Abstract
The discovery of 3-deazathiamine diphosphate (deazaThDP) as a potent inhibitor analog of the cofactor thiamine diphosphate (ThDP) has highlighted the need for an efficient and scalable synthesis of deazaThDP. Such a method would facilitate development of analogs with the ability to inhibit individual ThDP-dependent enzymes selectively. Toward the goal of developing selective inhibitors of the mycobacterial enzyme 2-hydroxy-3-oxoadipate synthase (HOAS), we report an improved synthesis of deazaThDP without use of protecting groups. Tribromo-3-methylthiophene served as a versatile starting material whose selective functionalization permitted access to deazaThDP in five steps, with potential to make other analogs accessible in substantial amounts.
The emergence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) strains of M. tuberculosis poses a global threat.1,2 New anti-infectives are needed to combat tuberculosis and other infectious diseases that are becoming resistant to the current arsenal of drugs. An estimated one-third of the global population is infected with Mycobacterium tuberculosis. Although most hosts do not develop active disease, tuberculosis kills almost two million people worldwide every year.3 M. tuberculosis adapts its metabolism to survive under adverse conditions in the host, such as nutrient deprivation, hypoxia, acidification and oxidative stress.4 One approach to finding new anti-infectives against tuberculosis is to design inhibitors that target biochemical pathways used by the pathogen to survive in the host.
It was recently demonstrated that M. tuberculosis organizes a tricarboxylic acid (TCA) cycle that lacks the α-ketoglutarate dehydrogenase complex (KDH).5 The TCA cycle is an essential pathway used by aerobic organisms to metabolize carbohydrates, amino acids and fatty acids to produce energy, reducing power and biosynthetic precursors. In the canonical TCA cycle, KDH carries out decarboxylation of α-ketoglutarate, producing succinyl-CoA, which is then converted to succinate. Lack of KDH activity is a TCA cycle variant common among anaerobic bacteria and microaerophiles.6 It was initially proposed that the M. tuberculosis enzyme Rv1248c converts α-ketoglutarate to succinate via an α-ketoglutarate decarboxylase (Kgd)-mediated decarboxylation of α-ketoglutarate to succinic semialdehyde (SSA), followed by oxidation of SSA to succinate by a succinyl semialdehyde dehydrogenase, GabD1 or GabD2.5 Recent studies demonstrated that production of SSA is too slow to support this as a means for joining the oxidative and reductive branches of the TCA cycle. Instead, Rv1248c couples the decarboxylation of α-ketoglutarate to carboligation with glyoxylate, forming 2-hydroxy-3-oxoadipate (HOA), which spontaneously decarboxylates to 5-hydroxylevulinate. Thus, Rv1248c has been re-named HOA synthase (HOAS).7
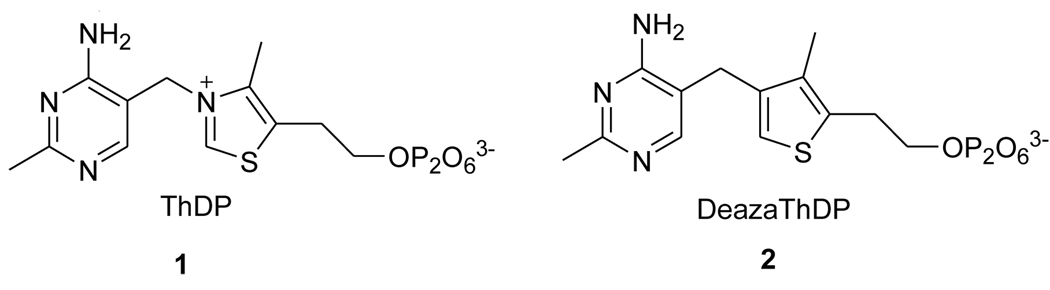
The HOAS reaction in M. tuberculosis is strictly dependent upon ThDP (1) (Figure 1).7 ThDP is an enzyme cofactor in a broad range of biosynthetic pathways and reactions, usually involving the cleavage and formation of C-C bonds adjacent to a carbonyl group.8,9 Humans cannot synthesize thiamine, thus making it an essential vitamin, whereas in bacteria, ThDP biosynthesis is regulated by riboswitches, metabolite-sensing domains found in messenger RNAs (mRNAs) of metabolite-synthesis proteins.10 The ThDP riboswitch detects ThDP with 1000-fold higher affinity than thiamine monophosphate or thiamine.11 Ligand-binding to the ThDP riboswitch promotes a conformational change in the mRNA that leads to termination of transcription and/or inhibition of translation.11–14 When intracellular ThDP concentrations are in excess, the expression of related metabolite-synthesis Pergamon proteins is repressed, and the repression is relieved when availability of metabolite falls below threshold.10 Because HOAS is predicted to be essential for survival of M. tuberculosis during infection15 and depends on ThDP, and because it appears that humans possess neither HOAS nor riboswitches, ThDP analogs selective for HOAS or selective for riboswitches might block survival of M. tuberculosis in infected individuals.
Figure 1.
Structures of Thiamine Diphosphate and 3-Deazathiamine Diphosphate.
Many ThDP analogs have been chemically synthesized16 and used to explore the biophysical significance of ThDP functional groups by serving as probes for spectroscopic and mechanistic studies. They have also been applied as putative transition or intermediate state analogs in protein crystallography.9,16 Some ThDP analogs are effective inhibitors of ThDP-dependent enzymes16 and riboswitches.13,16,17 DeazaThDP (2) is a ThDP analogue in which the N-3 atom of ThDP is replaced by a carbon, converting the charged thiazolium ring to a neutral thiophene. The absence of a positive charge at the 3-position prevents formation of the reactive ylid required for catalysis.16 Another result of this modification is that deazaThDP binds to target enzyme with greater affinity and speed than the natural coenzyme16,18 despite maintaining the size and steric profile of ThDP. Studies of Zymomonas mobilis pyruvate decarboxylase and the E.coli KDH E1 subunit suggest that deazaThDP binds these enzymes 25,000- and 500-times more tightly than ThDP, respectively.18
Given the established utility of deazaThDP as a tool to understand ThDP biochemistry, as well as its potential value as a precursor to development of an anti-infective targeting HOAS in M. tuberculosis, we sought to improve the efficiency and scalability of deazaThDP and deazaTh syntheses as further outlined below. We have developed a concise and general synthetic route to deazaTh. This report is a formal synthesis of deazaThDP since the transformation of the former to the latter has been described previously18,19 and worked in our hands.
Previously reported syntheses20,21 require amongst other things the construction of the thiophene ring itself. With a view toward a scalable synthesis, we reasoned to start with the readily available 2,3,5-tribromo-4-methylthiophene. We hoped to take advantage of different reactivities of the three bromine atoms in 2,3,5-tribromo-4-methyl thiophene 3 to instill a sequence of alkylating events that may, in principle, be conducted in one pot, and allow installation of desired functional groups in a predictable order. The three bromine atoms rank in reactivity from most reactive, Br-2, to the least reactive, Br-3. A wealth of possible transformations could be applied to construct thiophene-based chemical libraries. For example, Br-2 could be exchanged for a proton or metal-exchanged (umpolung) to react with an electrophile. Alternatively, these could also be substituted directly or through transition-metal catalyzed couplings to provide further diversity. These examples give a glimpse of the versatility of such a starting material.22
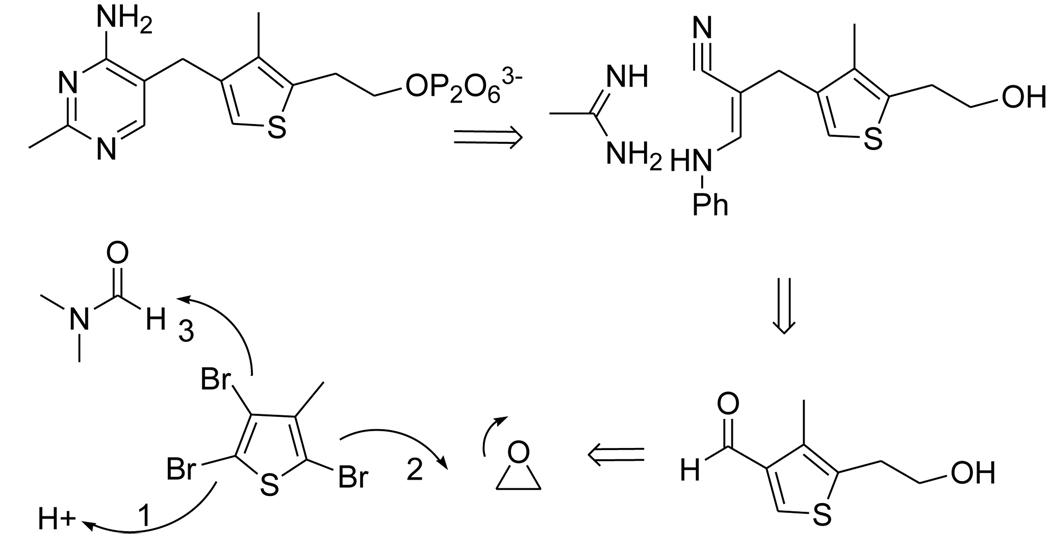
The retro-synthetic analysis profile of deazaThDP is as follows (Scheme 1). The pyrimidine ring should form easily from condensation of a cyanimine such as intermediate 7 and acetamidine. In turn, the cyanimine would result from reacting 3-anilinopropionitrile with aldehyde 6. The latter would be the result of an orchestrated sequence of transformations starting with halogen-proton exchange at Br-2, followed by metallation at position 5 and alkylation of ethylene oxide, and lastly, another metal-halogen exchange at Br-3, and treatment of the resulting anion with DMF.
Scheme 1.
Retrosynthetic Analysis Profile of 3-DeazaThDP.
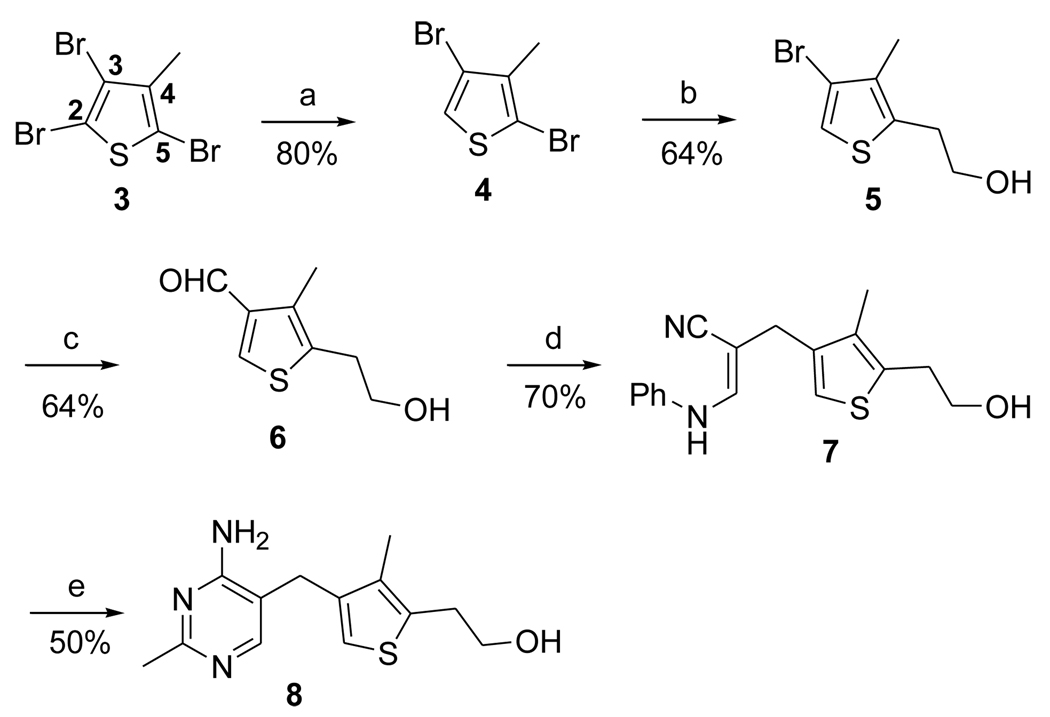
For the synthesis at hand, Br-2 in 2,3,5-tribromo-4-methyl thiophene was reductively cleaved with zinc powder in refluxing acetic acid22a,23 (Scheme 2) to give dibromide 4, which set the stage for the reaction of the next reactive bromide, Br-5. This was selectively translithiated and the resulting anion was condensed with ethylene oxide to introduce the hydroxyethyl side chain of 5 in good yield.
Scheme 2.
Synthesis of 3-Deazathiaminea
aReagents and conditions: (a) Zn powder, AcOH, reflux overnight. (b) n-BuLi, 1.0eq, ethylene oxide, BF3-Et2O, −78°C to 0°C, 3 h. (c) n-BuLi, 2.0eq, DMF, −78°C to rt, 3 h. (d) β-anilinopropionitrile, NaOMe/MeOH, DMSO, 40°C, 2 h. (e) acetamidine hydrochloride, NaOEt/EtOH, reflux, 48 h.
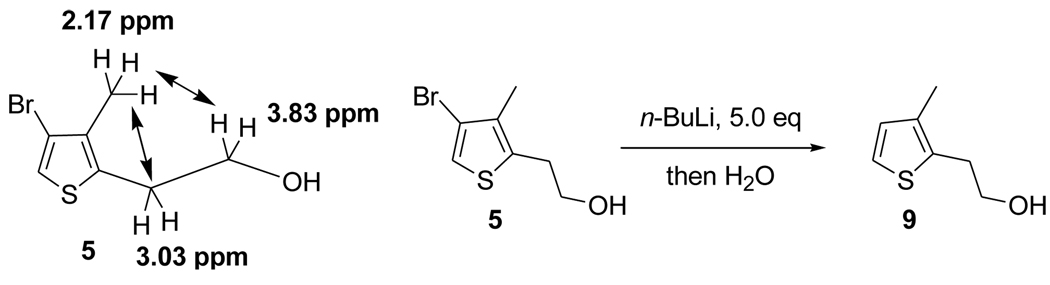
The structure of compound 5 was further confirmed by an NOE experiment (Table 1) and exhaustive lithiation, followed by quenching with water to ensure that regiochemistry occurred as intended, which was the case (Scheme 3). For instance, the methyl group showed a strong NOE with the two methylene groups on the side chain, but had no interaction with the aromatic proton at C2, providing additional proof of regiochemistry as drawn. Treatment of compound 5 with excess of n-butyllithium followed by quenching with water gave thiophene 9, which was easily recognized through the characteristic NMR signature which showed two aromatic hydrogens appearing as expected as two coupled doublets.
Table 1.
NOE data of 5
| Irradiated peaks (ppm) | Enhanced peaks (ppm) |
|---|---|
| 3.03 | 3.83, 2.17 |
| 2.17 | 3.83, 3.03 |
Scheme 3.
NOE and Exhaustive Lithiation of 5
The remaining bromine in 5 was transmetallated with n-butyllithium to give the corresponding lithiated intermediate, which was reacted with DMF to produce aldehyde 6 in 64 % isolated yield.24,25 Compound 6 thus obtained displayed proton and 13C NMR spectra identical to the described values.20 Finally, formation of the pyrimidine ring was accomplished in two steps.19,26 The aldehyde was condensed with β-anilinopropionitrile to give α,β-unsaturated nitrile 7, as a Z isomer. Base-catalyzed heterocyclization with acetamidine proceeded smoothly in refluxing ethanol to afford the desired deazathiamine 8.
As a preliminary indication of anti-infective potential, we tested the enzyme inhibitory activity of compound 8 in vitro (Figure 2). DeazaTh and deazaThDP inhibited HOAS-catalyzed reduction of ferricyanide (Figure 2) with IC50 values of 144 ± 11 µM (nH = 0.91 ± 0.05) and 6.7 ± 0.8 µM (nH = 1.28 ± 0.14), respectively. Correction for the competitive nature of the inhibition and concentration of TDP allowed estimation of inhibition constants KicalcdeazaTh = 76 µM and KicalcdeazaThDP = 3.5 µM.
Figure 2.
Inhibition curves for deazaTh and deazaThDP. Experiments were performed in phosphate buffer pH 7.4 at 37°C. DeazaTh (closed circles) was varied from 30 to 600 µM and deazaThDP (open circles) was varied from 1 to 200 µM. Symbols represent data and the solid curve represents the fit to ν = ν0/[1+(I/IC50)nH].
Conclusion
In summary, we have completed an efficient synthesis of deazaThDP. Readily available 2,3,5-tribromo-4-methyl thiophene allowed us to avoid construction of the thiophene ring, which greatly shortened the synthesis. To date, this five-step synthesis appears to be the most concise among reported routes. Metallations of the bromide allow for considerable flexibility, since it has been demonstrated that more than one electrophile couples with the lithiated intermediate. The ease of the C2 carbon modification, the active site in catalysis, offered by this method has the potential to provide general access to many deazaThDP analogs and to thiophene derivatives as well.
As with other ThDP-dependent enzymes, substitution of the nitrogen from the thiazolium ring of thiamine by a carbon leads to the generation of a good mimic of neutral or zwitterionic intermediates, which exist along the reaction pathway of ThDP-dependent enzymes. Some of these intermediates display greater affinity for these enzymes than ThDP itself. DeazaTh, which lacks the diphosphate portion of the cofactor, binds HOAS with affinity similar to ThDP, and deazaThDP binds 32-fold more tightly to HOAS than ThDP. These results indicate that deazaTh analogs are good starting points for rational inhibitor development or as chemical baits for click chemistry or other tethering approaches.
Supplementary Material
Acknowledgments
We thank Dr Samuel J. Danishefsky for critical reading of the manuscript. We also thank Drs. Hartmuth Kolb, Ramulu Poddutoori, and Avdhoot Velankar (UCLA) for early efforts in this project and Prof. Tadgh Begley for stimulating discussions. We are grateful for help with analytical data collection as well as interpretation provided by Drs. George Sukenick, Hui Liu, Silvy Ruesli, Ms Hui Fang from the NMR Analytical Core Facility at MSKCC, and Dr. Cliff Soll from the CUNY Mass Spectrometry Facility at Hunter College. The authors also thank Dr Cynthia Jung for valuable assistance with preparation of the manuscript. This work was supported by NIH grants AI64768 (CN) and P30 CA 008748-43 (core grant). The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Dorman SE, Chaisson RE. Nat. Med. 2007;13:295–298. doi: 10.1038/nm0307-295. [DOI] [PubMed] [Google Scholar]
- 2.Shenoi S, Friedland G. Annu. Rev. Med. 2009;60:307–320. doi: 10.1146/annurev.med.60.053107.103955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. Nat. Rev. Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook GM, Berney M, Gebhard S, Heinemann M, Cox RA, Danilchanka O, Niederweis M. Adv. Microb. Physiol. 2009;55:81–182. 318–319. doi: 10.1016/S0065-2911(09)05502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. Proc. Natl. Acad. Sci. U S A. 2005;102:10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guest JR. Philos. Trans. R Soc. Lond. B Biol. Sci. 1995;350:189–202. doi: 10.1098/rstb.1995.0152. [DOI] [PubMed] [Google Scholar]
- 7.de Carvalho LP, Zhao H, Dickinson CE, Arango NM, Lima CD, Fischer SM, Ouerfelli O, Nathan C, Rhee KY. Chem. Biol. 2010;17:323–332. doi: 10.1016/j.chembiol.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller M, Gocke D, Pohl M. Febs J. 2009;276:2894–2904. doi: 10.1111/j.1742-4658.2009.07017.x. [DOI] [PubMed] [Google Scholar]
- 9.Tittmann K. Febs J. 2009;276:2893. doi: 10.1111/j.1742-4658.2009.07016.x. [DOI] [PubMed] [Google Scholar]
- 10.Mandal M, Breaker RR. Nat. Rev. Mol. Cell. Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 11.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 12.Thore S, Leibundgut M, Ban N. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 13.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 15.Sassetti CM, Rubin EJ. Proc. Natl. Acad. Sci. U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Agyei-Owusu K, Leeper FJ. Febs J. 2009;276:2905–2916. doi: 10.1111/j.1742-4658.2009.07018.x. [DOI] [PubMed] [Google Scholar]; b) Fobare WF, Solvibile WR, Robichaud AJ, Malamas MS, Manas E, Turner J, Hu Y, Wagner E, Chopra R, Cowling R, Jin G, Bard J. Bioorg. Med. Chem. Let. 2007;17:5353–5356. doi: 10.1016/j.bmcl.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Edwards TE, Ferre-D'Amare AR. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Mann S, Perez Melero C, Hawksley D, Leeper FJ. Org. Biomol. Chem. 2004;2:1732–1741. doi: 10.1039/b403619k. [DOI] [PubMed] [Google Scholar]
- 19.Erixon KM, Dabalos CL, Leeper FJ. Chem. Commun. (Camb) 2007:960–962. doi: 10.1039/b615861g. [DOI] [PubMed] [Google Scholar]
- 20.Hawksley D, Griffin DA, Leeper FJ. J. Chem. Soc., Perkin Trans. 2001;1:144–148. [Google Scholar]
- 21.Thomas AA, De Meese J, Le Huerou Y, Boyd SA, Romoff TT, Gonzales SS, Gunawardana I, Kaplan T, Sullivan F, Condroski K, Lyssikatos JP, Aicher TD, Ballard J, Bernat B, DeWolf W, Han M, Lemieux C, Smith D, Weiler S, Wright SK, Vigers G, Brandhuber B. Bioorg. Med. Chem. Lett. 2008;18:509–512. doi: 10.1016/j.bmcl.2007.11.098. [DOI] [PubMed] [Google Scholar]
- 22.(a) Mitchell RH, Iyer VS. J. Am. Chem. Soc. 1996;118:722–726. [Google Scholar]; (b) Piller FM, Knochel P. Org. Lett. 2009;11:445–448. doi: 10.1021/ol802513q. [DOI] [PubMed] [Google Scholar]; (c) Dapperheld S, Feldhues M, Litterer H, Sistig F, Wegener P. Synthesis. 1990:403–405. [Google Scholar]
- 23.Hull JW, Jr, Romer DR, Podhorez DE, Ash ML, Brady CH. Beilstein J. Org. Chem. 2007;3(No. 23) doi: 10.1186/1860-5397-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To insure absence of any regioisomer, we recommend the bidistillation of dibromide 4 and purification of alcohol 5. Should any regioisomer be present, we have found the desired aldehyde 6 to be chromatographically separable at this stage (please see supplementary material section).
- 25.Slocum DW, Gierer PL. Chem. Commun. 1971;7:305–306. [Google Scholar]
- 26.Roth B, Aig E, Rauckman BS, Strelitz JZ, Phillips AP, Ferone R, Bushby SR, Sigel CW. J. Med. Chem. 1981;24:933–941. doi: 10.1021/jm00140a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.