Abstract
Human immunodeficiency virus (HIV) infections are rarely acquired via an oral route in adults. Previous studies have shown that human whole saliva inhibits HIV infection in vitro, and multiple factors present in human saliva have been shown to contribute to this antiviral activity. Despite the widespread use of simian immunodeficiency virus (SIV)-infected rhesus macaques as models for HIV pathogenesis and transmission, few studies have monitored SIV in the oral cavity of infected rhesus macaques and evaluated the viral inhibitory capacity of macaque saliva. Utilizing a cohort of rhesus macaques infected with SIVMac251, we monitored virus levels and genotypic diversity in the saliva throughout the course of the disease; findings were similar to previous observations in HIV-infected humans. An in vitro infectivity assay was utilized to measure inhibition of HIV/SIV infection by normal human and rhesus macaque whole saliva. Both human and macaque saliva were capable of inhibiting HIV and SIV infection. The inhibitory capacity of saliva samples collected from a cohort of animals postinfection with SIV increased over the course of disease, coincident with the development of SIV-specific antibodies in the saliva. These findings suggest that both innate and adaptive factors contribute to inhibition of SIV by whole macaque saliva. This work also demonstrates that SIV-infected rhesus macaques provide a relevant model to examine the innate and adaptive immune responses that inhibit HIV/SIV in the oral cavity.
Introduction
Human immunodeficiency virus (HIV) infection and AIDS remain significant public health challenges worldwide. The majority of new HIV-1 infections are acquired across mucosal surfaces through high risk sexual contact with infected individuals.1 Epidemiologic and clinical reports agree that transmission of virus rarely occurs via an oral route in adults, despite the presence of detectable levels of virus in the saliva, salivary glands, and oropharyngeal tissues of HIV-infected patients.2–4
The level and potential infectivity of virus present in the oral cavity of HIV-positive individuals have been evaluated using various methodologies.2,5 Studies using culture-based methods have reported low frequencies of HIV-1 detection in saliva, while more quantitative methods, such as polymerase chain reaction (PCR), have led to higher frequencies of detection.5 One study detected HIV-1 RNA by PCR in 96% of cell-free whole saliva samples, with a median viral load of 162 copies/ml saliva (range 0–72,080 copies/ml).4 Another study examined virus levels in saliva by the Nuclisens (nucleic acid sequence-based amplification) assay and detected HIV-1 RNA in 42% of salivary secretions, at a mean level of 794 copies/ml (range 79–794,328 copies/ml).6 HIV-1 RNA levels in saliva are typically lower than those of matched plasma samples; however, discordant viral loads in saliva and plasma have been observed, leading investigators to suggest that the oral cavity may be a viral reservoir.4,7 Genotypic analyses of HIV-1 envelope diversity in saliva and plasma have demonstrated homogeneous viral populations in these compartments.8,9 Homogeneity of the HIV-1 populations in saliva and plasma suggests that viral variants in saliva and plasma originate from a common source and raises the possibility that the oral cavity may be used as a noninvasive site to monitor viral evolution and disease progression in HIV-infected individuals.8,9
Attempts to culture infectious virus from the saliva of HIV-infected individuals have been largely unsuccessful, with recovery rates ranging from 0% to 5%, despite detectable and, in some cases, high levels of viral RNA in saliva.10 In a number of studies, human saliva collected from HIV seronegative and seropositive individuals has been shown to inhibit HIV infection in vitro.11–15 Multiple endogenous inhibitory factors have been identified in saliva, which contribute to this antiviral effect, including mucins, cystatins, defensins, secretory leukocyte protease inhibitor, lactoferrin, anti-HIV antibodies, and other factors.2,10 The innate inhibitory capacity of saliva is, at least in part, responsible for limiting viral expression and transmission of HIV in the oral cavity.2,3,16
Simian immunodeficiency virus (SIV)-infected rhesus macaques are a widely accepted model for the study of HIV transmission and pathogenesis. Experimental infection of macaques with SIV leads to immune dysfunction and progression through advanced disease similar to HIV infection in humans.17 Although SIV in the oral cavity has not been fully investigated, one recent report quantified SIV RNA levels in the saliva and peripheral blood of five SIVMac251-infected rhesus monkeys over the first 90 days postinfection (dpi).18 Levels of SIV RNA in saliva were variable between the five animals. Saliva and peripheral blood viral RNA levels peaked at 14 dpi, and median levels were 105 and 107 viral RNA copies/ml, respectively. Following 35 dpi, only occasional saliva samples had detectable levels of SIV RNA, which were observed mainly in animals with high set point plasma viral RNA levels.
Rhesus macaques may also provide a good model to evaluate the innate protective factors present in the oral cavity, including the potential SIV-inhibitory capacity of macaque saliva. Initial reports demonstrated that both human and chimpanzee saliva were able to inhibit HIV-1 infection of peripheral blood mononuclear cells in vitro.11 However, human submandibular saliva had little to no inhibitory capacity against HIV-2ROD or SIVMac239 infection in vitro.13 To evaluate the utility of the SIV-infected macaque model for the study of oral HIV pathogenesis, we examined oral SIV loads and genotypes in saliva collected from a cohort of SIVMac251-infected rhesus macaques throughout the course of disease and evaluated the inhibitory capacity of rhesus macaque whole saliva against SIV in vitro.
Materials and Methods
Cohort and sample collection
All samples were collected using protocols approved by LSUHSC IRB and LSUHSC and Tulane IACUC committees. Whole saliva samples were collected from eight normal human volunteers. Volunteers were asked to abstain from food and drink only water for 1 h prior to sample collection. Saliva (5 ml) was collected from each volunteer by expectoration into 15-ml polypropylene tubes on ice. Whole saliva samples were clarified by centrifugation (12,000 × g for 1 min) and aliquots of supernatants were stored at −80°C until use.
Whole saliva samples were collected from eight normal, untreated juvenile male rhesus macaques (Macaca mulatta) housed at the LSU Health Sciences Center animal facility. Whole saliva samples were also collected from 16 juvenile male rhesus macaques infected intravenously with SIVMac251 and housed at the Tulane National Primate Research Center in Covington, LA, at several time points post-SIV infection (1, 2, 4, 6, 8, 12, 16, 30, or 32, and 48 weeks postinoculation). All animals in this report were enrolled in larger studies at LSUHSC with similar treatment protocols. Whole saliva samples were collected from anesthetized macaques by placement of two Weck-Cel surgical sponges (Solon-Medtronics, Solon, OH) in the buccal cavity for 5–10 min.19 Sponges soaked in saliva were placed in 15-ml polypropylene tubes on ice immediately following collection. Whole saliva was recovered from the sponges by centrifugation (12,000 × g for 1 min) through a sterile 0.5-ml tube containing small holes at the bottom into a 1.5-ml collection tube. Whole fluid was then clarified by centrifugation, and supernatants were stored at −80°C until analyzed.
As varying volumes of saliva were recovered, the quality of macaque whole saliva samples was assessed for total protein levels by BCA assay (Thermo Pierce), according to manufacturer's instructions. Typical total protein levels in macaque whole saliva ranged from 2000 to 4000 μg/ml.
Peripheral blood samples were collected in tubes containing EDTA from 16 rhesus macaques at various time points following SIVMac251 infection, coincident with the time points of saliva collection. Plasma aliquots were stored at −80°C until analyzed.
Quantitation of SIV RNA
Levels of SIV RNA in rhesus macaque plasma and saliva samples were quantified by real-time TaqMan, reverse-transcriptase PCR (RT-PCR) using SIV gag primers and probe, as previously described.20 Briefly, viral RNA was isolated from 1 ml plasma using the Trizol reagent (Life Technologies, Rockville, MD) and reverse transcribed to cDNA. Products were PCR amplified in duplicate reactions using 1/10 of the total volume of cDNA.
Whole saliva SIV RNA levels were quantified by the same method, with some modifications for accurate measurement of viral RNA in mucosal secretions. Briefly, RNA was isolated from 50 μl of whole saliva supernatant using the Trizol-LS reagent (Life Technologies, Rockville, MD) and reverse transcribed into cDNA using Multiscribe Reverse Transcriptase (Applied Biosystems, Foster City, CA). PCR amplification was performed in duplicate reactions utilizing the total amount of cDNA. Due to the limited volumes of saliva available for assessment, the assay had a quantitation limit of 100 copies/ml of saliva.
Characterization of SIV genotypes
Viral genotypes expressed in plasma and saliva over the course of disease were evaluated in SIV-infected animals by heteroduplex tracking assays (HTA) utilizing SIV envelope V1 sequences amplified by RT-PCR from time-matched plasma and saliva samples, as previously described.21,22 Briefly, viral RNA was isolated using Trizol reagent (Life Technologies, Rockville, MD) and reverse transcribed into cDNA using Multiscribe Reverse Transcriptase (Applied Biosystems, Foster City, CA). Using conserved primer pairs, a 480-bp fragment of SIV envelope V1/V2 sequences was amplified from cDNA by nested PCR. Products from independent V1 PCR amplifications, at least three per plasma sample and two per saliva sample, were pooled for more accurate analysis of viral diversity by HTA.
A single-stranded 32P-labeled probe was constructed by end point dilution of SIVMac251 stock and PCR amplification of the V1 region from the predominant envelope genotype present in the stock. Pooled V1 PCR products generated from plasma and saliva samples were mixed with 1 μl 32P-labeled probe and 2 μl annealing buffer in a total volume of 20 μl. This mixture was heated to 90°C for 3 min, chilled on ice for 3 min, and then separated by electrophoresis on a 12% polyacrylamide gel for 1600 V-h. DNA migration patterns were visualized and evaluated by phosphorimaging and ImageQuant software (GE Healthcare).
Culture and coculture of SIV from whole saliva
CEMx174-R5 and MT4-R5 cell lines and rhesus macaque peripheral blood mononuclear cells (RhPBMCs) were used for SIV culture/coculture experiments and expansion of virus stocks. CEMx174-R5 and MT4-R5 T cell lines were a gift from James Robinson, Tulane University School of Medicine.23 Both cell lines express CCR5 cell surface receptors under puromycin selection. RhPBMCs were isolated from normal (SIV-seronegative) rhesus macaque blood using Ficoll-Hypaque gradient separation and stimulated with phytohemagglutinin (PHA) in interleukin (IL)-2-containing media for 72 h prior to experiments. PBMC and T cell line cultures were evaluated for cytotoxic effects of whole saliva by cell viability counts using Trypan blue staining. At a 1:20 saliva dilution, cells did not show any evidence of decreased viability over 14 days in culture.
Acute (2 wpi) and chronic (18 months postinoculation) whole saliva samples from SIVMac251-infected rhesus macaques were clarified by centrifugation. The supernatant fraction was used for SIV culture studies, and the cell fraction was used for SIV coculturing studies. Cultures and cocultures were maintained at 37°C in six-well plates and contained 1 × 106 RhPBMC, CEMx174-R5, or MT4-R5 cells and 1:20 dilution of cell-free whole saliva supernatant or whole saliva cell fraction in 2 ml appropriate culture media. Culture/coculture aliquots were assessed at 4, 7, 10, and 14 days for SIV RNA and DNA levels by real-time RT-PCR, as described above.
Viral isolates
HIV isolate, HIV-1Bal, and HIV-1JRFL, were obtained from the NIH Reference and Reagent Program. The SIVMac251 virus was generously provided by Preston Marx, Tulane Regional Primate Center, Covington, LA. Viral isolates were expanded on CEMx174-R5 or MT4-R5 cell lines in RPMI media containing 10% fetal calf serum for 3 weeks at 37°C. Culture supernatants were clarified by centrifugation (250 rcf for 10 min) and vacuum filtered using the Steriflip apparatus (Millipore).
An SIVMac239 molecular infectious clone, containing the full length proviral DNA genome in a modified pBR322 plasmid vector, was a gift from Toshi Kodama, University of Pittsburgh School of Medicine, Pittsburgh, PA (GenBank accession: M33262).24,25 To examine the inhibitory capacity of saliva against a neutralization-sensitive SIV envelope, we replaced the gp160 region of the SIVMac239 molecular clone with the predominant viral envelope of the SIVDeltaB670 quasispecies, an isolate shown to be sensitive to plasma neutralization in vitro.26 SIVMac239-Cl3env is a molecular infectious clone that contains the SIVDeltaB670-clone 3 envelope (GenBank accession: FJ842859) in an SIVMac239 background.
To obtain virus stocks, plasmids containing the proviral genomes of SIVMac239 and SIVMac239-Cl3env (3 μg DNA) were transfected, using the Fugene 6 transfection reagent (Roche), into 1 × 106 HEK293T cells seeded overnight in T25 cell culture flasks at 37°C. Culture supernatants were harvested after 72 h at 37°C and were used to infect 5 × 105 MT4-R5 cells in RPMI containing 10% fetal calf serum for expansion of SIVMac239 and SIVMac239-Cl3env virus stocks. After 3 weeks, virus culture supernatants were prepared as described above. All virus stocks were aliquoted and stored at –80°C. The 50% tissue culture infectious dose (TCID50) was determined by serial dilution of virus stocks and infection of TZM-bl cells as described.27
Inhibition assay
Inhibition of HIV and SIV isolates by human and rhesus macaque whole saliva was evaluated using an in vitro neutralization assay previously described for evaluation of HIV plasma neutralizing antibody responses.27–29 This assay utilizes TZM-bl cells, a HeLa cell line that stably expresses CD4, CXCR4, and CCR5 receptors and contains a tat-responsive firefly luciferase (Luc) reporter gene. This cell line requires only a single round of infection to accurately measure in vitro neutralization of virus. Briefly, human or macaque whole saliva samples were diluted in DMEM with 10% fetal calf serum (FCS) and incubated with 100 TCID50s of cell-free virus at 37°C in 96-well plates for 1 h prior to addition of cells. TZM-bl cells were counted and diluted in DMEM containing 10% FCS and 75 μg/ml DEAE-Dextran (Sigma); 8000 cells were added to each reaction in a final volume of 250 μl per well. Plates were then incubated at 37°C for 48 h. Luminescence for each reaction was assessed using the Bright-Glo Luciferase Assay System (Promega) and quantified in 96-well black plates using a Hidex Oy CHAMELEON V plate reader. Inhibition of virus by saliva samples was expressed as the reduction in relative light units (RLU) as compared to virus controls and reported as the percent reduction in viral infection in vitro. Using 100 TCID50 doses, virus only control wells produced RLU that were on average 10 times those of background. All virus stocks used in these studies produced similar RLU in control wells. Saliva samples were assayed in duplicate; cell only (negative) and virus only (positive) controls were included in replicates of eight within each plate. Only samples that displayed a 50% or more reduction in RLU as compared to the average of control wells were considered to have quantifiable inhibitory effects.
To evaluate potential cytotoxic effects of whole saliva on TZM-bl cells, viability and metabolic activity of cells treated with 1:10 and 1:20 dilutions of whole saliva were assessed and compared to untreated controls. Cell viability counts using Trypan blue staining were similar in treated and control cultures, as were measurements of background RLU following analysis in the Bright-Glo Luciferase Assay. Similarly, no differences in metabolic activity were observed following culture of TZM-bl cells in the presence of saliva as compared to mock-treated control wells when assessed by a cell proliferation MTS assay (Promega CellTiter 96 AQueous One Solution Cell Proliferation Assay), according to manufacturer protocols.30
Measurement of total and gp130 (SIV-envelope-specific) antibody levels by ELISA
Total IgG and IgA levels in macaque saliva were measured by ELISA as previously described.31 Briefly, Fisherbrand microtiter plates were coated overnight at 4°C with 100 μl PBS, pH 7.4 containing 0.4 μg/ml goat F(ab′)2 to monkey IgG (1.2 μg/ml total protein; MP Biochemicals, Solon, OH) or containing 0.5 μg/ml goat antimonkey IgA (Alpha Diagnostics, San Antonio, TX). Blocking was done at room temperature for 30 min in PBS (1×) containing 0.05% Tween-20 and 2% goat serum (Equitech-Bio Inc., Kerrville, TX). Whole saliva was diluted 1:1000 in blocking buffer, added to the first row of wells, and then further diluted in a 2-fold series down the plate. The standard was pooled normal monkey serum containing known amounts of IgA and IgG.32 After overnight storage at 4°C, plates were developed by consecutive treatment with biotin-conjugated goat antihuman IgG (SouthernBiotech, Birmingham, AL) or antimonkey IgA (Alpha Diagnostics) for 1 h at 37°C, then 0.5 μg/ml streptavidin-peroxidase (Sigma) for 30 min at room temperature. Tetramethylbenzidine Supersensitive substrate (Sigma) was added for 30 min, followed by 1 M H2SO4 stop solution. Absorbance was read at 450 nm in a SpectraMax 5 plate reader (Molecular Devices, Sunnyvale, CA). The SoftMax Pro computer program (Molecular Devices) was then used to construct four-parameter standard curves and calculate the total IgG and IgA in saliva samples.
SIV envelope-specific IgG concentrations were similarly measured by ELISA using recombinant SIVmac251 gp130 (ImmunoDiagnostics, Woburn, MA) as coating reagent, 1:50 starting dilutions of saliva, and a standardized pool of plasma from SIV-infected macaques. Plates were developed with secondary antibody to IgG as described above.
Statistical analyses
All statistical comparisons in this study were performed by the Mann–Whitney U test. p values <0.05 were considered significant.
Results
SIV levels in the oral cavity of rhesus macaques
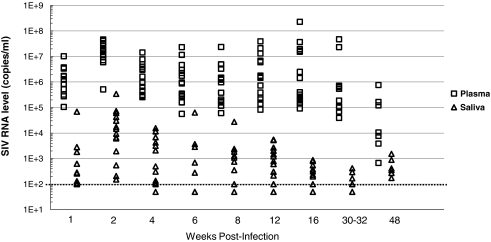
Whole saliva and peripheral blood samples were obtained from 16 SIVMac251-infected rhesus macaques over a 48-week period following inoculation. SIV RNA levels were quantified in plasma and saliva by real-time TaqMan RT-PCR and compared (Fig. 1). As previously shown in HIV-infected individuals, detectable levels of viral RNA were present in the saliva of SIV-infected rhesus macaques.4,6 The level of viral RNA present in saliva of each animal was approximately 2–3 logs lower than time-matched plasma viral RNA levels. Only 20 of the 111 total saliva samples evaluated (18%) contained undetectable levels of SIV RNA (less than 100 viral RNA copies/ml of saliva). These 20 samples were collected from 10 of the 16 rhesus macaques between 4 and 32 weeks following infection, representing 1–2 time points per animal. Therefore, during the majority of the disease course, each animal in the cohort had detectable levels of virus in their saliva, with several time points having >1000 copies/ml. The highest levels of SIV RNA in macaque saliva were observed at 2 weeks postinoculation (wpi), ranging between 150 and 340,000 copies/ml of saliva (Fig. 1).
FIG. 1.
Levels of SIV RNA in time-matched plasma (□) and saliva (▵) samples collected from SIVMac251-infected rhesus macaques (n = 16) over 48 weeks following inoculation. The real-time RT-PCR assay quantitation limit of 100 copies/ml is indicated on the chart by the dotted line. Samples shown that are below the dotted line contain less than 100 copies of SIV RNA/ml fluid.
Despite the detectable levels of SIV RNA measured in macaque saliva, we were unable to culture virus from the saliva of SIV-infected macaques collected during the acute (2 wpi) or chronic (18 months pi) stages of SIV disease using rhesus macaque PBMCs or CEMx174-R5 and MT4-R5 T cell lines. We were also unable to detect productive SIV infection through coculture of the cellular fraction of the same acute and chronic stage whole saliva samples.
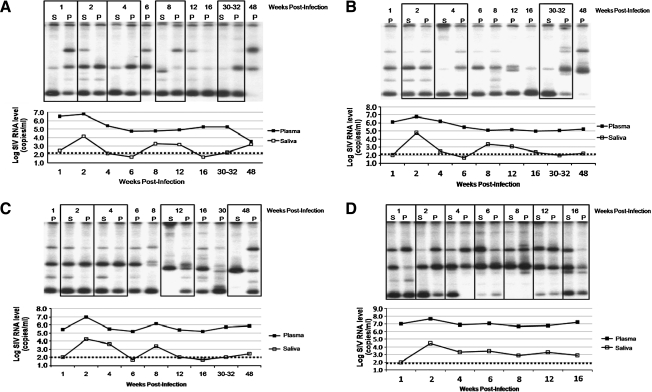
To examine the genotypic diversity of SIV present in saliva, we amplified the V1/V2 region of the SIV envelope by RT-PCR in matched macaque plasma and saliva samples collected over 48 weeks following infection and examined the resulting amplicons by HTA. Samples were evaluated from each of the 16 SIV-infected macaques over the course of disease; four animals were selected as representative examples of the cohort, and these data are shown in Fig. 2, along with the longitudinal SIV RNA levels in plasma and saliva. We were unable to amplify envelope V1 PCR products from all saliva samples, particularly samples with unquantifiable viral RNA levels. HTA was performed on all PCR-positive saliva samples, as shown in Fig. 2, with amplicons from time-matched plasma samples (boxes). Plasma samples collected from SIVMac251-infected macaques at 1 or 2 wpi contained four predominant envelope genotypes, which were also present in saliva. These four genotypes are representative of the SIVMac251 stock used for inoculation of the animals (data not shown). Some of the inoculum envelope genotypes persisted in plasma and saliva of each animal throughout the disease course, but new genotypes appeared in both compartments at various time points (typically appearing from 8 to 12 wpi). Late in the disease course (30–48 wpi), saliva samples from animals A, B, and C contained only one or two of the predominant V1 genotypes found in more diverse plasma samples. The loss of genotypic heterogeneity observed in saliva over the disease course may reflect limitations in assay sensitivity for genotypes present at very low levels, lower viral levels, and/or the presence of antiviral salivary factors. Animal D had high levels of saliva SIV RNA and succumbed early to disease at 4 months following infection. At end-stage disease in animal D (16 wpi), four envelope genotypes were identified in both plasma and saliva.
FIG. 2.
SIV envelope V1 genotyping by heteroduplex tracking assay (HTA) on time-matched plasma and saliva samples collected from four cohort-representative SIVMac251-infected macaques over 48 weeks following inoculation. (A, B, C, and D) 32P-labeled V1 DNA banding patterns in the upper portion and time-coincident SIV RNA levels in the lower portion for plasma and saliva samples over the SIV disease time course collected from each of the four representative animals. Time-matched plasma and saliva samples are outlined in boxes. The dotted lines indicate the lower assay limit for measurement of SIV RNA levels.
Among the cohort of macaques, the SIV envelope genotypes present in saliva reflected the genotypes present in plasma. In a few instances, we did observe unique envelope V1 genotypes present in saliva, which were not found in matched plasma samples. However, the majority of saliva SIV genotypes were observed in plasma samples at concurrent or earlier time points in the disease course.
In vitro measurement of SIV inhibition by human and rhesus macaque whole saliva
To determine whether whole human or rhesus macaque saliva was capable of inhibiting SIV infection in vitro, we collected whole saliva samples from eight normal human volunteers and eight normal rhesus macaques. The inhibitory capacity of each normal human and macaque saliva (diluted 1:20) against two isolates of SIV, SIVMac239 and SIVMac239-Cl3env, was evaluated by their ability to inhibit infection of MT4-R5 T cells. Both normal human and rhesus macaque whole saliva samples from this collection were capable of near complete inhibition of SIVMac239-Cl3env and variable levels of SIVMac239 inhibition, demonstrating that whole saliva can inhibit SIV infection in vitro. However, infection levels in the MT4-R5 cell line by both SIV isolates were variable across replicate wells, making standardization of this assay difficult.
To develop a more sensitive, standardized assay to evaluate the virus-inhibitory capacity of saliva in vitro, we adapted an assay commonly used to measure antibody neutralization of HIV. Utilizing the TZM-bl cell line, we evaluated the inhibitory capacity of a set of normal human and rhesus macaque whole saliva against five viral isolates. Comparisons of human and monkey saliva against a specific virus were conducted in a single experiment. The results are shown in Fig. 3.
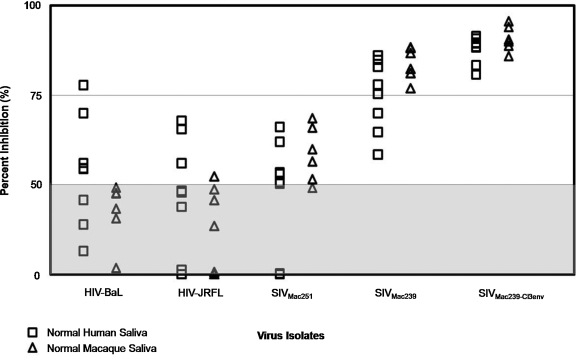
FIG. 3.
Inhibition of HIV and SIV infection by normal human (n = 8) and rhesus macaque (n = 8) whole saliva was evaluated using the TZM-bl assay. Saliva samples were diluted 1:20 and measured for inhibitory capacity against five viral isolates: HIV-1BaL, HIV-1JRFL, SIVMac251, SIVMac239, and SIVMac239-Cl3env. Percent inhibition of each virus is shown for normal human (□) and normal macaque (▵) saliva samples. Gray area indicates saliva samples that were negative for virus inhibition in this assay (less than 50% inhibition of viral isolate).
Percent inhibition of HIVBaL, an HIV-1 isolate commonly used to examine human salivary inhibition, varied widely among the eight normal human saliva samples tested, ranging from <10% to 75% inhibition. Human whole saliva was also capable of inhibiting HIVJRFL infection to a similar level. Rhesus macaque whole saliva was less inhibitory to the HIV isolates tested as compared to human saliva, with only one rhesus sample capable of inhibiting either HIV isolate by more than 50% in this assay. All macaque saliva samples (eight of eight) and six of eight human saliva samples tested were capable of inhibiting SIVMac251 infection. All normal human and macaque saliva samples were capable of inhibiting SIVMac239 and SIVMac239-Cl3env infection by greater than 50%. SIVMac239-Cl3env was the isolate most sensitive to inhibition by human and macaque saliva, with all saliva samples in this collection inhibiting SIVMac239-Cl3env infection by greater than 75% in this assay. These data indicate that both normal human and rhesus macaque whole saliva can inhibit in vitro infection of SIV, and that SIVMac239-Cl3env is highly sensitive to inhibition in vitro, providing a tool to evaluate the inhibitory capacity of macaque whole saliva. Samples which exhibited <50% inhibition showed varying levels of inhibition ranging between 0 and 50% in replicate experiments, and quantification of this amount was not reproducible with values below 50%. Therefore, these saliva samples were categorized as negative for virus inhibition.
Volumes of whole saliva obtained from anesthetized SIVMac251-infected macaques in this study were limited, and we were unable to perform repeated SIV inhibition measurements on individual saliva samples collected from many animals in the cohort. To assess the variability in SIV inhibitory capacity of multiple saliva samples collected from a single animal, saliva samples (n = 7) were collected over 3 months from two animals and SIVMac239-Cl3env inhibition was measured using the TZM-bl assay. We observed consistent, reproducible levels of SIV inhibition by replicate saliva samples collected from both animals; average percent inhibition by saliva was 89 ± 4.5 (standard deviation) and 82 ± 6.4.
Because the volume of saliva collected varied among animals, we assessed the quality of samples collected by measuring protein levels. Total protein levels were measured by a BCA assay in all macaque whole saliva samples collected. Levels of total salivary proteins among individual macaques ranged between 2000 and 4000 μg/ml saliva and were not associated with same-sample measures of SIV inhibitory capacity in vitro. These data suggest that the quality of saliva samples collected from macaques in this study was comparable.
The small volumes of saliva available from monkeys also limited our ability to analyze the SIV-inhibitory capacity of saliva at different dilutions. To determine the range of saliva dilutions capable of inhibiting SIV in vitro, a collection of samples with larger volumes available was diluted 1:20, 1:80, and 1:200 and evaluated for levels of SIVMac239-Cl3env inhibition using the TZM-bl assay. Based on assessment of replicate experiments, we determined that saliva samples diluted 1:20 that demonstrated >75% inhibition were also capable of inhibiting infection by >50% at a dilution of 1:80, whereas saliva samples with inhibitory levels between 50% and 75% of controls were not capable of inhibiting SIV infection at a higher dilution of saliva. Therefore to classify the SIV inhibitory capacity of volume-limited macaque saliva following analysis at a single dilution of saliva (1:20), we used the designations of less than 50% inhibition of SIVMac239-Cl3env infection as having little to no SIV-inhibitory capacity, greater than 50% inhibition as having SIV-inhibitory capacity, and greater than 75% as having a higher titer of inhibitory factors (≥1:80).
Inhibitory capacity of rhesus macaque saliva over the SIV disease course
The SIV-inhibitory capacity of whole saliva collected from the cohort of 16 SIVMac251-infected macaques was evaluated over 48 weeks following inoculation, as shown in Fig. 4. During the first 8 weeks following SIV infection, a wide range of inhibitory capacity among individual animals was observed; however, the majority of macaque saliva samples tested demonstrated >50% inhibition of SIV infection. Saliva collected from eight normal, untreated macaques all demonstrated levels of inhibition of >75% against SIVMac239-Cl3env (Fig. 3). The wider range of SIV inhibition observed by saliva from the SIV-infected macaque cohort may be due to SIV infection itself or may represent the variability of saliva inhibitory capacity among a larger group of macaques.
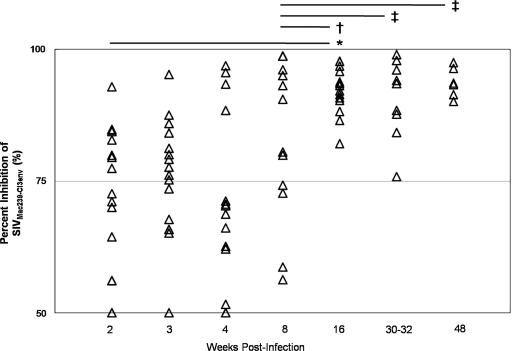
FIG. 4.
Saliva samples (▵) were collected from 16 SIVMac251-infected rhesus macaques at the indicated time points following inoculation. Saliva was diluted 1:20 and inhibitory capacity against SIVMac239-Cl3env was measured by TZM-bl assay for each animal over the course of disease. Changes in salivary inhibitory capacity were evaluated for statistical significance by the Mann–Whitney U test: (*) p < 0.001; (†) p < 0.01; (‡) p < 0.05.
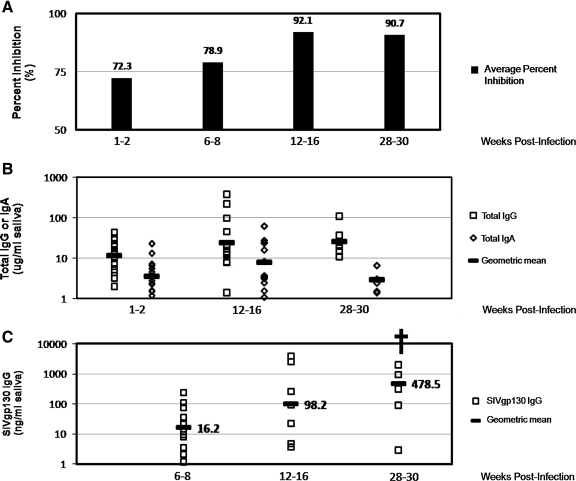
At 2 wpi, 14 of 16 of animals had positive saliva SIV-inhibitory capacities (>50% inhibition), 8 of which were >75% inhibition, and 2 of 16 animals had negative saliva SIV-inhibitory capacities (<50% inhibition). Averages among the cohort are shown in Fig. 5A. At 8 wpi, all animals had positive saliva SIV-inhibitory capacities, with half of the animals demonstrating >75% inhibition of SIV infection. By 16 wpi, all saliva samples exhibited positive SIV-inhibitory capacities of >75% inhibition. Although we did not observe any significant changes in inhibitory capacity during the acute stage of SIV disease (2–8 wpi), there was a significant improvement in the SIV-inhibitory capacity of saliva later in the disease course (16–48 wpi). The improvement in levels of SIV inhibition by infected macaque saliva was statistically significant between 2 and 16 wpi (p < 0.001) and between 8 and 16 wpi (p < 0.01). The SIV-inhibitory capacity of infected macaque saliva remained statistically higher (p < 0.05) than the 8 wpi time point throughout the disease course. There was no correlation between salivary SIVMac239-Cl3env inhibitory capacity and salivary SIV RNA levels over the disease course; however, at 16 wpi, virus levels in the saliva of all animals were less than 1000 copies/ml and remained low to undetectable (less than 100 copies/ml saliva) through 48 weeks. These data suggest that the development of effective adaptive host immune responses in the oral cavity between 8 and 16 weeks following infection may contribute to limiting expression of virus and to the SIV-inhibitory capacity of saliva.
FIG. 5.
Comparison of total and SIV envelope-specific antibody levels in saliva collected from 16 SIVMac251-infected rhesus macaque saliva at various time points over the course of disease to SIV-inhibitory capacity in vitro. (A) The average percent inhibition of SIVMac239-Cl3env infection by all saliva samples over time. Salivary antibody levels in each saliva sample were measured by ELISA, and total IgG and IgA levels in saliva (B) and SIVgp130 IgG (binding antibody) levels in saliva (C) are shown for select time points over the disease course. Changes in antibody levels were assessed for statistical significance by the Mann–Whitney U test; (†) p < 0.01.
To evaluate humoral immune responses in saliva of SIV-infected macaques, we examined total IgG and IgA antibody levels in macaque saliva as well as SIVgp130-specific IgG antibody levels. Figure 5 shows the comparison between salivary inhibition of SIVMac239-Cl3env and antibody levels over the course of disease. There were no significant changes in the levels of total IgG and IgA antibodies in macaque saliva corresponding to the increase in saliva inhibition of SIV infection in vitro over the course of disease. However, we did observe a statistically significant increase in SIVgp130-specific IgG levels in SIVMac251-infected macaque saliva between 6–8 wpi and 28–30 wpi (p < 0.01). The increases in SIV-specific humoral immune responses are coincident with the improvement in the SIV-inhibitory capacity of saliva observed later in the disease course of this cohort and suggest that SIV-specific antibody responses in the oral cavity of macaques contribute to the inhibition of SIV infection in vitro by saliva.
Discussion
In this study, we have demonstrated that SIV levels in the oral cavity of rhesus macaques are highly similar to previously reported viral loads in HIV-infected humans. SIVMac251-infected macaques (n = 16) had detectable levels of SIV RNA present in saliva over the course of disease. Among individual animals, saliva SIV RNA levels were 2–3 logs lower than the corresponding plasma levels and reflected fluctuations in the plasma SIV RNA levels over the disease course. However, among the animal cohort, we did not observe a direct correlation between viral RNA levels in plasma and saliva at any time point, in agreement with a recent report by Whitney et al.18 In that report, viral RNA was detected over a prolonged period of time in the blood and saliva of five SIVMac251-infected macaques, similar to our observations. Analysis of the genotypic diversity of virus populations in matched macaque plasma and saliva samples indicated the presence of identical SIV envelope variants in both fluids, similar to the observations in HIV-infected humans.8,9 Attempts to culture or coculture SIV from infected macaque saliva were unsuccessful, indicating that like human saliva, macaque saliva may contain inhibitory factors that reduce the viability of virus in the oral cavity.5
Multiple endogenous human salivary factors with specific and nonspecific mechanisms of anti-HIV activity have been described since Fultz first demonstrated inhibition of HIV infection in vitro by whole human and chimpanzee saliva.10,33,34 Despite the widespread use of SIV-infected rhesus macaques as an animal model for HIV transmission and pathogenesis, few studies have utilized this model to evaluate the inhibitory components of macaque saliva. One study, by Nagashunmugam et al., was unable to demonstrate any inhibitory activity of human submandibular saliva against HIV-2ROD or SIVMac239 infection of HUT78 cells in vitro, as measured by reverse transcriptase activity at 7 days postinfection.13 In this study, we first examined the SIV-inhibitory capacity of normal human and macaque saliva against SIVMac239 and SIVMac239-Cl3env using an MT4-R5 T cell inhibition assay. We were able to confirm saliva inhibition of SIV infection in vitro using the MT4-R5 cell line. However, variable levels of infection in positive controls were observed by both SIV isolates, making standardized and sensitive assessment of SIV inhibition by saliva difficult. To address similar difficulties with assessment of plasma neutralization of HIV isolates in vitro, rapid and sensitive assays for measurement of HIV-1 neutralization have recently been standardized using the TZM-bl reporter cell line.28,29 We utilized this assay to examine inhibition of viral infection by normal human and rhesus macaque saliva, and both were capable of inhibiting in vitro infection of HIV and SIV isolates to varying degrees using the TZM-bl assay. Therefore, lack of assay sensitivity may have played a role in the previous inability to measure saliva inhibition of SIV infection.13
The virus isolate used in previous studies may have also contributed to the inability of saliva to inhibit SIV infection in vitro. SIVMac239 is a lab-adapted isolate of SIV and has been characterized as resistant to in vitro neutralization by plasma antibodies.35 It has been reported that characteristics of SIVMac239 determined in vitro, such as cell tropism, were not predictive of in vivo tropism or pathogenesis of the virus, indicating that SIVMac239 may not be an ideal isolate to examine saliva inhibition against SIV in vitro.36 For assessment of in vitro neutralization of primary or biological HIV-1 isolates, investigators have utilized pseudotyping and molecular infectious cloning methods to limit T cell line passage and allow for more accurate measurement of antibody-mediated neutralization of heterologous HIV-1 envelopes.37 We hypothesized that the use of an SIV molecular infectious clone containing a plasma neutralization-sensitive envelope would allow for more accurate measurement of inhibition of SIV infection in vitro by macaque saliva. SIVMac239-Cl3env, used in this study, is a replication-competent virus created from an SIVMac239 molecular infectious clone and contains a neutralization-sensitive envelope derived from the SIVDeltaB670 quasispecies.24,38 In this study, SIVMac239-Cl3env was the viral isolate most sensitive to inhibition by saliva using the TZM-bl assay.
Utilizing a cohort of 16 SIV-infected macaques, at 2 wpi, we observed varying levels of SIV-inhibitory capacity by saliva, ranging from <50 to >90% inhibition, with average levels of 72%. In contrast, the collection of normal macaque saliva (shown in Fig. 3) exhibited more uniform levels of inhibition against SIVMac239-Cl3env, where each of the eight samples had >75 % inhibition. Samples were not available from time points prior to SIV infection of the 16 animal cohort to determine if SIV infection had an impact on the inhibitory capacity of saliva. It is also possible that these results represent the variation found among different animals, which was not reflected in the small cohort of the eight normal macaques evaluated.
Among the SIV-infected cohort, we observed a significant increase in the SIV-inhibitory capacity of saliva collected 16 wpi, corresponding to a statistically significant increase in levels of SIV-specific IgG in saliva and reduced salivary SIV RNA levels (less than 1000 copies/ml saliva). The development of SIV-specific humoral immune responses may play a key role in the control of viral replication in the oral cavity of infected macaques and together with endogenous antiviral factors in saliva, these responses may serve to limit virus in this compartment.
Many endogenous factors have been identified that contribute to the antiviral activity of human saliva in vivo and in vitro.33 The current study demonstrates that both normal human and macaque whole saliva are capable of inhibiting SIV and HIV infection in vitro using a sensitive, standardized assay. These data suggest that the same innate antiviral components identified in human saliva may also be present in macaque saliva, and that the inhibitory activities of these salivary components are not limited to HIV-1. Utilization of the rhesus macaque model to study viral pathogenesis in the oral cavity may be of particular importance in HIV vaccine and treatment studies. Recent reports indicate that frequent or high-risk contact between HIV serodiscordant individuals elicits protective, HIVgp160-specific neutralizing antibody responses at multiple mucosal surfaces, including IgA responses in the genital tract secretions of HIV-negative female Kenyan sex workers and in the parotid saliva of HIV-negative babies exposed to breast milk from HIV-infected mothers.39–41 Further examination of these findings is necessary to determine if these mucosal HIVgp160-specific antibodies may truly be protective against HIV infection in vivo or have potential as a prophylactic therapy. Although more work is needed to identify the specific protective factors present in macaque saliva and to determine similarities of these factors to human salivary molecules with anti-HIV activity, our findings demonstrate the relevance of the SIV-infected rhesus macaque for studying HIV pathogenesis in the oral cavity, as well as for identifying innate and acquired responses protective against HIV at mucosal surfaces.
Acknowledgments
We would like to thank the LSU Comprehensive Alcohol Research Center, Core Laboratories, Jason Dufour, DVM (Tulane Regional Primate Center), Ashok Aiyar, Ph.D. (LSUHSC), and Patricia Molina, M.D., Ph.D. (LSUHSC), for support, technical help, and macaque specimens. We thank Jane Schexnayder for technical assistance and James Robinson, M.D. (Tulane HSC) for technical advice and cell lines. This work was supported, in part, by the Louisiana Vaccine Center and the South Louisiana Institute for Infectious Disease Research sponsored by the Louisiana Board of Regents and by NIH Grants AA009803 (S. Nelson), AA007577 (G.J. Bagby), and AI058896 (P.A. Kozlowski).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hall HI. Song R. Rhodes P, et al. Estimation of HIV incidence in the United States. JAMA. 2008;300(5):520–529. doi: 10.1001/jama.300.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campo J. Perea MA. del Romero J. Cano J. Hernando V. Bascones A. Oral transmission of HIV, reality or fiction? An update. Oral Dis. 2006;12(3):219–228. doi: 10.1111/j.1601-0825.2005.01187.x. [DOI] [PubMed] [Google Scholar]
- 3.Moutsopoulos NM. Greenwell-Wild T. Wahl SM. Differential mucosal susceptibility in HIV-1 transmission and infection. Adv Dent Res. 2006;19(1):52–56. doi: 10.1177/154407370601900111. [DOI] [PubMed] [Google Scholar]
- 4.Liuzzi G. Clementi M. Bagnarelli P. Valenza A. Cataldo PT. Piazza M. Analysis of HIV-1 load in blood, semen and saliva: Evidence for different viral compartments in a cross-sectional and longitudinal study. AIDS. 1996;10(14):F51–F56. doi: 10.1097/00002030-199612000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Rothenberg RB. Scarlett M. del Rio C. Reznik D. O'Daniels C. Oral transmission of HIV. AIDS. 1998;12(16):2095–2105. doi: 10.1097/00002030-199816000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Shugars DC. Slade GD. Patton LL. Fiscus SA. Oral and systemic factors associated with increased levels of human immunodeficiency virus type 1 RNA in saliva. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;89(4):432–440. doi: 10.1016/s1079-2104(00)70124-7. [DOI] [PubMed] [Google Scholar]
- 7.Shugars DC. Wahl SM. The role of the oral environment in HIV-1 transmission. J Am Dent Assoc. 1998;129(7):851–858. doi: 10.14219/jada.archive.1998.0349. [DOI] [PubMed] [Google Scholar]
- 8.Freel SA. Williams JM. Nelson JAE, et al. Characterization of human immunodeficiency virus type 1 in saliva and blood plasma by V3-specific heteroduplex tracking assay and genotype analyses. J Virol. 2001;75(10):4936–4940. doi: 10.1128/JVI.75.10.4936-4940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freel SAFS. Pilcher CD. Menezes P. Giner J. Patrick E. Lennox JL. Hicks CB. Eron JJ., Jr Shugars DC. Envelope diversity, coreceptor usage and syncytium-inducing phenotype of HIV-1 variants in saliva and blood during primary infection. AIDS. 2003;17(14):2025–2033. doi: 10.1097/00002030-200309260-00003. [DOI] [PubMed] [Google Scholar]
- 10.Shugars DC. Endogenous mucosal antiviral factors of the oral cavity. J Infect Dis. 1999;179(Suppl 3):S431–S435. doi: 10.1086/314799. [DOI] [PubMed] [Google Scholar]
- 11.Fultz PN. Components of saliva inactivate human immunodeficiency virus. Lancet. 1986;2(8517):1215. doi: 10.1016/s0140-6736(86)92218-x. [DOI] [PubMed] [Google Scholar]
- 12.Shine N. Konopka K. Duzgunes N. The anti-HIV-1 activity associated with saliva. J Dent Res. 1997;76(2):634–640. doi: 10.1177/00220345970760020301. [DOI] [PubMed] [Google Scholar]
- 13.Nagashunmugam T. Davis C. Kennedy S. Goldstein LT. Malamud D. Human submandibular saliva specifically inhibits HIV type 1. AIDS Res Hum Retroviruses. 1997;13(5):371–376. doi: 10.1089/aid.1997.13.371. [DOI] [PubMed] [Google Scholar]
- 14.Nagashunmugam T MD. Davis C. Abrams WR. Friedman HM. Human submandibular saliva inhibits human immunodeficiency virus type 1 infection by displacing envelope glycoprotein gp120 from the virus. J Infect Dis. 1998;178:1635–1641. doi: 10.1086/314511. [DOI] [PubMed] [Google Scholar]
- 15.Malamud D. Berthold P. Roth E. Friedman HM. Human submandibular saliva aggregates HIV. AIDS Research and Human Retroviruses. 1993;9:633–637. doi: 10.1089/aid.1993.9.633. [DOI] [PubMed] [Google Scholar]
- 16.Challacombe SJ. Sweet SP. Oral mucosal immunity and HIV infection: Current status. Oral Dis. 2002;8(2):55–62. doi: 10.1034/j.1601-0825.2002.00013.x. [DOI] [PubMed] [Google Scholar]
- 17.Lackner AA. Veazey RS. Current concepts in AIDS pathogenesis: Insights from the SIV/macaque model. Annu Rev Med. 2007;58:461–476. doi: 10.1146/annurev.med.58.082405.094316. [DOI] [PubMed] [Google Scholar]
- 18.Whitney JB. Luedemann C. Bao S, et al. Monitoring HIV vaccine trial participants for primary infection: Studies in the SIV/macaque model. AIDS. 2009;23(12):1453–1460. doi: 10.1097/QAD.0b013e32832b43d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlowski PA. Lynch RM. Patterson RR. Cu-Uvin S. Flanigan TP. Neutra MR. Modified wick method using Weck-Cel sponges for collection of human rectal secretions and analysis of mucosal HIV antibody. J Acquir Immune Defic Syndr. 2000;24(4):297–309. doi: 10.1097/00126334-200008010-00001. [DOI] [PubMed] [Google Scholar]
- 20.Bagby GJ. Zhang P. Purcell JE. Didier PJ. Nelson S. Chronic binge ethanol consumption accelerates progression of simian immunodeficiency virus disease. Clin Exp Res. 2006;30(10):1781–1790. doi: 10.1111/j.1530-0277.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- 21.Amedee A. Rychert J. Lacour N. Fresh L. Ratterree M. Viral and immunological factors associated with breast milk transmission of SIV in rhesus macaques. Retrovirology. 2004;1(1):17. doi: 10.1186/1742-4690-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rychert J. Lacour N. Amedee AM. Genetic analysis of simian immunodeficiency virus expressed in milk and selectively transmitted through breastfeeding. J Virol. 2006;80(8):3721–3731. doi: 10.1128/JVI.80.8.3721-3731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krowicka H. Robinson JE. Clark R. Hager S. Broyles S. Pincus SH. Use of tissue culture cell lines to evaluate HIV antiviral resistance. AIDS Res Hum Retroviruses. 2008;24(7):957–967. doi: 10.1089/aid.2007.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kestler HW., 3rd Naidu YN. Kodama T, et al. Use of infectious molecular clones of simian immunodeficiency virus for pathogenesis studies. J Med Primatol. 1989;18(3–4):305–309. [PubMed] [Google Scholar]
- 25.Kodama T. Burns DP. Kestler HW., 3rd Daniel MD. Desrosiers RC. Molecular changes associated with replication of simian immunodeficiency virus in human cells. J Med Primatol. 1990;19(3–4):431–437. [PubMed] [Google Scholar]
- 26.Cole KS. Rowles JL. Jagerski BA, et al. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J Virol. 1997;71(7):5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protocall Immunol. 2005:11. doi: 10.1002/0471142735.im1211s64. Chapter 12:Unit 12. [DOI] [PubMed] [Google Scholar]
- 28.Mascola JR. D'Souza P. Gilbert P, et al. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79(16):10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polonis VR. Brown BK. Rosa Borges A, et al. Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology. 2008;375(2):315–320. doi: 10.1016/j.virol.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Cory AH. Owen TC. Barltrop JA. Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3(7):207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 31.Kozlowski PA. Chen D. Eldridge JH. Jackson S. Contrasting IgA and IgG neutralization capacities and responses to HIV type 1 gp120 V3 loop in HIV-infected individuals. AIDS Res Hum Retroviruses. 1994;10(7):813–822. doi: 10.1089/aid.1994.10.813. [DOI] [PubMed] [Google Scholar]
- 32.Wang SW. Kozlowski PA. Schmelz G, et al. Effective induction of simian immunodeficiency virus-specific systemic and mucosal immune responses in primates by vaccination with proviral DNA producing intact but noninfectious virions. J Virol. 2000;74(22):10514–10522. doi: 10.1128/jvi.74.22.10514-10522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shugars DC. Alexander AL. Fu K. Freel SA. Endogenous salivary inhibitors of human immunodeficiency virus. Arch Oral Biol. 1999;44(6):445–453. doi: 10.1016/s0003-9969(99)00003-5. [DOI] [PubMed] [Google Scholar]
- 34.Cohen MS. Shugars DC. Fiscus SA. Limits on oral transmission of HIV-1. Lancet. 2000;356(9226):272. doi: 10.1016/S0140-6736(00)02500-9. [DOI] [PubMed] [Google Scholar]
- 35.Choi WS. Collignon C. Thiriart C, et al. Effects of natural sequence variation on recognition by monoclonal antibodies neutralize simian immunodeficiency virus infectivity. J Virol. 1994;68(9):5395–5402. doi: 10.1128/jvi.68.9.5395-5402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borda JT. Alvarez X. Kondova I, et al. Cell tropism of simian immunodeficiency virus in culture is not predictive of in vivo tropism or pathogenesis. Am J Pathol. 2004;165(6):2111–2122. doi: 10.1016/S0002-9440(10)63261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Follis KE. Trahey M. LaCasse RA. Nunberg JH. Continued utilization of CCR5 coreceptor by a newly derived T-cell line-adapted isolate of human immunodeficiency virus type 1. J Virol. 1998;72(9):7603–7608. doi: 10.1128/jvi.72.9.7603-7608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole KS. Alvarez M. Elliott DH, et al. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology. 2001;290(1):59–73. doi: 10.1006/viro.2001.1144. [DOI] [PubMed] [Google Scholar]
- 39.Hirbod T. Reichard C. Hasselrot K, et al. HIV-1 neutralizing activity is correlated with increased levels of chemokines in saliva of HIV-1-exposed uninfected individuals. Curr HIV Res. 2008;6(1):28–33. doi: 10.2174/157016208783571964. [DOI] [PubMed] [Google Scholar]
- 40.Hasselrot K. Saberg P. Hirbod T, et al. Oral HIV-exposure elicits mucosal HIV-neutralizing antibodies in uninfected men who have sex with men. AIDS. 2009;23(3):329–333. doi: 10.1097/QAD.0b013e32831f924c. [DOI] [PubMed] [Google Scholar]
- 41.Farquhar C. VanCott T. Bosire R, et al. Salivary human immunodeficiency virus (HIV)-1-specific immunoglobulin A in HIV-1-exposed infants in Kenya. Clin Exp Immunol. 2008;153(1):37–43. doi: 10.1111/j.1365-2249.2008.03664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]