Abstract
Compared to biochemical reactions taking place in relatively well-defined aqueous solutions in vitro, the corresponding reactions happening in vivo occur in extremely complex environments containing only 60–70% water by volume, with the remainder consisting of an undefined array of bio-molecules. In a biological setting, such extremely complex and volume-occupied solution environments are termed ‘crowded’. Through a range of intermolecular forces and pseudo-forces, this complex background environment may cause biochemical reactions to behave differently to their in vitro counterparts. In this review, we seek to highlight how the complex background environment of the cell can affect the diffusion of substances within it. Engaging the subject from the perspective of a single particle’s motion, we place the focus of our review on two areas: (1) experimental procedures for conducting single particle tracking experiments within cells along with methods for extracting information from these experiments; (2) theoretical factors affecting the translational diffusion of single molecules within crowded two-dimensional membrane and three-dimensional solution environments. We conclude by discussing a number of recent publications relating to intracellular diffusion in light of the reviewed material.
Keywords: ‘Crowded’ solutions, Brownian motion, Single particle, intracellular diffusion, In vitro reactions, Bio-molecules
Introduction
In colloidal and polymer chemistry, the theoretical description of diffusion in non-turbulent fluids1 has reached a high level of predictive capability primarily due to the ability to test and refine the theory using relatively well-defined experimental systems (Doi and Edwards 1999; Batchelor 2000; Byron Bird et al. 2002). In contrast, a predictive theory of diffusion in cell biology has yet to be attained due to the general lack of system definition associated with the type of reaction vessels, i.e. there are many different cell types, with each cell’s internal composition capable of dramatic variation over the very small spatial and temporal scales associated with diffusive motion. Indeed, as indicated by Fig. 1, we are often largely ignorant of the exact chemical surroundings in which our diffusing probe is moving, making the application of complex theory problematic. As a result, our measurements become largely phenomenological in nature and lack ready transferability to other situations. Deciphering observations of diffusive motion made in the complex patchwork collection of microstructures within the cell requires that the discussion of diffusion be conducted within a reductionist framework. In such an approach, general physical principles espoused from the consideration of more highly resolved situations are used to help interpret observations made from small regions of the intracellular molecular landscape (Baumeister 2004). It is towards this viewpoint that we have tailored this review. In the belief that the next step towards a quantitative accounting of intracellular diffusion will be assisted by benchmark conceptualizations of the process, we have reviewed some of the general physical features associated with diffusion in crowded solutions from the perspective of a single molecule’s Brownian motion, giving a simple introduction to the factors responsible for the phenomenon and highlighting methods capable of extracting maximum information from the data. The subject of diffusion is a perennial favourite for review, with many excellent examples on different aspects of the topic (Berg 1993; Saxton and Jacobson 1997; Peskir 2003; Frey and Kroy 2005; Philibert 2006; Dix and Verkman 2008; Saxton 2008). Aware of this fact, we have tried our best to approach the subject matter in a manner that will hopefully prove useful as both a practical and conceptual aid for those actually engaging with the complexities of diffusion in the cell using single particle tracking methods as their experimental avatar.
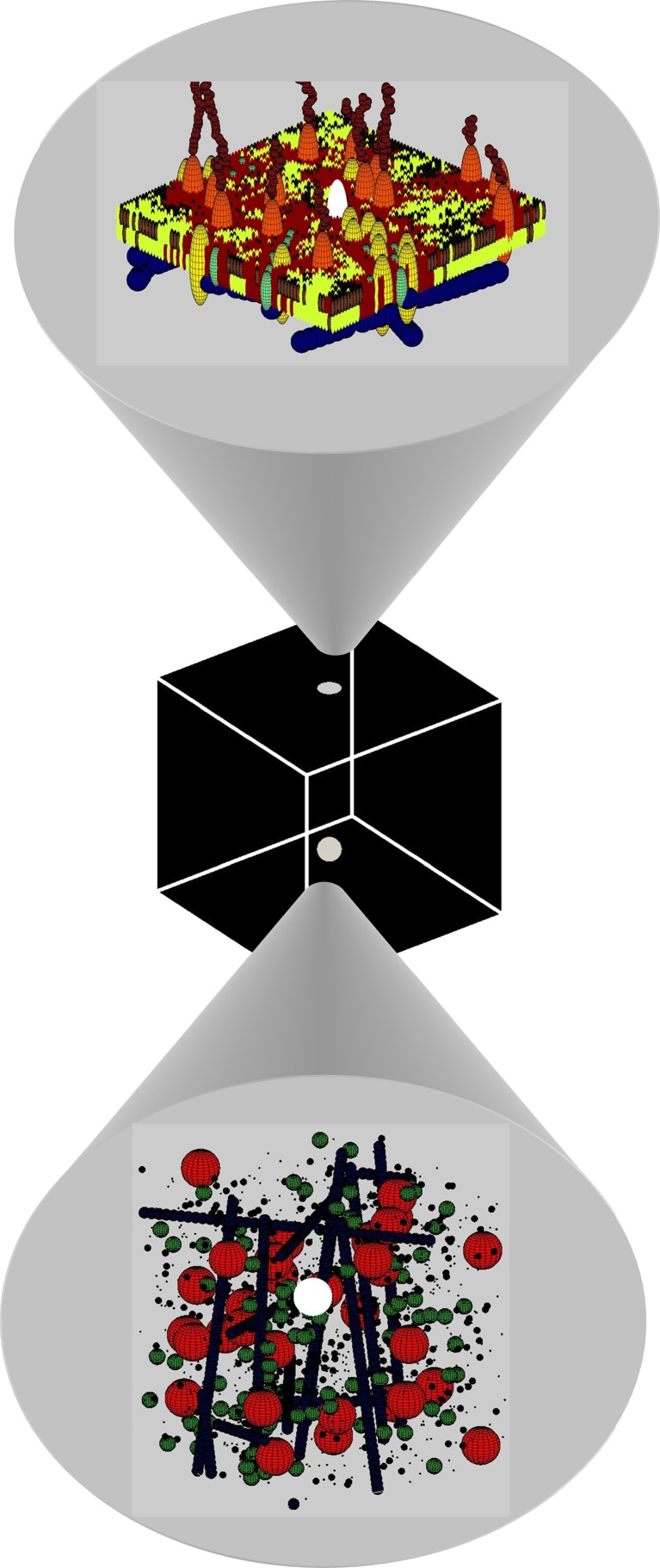
Fig. 1.
Interrogating Brownian motion in the ‘black box’ of the cell. A labeled (white) particle’s motion is followed in (top) the two-dimensional (2D) plane of the cell membrane (shown here as composed of various lipid domains and integral membrane proteins in contact extracellularly with polysaccharides and intracellularly with the cortical cytoskeleton) and (bottom) in the three-dimensional (3D) space of the cell cytosol (shown here as composed of various soluble proteins, cytoskeletal elements, nucleic acids, lipids, carbohydrates and inorganic ions)
Some diffusion basics
Particles in fluids undergo near continual random displacements due to collisions and subsequent momentum transfer with the surrounding solvent molecules—a situation termed ‘Brownian motion’.2 For a large number of particles initially located near to the same region in space, such Brownian motion has the consequence that the particles will tend to disperse over time, this situation being known as diffusion (Fig. 2). At the macroscopic level, the dispersal of the concentration profile of an arbitrary ideal solute component i, Ci(r,t), can be described by Fick’s second law of diffusion3 (Eq. 1a), which is an equation relating the partial time derivative of the concentration to its second partial spatial derivative using a phenomenological4 coefficient of diffusion, Di (units of m2s-1). At the time of its development, the diffusion coefficient was interpreted as a simple constant denoting a shared characteristic of the diffusing component i and the operative solution conditions under which the experiment was conducted. Throughout the period from 1905 to 1908, three scientists, Einstein (Einstein 1956), Smoluchowski (Fulinski 1998) and Langevin (Langevin 1908), all using different approaches, were able to provide a theoretical link between the phenomenological diffusion coefficient utilized by Fick and the system properties governing the individual particle displacements, ∆r i = [∆x, ∆y, ∆z] T, occurring over time intervals, ∆t, first observed by Brown (Eq. 1b)5 (Fig. 2).
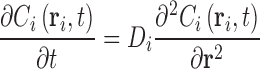 |
1a |
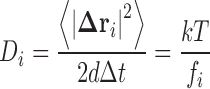 |
1b |
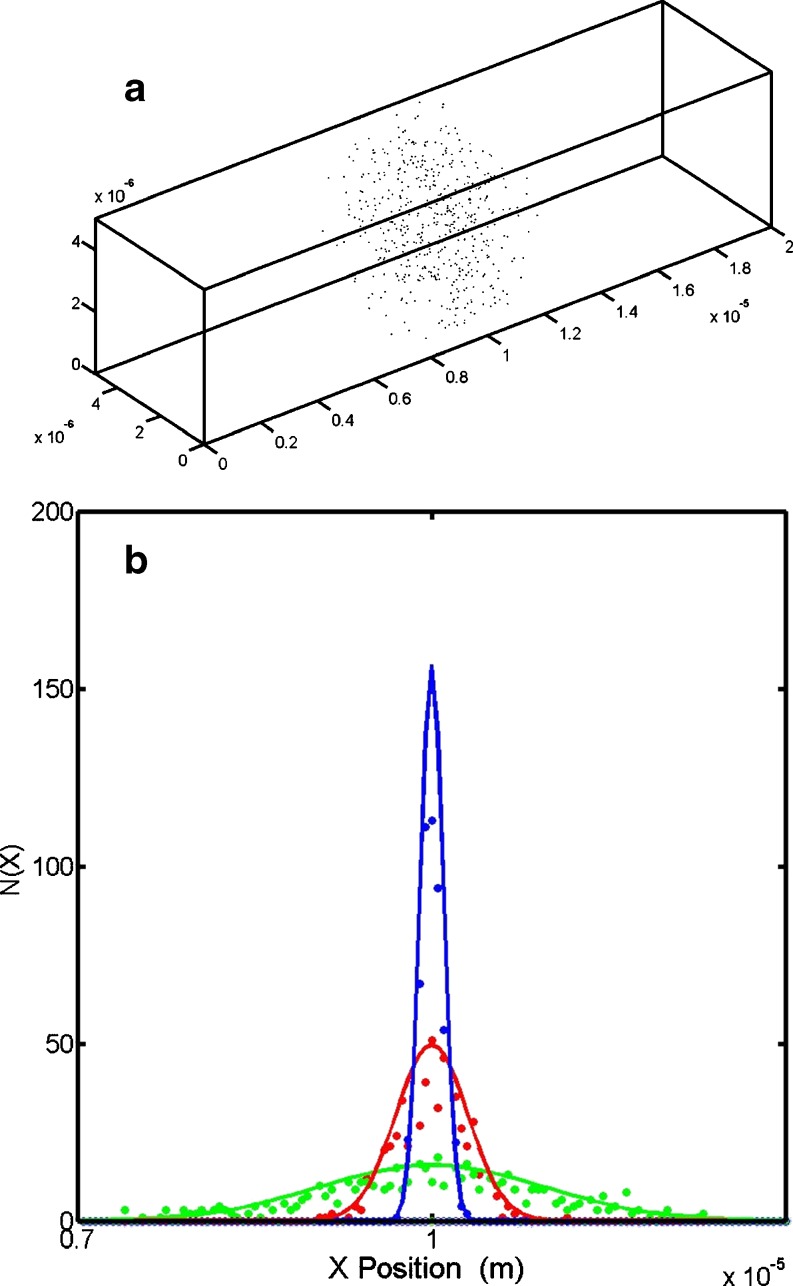
Fig. 2.
Diffusion occurs as the result of the Brownian motion of a large number of particles. a Time snapshot of the dispersal of 350 solute particles undergoing one-dimensional (1D) Brownian movement. Particles were initially located along the central plane of the box. The conditions were T = 37°C, η = 1 × 10−4 kgm−1 s−1, particle radii = 5 nm, snapshot time =0.001 s. b Dispersal profile described in terms of the number particle linear density as calculated using Eq. 1a (solid lines) and via stochastic simulation of the individual particles trajectories (dots) using Eq. 1b (circles). For solutions carried out using Eq. 1a, DI was estimated from the theoretical relation given in Eq. 1b. Simulation times were t = 0.001 s (green), t = 0.0001 s (red), and t = 0.00001 s (blue)
In this description, k is Boltzmann’s constant, T is the absolute temperature, fi is the particle’s frictional constant, and d is the dimensionality of the Cartesian space involved in the diffusion process. These theoretical predictions relating the diffusion coefficient to the sampling observation time and the particle mean squared displacement (MSD) were confirmed experimentally in the following years by optical microscopy-based observations of colloidal behaviour made by Perrin and co-workers (Perrin 1910) and sedimentation dispersion profiles measured by Svedberg and colleagues in the newly invented analytical ultracentrifuge (Svedberg and Pederson 1940). Importantly, both the theoretical assumptions used to derive Eqs. 1a and 1b as well as the experimental requirements necessary for achieving agreement with the theory, require that the solute particles be dilute and non-interacting—situations which are far removed from the reality inside the cell.
Experimental measurement of single particle motion within the cell
In correspondence with the macroscopic and particle level descriptions of diffusion embodied by Eqs. 1a and 1b, there are two general approaches for experimental determination of the diffusion coefficient: methods that measure the time-dependent behaviour of a large number (virtual continuum) of particles over relatively large regions of space and methods that involve observing the properties of individual particles over a small region of space. Experimental techniques belonging to the former category include pulsed gradient nuclear magnetic resonance (NMR) (Stejskal and Tanner 1965; Morris and Johnson Jr. 1992; Regan and Kuchel 2003), electron spin resonance (ESR) procedures (Regan and Kuchel 2003; Mirosavljević and Noethig-Laslo 2008), optical microscopy-based observation of light-absorbing or light-emitting solutes (Guo et al. 2008; Lorén et al. 2009), neutron scattering (Doster and Longeville 2007) and single- and multi-wavelength dynamic light scattering experiments (Peetermans et al. 1987). Experimental methods belonging to the later category, i.e. methods capable of observing intracellular single molecule diffusive motion at the microsecond and 100-nm level of precision (approximately) are largely due to technological advances in optical microscopy (Diaspro 2002; Elson 2004; Murcia et al. 2007; Guo et al. 2008). Particular promising from the viewpoint of deciphering the effects of underlying internal cellular structure are the single particle molecule tracking (SPT) techniques (Saxton and Jacobson 1997; Kusumi et al. 2005; Murcia et al. 2007) and the confocal based fluctuation microscopy approaches (Elson 2004; Weiss 2008). As the confocal based fluctuation approaches have been recently reviewed, both in this journal and elsewhere (Elson 2004; Weiss 2008; Chiantia et al. 2009; Jameson et al. 2009), we place our focus in this review on the SPT approach.
Optical microscope based SPT experiments involve recording the trajectory of a single molecule possessing either some naturally occurring detectable feature or an attached label (Fig. 3). The most common formats for realization of this procedure involve tagging the diffusing component of interest with a fluorescent compound having a high quantum yield (e.g. fluorescent proteins attached via molecular biology procedures (Hibino et al. 2009) or modern fluorescent ‘laser’ dyes and quantum dots (Alcor et al. 2009) which can be visualized using various implementations of fluorescence microscopy. Alternatively, differentially polarizable materials (e.g. small metal or latex colloidal particles) which are capable of being observed using standard bright field or interference based microscopy approaches (de Brabander et al. 1991; Geerts et al. 1991; Kusumi et al. 2005) can be attached to the molecule of interest. The choice of format for particle detection is usually dictated by the experimental problem at hand in conjunction with the photophysical properties (e.g. fluorescence excitation/emission bands, fluorescence lifetime, ‘blinking’ artefacts) and physical compatibility (size and chemical properties of the label in relation to the tracer molecule of interest). In the cases where a molecular biology based approach is not adopted, the labelled tracer can be introduced into the cell via passive uptake, liposome fusion, electroporation or micro-injection (Kusumi et al. 2005; Alcor et al. 2009). Once in place, a video or digital movie camera attached to the microscope is used to record the Brownian motion of the individual particles. The coupling of highly sensitive camera equipment (CCD or CMOS) with modern microscope developments, such as cylindrical objective lenses with multi-focus capabilities (Kao and Verkman 1994), confocal and multiphoton methods (Diaspro 2002; Hellriegel and Gratton 2009) and evanescent wave-based total internal reflection illumination procedures for recording close to surfaces, has meant that time-dependent single particle tracking (SPT) experiments are able to be performed in both the two- and three-dimensional (2D and 3D, respectively) intracellular spaces (e.g. the cell membranes and cytosol) at relatively high temporal and spatial frequencies.
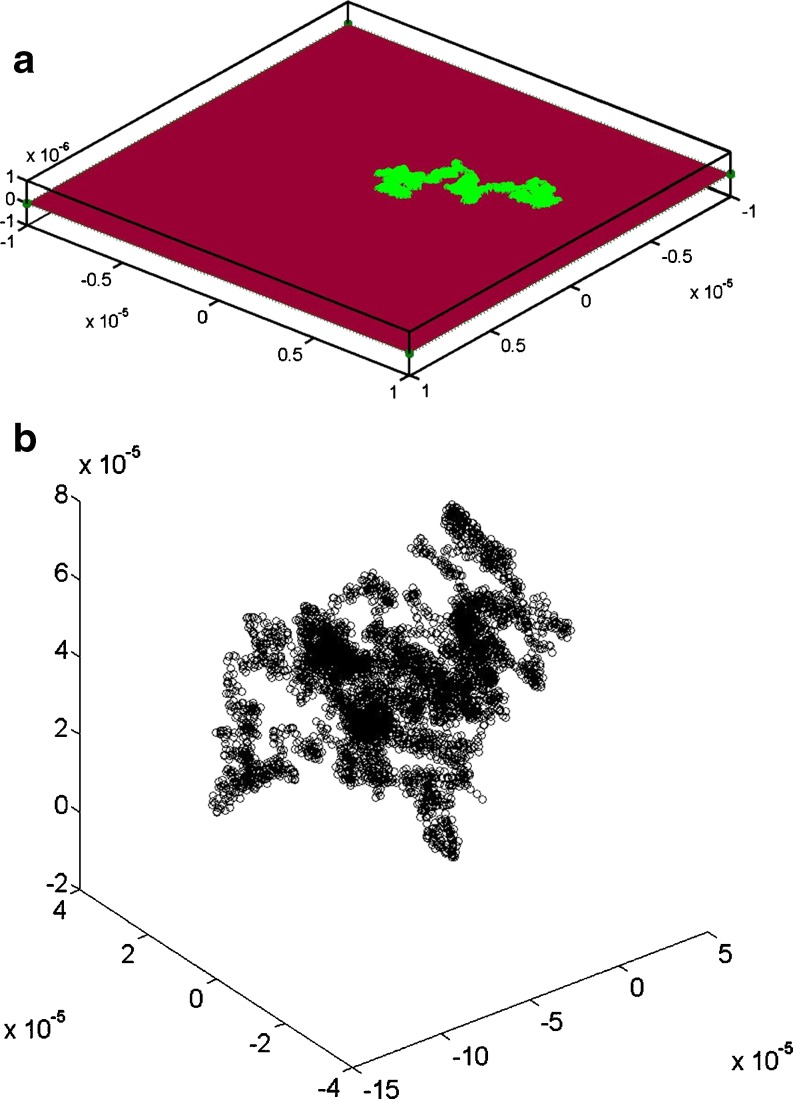
Fig. 3.
Single particle trajectories for tracer diffusion in simple (a) 2D (Di = 5.4 μm2 s-1) and (b) 3D continuum fluids 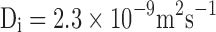 . Ri = 1 nm, Hi = 5 nm,
. Ri = 1 nm, Hi = 5 nm, 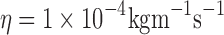 ,
, 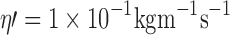 , T = 37°C, total simulation time was 1 s. Particles were initially located at the central position. Particle trajectories were simulated using time steps of 25 μs
, T = 37°C, total simulation time was 1 s. Particles were initially located at the central position. Particle trajectories were simulated using time steps of 25 μs
Extracting information about the diffusive properties of the observed tracer particle involves analysing the particle’s position trajectory, r(t) (Fig. 3), by repetitive calculation of the position difference vector, ∆r, over an arbitrary time interval, n∆t (Eq. 2).
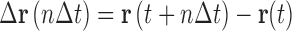 |
2 |
For a given integer multiplier, n, of the smallest recorded sampling time interval, ∆t, the diffusion tensor of component i, D i, (Batchelor 2000; Fernandes and de la Torre 2002) may be calculated by sufficiently averaging the successive product of the position difference vector with its transpose, both throughout the trajectory and, where possible, by averaging from multiple particles migrating through the same region of space. (Eq. 3).
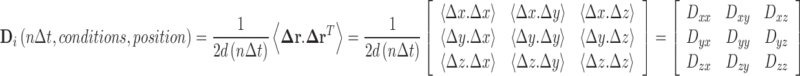 |
3 |
The diffusion tensor as calculated by Eq. 3 provides a measure of the isotropy6 of the system. For isotropic systems, all diagonal elements of D will be equal, and all non-diagonal elements will average to zero, thereby allowing a description of the system by a single diffusion constant D = Dxx = Dyy = Dzz. When this condition is not met, a rotational matrix, R, can be applied to the diffusion tensor with the condition of producing a transformed diffusion tensor D′ that is diagonal in nature and necessarily defined in terms of a Cartesian coordinate system x′,y′,z′ that lies along the principle axes of the anisotropy (Eq. 4)
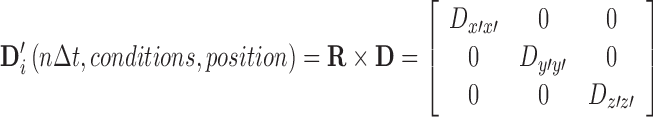 |
4 |
The system isotropy can be visualized graphically by making the diagonal terms of the diffusion tensor the characteristic measures of an ellipsoid lying along the axes specified by the rotational transform (Fig. 4). For the isotropic case in which diffusion along all three axes will be equally likely the representation will be a sphere.
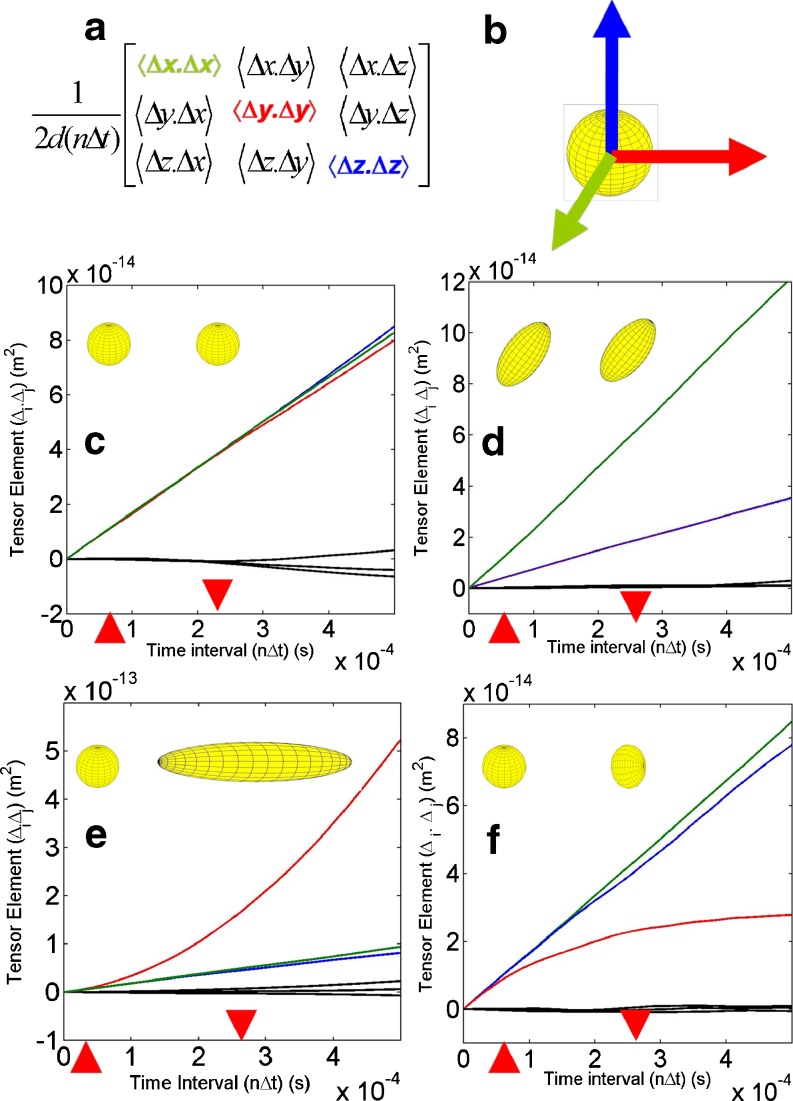
Fig. 4.
Analysis of the mean squared displacement components of the diffusion tensor as a function of the sampling interval time for 3D tracer diffusion. a Colour-coded averaged tensor elements. b Ellipsoid representation of the diffusion tensor using the diagonal components of the diffusion tensor to represent the characteristic lengths of the ellipsoid. c Isotropic ‘simple’ Brownian motion. All diagonal components equal (Dxx = Dyy = Dzz), and the mean squared displacement (MSD) is directly proportional to the sampling interval (α = 1). d Anisotropic ‘simple’ diffusion: Dxx = 3Dyy = 3Dzz (α = 1). e Anisotropic ‘super’ diffusion produced by Brownian diffusion with a weak constant drift velocity in the y direction [Dyy (α > 1)] > [Dxx = Dzz (α = 1)]. f Anisotropic ‘anomalous’ diffusion produced by a weak harmonic potential restoring force acting along the y direction only [Dyy (α < 1)] > [Dxx = Dzz (α = 1)]. The red arrow blocks refer to the time intervals at which the ellipsoids representing D were constructed
Calculation of the tensor elements at different sampling time intervals, n∆t, provides additional information on the so-called anomalous nature of the diffusion constant. In this approach, each tensor element calculated at the limiting sampling time interval is effectively modified by an empirical function, f(n∆t), of the sampling time interval. A common form of the function, f(n∆t) = (n∆t)(α), is shown in Eq. 5 in which the parameter α is termed the anomalous diffusion coefficient.
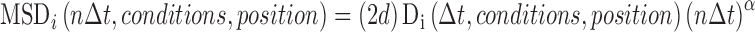 |
5 |
For normal diffusion, the modifying functional parameter, α, is equal to 1 (Fig. 4c. d). For the case of super-diffusion (Di increasing with time), α is ≥ 1 (Fig. 4e), and in the case of sub-diffusion (Di decreasing with time), α is ≤1 (Fig. 4f). Although the problem is essentially an inverse one, many researchers have used an iterative process of model building and simulation to provide an interpretation of experimentally observed anomalous diffusion in terms of cellular structural characteristics or local solution conditions (Goulian and Simon 2000; Jin and Verkmann 2007; Sanabria et al. 2007; Saxton 2007, Saxton 2008; Weiss 2008). We cite it here as an important descriptor of the diffusion process in crowded solutions (Banks and Fradin 2005; Ridgway et al. 2008; Weiss 2008). It also serves as an important reminder of the importance of asserting the sampling interval time when comparing tracer diffusion coefficients. Indeed, the two limiting forms corresponding to short time n∆t → 0 and long time n∆t →∞ are commonly used for comparative purposes (Bernadó et al. 2004).
Theoretical description of Brownian motion within the cell
Parallel to the advances in technology that have made high spatial, high temporal frequency single particle tracking a relatively straightforward technique, corresponding developments in computing power have meant that coarse grained particle modelling of cell like situations have started to become a possible, if not routine, addition to the biophysicist’s bag of tools, providing a much required extra level of discrimination in the construction and testing of cellular level biological hypotheses. In general the particle based models available for simulating intracellular diffusion involve the specification of a set number of solute particles within a boundary enclosing volume representing the cell wall (Andrews and Bray 1994; Jeschke 2008; Czech et al. 2009; Moraru et al. 2009; Takahashi et al. 2005; Wils and De Schutter 2009). If one considers the average cell size to be defined by a length scale approximately 10 μm and the average protein radius approximately 2 nm, then it becomes obvious that a full particle representation of the components of the cell is beyond current computing capabilities as it would involve on the order of 1 × 1010 particles. To subvert this problem models of intracellular diffusion are ‘coarse grained’ i.e. they are simplified by reducing the number of components and the level of detail. Such modelling approaches often involve simplifications in which Brownian motion characteristics are considered to be independent of the surrounding local environment, and particle interactions (if considered) are incorporated at the level of like/dislike rule based algorithms. Contrarily, much of the higher order theory already developed for the description of diffusion in concentrated solution environments cannot feasibly be employed due to the problems discussed in the introduction relating to general ignorance of the cellular solution composition. In the following section we discuss some of the factors affecting the Brownian motion of single particles in crowded environments and then we review some different levels of approximation for carrying out particle model simulations at different time and length scales commensurate with the level of knowledge of the fine structure of the cell.
Factors affecting the Brownian motion of single particles in crowded environments
The Langevin approximation (Langevin 1908; Dhont 1996; Snook 2006) of Brownian motion formalises the particle model approach by considering the solution as an implicit background of solvent containing a number of explicitly recognized particles. As such the Newtonian acceleration of the ith particle can be calculated as the net force due to the combination of intermolecular potentials acting between explicitly considered particles, (F p)i, a frictional forces arising from the tendency of the solvent to resist directed motion, (F f)i, and random forces resulting from unbalanced collisions with the solvent molecules, (F r)i. When the number of explicitly recognised particles is low the Langevin description approaches its dilute limiting form (Eq. 6a). However at elevated ‘crowded’ conditions the intermolecular potentials become significant and must be accounted for (Eq. 6b).
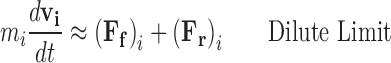 |
6a |
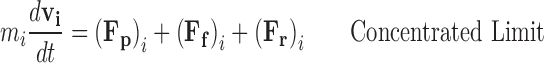 |
6b |
Although the Langevin equation has limited potential in the simulation of diffusion in crowded systems due to its requirement for short time scales during numerical integration, it provides a very useful pedagogical aid for developing a mechanical understanding of the subject. For now we note that in the concentrated limit the Langevin equation is composed of three force terms, F p, F f and F r, each of which we will discuss in the sections below.
Forces resulting from intermolecular potentials (Fp)
Although the term macromolecular crowding is used in biology to describe the special role played by the cell’s crowded microenvironment on the biochemical reactions occurring within it, the effects of crowding are, as for all chemical reactions, physically manifested through the existence of intermolecular potentials acting between individual particles. To a first approximation, the properties of solutions can be described on the basis of the summation of external forces acting on each of the individual particles and the summed contribution of the individual particle-particle pairwise interaction potentials (Elimelech et al. 1995; Leach 2001) (Eq. 7).
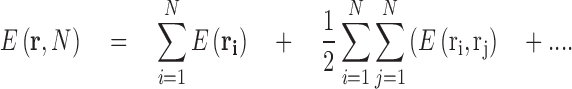 |
7 |
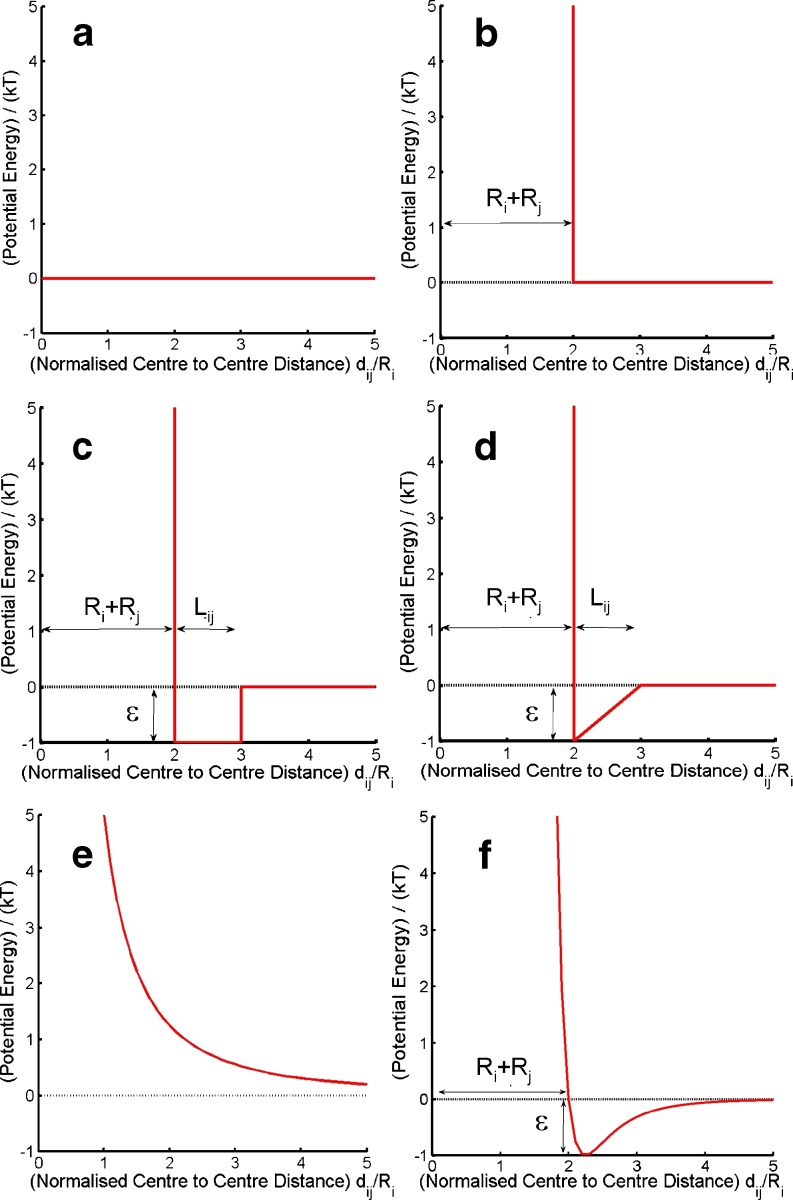
The types of potential specified depend upon the level of detail included in the description of tracer and background molecules (Elimelech et al. 1995; Leach 2001; Elcock 2003; McGuffee and Elcock 2006; Qin and Zhou 2009). Figure 5 and Table 1 describe six simple interparticle potentials commonly used in polymer and colloidal physical chemistry (Minton 1989; Elimelech et al. 1995; Doi and Edwards 1999; Zhou et al 2008) to account for the effects of concentrated solution environments. In this review, we regard the tracer particle (particle i) and crowding background molecule (particle j) as spheres of respective radii Ri and Rj. In general, the intermolecular potentials all feature some form of repulsive interaction at short range that physically arises from electronic repulsion. At longer distances, the interaction may take on attractive or repulsive characteristics empirically defined in terms of a potential energy depth ε and a screening length, Lij. The potentials described in Fig. 5 and Table 1 are assumed to have spherical symmetry and hence are functions of intermolecular distance, dij, only 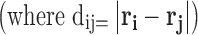 .
.
Fig. 5.
Pair potential functions useful for describing particle interactions at the coarse grained level. a Free overlap, b hard particle, c square well, d saw tooth, e soft sphere, f Lennard Jones. The associated mathematical descriptions are included in Table 1
Table 1.
Intermolecular potentials commonly used to describe particle interactions and their associated mathematical functions
| Pair-potential function | Potential energy function (ΔEij(dij)) | Force derivation [(Fp)i (dij)]a, b |
|---|---|---|
| Free Overlap | ∆E ij = 0 | (F p)i = 0 |
| Hard Particle |
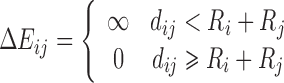
|
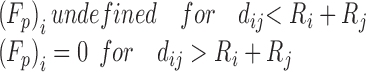
|
| Square Well |
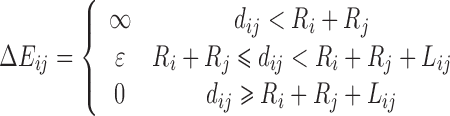
|
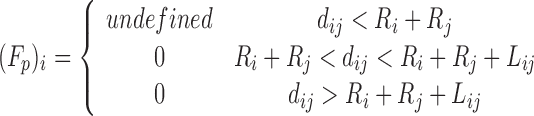
|
| Sawtooth |
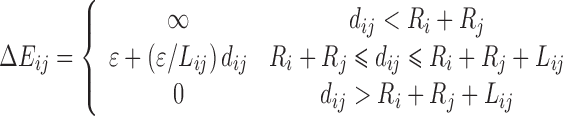
|
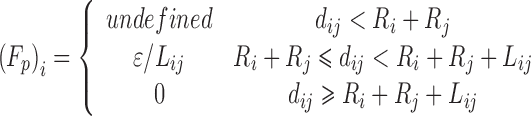
|
| Soft Sphere |
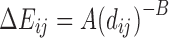
|
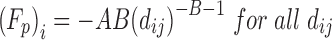
|
| Lennard-Jones |
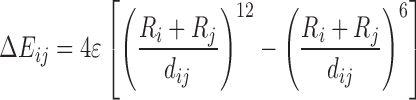
|
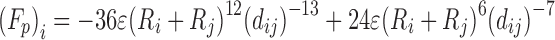
|
aThe assumption of spherical symmetry means that the calculated force acts in a direction given by the unit vector 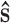 acting along the centre-to-centre line between the two particles.
acting along the centre-to-centre line between the two particles.
bIn some cases the pair potential functions are not differentiable because of discontinuities. Such functions are generally more suited to position Langevin and overdamped Brownian dynamics simulations. Alternatively, they can be used with an arbitrary specification of rules for dealing with physically forbidden overlaps.
The force acting on particle i, (F
p)i, is calculated as the sum of the gradients of the intermolecular potentials existing between the particle under consideration and all of the surrounding explicit species, Eq. 8. The assumption of spherical symmetry requires that the force calculated from each pair potential acts in a direction given by the unit vector 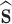 acting along the centre-to-centre line between the two particles.
acting along the centre-to-centre line between the two particles.
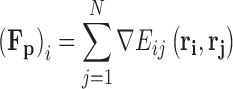 |
8 |
Although the described potential functions and the forces derived from them are not realistic, they serve both heuristic and practical purposes in that they are (1) easy to comprehend, (2) easy to incorporate into simulations and (3) represent an advance to just ignoring these features. Additional features, such as the dependence of the interparticle potentials on such things as ionic strength, can be incorporated phenomenologically by the inclusion of scaling terms in the potential depth and screening length terms (Elimelech et al. 1995).
Forces resulting from friction exerted by the solvent (Ff)
At limiting dilution in non-turbulent flows, a tracer particle of characteristic size <10 μm undergoing motion relative to a viscous solvent environment will experience a frictional force opposing its motion that is equal to the negative product of the velocity of the particle, v i (relative to the velocity of the surrounding fluid), and a frictional constant, fi, characteristic of the shape of the particle (Dhont 1996; Byron Bird et al. 2002) (Eq. 9a). For spherical particles undergoing translational motion in a 3D solvent environment, fi is given by Stokes law (Eq. 9b: Jones 2002; Byron Bird et al. 2002). Proteins and lipids in the two dimensions of the cell membrane are usually modelled as upright cylinders (of characteristic radius Ri and cylinder height, Hi) with the frictional coefficient of such a cylinder given by Eq. 9c (Saffman and Delbrűck 1975). In Eqs. 9b and c, η and η′ are the dynamic viscosities (units kg m-1s-1) of the solvent or membrane, respectively. Typical values for η and η′ at 37°C are ∼1 × 10-4 and ∼1 × 10-1 kg m-1 s-1, respectively (Byron Bird et al. 2002; Spooner et al. 2000;).
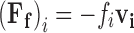 |
9a |
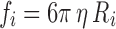 |
9b |
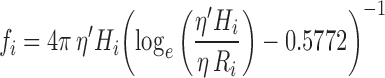 |
9c |
At non-dilute concentrations, the simple expressions for the frictional force described in Eqs. 9 becomes suspect due to the fact that all particles undergoing directed motion7 through a solvent environment must necessarily exert force on the surrounding solvent, which in turn communicates that force to other nearby regions of fluid. This means that the relative velocity between the particle and the surrounding solution environment can no longer be simply equated with the velocity of the particle with respect to an ideal consideration of its local streamline. This additional component to the frictional force, known as the hydrodynamic interaction (HI) force, becomes more pronounced with increasing concentration and level of volume occupation. Consequently, recognition of these HI effects in studies on macromolecular crowding is desirable. Within the Langevin framework, hydrodynamic interactions can be incorporated by including supporting functions containing various degrees of information on the positions, velocities and precise geometries of all N explicitly considered simulation components (Eq. 10a) (Rotne and Prager 1969; Ermak and McCammon 1978; Batchelor 1982; Fernandes and de la Torre 2002; Padding and Louis 2006; Geyer and Winter 2009). As such a precise level of system definition is often lacking when conducting investigations of diffusion within the cell, coarser methods for factoring in HI forces based on effective viscosity have been attempted (Heyes 1995; Wade 1996; Urbina-Villalba et al. 2003; Sun and Weinstein 2007). In this approach, a localized mean field approximation is used to calculate an appropriately averaged scalar value of the frictional coefficient, <f i>, over the displacement ∆r (Eq. 10b)
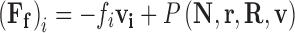 |
10a |
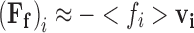 |
10b |
In practice the average frictional coefficient, <f i>, is approximated from the known value of the short-time diffusion constant8 of a tracer particle in a hard particle fluid, with the value either derived from experimental data (Urbina-Villalba et al. 2003) or made using a higher order theoretical method for implementation of hydrodynamic interactions (e.g. Fernandes and de la Torre 2002; Falck et al. 2004; Geyer and Winter 2009). For calculations of tracer diffusion within a solution of identical particles, these averaged values for <fi > in three dimensions (written in terms of fractional volume ϕ) or two dimensions (written in terms of fractional area θ) are presented as Eqs. 11a and 11b, respectively.
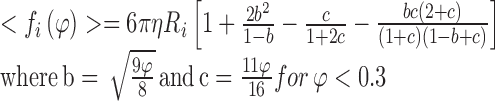 |
11a |
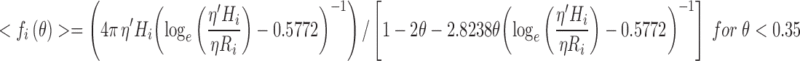 |
11b |
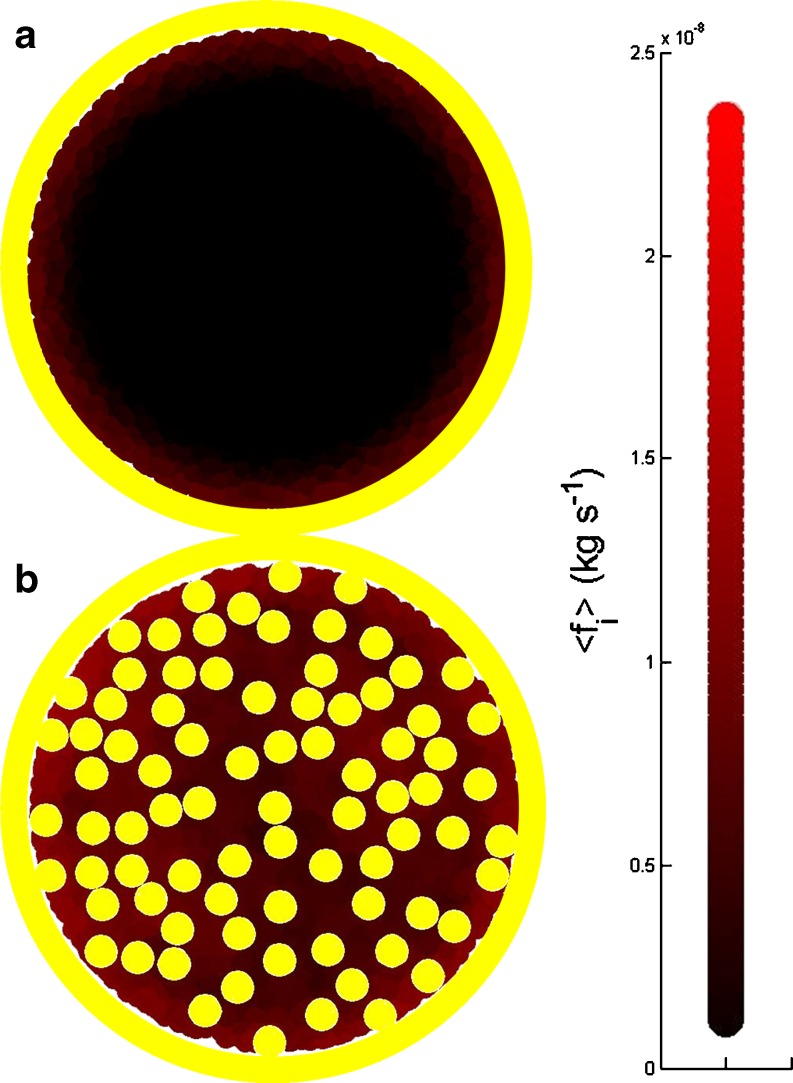
In Eqs. 11a and 11b, ϕ and θ refer to the local fraction of volume and area, respectively, occupied by all other explicitly recognized particles9 within a sphere or circle of radius 4Ri centred at position r i . As an example, Fig. 6 describes the altered frictional force resulting from HI effects experienced by a tracer protein Ri = 2 nm) in an enclosed 2D membrane at either limiting dilution or under crowded conditions (θ = 0.4). The equations derived above should be seen as a first order inclusion of HI as they ignore differences in the perpendicular and parallel components of the viscosity predicted by higher order theory (Elimelech et al. 1995; Dhont 1996; Batchelor 2000).
Fig. 6.
Colour-coded description of frictional coefficient experienced by a tracer particle (R = 2 nm, H = 5 nm) in a 2D fluid membrane 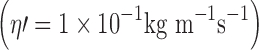 at 37°C bounded by a hard wall (a) at limiting dilution (b) at θ = 0.35 (as approximated by Eq. 11b). Note the stagnant regions adjacent to the walls and high local densities of particles shown in brighter red, indicating higher viscosity/higher local frictional coefficient
at 37°C bounded by a hard wall (a) at limiting dilution (b) at θ = 0.35 (as approximated by Eq. 11b). Note the stagnant regions adjacent to the walls and high local densities of particles shown in brighter red, indicating higher viscosity/higher local frictional coefficient
Forces resulting from random collisions with the solvent (Fr)
The final term required for specification of Eq. 5 is the stochastic force variable, [F r(∆t)]i, written here as a function of the time interval, ∆t, specifying the frequency with which it is updated. A simple conceptualization of the random force term that offers physical insight is that of a thermostat-regulating function (Langevin 1908; Snook 2006) through which it combats the tendency of the viscous frictional force to slow the system down (and hence cool it). The most common procedure for assigning a numerical value to [F r(∆t)]i involves requiring that it conforms to relations derived from application of the fluctuation dissipation theorem (Eq. 12) (Kubo 1966).
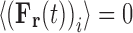 |
12a |
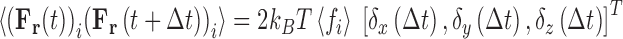 |
12b |
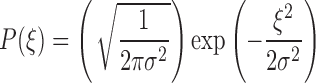 |
12c |
where δ is the Dirac delta function. It can be shown (Kubo 1966) that these requirements are satisfied by selecting the random component forces (ξ) from a Gaussian distribution (Eq. 12c) with a standard deviation, σ, given by 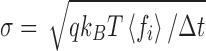 . The magnitude of σ can be seen to be dependent upon the sampling time,10 the temperature and the local frictional coefficient (Saxton 2008). The mathematics behind the result presented in Eq. 12 can be relatively difficult to follow so here we offer a simple argument which although only yielding an approximate result provides some useful insight into the physical origins of the random force and the natural time constant of the system. In this simplified model, the average kinetic energy of the system is first equated with the average thermal energy via the equipartition theorem, i.e.
. The magnitude of σ can be seen to be dependent upon the sampling time,10 the temperature and the local frictional coefficient (Saxton 2008). The mathematics behind the result presented in Eq. 12 can be relatively difficult to follow so here we offer a simple argument which although only yielding an approximate result provides some useful insight into the physical origins of the random force and the natural time constant of the system. In this simplified model, the average kinetic energy of the system is first equated with the average thermal energy via the equipartition theorem, i.e. 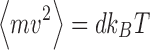 therefore,
therefore, 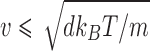 . Over time scales sufficiently small enough to preserve inertial memory, we can equate the average magnitude of the random force with the average absolute value of the frictional force (Eq. 13a), with this relationship being further developed by combination with the relation for the average particle velocity to determine the magnitude of the random force term (Eq. 13b).
. Over time scales sufficiently small enough to preserve inertial memory, we can equate the average magnitude of the random force with the average absolute value of the frictional force (Eq. 13a), with this relationship being further developed by combination with the relation for the average particle velocity to determine the magnitude of the random force term (Eq. 13b).
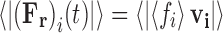 |
13a |
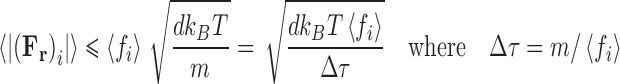 |
13b |
From a dimensional analysis of Eq. 13b, it can be noted that m/<fi> has units of time and can thus be interpreted as specifying a natural time constant for the system, such that 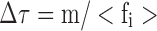 . For specification of larger time intervals ∆t >> ∆τ, such a direct physical interpretation of the random force term breaks down due to the fact that the path taken between the application of the intervening stochastic random forces is not linear, but rather occurs as the result of a random walk produced by many collisions, each occurring at or around the natural time constant of the system. In this time regime, the average motion of the particle is best described as a function of its mean squared displacement (Langevin 1908; Einstein 1956) (Eq. 1b). Using that definition of the diffusion coefficient, a phenomenological velocity can be determined over the time step and calculated displacement (Eq. 14)
. For specification of larger time intervals ∆t >> ∆τ, such a direct physical interpretation of the random force term breaks down due to the fact that the path taken between the application of the intervening stochastic random forces is not linear, but rather occurs as the result of a random walk produced by many collisions, each occurring at or around the natural time constant of the system. In this time regime, the average motion of the particle is best described as a function of its mean squared displacement (Langevin 1908; Einstein 1956) (Eq. 1b). Using that definition of the diffusion coefficient, a phenomenological velocity can be determined over the time step and calculated displacement (Eq. 14)
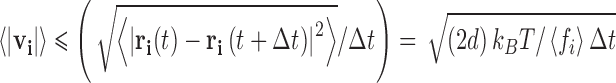 |
14 |
By the same argument as adopted for the small time step, the case above the magnitude of the random force term can be written by Eq. 15.
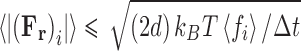 |
15 |
Using the Langevin equation to simulate diffusion at different time scales
The approximate relationships developed for the three force terms can be inserted into Eq. 6b and subsequently directly integrated over the time interval t-to = ∆t11, to yield Eq. 16.
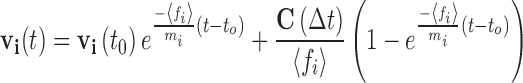 |
16a |
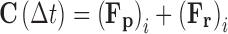 |
16b |
If Eqs. 16a and 16b are further integrated over a short time step (∆t < ∆τ) some of the information relating to the velocity previously held by the particle is retained (Eq. 17a). At longer time steps (∆t > ∆τ) the contributions from the exponential terms becomes relatively minor and a description of the over-damped Brownian dynamics algorithm (Ermack 1975; Ermack and McCammon 1978) is recovered (Eq. 17b).
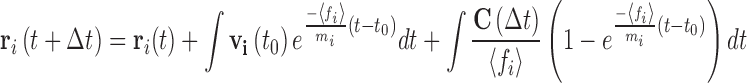 |
17a |
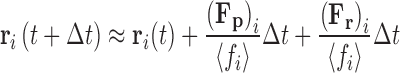 |
17b |
The upper limit for the time step, τB = ∆t, chosen for numerical integration in the Brownian dynamics regime corresponds to a value which prevents particles from passing through each other over the interval. For systems composed of particles of a single radius Ri, τB ∼ R2i <fi>/(2d kBT). Due to its simplicity and ability to access larger time scales, Eq. 17b is particularly suitable for examining diffusion in crowded solution environments.
For simulating Brownian motion in a homogenous phase over time scales, ∆t >> τB Eq. 17b can be first be used to produce a sufficiently long time simulation to allow for the desired time interval diffusion tensor, D
i(n∆t), to be either directly calculated from the data using the methods shown in Eq. 3 or alternatively estimated by extrapolation. This diffusion tensor can then be used in conjunction with a unit vector 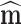 to simulate Brownian motion over the homogenous phase using relatively large time steps as a form of the position Langevin simulation method (Lax 1966; Zwanzig 1969; Hall 2008; Hall 2010) (Eq. 18a). In this approach each component in the displacement vector, ∆r
i(n∆t), is randomly chosen from a Gaussian distribution of zero mean and standard deviation given by the component term indicated by Eq. 18b.
to simulate Brownian motion over the homogenous phase using relatively large time steps as a form of the position Langevin simulation method (Lax 1966; Zwanzig 1969; Hall 2008; Hall 2010) (Eq. 18a). In this approach each component in the displacement vector, ∆r
i(n∆t), is randomly chosen from a Gaussian distribution of zero mean and standard deviation given by the component term indicated by Eq. 18b.
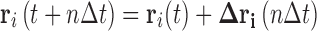 |
18a |
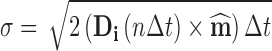 |
18b |
Discussion of recent experimental findings
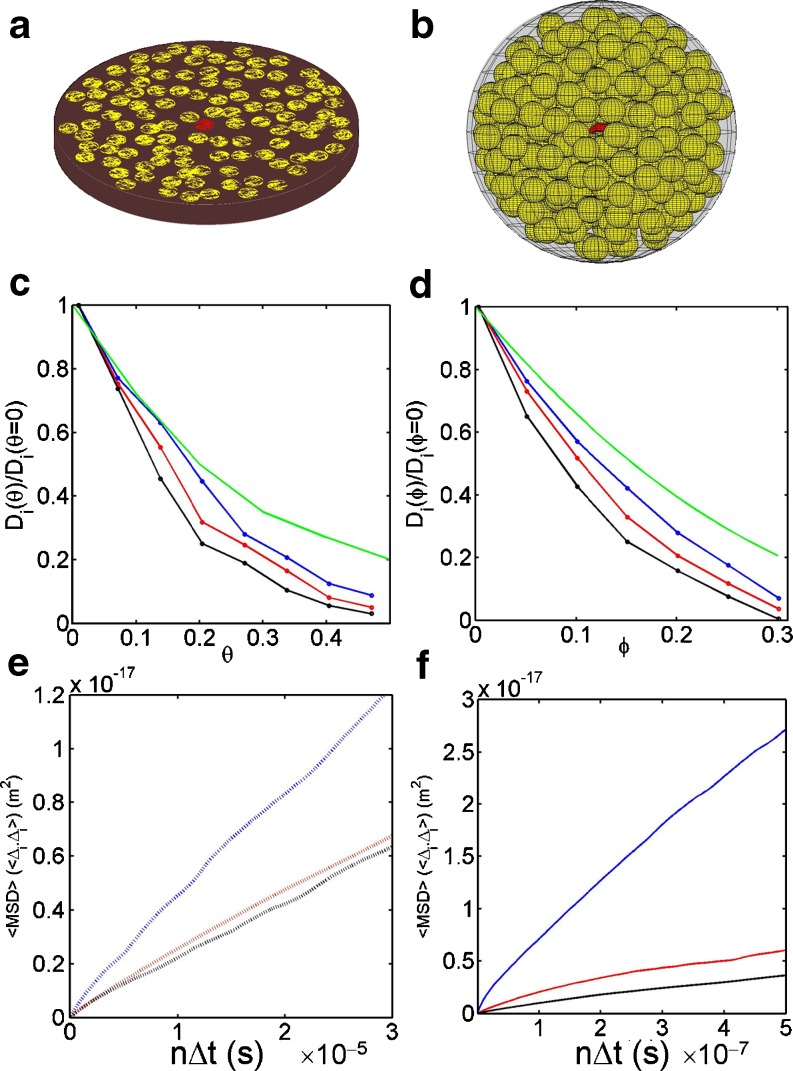
From the very earliest studies of diffusive motion in the 2D and 3D biological environments of the cell membrane and cell cytosol, strong differences to the simple behaviour predicted for continuum fluids have been observed (e.g. 3D: Jacobson and Wojcieszyn 1984; Gershon et al. 1985; Arrio-Dupont et al. 2000; Goulian and Simon 2000; e.g. 2D: Schlessinger et al. 1977; Sheetz et al. 1980; Haggie and Verkman 2002; Sakaki et al. 1982). Recently, rather than being interpreted as just unnecessary complications, these effects are now seen as probable design features of the cellular reaction environment that have resulted from the natural optimization process of evolution (Kurganov et al. 1985; Bray 1998; Luby-Phelps et al. 1988; Burdzy and Hołyst 2001; Haggie and Verkman 2002; Schnell and Turner 2004; Sear 2005; Ma'ayan et al. 2005; Weiss 2008). In this vein, the degree of anomalous diffusion in the cytoplasm has been interpreted as a means of tuning the search process for an interacting partner (Burz et al. 2006; Iwahara and Clore 2006;Tang et al. 2006) in crowded solution conditions by helping to both spatially determine the location of the interacting partner and also to enhance the probability of encounter (Weiss 2008; Szymanski and Weiss 2009). In cells lacking significant compartmentalization and structural organization (e.g. prokaryotes), the intracellular 2D and 3D fluids might be approximated by concentrated macromolecular environments (Zimmerman and Trach 1991; Konopka et al. 2006). Using the Brownian dynamics algorithm presented in Eq. 17b, we provide example simulations of 2D and 3D diffusion in a crowded solution of particles of identical size (Fig. 7). Estimates of the long time tracer diffusion coefficient12 indicate that for intermediate crowding regimes, the effect of a repulsive potential is to increase the magnitude of the short-time diffusion coefficient by helping to keep the tracer particle out of the ‘stagnant’ region associated with high local densities of background particles. Not surprisingly, the effect of a weakly attractive potential shows the reverse behaviour, with the tracer diffusion coefficient decreasing due to a combination effect arising from the attractive intermolecular potential (i.e. larger particles diffuse more slowly) and the HI forces (i.e. the tracer spends more time located in higher viscosity regions). The purely hard sphere tracer case with no associated potential was also simulated and is intermediate between the two13. The time dependence of the diffusion coefficient was also analysed and is displayed in Fig. 7 e, f for the three different types of associated intermolecular potential. All three examples demonstrated anomalous diffusion characteristics (α ≈ 0.85) similar in magnitude to those previously measured (Banks and Fradin 2005; Szymanski and Weiss 2009).
Fig. 7.
Effect of crowding on diffusion Top panel (a and b) Schematic showing diffusion of a tracer particle in crowded and confined 2D and 3D fluids of same sized particles (tracer R = 2 nm, H = 5 nm, fluid vessel radius 30 nm). Central panel (c and d) Normalized long-time tracer diffusion coefficients as a function of fractional area, θ, or volume, ϕ: occupation for repulsive saw tooth (blue), attractive saw tooth (black) and hard particle (red) intermolecular potentials operative between tracer and crowder molecules (ε = +kBT, -kBT or 0, Lij = Ri) (all trajectories were ended before approaching the wall). Green line indicates reduction in long-time tracer diffusion for hard particle cases as calculated for unconfined 2D fluids by Bussell et al. 1995 and in unconfined 3D fluids by Tokuyama and Oppenheim 1994. Bottom panel (e and f) Averaged diagonal elements of the mean squared displacements from the diffusion tensor for repulsive (blue), attractive (black) and hard particle only (red) for the most crowded cases considered in 2D and 3D, respectively
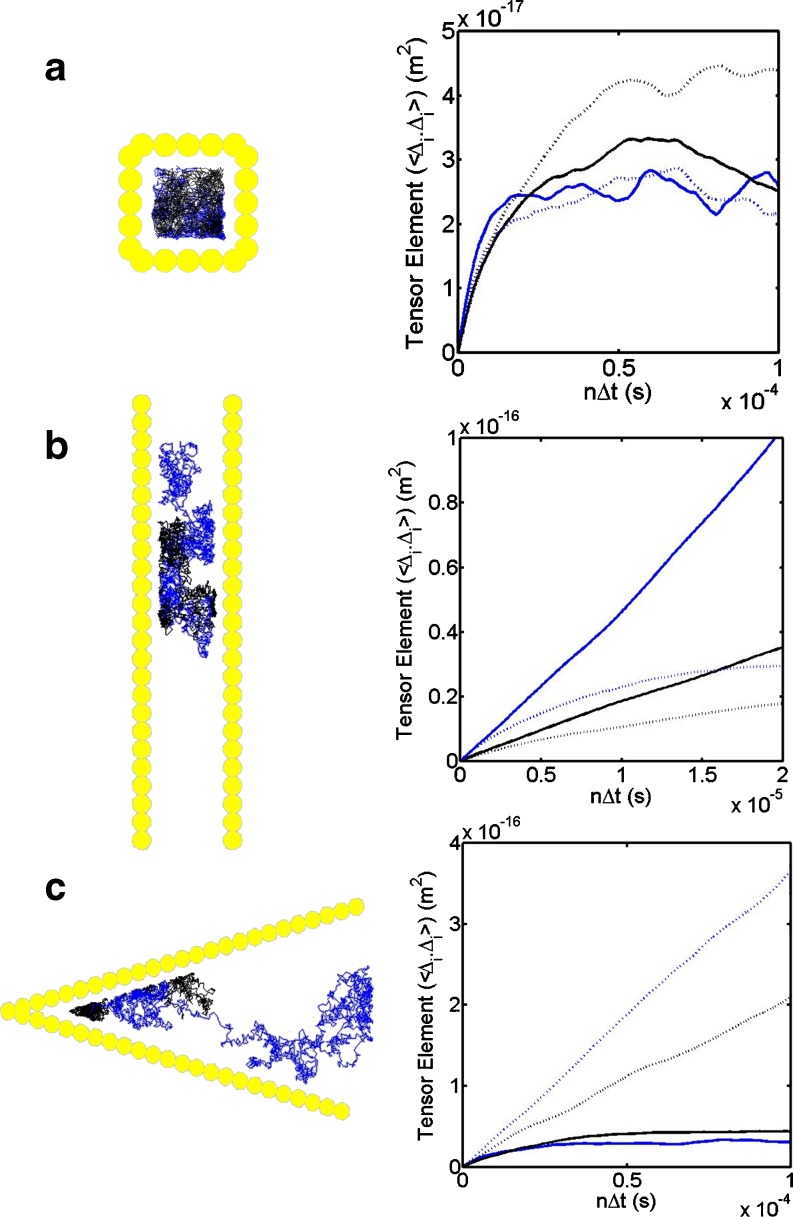
The existence within the cell of internal membrane compartments along with the numerous fibrous elements associated with the cellular cytoskeleton means that certain regions of volume (Blum et al. 1989; Provance et al. 1993; Goulian and Simon 2000; Sanabria et al. 2007; Klann et al. 2009) or area (Sheetz et al. 1980; Saxton 1995; Kusumi et al. 2005) may be effectively caged or confined. The partitioning of the cell membrane into such caged regions has been interpreted as a means for forming reaction modules (Fig. 8a) (Kurganov et al. 1985; Bray 1998; Kusumi et al. 2005; Ma'ayan et al. 2005). For the case of caging (Fig. 8a), our example simulations are in general agreement with the predictions and observations of hop diffusion (Saxton 1995; Saxton 2008; Kusumi et al. 2005), with the mean squared displacement attaining a horizontal asymptote that is dependent upon the permeability of the confining elements. Interestingly, only small differences are seen for molecules having different limiting case intermolecular potential behaviours, although this difference would be expected to increase with increasing potential magnitude. Additionally, the intrinsic asymmetry of the fibrous elements can impart orientation derived ordering effects similar to those observed in liquid crystal theory (Hentschke and Herzfeld 1989; Herzfeld 2004), leading to regions that are capable of generating altered diffusion behaviour that is direction-dependent (Blum et al. 1989; Han and Herzfeld 1993; Kang et al. 2007). We highlight some of these different findings in Fig. 8b and c using example simulations showing the general effects of confinement and ordering for the 2D cases of an open channel (Fig. 8b) and a confined wedge (Fig. 8c). Analysis of the diffusion tensor indicates that the structures have an intrinsic anisotropy generating function that is dependent upon both the width and angle of alignment in accordance with previous work (Blum et al. 1989; Han and Herzfeld 1993; Kang et al. 2007).
Fig. 8.
Effect of different types of confinement on the diffusion of a tracer particle (R = 2 nm) in 2D and subsequent analysis of its motion in terms of the diagonal diffusion tensor components (dotted lines <∆x∆x>, solid lines <∆y∆y>) for the cases of weak repulsive saw tooth (blue) and weak attractive saw tooth (black) intermolecular potentials operating between tracer and confining elements. Top panel Diffusion of tracer particle in a cage of diameter 20 nm and anomalous characteristic flattening of the MSD plot. Central panel Diffusion of tracer particle in a channel bounded in the x direction (width 10 nm) but unbounded in the y direction. Characteristic anomalous flattening of the MSD plot for the x component but ‘normal’ behaviour shown by the y component (with repulsive potential case, indicating greater diffusion rate in both y and x directions than attractive potential case). Bottom panel Diffusion of tracer particle in a sector shaped channel (angle =30°) unbounded in the positive x direction
Modelling approaches capable of accounting for such complex diffusive behaviour as that described above are required if we are to develop predictive models of intracellular diffusion. Towards the development of a such a predictive theory explicit accounting for the spatial dimension is an important first step. Methods for factoring in the dimensions of the cellular reaction vessels have been implemented in programs such as MCELL (Czech et al. 2009), VIRTUAL CELL (Moraru et al. 2009), E-CELL (Takahashi et al. 2005) and STEPS (Wils and De Schutter 2009) by incorporation of structures from confocal or EM image traces. However, as we have noted in this review diffusive, transport processes occurring in solution are not just affected by geometric considerations, but also by a combination of intermolecular potentials, hydrodynamic interactions and random forces effected by the solvent. Recent modelling and simulation efforts have started to address some of these issues (Bicout and Field 1996; Andrews and Bray 2004; Takahashi et al. 2005; Jeschke 2008; Czech et al. 2009; Moraru et al. 2009; Wils and De Schutter 2009). It is towards the goal of providing a readily digestible review of the general physical factors affecting Brownian behaviour that we have endeavoured to achieve here.
Concluding remarks
The general area of study relating to how the complex structured microenvironment of the cell affects the fate of biochemical reactions occurring within it has been termed as ‘macromolecular crowding and confinement’ or simply ‘crowding’ for short (Minton and Wilf 1981; Harris and Winzor 1985; Minton 1992; Zhou et. al. 2008). Despite the fascination that ‘biological chemistry’ holds for many, the study of intracellular crowding effects is a difficult subject area in which to both design and conduct experiments and to theoretically interpret the experimental results (Zimmerman and Minton 1993; Hall and Minton 2003; Zhou et al. 2008). In conventional biochemical investigations complications arising from unwanted side-reactions are made tractable via reduction of the number and extent of variable system components (Goto et al 1993; Hoshino et al 1997). However, even in in vitro model system approximations of crowding, such an option is limited when the objective is to examine the effect of a complex and highly concentrated background environment of the biochemical reaction at hand. The situation becomes more difficult when studying crowding in, in vivo, model systems due to a lack of definition/knowledge of both the composition (Zimmerman and Trach 1993; Ma'ayan et al. 2005) and position (Luby-Phelps et al. 1988; Diaspro 2002; Leis et al. 2009) of the system components. Often times, the irreducible nature of the experimental system can lead to the presence of multiple competing effects which can confound the ready application of theoretical methods. As such, the ability of modelling approaches for generating easily accessible guiding principles has generally been inversely proportional to their degree of realism (Zimmerman and Minton 1993; Schnell and Turner 2004; Sear 2005; Hall and Dobson 2006; McGuffee and Elcock 2006; Qin and Zhou 2009). Relatively simple models of diffusion in crowded solution have provided considerable insight into diffusive behaviour within cells (Muramatsu and Minton 1988; Minton 1989; Han and Herzfeld 1993; Hall 2008, Hall 2010). Recent advances in both single particle tracking (Saxton and Jacobson 1997, Kusumi et al. 2005; Murcia et al. 2007; Hellriegel and Gratton 2009) and single particle simulation methods (Bernadó et al. 2004; Sun and Weinstein 2007; Dix and Verkman 2008; Ridgway et. al. 2008; Weiss 2008) hold the promise that as we move into the new decade our potential to understand diffusive behaviour within the complex confines of the cell will continue to grow – perhaps putting a predictive theory of diffusion within the cell within eventual reach.
Acknowledgements
We would like to acknowledge Prof. M. Okuwaki, Prof. D. Stanley, Dr. N. Hirota and Dr. M. Nemoto for providing helpful comments on an early draft of this manuscript. We also greatly appreciate the insightful reviews provided by Dr. Allen P. Minton and the anonymous reviewer 2 which helped to significantly improve the paper. This research was supported by grants to D.H. from the Japanese Science and Technology Agency (JST) and the University of Tsukuba under the special coordinated scheme ‘Funds in Aid for the Promotion of Young Scientists’ Independent Research’ and to M.H. from ‘Grants-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science and Technology’.
Footnotes
This review is restricted to the discussion of diffusion in low Reynolds number fluid systems (the Reynolds number is a dimensionless parameter reflecting the relative importance of inertial to viscous forces), which are good approximations of the intracellular environment.
After the botanist, Robert Brown, who first noted it while observing pollen grains under a microscope in the early 1800s.
Adolf Fick published a series of papers on diffusion in 1855 (a modern analysis of his observations of the diffusion of salt is provided by Philibert 2006). By likening the diffusion of ions in solution to the flow of heat, Fick was able to adopt the mathematical methods developed by Fourier, thus explaining why the diffusion equation is near identical to equations developed for modelling heat flow.
Phenomenological as of the time of Fick.
Einstein was first, closely followed by Smoluchowski second; therefore, the relationship is alternately called the Einstein relation or the Einstein–Smoluchowski equation.
Isotropy refers to the equivalence of a given process along different directions through the medium.
In this instance directed motion refers to the short steps with transient direction associated with Brownian motion.
Reflective of HI only.
Not including the volume or area occupied by the i-th molecule under consideration.
The value of q = 1 at time scales small enough (∆t < m/<fi>) to preserve inertial memory, whereas q = 2 for time scales greater than this ((∆t > m/<fi>).
Note this integration is based on the assumption that all force terms are constant over the time interval, ∆t, for which the integration is carried out. Additionally we have introduced a factor q here to describe the different time interval ranges i.e. when ∆t < ∆τ, q = 1 and when ∆t >> ∆τ, q = 2d.
The predicted reduction in diffusion coefficient for the hard cylinder and sphere cases shown in Fig. 7 is slightly steeper than suggested by previous theoretical results for unbounded systems (3D: Tokuyama and Oppenheim, 1994; 2D: Bussell et al. 1995 superimposed in green), with the discrepancy due to our simulations being performed in a repulsive cylindrical/spherical container not featuring periodic boundary conditions.
References
- Alcor D, Gouzer G, Triller A. Single-particle tracking methods for the study of membrane receptors dynamics. Eur J Neurosci. 2009;30:987–997. doi: 10.1111/j.1460-9568.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- Andrews SS, Bray D. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys Biol. 1994;1:137–151. doi: 10.1088/1478-3967/1/3/001. [DOI] [PubMed] [Google Scholar]
- Arrio-Dupont M, Foucault G, Vacher M, Devaux PF, Cribier S. Translational diffusion of globular proteins in the cytoplasm of cultured muscle cells. Biophys J. 2000;78:901–907. doi: 10.1016/S0006-3495(00)76647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DS, Fradin C. Anomalous diffusion of proteins due to molecular crowding. Biophys J. 2005;89:2960–2971. doi: 10.1529/biophysj.104.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor GK. Sedimentation in a dilute polydisperse system of interacting spheres. Part 1. General theory. J Fluid Mech. 1982;119:379–408. doi: 10.1017/S0022112082001402. [DOI] [Google Scholar]
- Batchelor GK (2000) An introduction to fluid dynamics. Cambridge University Press, Cambridge
- Baumeister W. Mapping molecular landscapes inside cells. Biol Chem. 2004;385:865–872. doi: 10.1515/BC.2004.113. [DOI] [PubMed] [Google Scholar]
- Berg HC (1993) Random walks in biology. Princeton University Press, Princeton
- Bernadó P, García de la Torre J, Pons M. Macromolecular crowding in biological systems: hydrodynamics and NMR methods. J Mol Recognition. 2004;17:397–407. doi: 10.1002/jmr.694. [DOI] [PubMed] [Google Scholar]
- Bicout DJ, Field MJ. Stochastic dynamics simulations of macromolecular diffusion in a model of the cytoplasm of E. coli. J Phys Chem. 1996;100:2489–2497. doi: 10.1021/jp9525191. [DOI] [Google Scholar]
- Blum JJ, Lawler G, Reed M, Shin I. Effect of cytoskeletal geometry on intracellular diffusion. Biophys J. 1989;56:995–1005. doi: 10.1016/S0006-3495(89)82744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. Signalling complexes: biophysical constraints on intracellular communication. Annu Rev Biophys Biomol Struct. 1998;27:59–75. doi: 10.1146/annurev.biophys.27.1.59. [DOI] [PubMed] [Google Scholar]
- Burdzy K, Hołyst R. Mechanisms for facilitated target location and the optimal number of molecules in the diffusion search process. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;64:011914–011915. doi: 10.1103/PhysRevE.64.011914. [DOI] [PubMed] [Google Scholar]
- Burz DS, Dutta K, Cowburn D, Shekhtman A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR) Nat Methods. 2006;3:91–93. doi: 10.1038/nmeth851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell SJ, Koch DL, Hammer DA. Effect of hydrodynamic interactions on the diffusion of integral membrane proteins: tracer diffusion in organelle and reconstituted membranes. Biophys J. 1995;68:1828–1835. doi: 10.1016/S0006-3495(95)80359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron Bird R, Stewart WE, Lightfoot EN (2002) Chapter 2 of Transport phenomena, 2nd edn. John Wiley and Sons, London
- Chiantia S, Ries J, Schwille P. Fluorescence correlation spectroscopy in membrane structure elucidation. Biochim Biophys Acta. 2009;1788:225–233. doi: 10.1016/j.bbamem.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Czech J, Dittrich M, Stiles JR. Rapid creation, Monte Carlo simulation, and visualization of realistic 3D cell models. Methods Mol Biol. 2009;500:237–287. doi: 10.1007/978-1-59745-525-1_9. [DOI] [PubMed] [Google Scholar]
- de Brabander M, Nuydens R, Ishihara A, Holifield B, Jacobsen K, Geerts H. Lateral diffusion and retrograde movements of individual cell surface components on single motile cells observed with nanovid microscopy. J Cell Biol. 1991;112:111–124. doi: 10.1083/jcb.112.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhont JKG (1996) An introduction to dynamics of colloids. In: Möbius D, Miller R (eds) Studies of interface science. Elsevier, Amsterdam, pp 327–329
- Diaspro A (ed) (2002) Confocal and two-photon microscopy: foundations, applications and advances. Wiley Liss, London
- Dix J, Verkman A. Crowding effects on diffusion in solutions and cells. Annu Rev Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- Doi M, Edwards S (1999) Theory of polymer dynamics. Clarendon Press, Oxford
- Doster W, Longeville S. Microscopic diffusion and hydrodynamic interactions of hemoglobin in red blood cells. Biophys J. 2007;93:1360–1368. doi: 10.1529/biophysj.106.097956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein A. Investigations on the theory of the Brownian movement. New York: Dover Publ; 1956. [Google Scholar]
- Elcock AH. Atomic-level observation of macromolecular crowding effects. Proc Natl Acad Sci USA. 2003;100:2340–2344. doi: 10.1073/pnas.0535055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elimelech M, Gregory J, Jia X, Williams R (1995) Particle deposition and aggregation: measurement modelling and simulation. Butterworth and Heinemann, Oxford
- Elson EL. A quick tour of fluorescence correlation spectroscopy from its inception. J Biomed Opt. 2004;9:857–864. doi: 10.1117/1.1779234. [DOI] [PubMed] [Google Scholar]
- Ermak DL. A computer simulation of charged particles in solution I. Technique and equilibrium properties. J Chem Phys. 1975;62:4189–4196. doi: 10.1063/1.430300. [DOI] [Google Scholar]
- Ermak DL, McCammon JA. Brownian dynamics with hydrodynamic interactions. J Chem Phys. 1978;69:1352–1360. doi: 10.1063/1.436761. [DOI] [Google Scholar]
- Falck E, Lahtinen JM, Vattulainen I, Ala-Nissila T. Influence of hydrodynamics on many particle diffusion in 2D colloidal suspensions. Eur Phys J E. 2004;13:267–275. doi: 10.1140/epje/i2003-10075-9. [DOI] [PubMed] [Google Scholar]
- Fernandes MX, de la Torre JG. Brownian dynamics simulation of rigid particles of arbitrary shape in external fields. Biophys J. 2002;83:3039–3048. doi: 10.1016/S0006-3495(02)75309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey E, Kroy K. Brownian motion: a paradigm of soft matter and biological physics. Ann Phys. 2005;14:20–50. [Google Scholar]
- Fulinski A. On Marain Smoluchowski’s life and contribution to physics. Acta Phys Polonica B. 1998;29:1523–1537. [Google Scholar]
- Geerts H, de Brabander M, Nuydens R. Nanovid microscopy. Nature. 1991;331:450–453. doi: 10.1038/351765a0. [DOI] [PubMed] [Google Scholar]
- Gershon ND, Porter KR, Trans BL. The cytoplasmic matrix: its volume and surface area and the diffusion of molecules through it. Proc Natl Acad Sci USA. 1985;82:030–5034. doi: 10.1073/pnas.82.15.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer T, Winter U. An O(N2) approximation for hydrodynamic interactions in Brownian dynamics simulations. J Chem Phys. 2009;21:114905. doi: 10.1063/1.3089668. [DOI] [PubMed] [Google Scholar]
- Goto Y, Hagihara Y, Hamada D, Hoshino M, Nishii I. Acid-induced unfolding and refolding transitions of cytochrome c: a three-state mechanism in H2O and D2O. Biochemistry. 1993;32:11878–11885. doi: 10.1021/bi00095a017. [DOI] [PubMed] [Google Scholar]
- Goulian M, Simon SM. Tracking single proteins within cells. Biophys J. 2000;79:2188–2198. doi: 10.1016/S0006-3495(00)76467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Har JY, Sankaran J, Hong Y, Kannan B, Wohland T. Molecular diffusion measurement in lipid bilayers over wide concentration ranges: a comparative study. Chem Physchem. 2008;9:721–728. doi: 10.1002/cphc.200700611. [DOI] [PubMed] [Google Scholar]
- Haggie PM, Verkman AS. Diffusion of tricarboxylic acid cycle enzymes in the mitochondrial matrix in vivo. Evidence for restricted mobility of a multienzyme complex. J Biol Chem. 2002;277:40782–40788. doi: 10.1074/jbc.M207456200. [DOI] [PubMed] [Google Scholar]
- Hall D. Analysis and interpretation of two-dimensional single-particle tracking microscopy measurements: effect of local surface roughness. Anal Biochem. 2008;377:24–32. doi: 10.1016/j.ab.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Hall D (2010) Effect of heterogeneity on the characterization of cell membrane compartments: I. Uniform size and permeability. Anal Biochem (in press) [DOI] [PubMed]
- Hall D, Dobson CM. Expanding to fill the gap: a possible role for inert biopolymers in regulating the extent of the 'macromolecular crowding' effect. FEBS Lett. 2006;580:2584–2590. doi: 10.1016/j.febslet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Hall D, Minton AP. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim Biophys Acta. 2003;1649:127–139. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- Han J, Herzfeld J. Macromolecular diffusion in crowded solutions. Biophys J. 1993;65:1155–1161. doi: 10.1016/S0006-3495(93)81145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SJ, Winzor DJ. Effect of thermodynamic nonideality on the subcellular distribution of enzymes: adsorption of aldolase to muscle myofibrils. Arch Biochem Biophys. 1985;243:598–604. doi: 10.1016/0003-9861(85)90537-5. [DOI] [PubMed] [Google Scholar]
- Hellriegel C, Gratton E. Real-time multi-parameter spectroscopy and localization in three-dimensional single-particle tracking. J R Soc Interface. 2009;6(Suppl 1):S3–S14. doi: 10.1098/rsif.2008.0313.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschke R, Herzfeld J. Nematic behaviour of reversibly of reversibly polymerising proteins. J Chem Phys. 1989;90:5094–5101. doi: 10.1063/1.456692. [DOI] [Google Scholar]
- Herzfeld J. Crowding-induced organization in cells: spontaneous alignment and sorting of filaments with physiological control points. J Mol Recognition. 2004;17:376–381. doi: 10.1002/jmr.703. [DOI] [PubMed] [Google Scholar]
- Hibino K, Hiroshima M, Takahashi M, Sako Y. Single-molecule imaging of fluorescent proteins expressed in living cells. Methods Mol Biol. 2009;544:451–460. doi: 10.1007/978-1-59745-483-4_30. [DOI] [PubMed] [Google Scholar]
- Heyes DM. Mean-field hydrodynamics Brownian dynamics simulations of viscosity and self diffusion of near-hard-sphere colloidal liquids. J Phys Condens Matter. 1995;7:8857–8865. doi: 10.1088/0953-8984/7/47/006. [DOI] [Google Scholar]
- Hoshino M, Yumoto N, Yoshikawa S, Goto Y. Design and characterization of the anion-sensitive coiled-coil peptide. Protein Sci. 1997;6:1396–1404. doi: 10.1002/pro.5560060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- Jacobson K, Wojcieszyn J. The translational mobility of substances within the cytoplasmic matrix. Proc Natl Acad Sci USA. 1984;81:6747–6751. doi: 10.1073/pnas.81.21.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson DM, Ross JA, Albanesi JP. Fluorescence fluctuation spectroscopy: ushering in a new age of enlightenment for cellular dynamics. Biophys Rev. 2009;3:105–118. doi: 10.1007/s12551-009-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke M (2008) Multi-resolution spatial simulation for molecular crowding. In: Proc 40th Conf Winter Simulation, pp1384–1392
- Jin S, Verkmann AS. Single particle tracking of complex diffusion in membranes: simulation and detection of barrier, raft and interaction phenomena. J Phys Chem B. 2007;111:3625–3632. doi: 10.1021/jp067187m. [DOI] [PubMed] [Google Scholar]
- Jones RAL (2002) Soft condensed matter. Oxford University Press, Oxford
- Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Wilk A, Patkowski A, Dhont JK. Diffusion of spheres in isotropic and nematic networks of rods: electrostatic interactions and hydrodynamic screening. J Chem Phys. 2007;126(214501):1–17. doi: 10.1063/1.2737446. [DOI] [PubMed] [Google Scholar]
- Klann MT, Lapin A, Reuss M. Stochastic simulation of signal transduction: impact of the cellular architecture on diffusion. Biophys J. 2009;96:5122–5129. doi: 10.1016/j.bpj.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka MC, Shkel IA, Cayley S, Record MT, Weisshaar JC. Crowding and confinement effects on protein diffusion in vivo. J Bacteriol. 2006;188:6115–6123. doi: 10.1128/JB.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo R. The fluctuation dissipation theorem. Rep Prog. Phys. 1966;29:255–284. doi: 10.1088/0034-4885/29/1/306. [DOI] [Google Scholar]
- Kurganov BI, Sugrobova NP, Mil’man LS. Supramolecular organization of glycolytic enzymes. J Theor Biol. 1985;116:509–526. doi: 10.1016/S0022-5193(85)80086-2. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- Langevin P (1908) Sur la thėorie du mouvement brownien. C.R. Acad. Sci. (Paris) 146: 530–533 [As translated by Lemons DS, Gythiel A (1995) Paul Langevin’s 1908 paper “On the Theory of Brownian Motion”. Am J Phys 65:1079–1081]
- Lax M. Classical noise. IV. Langevin methods. Rev Mod Phys. 1966;38:541–566. doi: 10.1103/RevModPhys.38.541. [DOI] [Google Scholar]
- Leach AR (2001) Molecular modelling: principles and applications, 2nd edn. Pearson–Prentice Hall, Upper Saddle River
- Leis A, Rockel B, Andrees L, Baumeister W. Visualizing cells at the nanoscale. Trends Biochem Sci. 2009;36:60–70. doi: 10.1016/j.tibs.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Lorén N, Nydén M, Hermansson AM. Determination of local diffusion properties in heterogeneous biomaterials. Adv Colloid Interface Sci. 2009;150:5–15. doi: 10.1016/j.cis.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K, Lanni F, Taylor DL. The submicroscopic properties of cytoplasm as a determinant of cellular function. Annu Rev Biophys Biophys Chem. 1988;17:369–396. doi: 10.1146/annurev.bb.17.060188.002101. [DOI] [PubMed] [Google Scholar]
- Ma'ayan A, Blitzer RD, Iyengar R. Toward predictive models of mammalian cells. Annu Rev Biophys Biomol Struct. 2005;34:319–349. doi: 10.1146/annurev.biophys.34.040204.144415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffee SR, Elcock AH. Atomically detailed simulations of concentrated protein solutions: the effects of salt, pH, point mutations, and protein concentration in simulations of 1000-molecule systems. J Am Chem Soc. 2006;128:12098–12110. doi: 10.1021/ja0614058. [DOI] [PubMed] [Google Scholar]
- Minton AP. Lateral diffusion of membrane proteins in protein-rich membranes. A simple hard particle model for concentration dependence of the two-dimensional diffusion coefficient. Biophys J. 1989;55:805–808. doi: 10.1016/S0006-3495(89)82880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP. Confinement as a determinant of macromolecular structure and reactivity. Biophys J. 1992;63:1090–1100. doi: 10.1016/S0006-3495(92)81663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP, Wilf J. Effect of macromolecular crowding upon the structure and function of an enzyme: glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1981;20:4821–4826. doi: 10.1021/bi00520a003. [DOI] [PubMed] [Google Scholar]
- Mirosavljević K, Noethig-Laslo V. Spin labelling study of interfacial properties of egg-phosphatidylcholine liposomes as a function of cholesterol concentrations. Chem Phys Lipids. 2008;155:74–79. doi: 10.1016/j.chemphyslip.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Moraru II, Schaff JC, Slepchenko BM, Blinov ML, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM. Virtual Cell modelling and simulation software environment. IET Syst Biol. 2009;2:352–362. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KF, Johnson CS, Jr, Andrees L, Baumeister W. Diffusion-ordered two-dimensional nuclear magnetic resonance spectroscopy. J Am Chem Soc. 1992;114:3139–3141. doi: 10.1021/ja00034a071. [DOI] [Google Scholar]
- Muramatsu N, Minton AP. Tracer diffusion of globular proteins in concentrated protein solutions. Proc Natl Acad Sci USA. 1988;85:2984–2988. doi: 10.1073/pnas.85.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia MJ, Garg S, Naumann CA. Single-molecule fluorescence microscopy to determine phospholipid lateral diffusion. Methods Mol Biol. 2007;400:277–294. doi: 10.1007/978-1-59745-519-0_19. [DOI] [PubMed] [Google Scholar]
- Padding JT, Louis AA. Hydrodynamic interactions and Brownian forces in colloidal suspensions: coarse graining over time and length scales. Phys Rev E. 2006;74031402:1–29. doi: 10.1103/PhysRevE.74.031402. [DOI] [PubMed] [Google Scholar]
- Peetermans JA, Foy BD, Tanaka T. Accumulation and diffusion of crystallin inside single fiber cells in intact chicken embryo lenses. Proc Natl Acad Sci USA. 1987;84(7):1727–1730. doi: 10.1073/pnas.84.7.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin J (1910) Brownian movement and molecular reality (translated edition published by Taylor and Francis, London, 1910)
- Peskir G. On the diffusion coefficient: the Einstein relation and beyond. Stochastic Models. 2003;19:383–405. doi: 10.1081/STM-120023566. [DOI] [Google Scholar]
- Philibert J. One and a half century of diffusion: Fick, Einstein, before and beyond. Diffusion-Fundamentals. 2006;6:1–19. [Google Scholar]
- Provance DW, Jr, McDowall A, Marko M, Luby-Phelps K. Cytoarchitecture of size-excluding compartments in living cells. J Cell Sci. 1993;106:565–578. doi: 10.1242/jcs.106.2.565. [DOI] [PubMed] [Google Scholar]
- Qin S, Zhou HX. Atomistic modeling of macromolecular crowding predicts modest increases in protein folding and binding stability. Biophys J. 2009;97:12–19. doi: 10.1016/j.bpj.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan DG, Kuchel PW. Simulations of NMR-detected diffusion in suspensions of red cells: the effects of variation in membrane permeability and observation time. Eur Biophys J. 2003;32:671–675. doi: 10.1007/s00249-003-0331-x. [DOI] [PubMed] [Google Scholar]
- Ridgway D, Broderick G, Lopez-Campistrous A, Ru’aini M, Winter P, Hamilton M, Boulanger P, Kovalenko P, Ellison M. Coarse-grained molecularsimulation of diffusion and reaction kinetics in a crowdedvirtual cytoplasm. Biophys J. 2008;17(5):493–498. doi: 10.1529/biophysj.107.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotne J, Prager S. Variational treatment of hydrodynamic interaction in polymers. J Chem Phys. 1969;50:4831–4841. doi: 10.1063/1.1670977. [DOI] [Google Scholar]
- Saffman PG, Delbrűck M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T, Tsuji A, Chang CH, Ohnishi S. Rotational mobility of an erythrocyte membrane integral protein band 3 in dimyristoylphosphatidylcholine reconstituted vesicles and effect of binding of cytoskeletal peripheral proteins. Biochemistry. 1982;21:2366–2372. doi: 10.1021/bi00539a014. [DOI] [PubMed] [Google Scholar]
- Sanabria H, Kubota Y, Waxham MN. Multiple diffusion mechanisms due to nanostructuring in crowded environments. Biophys J. 2007;92:313–322. doi: 10.1529/biophysj.106.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Single-particle tracking: effects of corrals. Biophys J. 1995;69:389–398. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. A biological interpretation of transient anomalous subdiffusion. I. Qualitative model. Biophys J. 2007;92:1178–1191. doi: 10.1529/biophysj.106.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Modeling 2D and 3D diffusion. Methods Mol Biol. 2008;400:295–321. doi: 10.1007/978-1-59745-519-0_20. [DOI] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single Particle Tracking: applications and membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Elson EL, Webb WW, Yahara I, Rutishauser U, Edelmane GM. Receptor diffusion on cell surfaces modulated by locally bound concanavalin A. Proc Natl Acad Sci. 1977;74:1110–1114. doi: 10.1073/pnas.74.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell S, Turner TE. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Progr Biophys Mol Biol. 2004;85:235–260. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Sear RP. The cytoplasm of living cells: a functional mixture of thousands of components. J Phys Condens Matter. 2005;17:S3587–S3595. doi: 10.1088/0953-8984/17/45/052. [DOI] [Google Scholar]
- Sheetz MP, Schindler M, Koppel DE. Lateral mobility of integral membrane proteins is increased in spherocytic erythrocytes. Nature. 1980;285:510–511. doi: 10.1038/285510a0. [DOI] [PubMed] [Google Scholar]
- Snook I (2006) The Langevin and generalized Langevin approaches to the dynamics of atomic, polymeric and colloidal systems, 1st edn. Elsevier, Amsterdam
- Spooner P, Jr, Friesen RHE, Knol J, Poolman B, Watts A. Rotational mobility and orientational stability of a transport protein in lipid membranes. Biophys J. 2000;79(2):756–766. doi: 10.1016/S0006-3495(00)76333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42:288–292. doi: 10.1063/1.1695690. [DOI] [Google Scholar]
- Sun J, Weinstein H. Towards realistic modelling of dynamic processes in cell signalling: quantification of macromolecular crowding effects. J Chem Phys. 2007;127(155105):1–10. doi: 10.1063/1.2789434. [DOI] [PubMed] [Google Scholar]
- Svedberg T, Pederson KO. The Ultracentrifuge. Oxford: Clarendon Press; 1940. [Google Scholar]
- Szymanski J, Weiss M. Elucidating the origin of anomalous diffusion in crowded fluids. Phys Rev Lett. 2009;103:038102–1. doi: 10.1103/PhysRevLett.103.038102. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Arjunan SN, Tomita M. Space in systems biology of signaling pathways-towards intracellular molecular crowding in silico. FEBS Lett. 2005;579:1783–1788. doi: 10.1016/j.febslet.2005.01.072. [DOI] [PubMed] [Google Scholar]
- Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- Tokuyama M, Oppenheim I. Dynamics of hard-sphere suspensions. Phys Rev E. 1994;50:R16–R19. doi: 10.1103/PhysRevE.50.R16. [DOI] [PubMed] [Google Scholar]
- Urbina-Villalba G, García-Sucre M, Toro-Mendoza J. Average hydrodynamic correction for the Brownian dynamics calculation of flocculation rates in concentrated dispersions. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68:061408–1. doi: 10.1103/PhysRevE.68.061408. [DOI] [PubMed] [Google Scholar]
- Wade RC. Brownian dynamics simulations of enzyme substrate encounter. Biochem Soc Trans. 1996;24:254–259. doi: 10.1042/bst0240254. [DOI] [PubMed] [Google Scholar]
- Weiss M. Probing the interior of living cells with fluorescence correlation spectroscopy. Ann N Y Acad Sci. 2008;1130:21–27. doi: 10.1196/annals.1430.002. [DOI] [PubMed] [Google Scholar]
- Wils S, De Schutter E. STEPS: modelling and simulating complex reaction-diffusion systems with python. Front Neuroinformatics. 2009;3:15–23. doi: 10.3389/neuro.11.015.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-V. [DOI] [PubMed] [Google Scholar]
- Zwanzig R. Langevin theory of polymer dynamics in dilute solution. Adv Chem Phys. 1969;15:325–331. doi: 10.1002/9780470143605.ch17. [DOI] [Google Scholar]