Abstract
In this study, HIV strains circulating among military personnel were characterized, in Malabo, the capital city of Equatorial Guinea. One sample was found to be HIV-2 group A while a high degree of genetic diversity was recorded in the pol region of 41 HIV-1-positive samples. CRF02_AG accounted for 53.7% of the strains, and 11 different variants were obtained in the remaining 19 samples: subtype G (n = 3), A3 (n = 2), C (n = 2), CRF26_A5U (n = 2), F2 (n = 1), CRF06 (n = 1), CRF09 (n = 1), CRF11 (n = 1), CRF22 (n = 1), and divergent subtype A (n = 1) and F (n = 1). One strain could not be classified and three were unique recombinants. Analysis of antiretroviral drug resistance mutations revealed two patients each harboring one major mutation, M46I in protease and D67N in reverse transcriptase sequences, respectively. The high genetic diversity and emerging ARV resistance mutations call for frequent surveys and appropriate monitoring of ARV considering the increasing access to ARV in the country.
Sub-Saharan Africa remains the most affected area in the global AIDS epidemics, and accounted for 70% of all HIV infections in 2008 (http://unaids.org). It is now widely accepted that the emergence of human immunodeficiency viruses, HIV-1 and HIV-2, resulted from cross-species transmission from simian immunodeficiency viruses (SIV). At least eight interspecies transmissions implicating SIVsmm (from sooty mangabey monkeys) have occurred, resulting in eight HIV-2 groups, and four independent cross-transmissions from SIVcpzPtt and/or SIVgor infecting chimpanzees and gorillas, respectively, which have given rise to the four groups of HIV-1 (M, N O, and P).1,2 Only HIV-1 group M spread globally and, to date, the remarkable viral diversity of HIV-1 M strains has resulted in the classification of the virus into nine subtypes (A–D, F–H, J–K), subsubtypes (A1–A5, B–D, F1–F2), and at least 43 circulating recombinant forms (CRFs) (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). In addition, there are large numbers of unique recombinants (URFs) and a proportion of viruses that still remains unclassified (U), especially in Central Africa.
The classification of HIV-1/M does not reflect the evolutionary history of the virus, since it followed consecutive samplings of HIV-1 strains, but evidences a considerable heterogeneity in the prevalence and geographic distribution of the various forms of the virus. The classification of HIV strains, however, does allow for the course of the HIV pandemic to be tracked over time. Whereas subtype C predominates in Southern Africa where the highest HIV/AIDS country prevalences are recorded, subtypes A, C, and D predominate in East Africa and CRF02_AG and CRF06_cpx in West Africa.3,4 The highest genetic diversity with cocirculation of all HIV-1 groups, subtypes, and numerous CRFs and URFs of HIV-1 M strains has been reported in Central Africa. This has particularly been seen in the Democratic Republic of Congo (DRC), a country located in the heart of a region suspected to be an early epicenter of the global epidemic.5,6 Less is known of the HIV diversity in Equatorial Guinea (GQ), a small relatively wealthy country in Western Equatorial Africa. Information on HIV diversity has mainly been derived from samples collected in 1999 or from immigrants in Spain.7–10
Here we report the genetic diversity of HIV-1 in individuals identified as HIV positive on the basis of voluntary screening during an HIV prevention and education program among the military services in Malabo, the capital city, from Equatorial Guinea, in March 2008. Participants' age ranged from 18 to 64 years with a median age of 35 years. There were about twice as many military men participating than there were military women (male/female ratio = 2.1).
Blood samples were collected in EDTA tubes using standard methods from informed consenting volunteers from the military services in Malabo, the capital city, and surroundings. HIV screening was done on plasma samples by an approved national algorithm including two rapid tests, Determine HIV1/2 (Abbott, Tokyo, Japan) and Immunocomb HIV 1 + 2 Bispot (PBS Orgenics, Yavne, Israel) followed by confirmation with the Murex HIV Ag/Ab (Abbott, Wiesbaden, Germany). Forty-nine HIV-1-positive samples, for which sufficient plasma was left, were further characterized. To discriminate among HIV types and HIV-1 groups, the plasma samples were first subjected to an indirect ELISA assay using V3 loop peptide antigens specific to HIV-1 groups O, M, N, and HIV-2, as previously described.11 The serological assays identified 48 samples as HIV-1 group M and one sample as HIV-2. RNA was extracted from plasma using the QIAamp Viral RNA extraction kit (Qiagen SA, Courtabeauf, France) followed by RT-PCR to amplify a 1865-base pairs (bp) fragment of the pol gene using a previously described protocol for HIV-1 genotypic drug resistance testing.12 For the HIV-2-infected sample a 344-bp fragment in pol integrase was amplified with previously described universal HIV/SIV primers to confirm the HIV-2 infection.13 The amplified fragments for both HIV-1 and -2 samples were purified using the Geneclean Turbo Kit (Q-Biogen, MPbiomedicals, France) and directly sequenced using BigDye Terminator version 3.1 (Applied Biosystems, Courtaboeuf, France) according to the manufacturer's instructions. Electrophoresis and data collection were done on an Applied Biosystems 3130XL Genetic Analyzer. The sequenced fragments from both strands were reconstituted using Seqman II from the DNAstar package v5.08 (Lasergene, USA).
The newly determined sequences were aligned with known representatives of the different groups, subtypes, subsubtypes, and CRFs. The authors paid particular attention to including all CRFs that circulate in Central and West Africa, including the most recently characterized.14 Positions with any gap between the sequences and areas of uncertain alignment were excluded from the analysis. Pairwise evolutionary distances were estimated with Kimura's two-parameter method. Phylogenetic trees were constructed by the neighbor-joining method, and the reliability of the tree topology was assessed by bootstrap analysis as implemented in Clustal X software. To clearly identify whether a sequence belonged to a subgroup corresponding to a particular subtype or CRF, phylogenetic analysis was done for each sequence individually, then a general tree was drawn, and the clustering of each new sequence between the trees was compared. For simplicity, the general tree was drawn with the minimal numbers of references, i.e., CRFs that are not known from EG and were not represented among the current study were excluded from the tree. The new pol sequences were also further investigated for recombination using Simplot 3.2 beta software (Stuart Ray, http://www.med.jhu.edu/deptmed/sray/). Similarity and bootscan plots were done by moving a window of 350–400 bp with 10–20 bp increments along the sequence alignment. In these two sets of analysis, the new sequences were aligned with consensus sequences (50% threshold) representing all the references from the same alignment used for the phylogenetic analysis.
Phylogenetic analysis showed that the HIV-2 sample belonged to subtype A (data not shown). Among the 48 HIV-1 group M samples, 41 were successfully amplified and sequenced. Insufficient DNA volume in the PCR negative samples did not allow further testing using different primer sets and conditions, but all were confirmed as HIV-1 group M positive in the peptide ELISA.
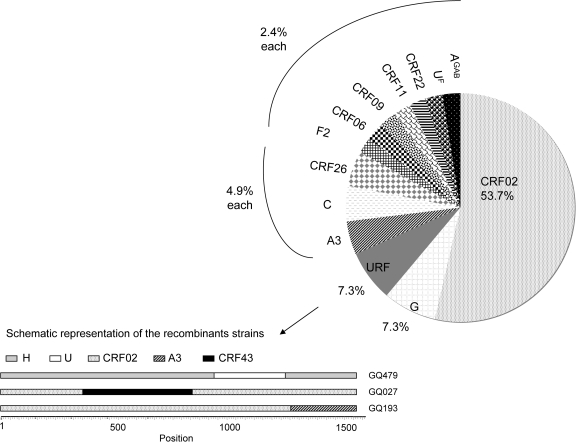
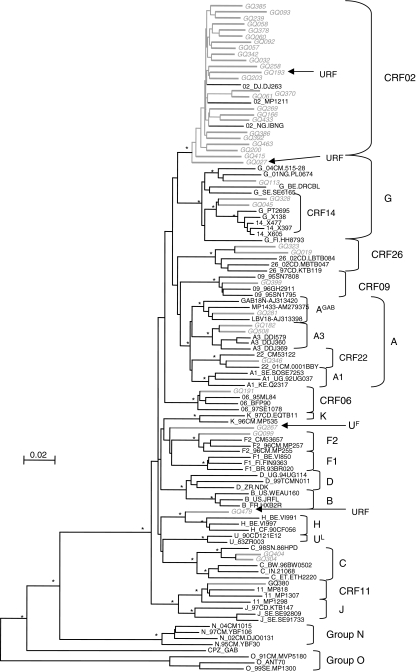
The subtype/CRF distribution is as follows in decreasing order of importance (see Fig. 1): CRF02_AG (n = 22, 53.7 %), subtype G (n = 3, 7.3%), CRF26_A5U (22) (n = 2, 4.9%), subsubtype A3 (n = 2, 4.9%), subtype C (n = 2, 4.9%), subsubtype F2 (n = 1, 2.4%), CRF06cpx (n = 1, 2.4%), CRF09cpx (n = 1, 2.4%), CRF11cpx (n = 1, 2.4%), CRF22cpx (n = 1, 2.4%), a divergent subtype F (UF) sample, and a divergent subtype A strain. This latter strain is closely related to subtype A strains described in Gabon and noted as AGAB in the phylogenetic tree (Fig. 2). Finally, more bootscan and similarity plots on all pol sequences revealed that three strains were unique recombinants: CRF02/CRF43/CRF02, CRF02/A3, and H/U/H (7.3%) and are noted as URF in Fig. 1. Among the three subtype G strains, two were closely related to the CRF14_BG strains, which have been characterized in intravenous drug users in Galicia (Spain) and Portugal.15 The authors postulated that this CRF was probably generated locally with subtype B strains spreading in Europe and subtype G originating from Africa. We therefore studied, in more detail, subtype G sequences from different geographic origins as well as the parental subtype G strains isolated also in Spain (X138) and Portugal (PT2695). The two sequences from Equatorial Guinea are located at the basal position of the CRF14_BG subcluster, suggesting that they may be representative of the original subtype G of African origin implicated in the CRF14_BG structure.
FIG. 1.
Schematic representation of the distribution of the HIV-1 subtypes/CRFs in the military population from Malabo, Equatorial Guinea. The profile of HIV-1 variants was calculated on the basis of 41 samples. Bottom: schematic representation of the structure of the three recombinant strains.
FIG. 2.
Phylogenetic relationships of the pol (protease + RT) sequences from the 41 amplified HIV-1 group M samples from the study population in Equatorial Guinea. The phylogenetic tree analysis was done using the neighbor joining (NJ) method with 100 bootstrap resamplings on 1529 unambiguously aligned nucleotides from the gap-stripped alignment. Sequences from Equatorial Guinea are highlighted in gray and bootstrap values above 90% are indicated with an asterisk (*). The strains identified as unique recombinants after Simplot and bootscan analysis were noted as “URF” and indicated with an arrow.
We also analyzed the amino acid sequences for the presence of drug resistance mutations according to the last update of the WHO list for the surveillance of drug resistance mutations in antiretroviral-naive patients (SDRM version 2009, http://hivdb.stanford.edu/pages/WHOResistanceList.html).16 The M46I major mutation-inducing resistance to protease inhibitors (PI) was detected in a CRF02-AG HIV-1 strain from a 24-year-old woman, in the absence of any other resistance-inducing mutation. A D67N mutation inducing nucleoside reverse transcriptase inhibitor (NRTI) resistance was identified in a 26-year-old man also infected with a CRF02-AG strain. The D67N change contributes to some degree of resistance to each of the NRTIs except lamivudine (3TC) and emtricitabine (FTC), whereas the M46I/L mutation may decrease susceptibility to many protease inhibitors such as atazanavir, indinavir, and lopinavir. when present with other mutations.
Overall, our data suggest that there is high genetic diversity among a small number of HIV-positive individuals belonging to the military services in Malabo, Equatorial Guinea who participated in a voluntary HIV testing campaign. Among the 49 HIV-positive samples, one was HIV-2 and the remaining 48 were all HIV-1 group M infections. HIV-2 infections have been previously reported among Equato-Guinean immigrants in Spain,17 but they belonged to group B whereas in the current study the sample belonged to group A. Phylogenetic analyses on HIV-1 group M sequences showed the cocirculation of 12 different variants, with CRF02_AG accounting for 53% of the sequences. This finding is in accordance with previous reports on subtypes in patients from Equatorial Guinea,7–10 and is similar to subtype distribution in neighboring Cameroon.12,18 Moreover, the subtype distribution has not changed significantly over time in both countries.7,18 This suggests that the HIV/AIDS epidemics in both countries are mature epidemics and follow a similar trend due to important exchanges between the two countries. Interestingly, subsubtype A3, traditionally known to circulate in Western Africa, was found in two of the participants. However, previous studies already reported the presence of this variant in Equatorial Guinea and the republic of Congo.19,20 Interestingly, we also found representatives of the recently characterized CRF26_A5U variant that has been reported to circulate in DRC and in previous samples from Equatorial Guinea.14 We also documented the presence of variants previously known only from Cameroon, such as subsubtype F2 and CRF22_A101. We also note that although subtype C strains represented 14% of infections in 1999,7 only two (4.8%) subtype C strains were identified in our study population.
In conclusion, the distribution of HIV variants in this study from Equatorial Guinea is very similar to that of neighboring Cameroon with which numerous exchanges occur; however, some strains seem to originate from other Central African countries such as CFR26 from DRC and divergent A strains from Gabon. HIV-2 and subsubtype A3 that are mainly restricted to West Africa have also been introduced in Equatorial Guinea. We have also demonstrated that rare resistance mutations to the protease inhibitors (PIs) and reverse transcriptase inhibitors can be found in this antiretroviral naive population, one mutation to PIs and one to NRTIs. Although our sample size is small, the prevalence of circulating drug-resistant strains is similar to that reported (0–5%) for other sub-Saharan countries where antiretroviral treatment has recently been scaled-up.21 Overall, these data suggest the need to extend these investigations to other parts of the country and to other population groups. Additionally, as access to antiretroviral treatment in Equatorial Guinea continues to improve, care must be taken to ensure treatment compliance to combat the further development of drug resistance.
Sequence Data
The sequences described in this article have been deposited in the EMBL Nucleotide Sequence Database under accession numbers FN557303 to FN557343.
Acknowledgments
We thank the Ministry of Public Health and the Ministry of National Defense of Equatorial Guinea for permission to undertake this study and the U.S. Embassy in Equatorial Guinea for their support. We owe sincere gratitude to Dr. Cruz Baca and Dr. Nemesio Eyi from the National AIDS Control Program and Mrs. Raquel Bondje of the National Reference HIV Laboratory for their active assistance in the field. C.F.D. was supported, in part, by funds from the National Institutes of Health (NIH) Fogarty International Center (FIC) AIDS International Training and Research Program (2 D 43 TW000010-16/17). N.D.W. is supported by the NIH Director's Pioneer Award (DP1-OD000370), the FIC International Research Scientist Development Award (5 K01 Tw000003-05), and the National Geographic Society. Additional support was provided by the Global Viral Forecasting Initiative (GVFI), The Institut de Recherche pour le Dévelopement (IRD), the WW Smith Charitable Trust, the U.S. Department of Defense HIV/AIDS Prevention Program (DHAPP), the Henry Jackson Foundation, Google.org, and the Skoll Foundation. We owe sincere gratitude to the personnel of IRD-UMR 145, the unit Director Pr. Eric Delaporte, in particular for the great support, and the GVFI Cameroon Program staff for their professional commitment.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hahn BH. Shaw GM. De Cock KM. Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287(5453):607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 2.Plantier JC. Leoz M. Dickerson JE, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 3.Peeters M. Toure-Kane C. Nkengasong JN. Genetic diversity of HIV in Africa: Impact on diagnosis, treatment, vaccine development and trials. AIDS. 2003;17(18):2547–2560. doi: 10.1097/01.aids.0000096895.73209.89. [DOI] [PubMed] [Google Scholar]
- 4.McCutchan FE. Understanding the genetic diversity of HIV-1. AIDS. 2000;14(Suppl 3):S31–44. [PubMed] [Google Scholar]
- 5.Vidal N. Peeters M. Mulanga-Kabeya C, et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74(22):10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worobey M. Gemme M. Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–665. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz M. Sanchez I. Gonzalez MP, et al. Molecular epidemiology of HIV type 1 subtypes in Equatorial Guinea. AIDS Res Hum Retroviruses. 2001;17(9):851–855. doi: 10.1089/088922201750252043. [DOI] [PubMed] [Google Scholar]
- 8.Ortiz M. Munoz L. Bernal A, et al. Molecular characterization of non-B HIV type 1 subtypes from Africa in Spain. AIDS Res Hum Retroviruses. 2000;16(18):1967–1971. doi: 10.1089/088922200750054693. [DOI] [PubMed] [Google Scholar]
- 9.Toro C. Jimenez V. Rodrıguez C, et al. Molecular and epidemiological characteristics of blood-borne virus infections among recent immigrants in Spain. J Med Virol. 2006;78:1599–1608. doi: 10.1002/jmv.20744. [DOI] [PubMed] [Google Scholar]
- 10.Holguín A. Rodés B. Soriano V. Protease gene analysis of HIV type 1 non-B subtypes in Spain. AIDS Res Hum Retroviruses. 2000;16(14):1395–1403. doi: 10.1089/08892220050140946. [DOI] [PubMed] [Google Scholar]
- 11.Vergne L. Bourgeois A. Mpoudi-Ngole E, et al. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology. 2003;17(9):851–855. doi: 10.1016/s0042-6822(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 12.Vergne L. Peeters M. Mpoudi-Ngole E, et al. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: Evidence of many minor drug resistance mutations in treatment-naive patients. J Clin Microbiol. 2000;38(11):3919–3925. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peeters M. Courgnaud V. Abela B, et al. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis. 2002;8(5):451–457. doi: 10.3201/eid0805.01-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal N. Bazepeo SE. Mulanga C. Delaporte E. Peeters M. Genetic characterization of eight full-length HIV type 1 genomes from the Democratic Republic of Congo (DRC) reveal a new subsubtype, A5, in the A radiation that predominates in the recombinant structure of CRF26_A5U. AIDS Res Hum Retroviruses. 2009;25(8):823–832. doi: 10.1089/aid.2008.0283. [DOI] [PubMed] [Google Scholar]
- 15.Delgado E. Thomson MM. Villahermosa ML, et al. Identification of a newly characterized HIV-1 BG intersubtype recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J AIDS. 2002;29:536–543. doi: 10.1097/00126334-200204150-00016. [DOI] [PubMed] [Google Scholar]
- 16.Bennett DE. Camacho RJ. Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug resistance: 2009 update. Plos One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heredia A. Vallejo A. Soriano V, et al. Evidence of HIV-2 infection in Equatorial Guinea (central Africa): Partial genetic analysis of a B subtype virus. AIDS Res Hum Retroviruses. 1997;13(5):439–440. doi: 10.1089/aid.1997.13.439. [DOI] [PubMed] [Google Scholar]
- 18.Brennan CA. Bodelle P. Coffey R, et al. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J AIDS. 2008;49(4):432–439. doi: 10.1097/QAI.0b013e31818a6561. [DOI] [PubMed] [Google Scholar]
- 19.Meloni ST. Sankalé JL. Hamel DJ, et al. Molecular epidemiology of human immunodeficiency virus type 1 sub-subtype A3 in Senegal from 1988 to 2001. J Virol. 2004;78(22):12455–12461. doi: 10.1128/JVI.78.22.12455-12461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niama FR. Toure-Kane C. Vidal N, et al. HIV-1 subtypes and recombinants in the Republic of Congo. Infect Genet Evol. 2006;6:337–343. doi: 10.1016/j.meegid.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Bennett DE. Myatt M. Bertognolio S, et al. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antiviral Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]