratio of the equilibrium dissociation constants for incorrect and correct nucleotides (Kd,incorrect/Kd,correct), is 250 on average for an exonuclease-deficient mutant of the replicative human mitochondrial DNA polymerase γ,6; 7 110 on average for the rat DNA repair polymerase β (rPol β),10 and 4 for the human DNA lesion bypass polymerase η.14
Human DNA polymerase λ (Pol λ), an X-family member which shares 34% sequence identity with Pol β,21 has been postulated to play a role in base excision repair (BER), nonhomologous end joining, and V(D)J recombination while the participation of Pol β in BER has been confirmed.22; 23; 24; 25; 26; 27; 28; 29; 30; 31 Intriguingly, Pol λ exhibits tight nucleotide binding affinity for both correct and incorrect dNTPs in which the discrimination factor is 3 on average for nucleotide incorporation into a single-nucleotide gap DNA substrate.12 Despite the varying discrimination factors (110 versus 3) between these two similar DNA repair enzymes, the base substitution fidelity of rPol β and human Pol λ is within the same range: 10−4 to 10−5.10; 12
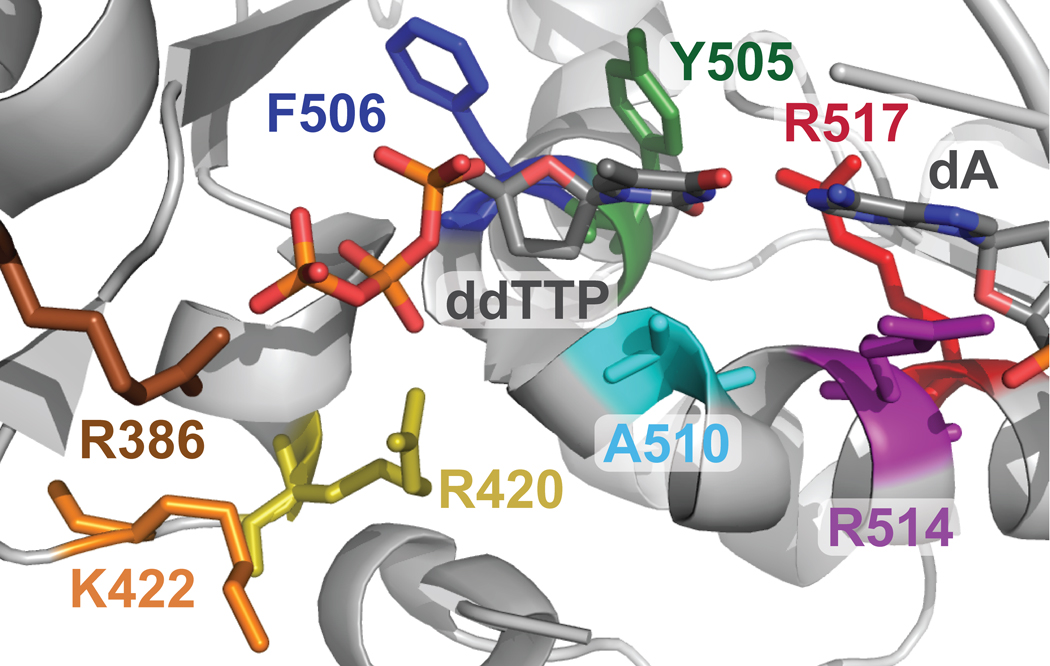
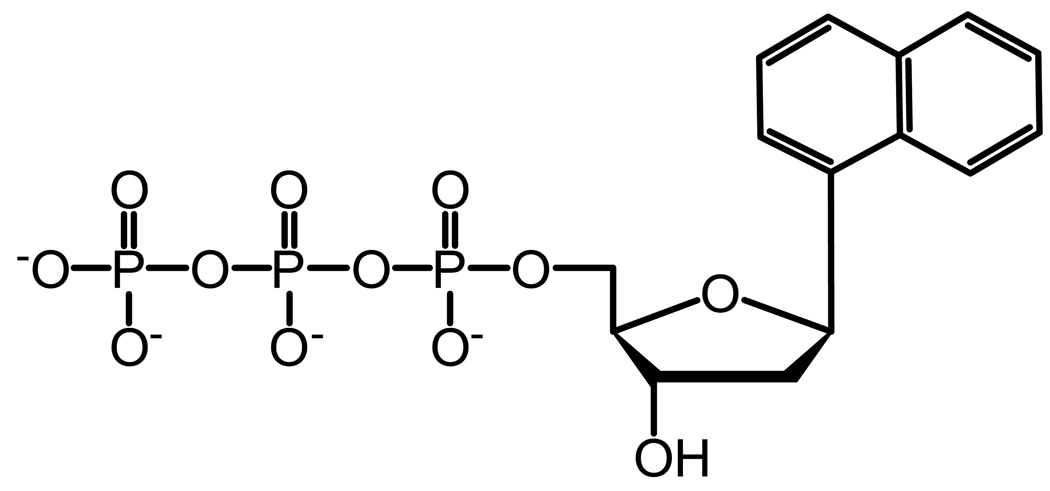
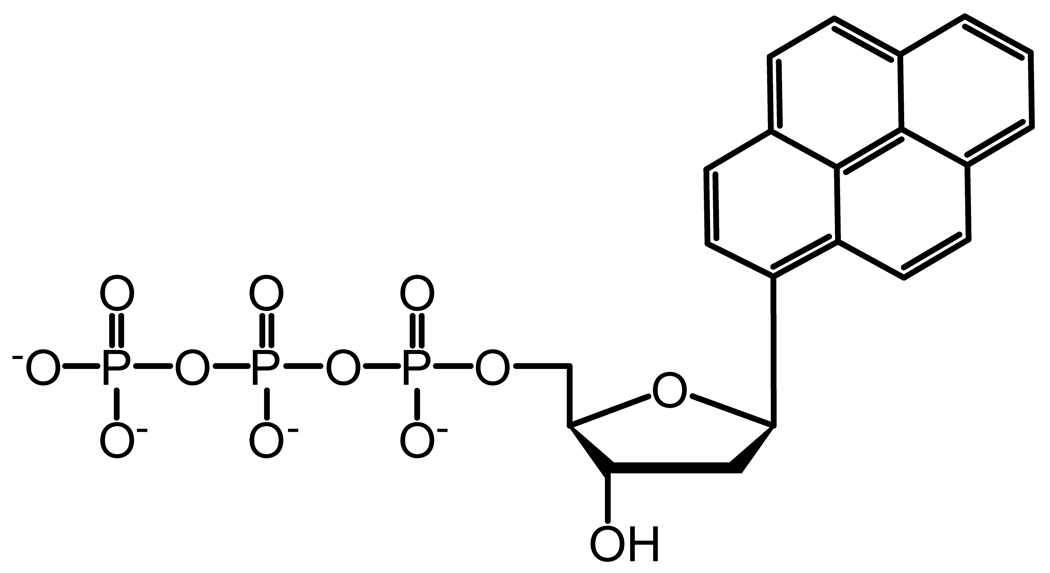
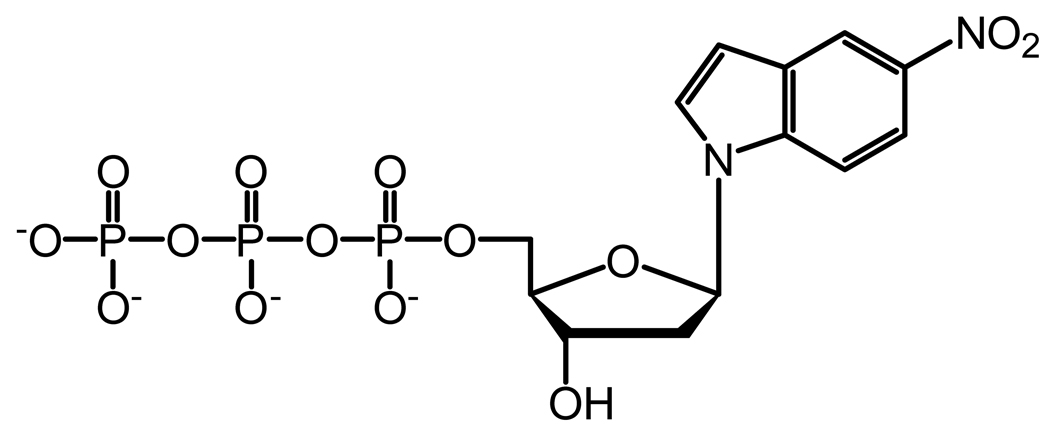
Based on the crystal structure of Pol β, the tight binding of correct nucleotides by Pol β in the presence of single-nucleotide gap DNA is predicted to be partly due to the contribution of its 5′-deoxyribose-5-phosphate lyase (dRPase) domain which interacts strongly with the downstream primer and the polymerase domain.32 This reasoning does not apply to Pol λ because all nucleotides have similarly high affinity to Pol λ, even with a non-gapped DNA substrate.33 Additionally, the N-terminal BRCT and Proline-rich domains of Pol λ are not responsible because the C-terminal Pol β-like domain34 binds to all nucleotides as tightly as Pol λ.12 To better understand the kinetic basis for the tight binding of all nucleotides, we have created site-specific mutants of Pol λ that are designed to target key interactions between the polymerase and its substrates (Figure 1). Furthermore, we investigated the kinetic contributions from the intrinsic properties of a nucleotide, such as hydrogen bonding, base stacking, and steric bulkiness, using three non-natural nucleotide analogs (Figure 2): 1-naphthalene 5′-triphosphate (1-dNaTP), pyrene 5′-triphosphate (dPTP), and 5-nitroindole 5′-triphosphate (5-dNITP).
Figure 1.
Active site of truncated Pol λ. Residues selected for mutagenesis and kinetic characterization are shown as sticks: R386 (brown), R420 (yellow), K422 (orange), F506 (dark blue), Y505 (green), A510 (cyan), R514 (purple), and R517 (red) (PDB 1XSN). The incoming ddTTP and DNA template base dA are shown as gray sticks with the important atoms in color. The remainder of the protein and DNA substrate is a solid, light gray ribbon.
Figure 2.
Chemical structure of nucleotide analogs. (A) 1-dNaTP, (B) dPTP, and (C) 5-dNITP.
RESULTS
The kinetic basis of Pol λ’s tight nucleotide binding was examined by creating several single-point mutants which disrupt specific interactions between each of the amino acid residues and the incoming dNTP or the template base. Based on an X-ray crystal structure of human Pol λ’s C-terminal Pol β -like domain in complex with gap DNA and a correct incoming ddTTP,35 the following amino acid residues were examined: R386, R420, K422, Y505, F506, A510, R514, and R517 (Figure 1). These amino acids are in close proximity to the triphosphate moiety (R386, R420, and K422), sugar moiety (Y505 and F506), or base of an incoming dNTP (A510) and the DNA template (R514, and R517) (Figure 1). Most of these residues are conserved in both Pol λ and Pol β. However, K422, A510, and R514 in Pol λ are A185, D276, and K280 in Pol β, respectively. Following the successful completion of site-directed mutagenesis and purification of the Pol λ mutants to greater than 90% purity (Figure S1), we examined whether the structure was altered due to the mutation(s). Therefore, we employed circular dichroism (CD) spectroscopy (see Materials and Methods). These data showed that all of the tested mutants had secondary structure similar to wild-type (WT) Pol λ which suggested that the mutations did not induce major structural changes (Figure S2). Pol λ R386E, R420A, and Y505G were not examined using CD spectroscopy due to their low purification yields.
Measurement of DNA binding affinity
Next, we determined whether the mutants were able to form a binary complex (Pol λ•DNA) by using a fluorescence titration assay (see Materials and Methods) to measure the equilibrium dissociation constant (KdDNA). The F-DNA substrate contains 2-aminopurine, an analog of adenine, at the template base position (Table 1). Increasing concentrations of Pol λ were titrated into F-DNA, and then a plot of the corrected 2-aminopurine intensity and concentration of Pol λ were fit to Eq. 1 which resolved a KdDNA of 110 ± 20 nM (Figure S3 and Table 2). Similar binding constants (70 – 300 nM) were measured for the Pol λ mutants, indicating that these enzymes have a folded structure that can recognize and bind a single-nucleotide gap DNA substrate similar to WT Pol λ.
Table 1.
DNA substrates
| F-DNAa | 5′–CGCAGCCGTCCAACCAACTCAG GTCGATCCAATGCCGTCC–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTCFCAGCTAGGTTACGGCAGG–5′ | |
| D-DNAb | 5′–CGCAGCCGTCCAACCAACTCA CGTCGATCCAATGCCGTCC–3′ |
| 3′–GCGTCGGCAGGTTGGTTGAGTXGCAGCTAGGTTACGGCAGG–5′ |
The ‘F’ in the 41mer template represents 2-aminopurine. The downstream 18-mer is 5′-phosphorylated.
The ‘X’ in the 41mer template represents dA for D-1 DNA and dT for D-7 DNA. The downstream 19-mer is 5′-phosphorylated.
Table 2.
Equilibrium dissociation constants of the Pol λ•DNA complex at 37 °C.
| Enzyme | KdDNA (nM) |
|---|---|
| WT | 110 ± 20 |
| R386A | 90 ± 20 |
| R386E | 300 ± 100 |
| R420A | 200 ± 100 |
| K422A | 150 ± 40 |
| Y505G | 150 ± 70 |
| Y505A | 150 ± 50 |
| F506A | 140 ± 40 |
| A510E | 120 ± 60 |
| R514A | 70 ± 10 |
| R517A | 80 ± 30 |
| R386A/A510E | 120 ± 40 |
| R386A/R514A | 80 ± 30 |
Measurement of nucleotide binding and incorporation rate catalyzed by Pol λ mutants
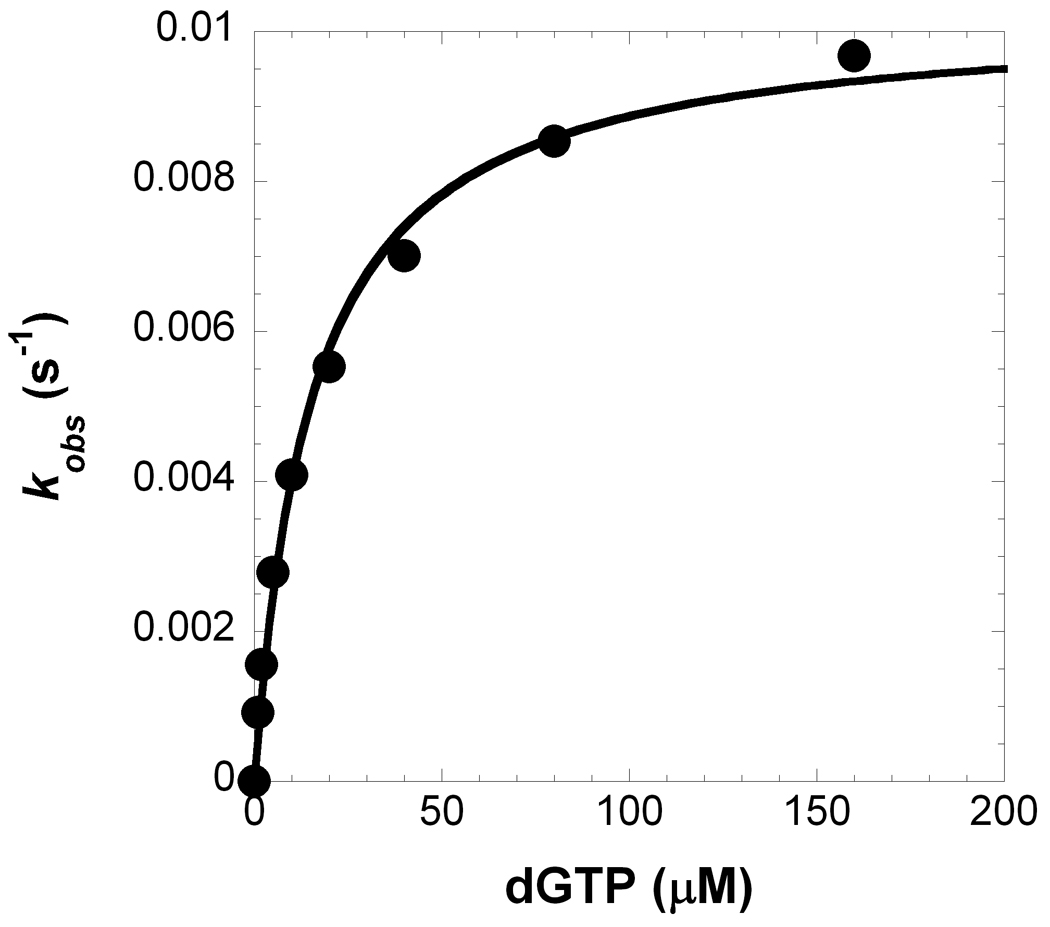
The single-point Pol λ mutants were kinetically characterized by measuring the maximum rate of nucleotide incorporation (kp) and the equilibrium dissociation constant (Kd) of an incoming nucleotide under single-turnover conditions. Single-turnover conditions prevent any complications from the steady-state phase that may originate from the rate of DNA dissociation being comparable to the rate of nucleotide incorporation for Pol λ .12; 34; 36 Recent studies with T7 DNA polymerase, an A-family enzyme, reveal that nucleotide binding to a polymerase complex with DNA induces several conformational changes preceding the incorporation step which may mask the measured Kd value under single-turnover reaction conditions.37 Since similar conformational changes have not been detected with Pol λ during nucleotide binding and incorporation35; 38 and the kinetic mechanisms of Pol λ with gap DNA and T7 DNA polymerase with non-gap DNA may be different,3; 39 the Kd values measured in this paper likely reflect the true nucleotide binding affinity. To perform the single-turnover kinetic assay (see Materials and Methods), a pre-incubated solution of Pol λ A510E and 5′-[32P]-labeled D-1 DNA (Table 1) was reacted with increasing concentrations of mismatched dGTP (1–160 µM). After quantitating DNA product formation at each nucleotide concentration (Figure 3), the pre-steady state kinetic parameters were determined: a kp of 0.0102 ± 0.0003 s−1 and a Kd of 15 ± 2 µM for Pol λ A510E. The pre-steady state kinetic parameters for all Pol λ mutants incorporating incorrect dGTP (Table 3) or correct dTTP (Table 4) into single-nucleotide gap D-1 DNA (Table 1) were determined using this single-turnover kinetic methodology. The kp and/or Kd values were then used to calculate the substrate specificity constant (kp/Kd), Kd ratio, and fidelity as defined in Tables 3 and 4. In this study, fidelity describes the probability of Pol λ making a base substitution error for the dGTP:dA mismatch. Our previous work determined the kinetic parameters for WT Pol λ.12
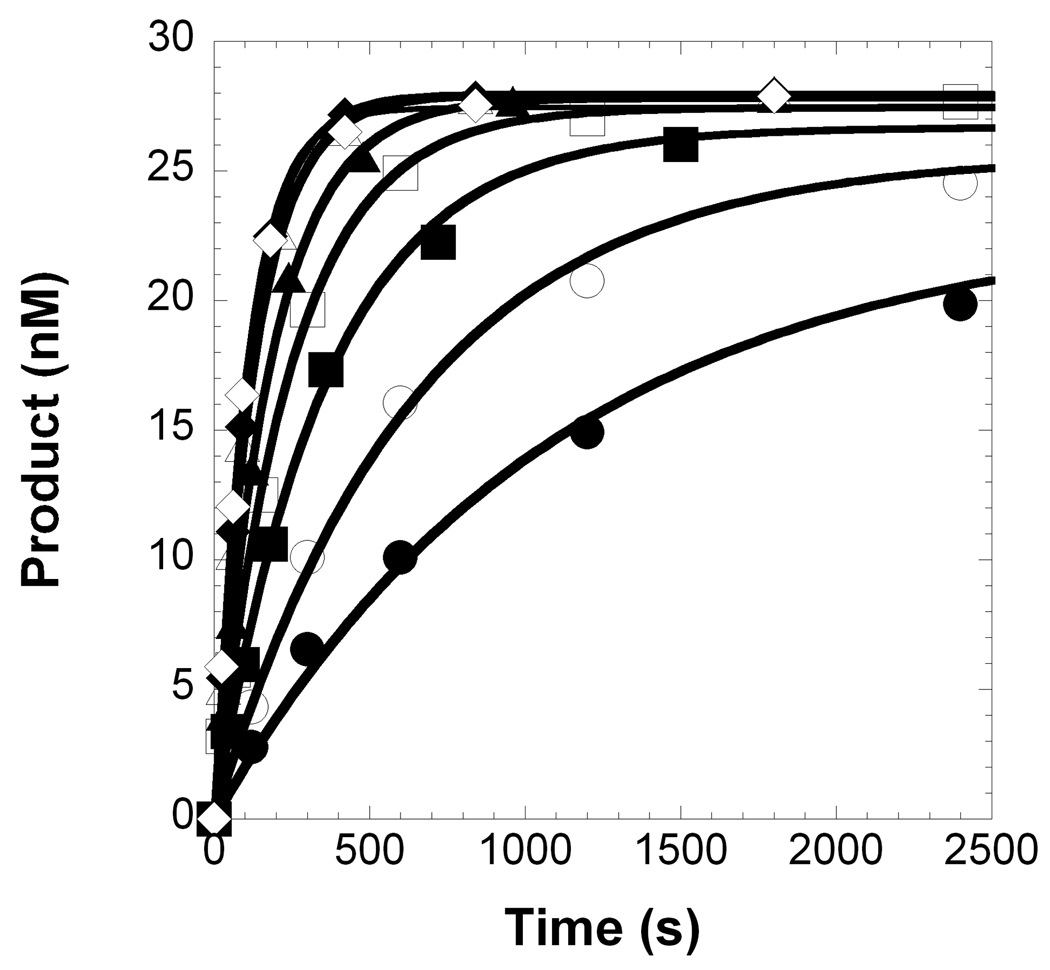
Figure 3.
Concentration dependence on the pre-steady state rate constant of nucleotide misincorporation. (A) A pre-incubated solution of Pol λ A510E (300 nM) and 5′-[32P]-labeled D-1 DNA (30 nM) was rapidly mixed with increasing concentrations of dGTP•Mg2+ (1 µM, ●; 2 µM, ○; 5 µM, ■; 10 µM, □; 20 µM, ▲; 40 µM, Δ; 80 µM, ◆; 160 µM, ◇) for various time intervals. The solid lines are the best fits to a single-exponential equation which determined the observed rate constants, kobs. (B) The kobs values were plotted as a function of dGTP concentrations. The data (●) were then fit to a hyperbolic equation, yielding a kp of 0.0102 ± 0.0003 s−1 and an apparent Kd of 15 ± 2 µM.
Table 3.
Kinetic parameters for incorrect nucleotide incorporation (dGTP) into single-nucleotide gapped DNA (D-1) catalyzed by human Pol λ at 37 °C.
| Pol λ mutant |
kp (s−1) |
Kd (µM) |
kp/Kd (µM−1s−1) |
Kd ratioa |
Fidelityb |
|---|---|---|---|---|---|
| WTc | (4.0 ± 0.2) × 10−4 |
3.2 ± 0.5 | 1.3 × 10−4 | - | 8.3 × 10−5 |
| R386A | (1.55 ± 0.06) × 10−3 |
50 ± 10 | 3.1 × 10−5 | 16 | 3.6 × 10−4 |
| R386E | No observed incorporation | ||||
| R420A | (1.1 ± 0.2) × 10−4 |
800 ± 200 | 1.4 × 10−7 | 250 | 8.1 × 10−3 |
| K422A | (6.3 ± 0.3) × 10−4 |
8 ± 1 | 7.9 × 10−5 | 3 | 3.4 × 10−4 |
| Y505G | (1.23 ± 0.07) × 10−3 |
60 ± 10 | 2.1 × 10−5 | 19 | 2.0 × 10−5 |
| Y505A | (7.5 ± 0.2) × 10−4 |
30 ± 3 | 2.5 × 10−5 | 9 | 1.9 × 10−5 |
| F506A | (2.9 ± 0.3) × 10−4 |
19 ± 6 | 1.5 × 10−5 | 6 | 6.0 × 10−6 |
| A510E | (1.02 ± 0.03) × 10−2 |
15 ± 2 | 6.8 × 10−4 | 5 | 1.4 × 10−3 |
| R514A | (8.2 ± 0.8) × 10−4 |
80 ± 20 | 1.0 × 10−5 | 25 | 1.1 × 10−5 |
| R517A | (1.5 ± 0.1) × 10−3 |
1.0 ± 0.3 | 1.5 × 10−3 | 0.3 | 1.1 × 10−1 |
| R386A/A510E | (6.9 ± 0.6) × 10−3 |
170 ± 40 | 4.1 × 10−5 | 53 | 2.3 × 10−3 |
| R386A/R514A | (7 ± 1) × 10 4 |
320 ± 90 | 2.2 × 10−6 | 100 | 6.4 × 10−5 |
Calculated as (Kd) Mutant/( Kd )WT.
Calculated as (kp/Kd)Incorrect/[(kp/Kd)Correct + (kp/Kd)Incorrect].
The kinetic parameters for WT Pol λ are from the work of Fiala et al.12
Table 4.
Kinetic parameters for correct nucleotide incorporation (dTTP) into single-nucleotide gap DNA (D-1) catalyzed by human Pol λ at 37 °C.
| Pol λ mutant |
kp (s−1)a |
Kd (µM) |
kp/Kd (µM−1s−1) |
Kd Ratioa |
|---|---|---|---|---|
| WTb | 3.9 ± 0.2 | 2.6 ± 0.4 | 1.5 | - |
| R386Ac | 1.3 ± 0.1 | 15 ± 5 | 8.7 × 10−2 | 6 |
| R386Ec | 0.005 ± 0.001 | 1000 ± 410 | 5.0 × 10−6 | 380 |
| R420A | 0.0119 ± 0.0005 | 710 ± 60 | 1.7 × 10−5 | 270 |
| K422A | 1.62 ± 0.08 | 7 ± 1 | 2.3 × 10−1 | 3 |
| Y505Gd | 0.85 ± 0.02 | 0.81 ± 0.09 | 1.0 | 0.3 |
| Y505Ad | 1.30 ± 0.03 | 1.0 ± 0.1 | 1.3 | 0.4 |
| F506Ad | 7.1 ± 0.2 | 2.8 ± 0.4 | 2.5 | 1 |
| A510E | 10.2 ± 0.8 | 21 ± 4 | 4.9 × 10−1 | 8 |
| R514A | 4.0 ± 0.1 | 4.1 ± 0.7 | 9.8 × 10−1 | 2 |
| R517A | 0.0153 ± 0.0004 | 1.3 ± 0.1 | 1.2 × 10−2 | 0.5 |
| R386A/A510E | 3.0 ± 0.2 | 170 ± 20 | 1.8 × 10−2 | 65 |
| R386A/R514A | 2.4 ± 0.1 | 70 ± 10 | 3.4 × 10−2 | 27 |
Pol λ mutants targeting interactions near the triphosphate moiety of an incoming dNTP
Pol λ single-point mutants of residues R386, R420, and K422 demonstrated a kinetically significant (i.e. >2-fold relative to WT) weaker binding affinity (1/Kd) for both correct dTTP and incorrect dGTP incorporation opposite a template base dA (Tables 3 and 4). The Kd value increased dramatically (250- and 270-fold) for Pol λ R420A, a residue situated near the β-phosphate of an incoming dNTP (Figure 1). In contrast, the neutrally-charged R386A mutant, which is located near the γ-phosphate of an incoming dNTP, had a more mild effect, as the binding was weakened by a modest 6- and 16-fold for a correct and incorrect dNTP, respectively. K422 is another positively-charged residue in the vicinity of the triphosphate moiety, although, the alanine substitution mutant revealed a mild, 3-fold weaker binding affinity for both dTTP and dGTP. Interestingly, the fidelity of Pol λ R420A dropped by almost two orders of magnitude (Table 3 and Figure 4). This effect was mostly due to a large decrease in the rate for dTTP incorporation (330-fold) while the rate for dGTP misincorporation (∼4-fold) remained similar to WT Pol λ.
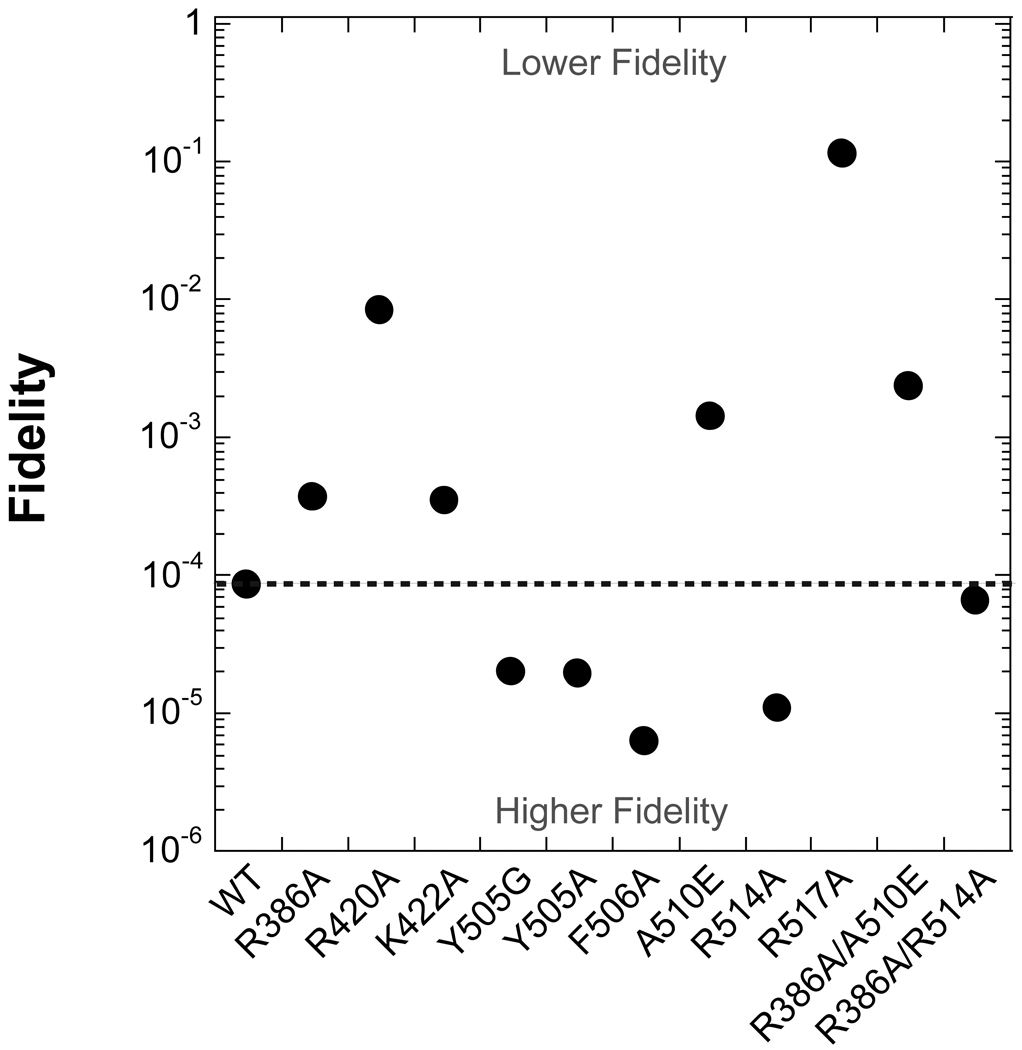
Figure 4.
Fidelity of mutant enzymes versus WT. The base substitution fidelity calculated in Table 3 is plotted for each enzyme. The dashed line coincides with the fidelity of WT Pol λ. Thus, mutants with a calculated fidelity value above the dashed line exhibit lower polymerase fidelity than WT Pol λ.
Pol λ mutants with altered interactions near the ribose of an incoming dNTP
For a misincorporation, the binding affinity of dGTP was weakened by 19-, 9-, and 6-fold for Y505G, Y505A, and F506A, respectively (Table 3). As a result, these mutants displayed a consistent increase in enzyme fidelity (4- to 13-fold) among the mutant enzymes examined in this work (Table 3 and Figure 4).
Pol λ mutants which disrupt interactions with bases of an incoming dNTP or DNA template
Replacing A510 with a glutamate resulted in modest 3- to 5-fold perturbations for the dTTP and dGTP Kd values (Tables 3 and 4). Additionally, the R517A mutant exhibited 2- to 3-fold tighter binding affinities for both dTTP and dGTP relative to WT Pol λ. However, the dTTP and dGTP Kd values for R514A were weakened by 2- and 25-fold, respectively. The fidelity of A510E dropped by approximately 17-fold, and this was caused by a 26-fold rate increase for a misincorporation (Table 3). In contrast, the fidelity of R517A dropped by three orders of magnitude which was due to a 250-fold rate decrease for a correct incorporation.
Kinetic characterization of double-point Pol λ mutants
Our above results suggested that the tight dNTP binding by Pol λ is a function of multiple amino acid residues. To examine if these residues behave individually or cooperatively, we created two double mutants: R386A/A510E and R386A/R514A. These three residues were selected because the Kd ratios for correct and incorrect dNTPs were modest so that, if binding was weaker for the double mutant, we could accurately measure the Kd. After determining the kp and Kd values under single-turnover conditions, the correct and incorrect Kd ratios for the double mutants ranged from 27- to 100-fold relative to WT Pol λ (Tables 3 and 4). Interestingly, the sum of the binding affinities for the single-substitution mutants did not equal the binding affinities for the double-substitution mutants. Thus, these dNTP binding data indicated that nucleotide binding was synergistic or cooperative for these sites tested. Similar to A510E, the fidelity for R386A/A510E dropped by 26-fold. This loss of fidelity was mostly due to a 17-fold increase in the rate of dGTP misincorporation. In contrast, the fidelity for R386A/R514A was similar to WT Pol λ but slightly higher than the single-point mutant R386A and lower for R514A (Table 3 and Figure 4), suggesting that the effects of these two point mutations were either offset or compensated by repositioning other active site residues.
Effects of hydrogen bonding, base stacking, and steric interactions during DNA synthesis
Nucleotide binding and selection may be influenced by the properties of the substrates, such as Watson-Crick hydrogen bonding, base stacking, steric interactions, and hydrogen bonds between the enzyme and DNA minor groove. To discern the role of these factors, we have determined the pre-steady state kinetic parameters (Table 5) for the incorporation of three non-natural nucleotide analogs (Figure 2) into D-7 DNA, a single-nucleotide gap DNA substrate with dT as the template (Table 1). This allows us to compare the kinetic parameters for the analogs with dATP, the nucleotide with the strongest base-stacking energy among the four natural bases.40
Table 5.
Kinetic parameters for non-natural nucleotide analog incorporation into single-nucleotide gap D-7 DNA catalyzed by WT human Pol λ at 37 °C.
| dNTP |
kp (s−1)a |
Kd (µM) |
kp/Kd (µM−1s−1) |
Kd Ratioa |
|---|---|---|---|---|
| dATPb | 1.5 ± 0.18 | 0.9 ± 0.3 | 1.7 | |
| dGTPb | (1.0 ± 0.2) × 10−2 | 8.4 ± 0.6 | 1.4 × 10−3 | 8 |
| dTTPb | (2 ± 1) × 10−4 | 7 ± 4 | 1.4 × 10−3 | 8 |
| dCTPb | (1.0 ± 0.2) × 10−2 | 7 ± 4 | 1.4 × 10−3 | 8 |
| dPTP | (3.3 ± 0.2) × 10−4 | 22 ± 2 | 1.5 × 10−5 | 24 |
| 1-dNaTP | (3.6 ± 0.3) × 10−4 | 11 ± 3 | 3.3 × 10−5 | 12 |
| 5-dNITP | (1.31 ± 0.04) × 10−2 | 1.5 ± 0.2 | 8.7 × 10−3 | 2 |
Calculated as (Kd )Analog/(Kd )dATP.
The kinetic parameters for natural dNTPs are from the work of Fiala et al.12
The three non-natural nucleotide analogs (dPTP, 5-dNITP, and 1-dNaTP) lack the ability to form hydrogen bonds with a template base, possess stronger base stacking energy, and increased steric bulkiness compared to a natural nucleotide.40 Nonetheless, WT Pol λ inserted dPTP, 5-dNITP, and 1-dNaTP, although, the incorporation efficiency was reduced significantly, mostly due to a slower rate of incorporation rather than nucleotide binding. Compared to dATP, the nucleotide binding was weakened by 24-, 12-, and 2-fold for dPTP, 1-dNaTP, and 5-dNITP, respectively. Overall, these results suggested that (i) Watson-Crick hydrogen bonding between the incoming nucleotide and template base is not essential for nucleotides with strong base-stacking energy but the hydrogen bonds likely enhance incorporation efficiency, (ii) the ground-state binding affinity of a nucleotide depends moderately on the formation of Watson-Crick hydrogen bonds, and (iii) the active site of WT Pol λ can accommodate an oversized base, although, the bulky pyrene moiety likely leads to a higher Kd.
DISCUSSION
Tight nucleotide binding depends partially on the properties of nascent base pair
We have previously demonstrated that all dNTPs, correct and incorrect, have tight and peculiarly similar binding affinities to Pol λ and single-nucleotide gap DNA.12 The biological relevance of tight nucleotide binding affinity likely facilitates efficient DNA repair synthesis by Pol λ, such as nonhomologous end joining, when cellular dNTP pools are low outside of S phase.34; 41 The tight binding of all dNTPs is rare because most kinetically characterized DNA polymerases generally discriminate against incorrect nucleotides via weak binding and slow incorporation.42 For example, incorrect dNTPs have 35- to 340-fold weaker binding affinity than correct dNTPs to the binary complex of Pol β and single-nucleotide gap DNA.10 To identify the major factors contributing to Pol λ’s tight nucleotide binding, we examined the physical properties of nascent base pairs including hydrogen bonding, base stacking, and base size. Previously, the importance of those factors for other DNA polymerases has been probed using non-natural nucleobases (e.g. difluorotoluene and pyrene) substituted in an incoming dNTP or a DNA template.43 The A- and Y-family DNA polymerases incorporate non-hydrogen bonding nucleotide analogs, albeit less efficiently than a correct, natural dNTP based on pre-steady state kinetic results.44; 45; 46 These findings suggested that Watson-Crick hydrogen bonds formed between a dNTP and a template base are not essential for nucleotide incorporation by the A- and Y-family DNA polymerases.43; 47; 48 Our kinetic data in Table 5 for the three nucleotide analogs (5-dNITP, dPTP, and 1-dNaTP) suggested that human Pol λ followed the same trend. Moreover, the moderately weaker binding affinities of dPTP, 5-dNITP, and 1-dNaTP suggested that Watson-Crick hydrogen bonds between canonical base pairs were not important for the tight binding of a nucleotide to Pol λ•DNA, but formation of these hydrogen bonds greatly accelerated the catalytic rate of Pol λ. Although base stacking contributed to the 7-fold tighter binding of 5-dNITP over 1-dNaTP, it is not the major factor to dictate nucleotide binding because correct and incorrect dNTPs exhibit tight binding affinities regardless of the base identity, e.g. purine versus pyrimidine.12
Active site residues coordinate unprecedentedly tight nucleotide binding
The X-ray crystal structure of truncated human Pol λ35 was used to rationally select residues to mutate and to determine the individual roles of eight active site residues (R386, R420, K422, Y505, F506, A510, R514, and R517) which interact with an incoming nucleotide or the template base (Figure 1). The common structural moieties between correct and incorrect nucleotides are triphosphate, ribose, and nucleobase. The Pol λ residues interact with specific structural moieties of a dNTP, therefore, the kinetic significance of binding each dNTP moiety can be dissected by averaging the Kd values for both correct and incorrect dNTPs using the mutants (Tables 3 and 4) that interact with the triphosphate (R386A, R420A, and K422A), ribose (Y505A and F506A), or nucleobase (A510E). The impact on dNTP binding generated the following ranking for each of the common structural moieties: triphosphate (Kd ,average = 265 µM) ≫ nitrogenous base (Kd,average = 18 µM) > ribose (Kd,average = 13 µM). Stabilizing the triphosphate group of an incoming nucleotide by R386, R420, and K422 was critical for tight nucleotide binding. For example, dramatic changes in nucleotide binding were observed when R386A or R386E disrupted the salt bridge between the guanidino group of R386 and the γ-phosphate of an incoming nucleotide.35; 38 Similarly, the electrostatic interactions between R420 and the β-phosphate of an incoming nucleotide maintain a low Kd. In comparison, the alanine mutations of Y505 and F506, which interact with the ribose of an incoming nucleotide,35 enhanced the ability of Pol λ to discriminate between correct and incorrect dNTPs by increasing the Kd value of an incorrect nucleotide by 6-to 9-fold (Tables 3 and 4). A510 of Pol λ is one of the few active site residues not conserved in Pol β. Previously, we34 and others41; 49 have proposed that Pol λ’s tight dNTP binding affinity may be due to a neutral residue A510 which is D276 in Pol β. Interestingly, the A510E substitution weakened the dNTP binding affinity of Pol λ by 5- to 8-fold (Tables 3 and 4) while mutating D276 to a neutral residue (e.g. valine and glycine) increases the correct nucleotide binding affinity of Pol β by 4- to 9-fold and incorrect nucleotide binding affinity by 3-fold.42; 50 Thus, a bulky, negatively-charged residue weakens the nucleotide binding affinity for both Pol λ A510E and Pol β. R514, which stacks against the template base dA, is another residue that enhanced the binding discrimination factor, for R514A weakened the binding of correct and incorrect nucleotides by 2- and 25-fold, respectively (Tables 3 and 4).
Several of the active site residues (R386, R420, K422, Y505, F506, A510, and R514) in Pol λ interact with a nascent base pair to cooperatively stabilize nucleotide binding in the ground state. Since the Kd values for the double-point mutants (R386A/A510E and R386A/R514A) were greater than the sum of the Kd values for the single-point mutants by 2- to 5-fold (Tables 3 and 4), these residues contribute to nucleotide binding in a synergistic manner. However, certain residues, e.g. R420, play a more important role in nucleotide binding than others. It is possible that other active site residues, which we did not examine here, may also participate in nucleotide binding. Importantly, the tight nucleotide binding affinity of Pol λ was not manifested in a single amino acid residue, indicating the mechanism of how Pol λ and Pol β discriminate at the ground-state nucleotide binding level is more complex. The different conformational dynamics of these enzymes preceding catalysis may play a role. For example, Pol β undergoes a conformational change during the binary (Pol β•DNA) to ternary (Pol β•DNA•dNTP) complex transition32; 51; 52 where as Pol λ only repositions selected active site residues.35; 38 Furthermore, the conformational change observed for correct and incorrect nucleotide incorporations is different for Pol β.32; 53 Thus, additional studies are needed to better understand the relationship between substrate recognition and polymerase conformational dynamics during the catalytic cycle.
Fidelity of Polλ is maintained by minor groove interactions
Among ten single-substitution Pol λ mutants, this work identified R517 as an important residue in maintaining polymerase fidelity. R517, which is R283 in Pol β, is involved in minor groove interactions with the DNA template (Figure 1).35 The R517A mutation did not significantly alter nucleotide binding affinity but dramatically decreased the kp value of correct dTTP by 250-fold, leading to a 1,360-fold loss of polymerase fidelity (Tables 3 and 4). The Pol β R283A mutant followed a similar kinetic trend as Pol λ R517A, whereby the rate of a correct dTTP incorporation dropped by 310-fold for an overall 260-fold loss of fidelity.54 Crystal structures have been solved for human Pol β R283A and truncated Pol λ R517A; both of these studies conclude that these arginine residues are essential for properly positioning the template strand through hydrogen bonding and/or van der Waal’s interactions.55; 56 Thus, these interactions provide Pol β and Pol λ with greater polymerization efficiency for correct incorporations.12; 54 In the absence of a large conformational change,35; 38 Pol λ relies, in part, on non-specific minor groove interactions to detect the proper nascent base pair geometry in a manner similar to that observed for A- and B-family DNA polymerases.57; 58; 59; 60
MATERIALS AND METHODS
Materials
These chemicals were purchased from the following companies: [γ-32P]ATP, MP Biomedicals; deoxyribonucleotides-5′-triphosphates (dNTP), GE Healthcare; 5-nitroindole 5′-triphosphate, TriLink Biotechnologies; Biospin columns, Bio-Rad Laboratories; OptiKinase™, USB Corporation; Quikchange XL site-directed mutagenesis kit, Stratagene; synthetic oligodeoxyribonucleotides, Integrated DNA Technologies. The mutagenesis, expression, and purification steps of full-length wild-type Pol λ (1–575) and the full-length mutants were performed as described previously.12; 61 The purity of the enzymes was above 90% (Figure S1). CD spectroscopic studies were performed for the Pol λ mutants as described previously.61 Pyrene 5′-triphosphate was synthesized as described previously.62 1-naphthalene 5′-triphosphate was synthesized according to the literature63 except that the epimerization followed a different procedure64 as did the formation of the triphosphate.65
DNA binding assay
The equilibrium dissociation constant (KdDNA) of the Pol λ•DNA binary complex was measured using a fluorescence titration assay. Increasing concentrations of Pol λ (10 – 2000 nM) were titrated into a fixed concentration of F-DNA (100 nM) in buffer L (50 mM Tris-Cl pH 8.4 at 37 °C, 5 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, and 10% glycerol). The F-DNA substrate (Table 1) was excited at a wavelength of 312 nm with emission and excitation slit widths of 5 nm. The emission spectra were collected at 1 nm intervals from 320 to 500 nm using a Fluoromax-4 (Jobin Jvon Horiba). Emission background from the buffer and intrinsic protein fluorescence were subtracted from each spectrum. A modified form of the quadratic equation (Eq. 1) was applied to a plot of the fluorescence intensity (F) measured at 370 nm versus enzyme concentration. Fmax and Fmin represent the maximum and minimum fluorescence intensity, respectively.
| (1) |
Measurement of the kp and Kd for single nucleotide incorporation
Kinetic assays were completed using optimized buffer L (50 mM Tris-Cl pH 8.4 at 37 °C, 5 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 10% glycerol, and 0.1 mg/ml BSA) as previously described.34 The purification, 5′-[32P] radiolabeling, and annealing of single-nucleotide gapped D-DNA substrates (Table 1) was performed as described previously.12; 34 All kinetic experiments described herein were performed at 37 °C and the reported concentrations are final after mixing all the components. A pre-incubated solution containing wild-type Pol λ (120 nM) or a Pol λ mutant (300 nM) and a single-nucleotide gap DNA substrate (30 nM) was mixed with increasing concentrations of nucleotide (0.25–1500 µM) in buffer L at 37 °C. Aliquots of the reaction mixtures were quenched at various times using 0.37 M EDTA. A rapid chemical-quench flow apparatus (KinTek) was utilized for fast nucleotide incorporations. Reaction products were resolved using sequencing gel electrophoresis (17% acrylamide, 8 M urea) and quantitated with a Typhoon TRIO (GE Healthcare) and ImageQuant software (Molecular Dynamics). The time course of product formation at each nucleotide concentration was fit to a single-exponential equation (Eq. 2) using a nonlinear regression program KaleidaGraph (Synergy Software) to yield an observed rate constant of nucleotide incorporation (kobs). The kobs values were then plotted as a function of nucleotide concentration and fit using the hyperbolic equation (Eq. 3) which resolved the kp and Kd values for nucleotide incorporation catalyzed by wild-type Pol λ or a mutant.
| (2) |
| (3) |
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health through Grants GM079403 (to Z.S.) and CA040463 (to Z.S. and J.T.). J.A.B. was supported by an American Heart Association Predoctoral Fellowship (Grant 0815382D) and an International P.E.O. Scholar Award. L.R.P. and S.A.N. were supported from a supplemental grant of the National Science Foundation Career Award (Grant MCB-0447899 to Z.S.). S.M.S. was supported by the National Institutes of Health Chemistry and Biology Interface Program at The Ohio State University (Grant T32-GM008512-13). J.D.F. was supported by an American Heart Association Predoctoral Fellowship (Grant 0615091B).
Abbreviations used are the following
- 1-dNATP
1-naphthalene 5′-triphosphate
- 5-dNITP
5-nitroindole 5′-triphosphate
- BER
base excision repair
- CD
circular dichroism
- dNTP
2′-deoxyribonucleotide-5′-triphosphate
- dPTP
pyrene 5′-triphosphate
- dRPase
5′-deoxyribose phosphate lyase
- Pol λ
human DNA polymerase λ
- Pol β
DNA polymerase β
- rPol β
rat DNA polymerase β
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 2.Fowler JD, Suo Z. Biochemical, structural, and physiological characterization of terminal deoxynucleotidyl transferase. Chem Rev. 2006;106:2092–2110. doi: 10.1021/cr040445w. [DOI] [PubMed] [Google Scholar]
- 3.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 4.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 5.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 6.Johnson AA, Johnson KA. Fidelity of nucleotide incorporation by human mitochondrial DNA polymerase. J Biol Chem. 2001;276:38090–38096. doi: 10.1074/jbc.M106045200. [DOI] [PubMed] [Google Scholar]
- 7.Lee HR, Johnson KA. Fidelity of the human mitochondrial DNA polymerase. J Biol Chem. 2006;281:36236–36240. doi: 10.1074/jbc.M607964200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Rhee C, Bebenek A, Drake JW, Wang J, Konigsberg W. The L561A substitution in the nascent base-pair binding pocket of RB69 DNA polymerase reduces base discrimination. Biochemistry. 2006;45:2211–2220. doi: 10.1021/bi052099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Brown JA, Newmister SA, Suo Z. Polymerization fidelity of a replicative DNA polymerase from the hyperthermophilic archaeon Sulfolobus solfataricus P2. Biochemistry. 2009;48:7492–7501. doi: 10.1021/bi900532w. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J, Kraynov VS, Zhong X, Werneburg BG, Tsai MD. DNA polymerase beta: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem J. 1998;331(Pt 1):79–87. doi: 10.1042/bj3310079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roettger MP, Fiala KA, Sompalli S, Dong Y, Suo Z. Pre-steady-state kinetic studies of the fidelity of human DNA polymerase mu. Biochemistry. 2004;43:13827–13838. doi: 10.1021/bi048782m. [DOI] [PubMed] [Google Scholar]
- 12.Fiala KA, Duym WW, Zhang J, Suo Z. Up-regulation of the fidelity of human DNA polymerase lambda by its non-enzymatic proline-rich domain. J Biol Chem. 2006;281:19038–19044. doi: 10.1074/jbc.M601178200. [DOI] [PubMed] [Google Scholar]
- 13.Washington MT, Prakash L, Prakash S. Yeast DNA polymerase eta utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107:917–927. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 14.Washington MT, Johnson RE, Prakash L, Prakash S. The mechanism of nucleotide incorporation by human DNA polymerase eta differs from that of the yeast enzyme. Mol Cell Biol. 2003;23:8316–8322. doi: 10.1128/MCB.23.22.8316-8322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA polymerase iota utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol Cell Biol. 2004;24:936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiala KA, Suo Z. Pre-Steady-State Kinetic Studies of the Fidelity of Sulfolobus solfataricus P2 DNA Polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 17.Cramer J, Restle T. Pre-steady-state kinetic characterization of the DinB homologue DNA polymerase of Sulfolobus solfataricus. J Biol Chem. 2005;280:40552–40558. doi: 10.1074/jbc.M504481200. [DOI] [PubMed] [Google Scholar]
- 18.Carlson KD, Johnson RE, Prakash L, Prakash S, Washington MT. Human DNA polymerase kappa forms nonproductive complexes with matched primer termini but not with mismatched primer termini. Proc Natl Acad Sci U S A. 2006;103:15776–15781. doi: 10.1073/pnas.0605785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell CA, Prakash S, Washington MT. Pre-steady-state kinetic studies of protein-template-directed nucleotide incorporation by the yeast Rev1 protein. Biochemistry. 2007;46:13451–13459. doi: 10.1021/bi701429v. [DOI] [PubMed] [Google Scholar]
- 20.Brown JA, Fowler JD, Suo Z. Kinetic basis of nucleotide selection employed by a protein template-dependent DNA polymerase. Biochemistry. 2010;49:5504–5510. doi: 10.1021/bi100433x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, Cocea L, Gupta N, Weill JC, Reynaud CA. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5'-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 23.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 24.Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamtich J, Sweasy JB. DNA polymerase Family X: Function, structure, and cellular roles. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 31.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 32.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 33.Duym WW, Fiala KA, Bhatt N, Suo Z. Kinetic effect of a downstream strand and its 5'-terminal moieties on single nucleotide gap-filling synthesis catalyzed by human DNA polymerase lambda. J Biol Chem. 2006;281:35649–35655. doi: 10.1074/jbc.M607479200. [DOI] [PubMed] [Google Scholar]
- 34.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43:6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 36.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. Enzymes. 1992;20:1–61. [Google Scholar]
- 37.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fowler JD, Brown JA, Kvaratskhelia M, Suo Z. Probing conformational changes of human DNA polymerase lambda using mass spectrometry-based protein footprinting. J Mol Biol. 2009;390:368–379. doi: 10.1016/j.jmb.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 40.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Diaz M, Bebenek K, Sabariegos R, Dominguez O, Rodriguez J, Kirchhoff T, Garcia-Palomero E, Picher AJ, Juarez R, Ruiz JF, Kunkel TA, Blanco L. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J Biol Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- 42.Vande Berg BJ, Beard WA, Wilson SH. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase beta. Implication for the identity of the rate-limiting conformational change. J Biol Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 43.Kool ET, Sintim HO. The difluorotoluene debate--a decade later. Chem Commun (Camb) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 44.Hwang H, Taylor JS. Role of base stacking and sequence context in the inhibition of yeast DNA polymerase eta by pyrene nucleotide. Biochemistry. 2004;43:14612–14623. doi: 10.1021/bi0489558. [DOI] [PubMed] [Google Scholar]
- 45.Potapova O, Chan C, DeLucia AM, Helquist SA, Kool ET, Grindley ND, Joyce CM. DNA polymerase catalysis in the absence of Watson-Crick hydrogen bonds: analysis by single-turnover kinetics. Biochemistry. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J Biol Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kool ET. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu Rev Biophys Biomol Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 48.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Diaz M, Bebenek K, Gao G, Pedersen LC, London RE, Kunkel TA. Structure-function studies of DNA polymerase lambda. DNA Repair (Amst) 2005;4:1358–1367. doi: 10.1016/j.dnarep.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Lavrik OI, Prasad R, Beard WA, Safronov IV, Dobrikov MI, Srivastava DK, Shishkin GV, Wood TG, Wilson SH. dNTP binding to HIV-1 reverse transcriptase and mammalian DNA polymerase beta as revealed by affinity labeling with a photoreactive dNTP analog. J Biol Chem. 1996;271:21891–21897. doi: 10.1074/jbc.271.36.21891. [DOI] [PubMed] [Google Scholar]
- 51.Zhong X, Patel SS, Werneburg BG, Tsai MD. DNA polymerase beta: multiple conformational changes in the mechanism of catalysis. Biochemistry. 1997;36:11891–11900. doi: 10.1021/bi963181j. [DOI] [PubMed] [Google Scholar]
- 52.Dunlap CA, Tsai MD. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase beta. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 53.Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Ahn J, Werneburg BG, Tsai MD. DNA polymerase beta: structure-fidelity relationship from Pre-steady-state kinetic analyses of all possible correct and incorrect base pairs for wild type and R283A mutant. Biochemistry. 1997;36:1100–1107. doi: 10.1021/bi961653o. [DOI] [PubMed] [Google Scholar]
- 55.Beard WA, Osheroff WP, Prasad R, Sawaya MR, Jaju M, Wood TG, Kraut J, Kunkel TA, Wilson SH. Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase beta. J Biol Chem. 1996;271:12141–12144. doi: 10.1074/jbc.271.21.12141. [DOI] [PubMed] [Google Scholar]
- 56.Bebenek K, Garcia-Diaz M, Foley MC, Pedersen LC, Schlick T, Kunkel TA. Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda. EMBO Rep. 2008;9:459–464. doi: 10.1038/embor.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 60.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci U S A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown JA, Fiala KA, Fowler JD, Sherrer SM, Newmister SA, Duym WW, Suo Z. A Novel Mechanism of Sugar Selection Utilized by a Human X-Family DNA Polymerase. J Mol Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiala KA, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor JS, Yang W, Suo Z. Mechanism of Template-independent Nucleotide Incorporation Catalyzed by a Template-dependent DNA Polymerase. J Mol Biol. 2007;365:590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren RXF, Chaudhuri NC, Paris PL, Rumney SIV, Kool ET. Naphthalene, Phenanthrene, and Pyrene as DNA Base Analogs: Synthesis, Structure, and Fluorescence in DNA. J. Am. Chem. Soc. 1996;118:7671–7678. doi: 10.1021/ja9612763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang YL, Stivers JT. Efficient epimerization of pyrene and other aromatic C-nucleosides with trifluoroacetic acid in dichloromethane. Tetrahedron Lett. 2003;44:85–88. [Google Scholar]
- 65.Matray TJ, Kool ET. A specific partner for abasic damage in DNA. Nature. 1999;399:704–708. doi: 10.1038/21453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.