Abstract
Transgenic mouse lines in which GFP expression is under the control of tissue-and stage specific promoters have provided powerful experimental tools for identification and isolation of cells at specific stage of differentiation along a lineage. In the present study we used primary cell cultures derived from the dental pulp from pOBCol3.6GFP and pOBCol2.3GFP transgenic mice as a model to develop markers for early stages of odontoblast differentiation from progenitor cells. We analyzed the temporal and spatial expression of 2.3-GFP and 3.6-GFP during in vitro mineralization. Using FACS to separate cells based on GFP expression, we obtained relatively homogenous sub-populations of cells and analyzed their dentinogenic potentials and their progression into odontoblasts. Our observations showed that these transgenes were activated before the onset of matrix deposition and in cells at different stages of polarization. The 3.6-GFP transgene was activated in cells in early stages of polarization whereas the 2.3-GFP transgene was activated at a later stage of polarization just before or at the time of formation of secretory odontoblast.
Keywords: Odontoblast differentiation, dental pulp, progenitors, type I collagen, green fluorescent protein
INTRODUCTION
Dentinogenesis is regulated by odontoblasts, highly specialized cells originating from the neural crest derived cells of the dental papilla. The differentiation of odontoblasts from the neural crest cells is a long process involving several intermediate steps that are dependent and regulated by epithelial signals [1–4]. The first epithelial signals from the early oral ectoderm lead to the induction of odontogenic potential in the cranial neural crest cells. The next step in the advancing differentiation within the odontoblast cell lineage is the formation of dental papilla during the transition from bud to cap stage of tooth development that is regulated by signals from epithelial bud and primary enamel knot. Next, dental papilla cells in close proximity to the epithelial–mesenchymal interface at the tip of the cusp differentiate into pre-odontoblasts, whereas the rest of dental papilla cells form the dental pulp [1–4].
The differentiation of pre-odontoblasts occurs at the bell stage of tooth development and is regulated by signals from inner enamel epithelium and secondary enamel knots [1–4]. Further differentiation proceeds in a graded fashion from cusp tips towards the inter-cuspal areas and cervical loops and include the withdrawal of pre-odontoblasts from cell cycle and the formation of polarized odontoblasts from pre-odontoblasts in close contact with the epithelial–mesenchymal interface. It has been suggested that pre-odontoblasts located away from the interface, becomes incorporated within the Höhl layer and during tooth injury leading to odontoblast death, differentiate into new odontoblasts for reparative dentinogenesis [5]. The formation of pre-odontoblasts is followed by the formation of polarizing odontoblasts, secretory/functional odontoblasts and finally the formation of mature and terminally differentiated odontoblasts [1–4].
Secretory/functional odontoblasts (also called young odontoblasts) are engaged in the secretion of unmineralized predentin matrix, composed primarily of type I collagen (Col1a1) [4, 6]. As these odontoblasts continue their differentiation, they secrete many non-collagenous proteins (NCPs) essential for the mineralization of collagen fibers and crystal growth [7]. The NCPs of dentin include proteins that are also found in bone such as decorin [8], biglycan [8], osteonectin [9, 10] and osteocalcin (OC) [10]. Other NCPs in dentin belong to the SIBLING (Small Integrin- Binding LIgand, N-linked Glycoprotein) family that includes osteopontin (OPN), bone sialoprotein (BSP), dentin matrix protein 1 (DMP1), dentin sialophosphoprotein (DSPP) and matrix extracellular phosphoglycoprotein (MEPE) [7]. High levels of expression of DSPP and DSP are the hallmark of odontoblast differentiation and are routinely used to distinguish differentiated odontoblasts from undifferentiated progenitors and osteoblasts [11–13]. DSPP expression is first detected in secretory odontoblasts and increases in terminally differentiated odontoblasts. After production of mineralized dentin, odontoblasts recede towards pulp and leave behind cell processes that extend into the mineralized dentin and give dentin matrix its characteristic tubular morphology [2–4]. In mice, the steps between the formation of pre-odontoblasts and mature odontoblasts are completed within 6–10 hours [14, 15].
The cellular and molecular mechanisms regulating odontoblast differentiation have been the subject of intense investigation using a variety of in vitro and in vivo approaches using cells derived from dental pulps [3, 11, 16]. The differentiation of odontoblasts in these cultures has been characterized by a number of standard morphological and functional methods including the expression of extracellular matrix components. Most often, published studies report end point assays in whole primary dental pulp cultures, which represent a heterogeneous population [3, 16] containing cells and nodules at various stages of odontoblast differentiation. The heterogeneity has made it difficult to fully characterize the intermediate steps in the odontoblast lineage and differentiation.
Key to the isolation of cells at intermediate stages during lineage progression is the ability to identify and isolate the cells at specific stages of differentiation. Techniques most often employed include isolation of cells by fluorescence-activated cell sorting (FACS), based on expression of cell surface antigens, and laser capture microscopy based on location of recognizable anatomical markers. In recent years, transgenic animals carrying Green Fluorescence Protein (GFP) coding sequences under the control of tissue-specific or stage-specific promoters and the utilization of FACS-mediated cell isolation and enrichment have provided powerful experimental tools for lineage studies [17–22].
Transgenic mouse lines in which GFP expression is under the control of tissue-and stage specific regulatory elements of genes involved in osteogenesis have provided valuable tools for examining the stepwise progression and differentiation of osteo-progenitors into pre-osteoblasts, osteoblasts and osteocytes [23–25]. Pre-osteoblasts could be identified by the expression pOBCol3.6GFP, osteoblasts by the expression of pOBCol2.3GFP, mature osteoblasts by OC-GFP and osteocytes by the expression of DMP1-GFP transgenes [23–25].
Col1a1 is the most abundant collagen in dentin (approximately 86–90%) [26–28]. The regulatory elements of the Col1a1 gene directing its expression to odontoblasts are similar to those in bone [29–32]. Furthermore, the stepwise progression of progenitor/stem cells into the odontoblast lineage is comparable with the process of osteoblasts differentiation, which starts from stem/progenitor cells differentiation into pre-osteoblasts and finally into terminally differentiated functional osteoblasts. These observations suggest that pOBCol3.6GFP and pOBCol2.3GFP transgenic animals in which 3.6- and 2.3-kb fragments of rat type I collagen promoter drive the expression of GFP, respectively, provide models for examination of the stepwise progression of progenitor/stem cells along the odontoblast lineage.
This possibility was supported by our in vivo studies in the developing teeth (molars and incisors) of these transgenic animals [12, 33, 34] that showed the expression of pOBCol3.6GFP (referred to as 3.6-GFP) and pOBCol2.3GFP (referred to as 2.3-GFP) transgenes at high intensity in secretory and differentiated odontoblasts expressing high levels of DSPP [12, 34]. Our transplantation studies in which pieces of dental pulp isolated from pOBCol2.3GFP mice were transplanted under the kidney capsule showed the formation of dentin-like and bone-like mineralized tissues by explanted dental pulps [12]. Dentin-like matrices were composed of tubular matrix (characterized by extended expression of 2.3-GFP into the tubular matrices) lined with cells expressing high levels of 2.3-GFP and DSPP. On the other hand, bone-like matrices were composed of atubular matrix with cells embedded within the matrix expressing high levels of 2.3-GFP and lacking the expression of DSPP [12]. These observations indicate that Col1a1-GFP transgenes (2.3-GFP and 3.6-GFP) provide excellent non-invasive markers for identification of odontoblasts and examination of the progression of odontoblast differentiation from progenitor cells.
In the present study, we have used 3.6-GFP and 2.3-GFP transgenic animals as an experimental model to examine the differentiation of odontoblast from progenitor cells in the dental pulp. We have analyzed the temporal and spatial expression of these transgenes during in vitro mineralization. With the ability of FACS to separate cells based on GFP expression, we also obtained relatively homogenous sub-populations of cells and analyzed their dentinogenic potentials and their progression into odontoblasts in vitro. Our observations show that these transgenes are activated before the onset of matrix deposition and in cells at different intermediate stages of odontoblast differentiation. The 3.6-GFP transgene was activated in cells in early stages of polarization whereas the 2.3-GFP transgene was activated at a later stage of polarization just before or at the time of formation of secretory odontoblast.
Materials and Methods
Primary dental pulp cultures
The coronal portions of the pulps from first and second molars were isolated from 5–7-day-old hemizygous pOBCol3.6GFP, pOBCol2.3GFP and non-transgenic mice and prepared for primary cultures as described previously [35, 36]. Briefly, 520 cells/mm2 (5×105 cells/well in 35-mm culture plates) were grown first in media containing Dulbecco's modified Eagles' medium (DMEM, Invitrogen), 20% fetal bovine serum (FBS, Hyclone, USA), 40µg/ml and 40U/ml of Pen-Strep antibiotics. Three days later media was changed to media containing 10% FBS that was switched to mineralization inducing media containing 50 µg/ml ascorbic acid, and 4 µM β-glycerophosphate in confluent cultures at day 7 [13, 35, 36]. Media was changed every 2 days for a period of 2 weeks.
Digital imaging and epifluorescence analysis in cell culture
GFP expression in cell culture was visualized using an Olympus IX50 inverted microscope equipped with an IX-FLA inverted reflected light fluorescence (Olympus America, Inc., Melville, NY, USA). A specific excitation wavelength was obtained using filters for GFPtpz (exciter: D500/20; dichroic: 525DCLP; emitter: D550/40) and GFPemd (exciter, D470/40; dichroic, 495LP; emitter, D525/50). Images were captured using a SPOTcamera (Diagnostic Instruments, Inc., Sterling Heights, MI).
Cell cultures were also examined with a Zeiss Axiovert 200 microscope (Carl Zeiss, Thornwood, NY) equipped with the motorized X-Y-Z platform, fluorescent cube, and AxioCam color digital camera controlled by a user-defined computation program using Openlab software (Improvision, Lexington, MA) as described before [37, 38]. Fluorescent expression of 3.6-GFP and 2.3-GFP in cultures was examined using a TOPAZ filter (YFP). The Zeiss Axiovert 200 microscope workstation allows the user to reproducibly record images of cultures at the same location and under the same conditions at different time points. Approximately 65% of the 35-mm culture dish can be repeatedly imaged and all the images taken can be concentrated into one image using AxioVision Rel 4.7 software.
Detection and Quantification of mineralization in cultures
Mineralized nodules in live cultures were visualized by Xylenol Orange (XO) staining as described previously [35]. Mineralization in fixed cultures was assayed and quantified using a modified von Kossa silver nitrate and modified Alizarin Red-S (AR-S) staining [36].Values represent mean ± SE of at least three independent experiment.
RNA extraction and analyses
Total RNA was prepared using TRI Reagent according to the manufacturer's instructions. For RT-PCR, isolated RNAs were reverse transcribed by Superscript II reverse transcriptase (Life Technologies) with oligo dT primers. Subsequent PCR amplifications were carried out using specific primers as described before [13], including primers for detection of GFP: 5′-GTCCGCCCTGAGCAAAGA and 3′-CTAGTGTACCAGGACGACCT [39]. RT-PCR products were resolved on 1% agarose gels, stained with ethidium bromide, and digitally photographed.
Flow cytometric analysis and Sorting (FACS)
Cultures were prepared for flow cytometric analysis by mild trypsin/EDTA digestion followed by centrifugation at +4°C. Cells were then re-suspended in 300–500 µl of staining medium containing 1µg/ml of propidium iodide (PI) and strained through a 70µm filter. Flow cytometry analysis was done collecting 20,000–100,000 cells on a BD FACScan/Calibur cytometer (Becton-Dickinson, San Jose, CA, USA) using a 488-nm excitation wavelength generated by a 15 mw argon ion laser. Emission was detected using a 500-nm long pass filter (GFP). The percentage of cells expressing GFP at high and low intensities was determined manually by setting a separation point at 102. Data were processed using Cell Quest software. Values represent mean ± S.E. determined from at least three independent experiments in which dental pulp cells obtained from non-transgenic mice were used as controls.
The expression of GFP was also examined in combination with various commercially available anti-mouse antibodies in 7 days old pulp cultures including CD45.2-Biotin; Sca-1-PE (D7); and CD90/Thy1.2-FITC (53-2.1) as described previously [13]. Approximately 0.5–1 ×106 cells was incubated with pre-titrated antibodies (1:50-1:800), in the presence of rat Ig (when necessary), washed and resuspended in 300µl of staining medium containing 1µg/ml of PI (propidium iodide) [39]. Between 20,000 and 100,000 cells was used for analysis.
For FACS sorting, seven day cultures were prepared by mild trypsin/EDTA digestion followed by centrifugation at 4°C. FACS sorting based on GFP expression was performed on 2.5×106 cells/ml by UCHC FACS facility on a FACS-Vantage BD cell sorter with a 130-µm nozzle at a speed of 3–5K cells/s. GFP was excited at 488 nm with an argon laser and a 550/30 emission filter was utilized. Upon separation, GFP+ and GFP− live cells were collected into DMEM with 20% FBS, and plated at 520 cells/mm2 (5×105 cells/well in 35-mm culture plates) density. Cultures were grown as described for unsorted population.
Immunocytochemistry and Confocal Microscopy
Dental pulp cells cultured on glass surface for 14 days were fixed and processed for immunocytochemistry as described previously [13]. Briefly, cells were fixed with 2% paraformaldehyde, washed and then incubated with 3% dry milk in PBS to block nonspecific staining. Cells were then incubated with 1:400 dilution of Anti-DSP antibody [LF-153 (kind gift from Dr. Larry Fisher)] for 1 hour at room temperature, and then with 1:800 dilution of Alexa Fluor 568 goat anti-rabbit antibody (Invitrogen, USA) for an additional hour at room temperature. For nuclear detection, samples were incubated with 1:500 dilution of TO-PRO-3 (Invitrogen, USA, 1:500 in PBS) for 30 min. After staining, cells were transferred to a slide with Prolong Gold anti-fade reagent (Invitrogen, A). A Zeiss LSM 510 confocal laser scanning microscope (Thornwood, NY) with a 63×, 1.4 numerical aperture oil immersion objective was used to collect images by simultaneous recording in the 568l and 647l channels. The expression of DSP was correlated with the expression of 3.6-GFP and 2.3-GFP.
Cell cycle analysis
To analyze the cell cycle and rate of proliferation in Col1a1-GFP+ and Col1a1-GFP− populations, three days cultures were washed with PBS, trypsinized and resuspended in a staining media (1×HBSS, 2% fetal calf serum, 10mM HEPES, pH7.2) containing 5µg/ml of a Hoechst 33342 nuclear dye (Molecular Probes, Invitrogen, USA) and incubated for 90 minutes at 37°C in a dark as described before [40]. Controls included cultures from the same animal but without addition of nuclear dye and cultures from wild type animals. Cells were analyzed in a BD LSRII flow cytometer (Becton-Dickinson). Data was analyzed using ModFitLT software (Verity Software House, Inc., Topshame, ME, USA) and FlowJo software. Values represent mean and ± SD of percentage of in G0+G1, G2+M and S phases in GFP+ and GFP− sub-populations in at least three independet experiments.
Statistical analysis
Unpaired, two-tailed t-tests were performed to determine statistically significant differences and p<0.05 was considered statistically significant.
RESULTS
Expression of 3.6-GFP and 2.3-GFP transgenes during mineralization in primary dental pulp cultures
We have previously characterized the sequence of mineralization in dental pulps from unerupted murine molars [13] and showed that when dissociated pulp cells are placed in primary culture, they proliferate rapidly and reach confluence at day 7. The proliferation phase is followed by the formation of distinct multi-layered individual nodules. The first sign of mineralization is around day 10, with increases thereafter so that by day 21 almost the entire culture dish is covered by a sheet of mineralized tissue [13].
To gain further insight into the stage of activation of 3.6-GFP and 2.3-GFP transgenes during odontoblast differentiation, we examined their expression during the mineralization in primary pulp cultures established from coronal portions of unerupted molars from hemizygous pOBCol3.6GFP and pOBCol2.3GFP transgenic mice (P5-P7). The pattern and intensity of GFP expression was correlated with the onset and subsequent growth of the mineralized nodules at various time points in real time by XO staining in the same cultures.
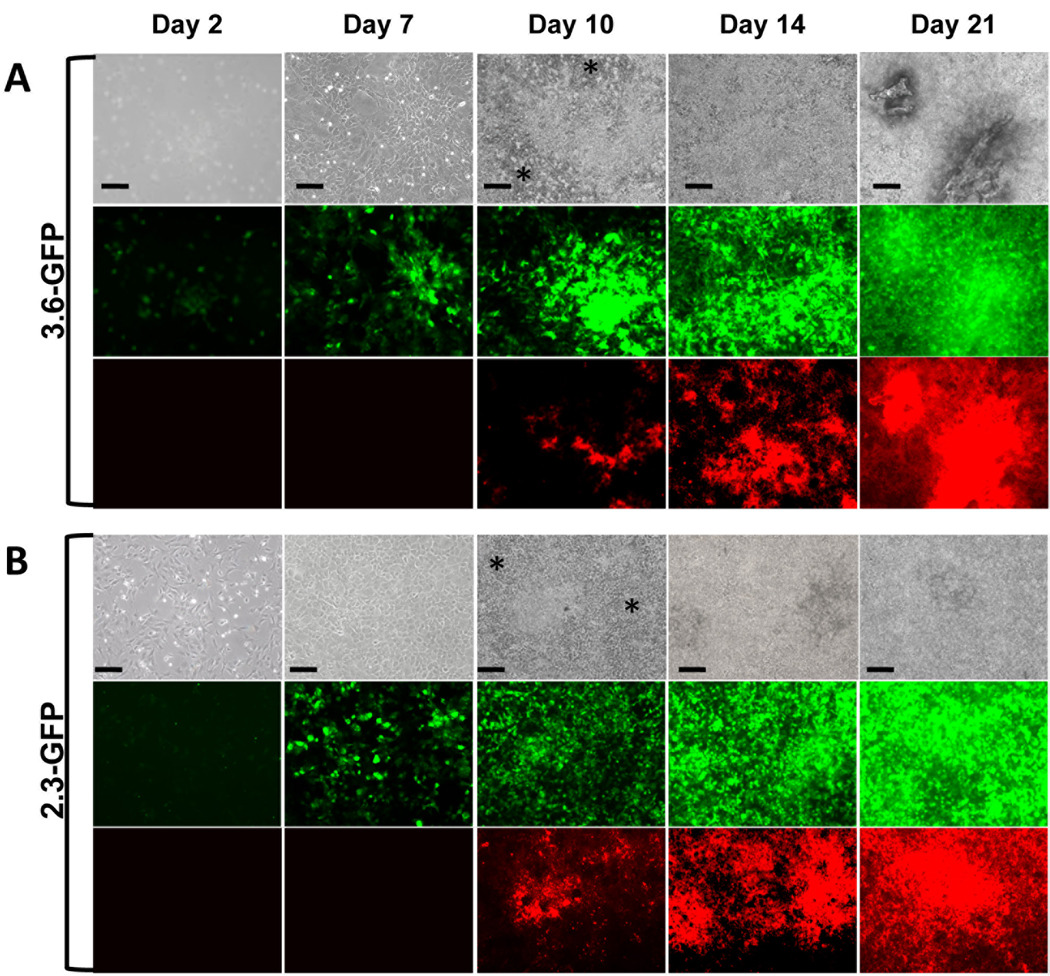
Epifluorescence analysis of primary pulp cultures from transgenic animals showed expression of the transgenes in scattered cells at day 2 (Figure 1A & B). At day 7, 3.6-GFP was detected in clusters of fibroblastic-like cells (Figure 2A) whereas 2.3-GFP was expressed in isolated cuboidal cells (Figure 2B). After addition of mineralization media, at day 10, 14 and 21, both transgenes were expressed at high intensity in multilayered and mineralized nodules (Figure 1A & B). At day 10, the areas expressing the transgenes at high intensity were larger than the areas of mineralization (XO staining) (Figure 1A & B). At days 14 and 21, the areas of XO staining overlapped more closely with areas expressing transgenes at high intensity. Cells expressing these transgenes at lower intensities were detected between nodules (Figure 1A & B).
Figure 1. Expression of 3.6-GFP and 2.3-GFP transgenes in primary dental pulp cultures during in vitro mineralization and dentinogenesis.
Primary dental pulp cultures obtained from pOBCol3.6GFP (A) and pOBCol2.3GFP (B) transgenic animals grown for 21 days in culture. Images of the same areas in cultures at different time points analyzed under phase contrast (upper rows in A and B), epifluorescent light using filters for GFPtpz and GFPemd for detection of GFP (middle rows in A and B) and epifluorescent light using TRITC Red filter for detection of XO staining (lower rows in A and B). Note the presence of GFP+ cells early in the culture (day 2), in cell clusters at day 7 and in differentiating and differentiated nodules between days 10–21. Note that areas of the cultures expressing GFP at day 10 extend beyond the areas of XO staining. There is a closer correlation of GFP expression and XO staining at days 14 and 21. Note the expression of low levels of 3.6-GFP and 2.3-GFP in the inter-nodular areas (indicated by asterisc). Scale bars=100µm.
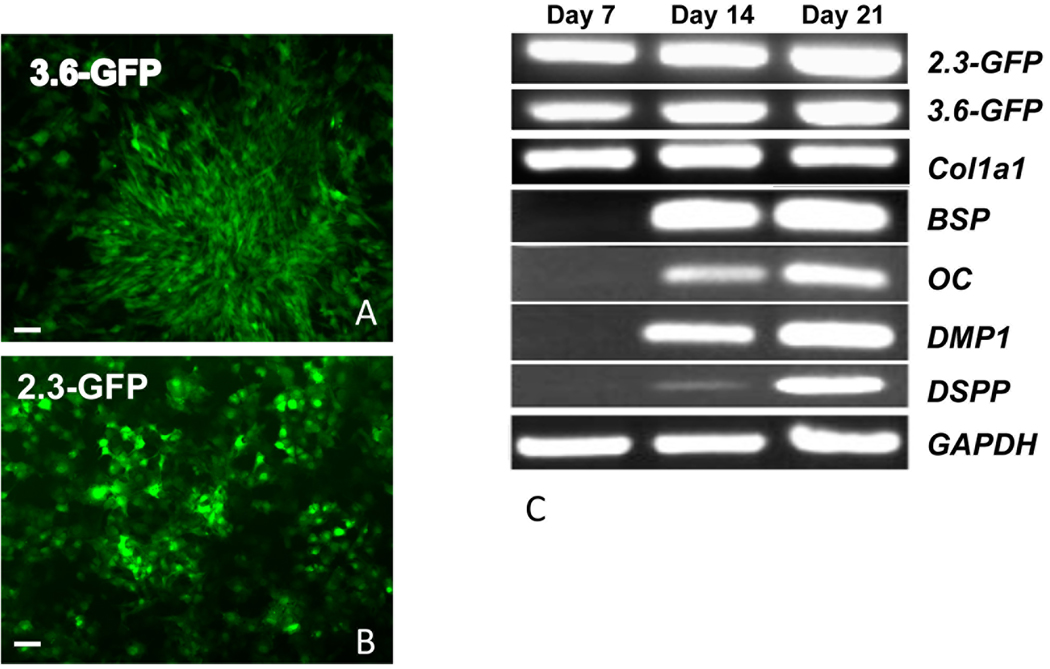
Figure 2. Expression of early and late markers of mineralization in primary pulp cultures and their correlation with 3.6- and 2.3-GFP expression.
A and B are epifluorescent images of cultures at day 7 showing the expression of 3.6-GFP in fibroblastic cells and 2.3-GFP in cuboidal cells. Scale bars=100µm.
(C) RT-PCR analysis of RNA extracted from primary dental pulp pulps at different time points from pOBCol3.6GFP and pOBCol2.3GFP mice. GFP and Col1a1 expression were detected at day 7 and increases at days 14 and 21. Markers of mineralization (BSP, OC and DMP1) and dentinogenesis (DSPP) were detected at day 14 and increased at day 21.
These results showed similarities in the temporal and spatial patterns of expression of these transgenes in primary pulp cultures. Both transgenes were expressed at early time points prior to mineralization. Cells undergoing mineralization expressed 3.6-GFP and 2.3-GFP transgenes at high intensity (Figure 1A & B).
Col1a1 transgenes are expressed before the expression of markers of mineralization
Our previous analyses of expression of these transgene in the developing teeth in vivo showed that they are expressed at high levels in secretory odontoblasts expressing DSPP [12, 34]. The expression of these transgenes at early time points (before mineralization) in primary pulp cultures was different from our in vivo studies and thus was further examined by RT-PCR analysis of primary pulp cultures from pOBCol2.3GFP and pOBCol3.6GFP transgenic animals.
Expression of both transgenes was detected at day 2 and 7 before the expression of various markers of mineralization including DSPP (Figure 2C and data not shown). In these cultures expression of DSPP, OC, BSP and DMP1 was detected at day 14 and increased at day 21 (Figure 2C).
Expression of Col1a1-GFP in odontoblasts and osteoblasts in primary dental pulp cultures
Our previous characterization of dental pulp cultures from unerupted murine molars showed dentinogenic and osteogenic potentials of these cells [13]. To correlate the expression of transgenes with dentinogenesis, DSP expression was examined in primary dental pulp and primary calvaria osteoblasts cultures from pOBCol3.6GFP and pOBCol2.3GFP transgenic mice. At day 14 expression of both transgenes was detected in mineralized nodules in primary dental pulp and primary calvaria osteoblasts cultures. In pulp cultures, DSP expression was detected in some but not all mineralized nodules expressing transgenes (supplemental Figure 1A–H and data not shown). DSP expression was not detected in mineralized nodules in primary calvaria osteoblasts (supplemental Figure 1I–L and data not shown). These results indicated that in primary pulp cultures these transgenes are expressed by both odontoblasts and osteoblasts.
Changes in Col1a1-GFP+ population and the intensity of GFP signal in primary dental pulp cultures
Changes in the percentage of 2.3-GFP and 3.6-GFP expressing cells (GFP+) and the signal strength within the GFP+ populations at different time points was examined by flow cytometry analysis (supplemental Figure 2 and Table 1).
Table 1.
Expression of GFP in primary pulp cultures obtained from pOBCol3.6GFP and pOBCol2.3GFP animals. FACS analysis of dental pulp cultures at different time points (days 2, 7, 10 and 14). Values represent means ± S.E. from at least three individual experiments. A) Percentage of GFP+ and GFP− populations. B) Distributions of cells expressing of GFP at high and low intensity within the GFP+ populations. Arrows in Figure 2 demonstrate the separation border between cells expressing high and low levels of GFP.
| A | |||
|---|---|---|---|
| Days in culture |
% of GFP+ cells |
% of GFP− cells |
|
| 3.6-GFP | 2 | 44.67 ± 2.7 | 55.3 ± 2.7 |
| 7 | 57.3 ± 1.5 | 42.7 ± 1.5 | |
| 10 | 61.3 ± 1.8 | 38.7 ± 1.8 | |
| 14 | 79.3 ± 1.5 | 20.7 ± 1.5 | |
| 2.3-GFP | 2 | 49.6 ± 1.8 | 50.4 ± 1.8 |
| 7 | 72.9 ± 11.8 | 27.1 ± 11.8 | |
| 10 | 83.6 ±10.5 | 16.4 ± 10.5 | |
| 14 | 92 ± 3.0 | 8.0 ± 3.0 | |
| B | |||
|---|---|---|---|
| GFP+ cells | |||
| Days in culture |
High intensity | Low intensity | |
| 3.6-GFP | 2 | 31.3 ± 4.3 | 13.3 ± 3.5 |
| 7 | 32.3 ± 2.2 | 25 ± 2.6 | |
| 10 | 39.7 ± 0.9 | 21.3 ± 4.5 | |
| 14 | 48.3 ± 2.0 | 30.3 ± 1.5 | |
| 2.3-GFP | 2 | 20.5 ± 3.7 | 29.1 ± 2.4 |
| 7 | 45.1 ± 9.7 | 27.7 ± 4.4 | |
| 10 | 65.0 ± 16.9 | 18.6 ± 6.5 | |
| 14 | 70.1 ± 12.8 | 21.9 ± 10.4 | |
Analysis of cultures from pOBCol3.6GFP mice showed that during mineralization there were increases in the percentage of GFP+ cells and about a 3 fold decrease in the GFP− cells (supplemental Figure 2 and Table 1A). At all time points 3.6-GFP was expressed at two levels of intensity (supplemental Figure 2 and Table 1B). Cells that expressed 3.6-GFP at the lower intensity (less or equal to 102) constituted 13% of the cells at day 2 and increased about 2.3 fold by day 14 (supplemental Figure 2 and Table 1B). Cells expressing 3.6-GFP at higher intensity (more than 102) increased about 1.5 fold between days 2 and 14 (supplemental Figure 2 and Table 1B). These changes were followed by increases in the mean intensity (strength) of GFP signal that were most significant between days 2 (approximately 230) and 7 (approximately 2116). No significant change in the mean intensity was seen in these cultures between days 7 and 14.
FACS analysis of primary cultures derived from pOBCol2.3GFP mice showed that mineralization in these cultures was also associated with increases in the percentage of 2.3-GFP+ cells and 6 fold decreases in the percentage of 2.3-GFP− cells (supplemental Figure 2 and Table 1A). Similar to 3.6-GFP, 2.3-GFP was also expressed at two intensities. Cells expressing 2.3-GFP at lower intensity constituted 29% of the cells at day 2 and decreased by day 14, whereas cells expressing 2.3-GFP at high intensity increased about 3.5 fold between days 2 and 14 (supplemental Figure 2 and Table 1B). There were also continuous increases in the mean intensity of 2.3-GFP between days 2 (450) and day 14 (>7420). At all time points the intensity of the 2.3-GFP exceeded the intensity of 3.6-GFP transgene.
Thus, during mineralization in primary pulp cultures there are increases in cells expressing 3.6-GFP transgene at low and high intensities and increases in the percentage of cells expressing 2.3-GFP at high intensity.
The increases in the percentage of the GFP+ cells in these cultures can be due to activation of transgenes in GFP− cells (did not express these transgenes before) and/or proliferation of existing GFP+ cells present at early time points in cultures. To distinguish between these possibilities, DNA content/cell cycle analysis was performed in three-day-old cultures using flow cytometry and Hoechst 33342 nuclear stain.
Cell cycle analysis in cultures from pOBCol3.6GFP mice showed proliferation in both populations. There were no significant differences in the rates of proliferation (G2M+S) between 3.6-GFP− (46.7%) and 3.6-GFP+ (42.8%) cells (Table 2). Analysis of cultures from pOBCol2.3GFP mice also showed similar rates of proliferation in 2.3-GFP− (47.5%) and 2.3-GFP+ (42.3%) cells (Table 2). The rates of proliferation (G2M+S) in 2.3-GFP+ and 2.3-GFP−cells were very similar to 3.6-GFP+ and 3.6-GFP− cells. These results indicated that increases in the number of GFP+ cells in cultures were due to proliferation of GFP+ as well as activation of these transgenes in cells that did not express GFP before.
Table 2. Analysis of cell proliferation using Hoecsht 33342 dye.
Three day old primary pulp cultures derived from pOBCol3.6GFP and pOBCol2.3GFP animals were trypsinized and incubated with Hoecsht 33342 for 90 minutes. Between 10,000–20,000 cells were FACS analyzed and results were obtained using ModFit Software. Results represent mean ± S.E. from at least three experiments.
| Phases of the cell cycle |
GFP− | GFP+ | |
|---|---|---|---|
| Col3.6GFP | G0G1 | 53.6 ± 1.8 | 56.8 ± 2.4 |
| S | 27.8 ± 1.8 | 21.8 ± 1.5 | |
| G2M | 18.6 ± 1.2 | 21.0 ± 1.2 | |
| Col2.3GFP | G0G1 | 52.6 ± 1.8 | 57.8 ± 2.4 |
| S | 6.9 ± 1.6 | 18.1 ± 1.9 | |
| G2M | 40.6 ± 0.4 | 24. 2 ± 0.7 |
Stage of activation of 3.6-GFP and 2.3-GFP transgenes in pulp cultures
The presence of a mixture of cells (GFP+ and GFP−) and the high rate of proliferation of GFP+ cells in pulp cultures made it difficult to study the stage of activation of this transgenes during mineralization and dentinogenesis. Therefore, as the next step, we studied the expression of these transgenes in Col1a1-GFP− populations using FACS based on GFP.
Primary pulp cultures were prepared from pOBCol3.6GFP and pOBCol2.3GFP transgenic mice and grown for 7 days. Following growth and expansion, FACS and re-analysis were used for separation of two populations with >97% purity (Figures 3A and 4A). Both populations were plated at same density as unsorted cells (5×105 cells/well in 35-mm culture plates). Cells were first grown for 7 days in media supporting their proliferation and then for an additional 7 to 14 days in mineralization inducing media. Controls included cultures from unsorted cells (primary cultures) and cultures from unsorted cells that were re-plated after 7 days (secondary culture).
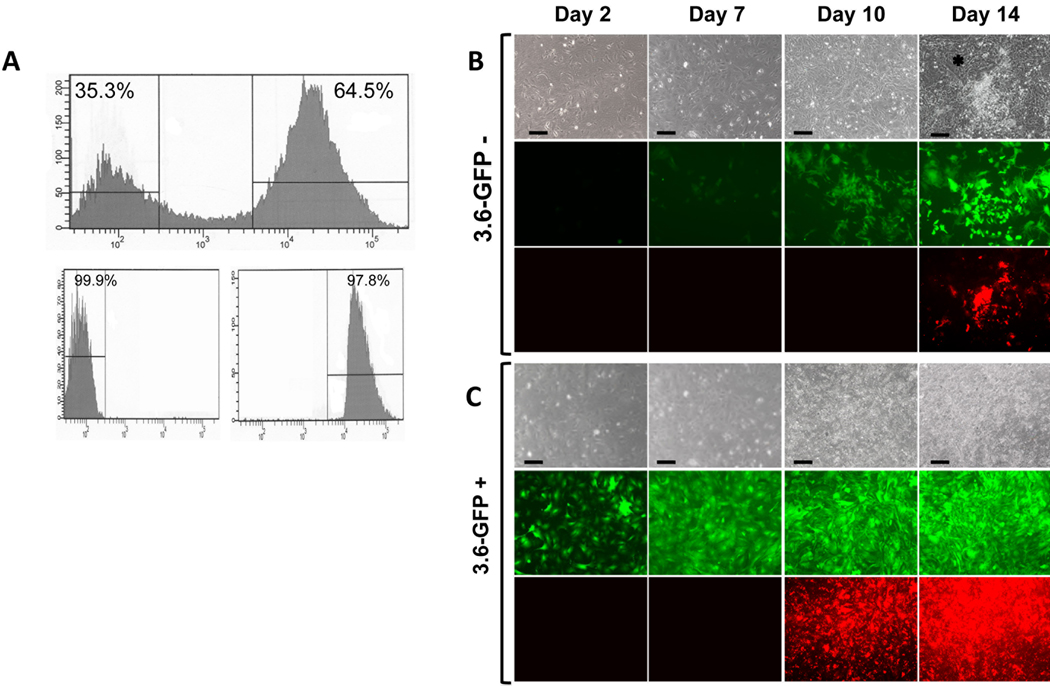
Figure 3. Comparison of behavior of 3.6-GFP+ and 3.6-GFP− populations.
Primary pulp cultures from pOBCol3.6GFP were grown for 7 days and processed for FACS sorting. (A) Histogram showing that FACS sorting resulted in clear separation of 3.6-GFP+ (approximately 64%) and 3.6-GFP− (approximately 35%) populations. Histogram of the FACS re-analysis shows that the purity of isolated cell populations was higher from 97%.
B and C represent images of the same areas in live cultures at different time points analyzed under phase contrast (upper rows), epifluorescent light using filters for GFPtpz (middle rows in B and C) and epifluorescent light using TRITC Red filter for detection of XO staining (lower rows in B and C). Note that in 3.6-GFP+ cultures, all cells continuously expressed this transgene. XO staining was detected at day 10 and increased at day 14. In 3.6-GFP− cultures, GFP expression was not detected at day 2. At day 7, 3.6-GFP was expressed in some cells at low intensity. 3.6-GFP was expressed in the cell clusters at day 10 and increased at day 14. XO staining was detected at day 14. Cells in the inter-nodular area are indicated by asterisk. Scale bars=100µm.
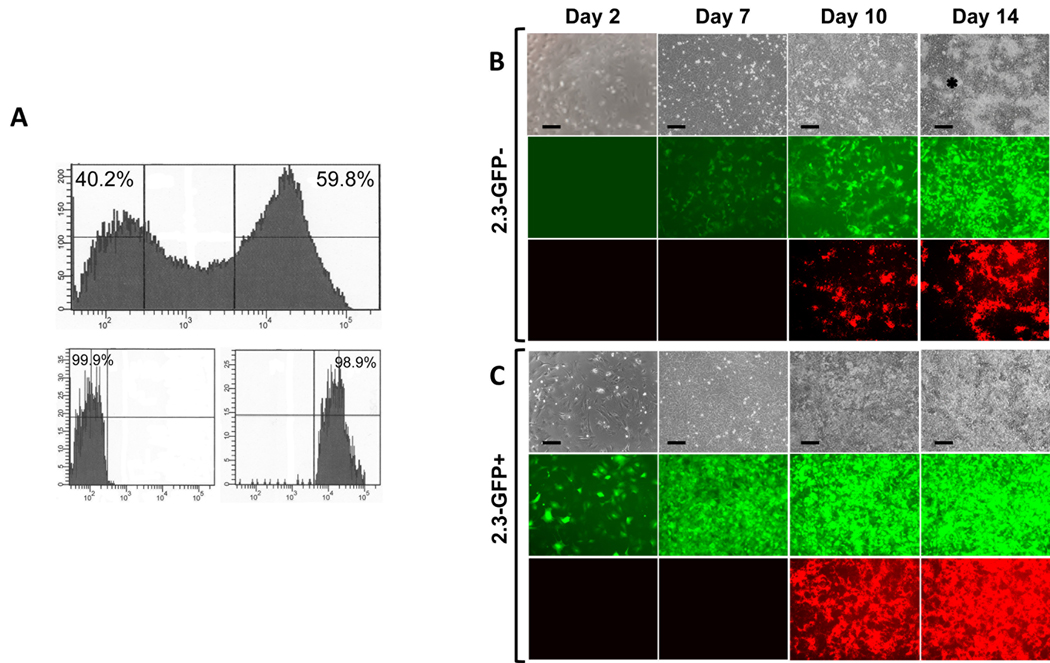
Figure 4. Comparison of behavior of 2.3-GFP+ and 2.3-GFP− sub-populations.
Primary pulp cultures derived from pOBCo12.3GFP mice were grown for 7 days and processed for FACS. (A) Histogram showing that FACS sorting resulted in clear separation of 2.3-GFP+ (approximately 60%) and 2.3-GFP− (approximately 40%) sub-populations. Histogram of FACS re-analysis on isolated cell sub-populations showed that the purity of both was higher than 98%.
B and C represent images of the same areas in live cultures at different time points analyzed under phase contrast (upper rows), epifluorescent light using filters for GFPemd (middle rows in B and C) and epifluorescent light using TRITC Red filter for detection of XO staining (lower rows in B and C). Note that in cultures from 2.3-GFP+, all cells continuously expressed this transgene. XO staining was detected at day 10 and increased at day 14.
In 2.3-GFP− cultures, expression of 2.3-GFP was not detected at day 2. At day 7, 2.3-GFP was expressed in some cells. Expression of 2.3-GFP was intensified at day 10 and 14. XO staining is detected at day 10 and 14. Cells in the inter-nodular area are indicated by asterisk. Scale bars=100µm.
Cultures from 3.6-GFP− cells remained healthy and proliferated (Figures 3B). In these cultures, expression of 3.6-GFP was not detected during the first 6 days (Figure 3B) and appeared in scattered cells around day 7. Expression of 3.6-GFP was detected in clusters of fibroblastic-like cells at day 10 and in cuboidal cells in mineralized nodules at day 14 and 21. XO staining showed that the onset of mineralization in 3.6-GFP− cultures was at day 14 and later than in primary and secondary unsorted cells (day 10).
The appearance of GFP signal in 2.3-GFP− cultures was similar to 3.6-GFP− cultures. In these cultures, expression of 2.3-GFP appeared at day 7 in isolated cuboidal cells and increased thereafter (Figure 4B). The onset of mineralization in 2.3-GFP− cultures was at day 10 and similar to primary and secondary unsorted cells. Expression of 2.3-GFP was detected in isolated cuboidal cells (Figure 4B).
Comparison of the whole cultures at days 14 and 21 showed that the intensity of the GFP in cultures from 2.3-GFP− and 3.6-GFP− populations was significantly lower than in unsorted cells (supplemental Figure 3). Mineralization in 3.6-GFP− and 2.3-GFP− cultures displayed nodular patterns.
These observations showed similarities in the stage of activation of these transgenes. Both transgenes were activated around days 6/7 prior to mineralization indicating that during odontoblasts differentiation these transgenes are activated prior to the formation of secretory odontoblasts in either pre-odontoblasts or polarizing odontoblasts.
Mineralization and dentinogenic potentials of Col1a1-GFP+ populations
To gain further insight into stage of activation of these transgenes, mineralization and dentinogenic potentials of cultures from different populations were examined and compared. Controls in these experiments also included primary and secondary pulp cultures.
Cultures from 3.6-GFP+ cells remained healthy and proliferated (Figures 3C & 4C). In these cultures GFP expression was maintained throughout the entire culture period. The first sign of mineralization in these cultures was at day 10 with increases thereafter (Figures 3C & 4C). At days 14 and 21, extensive mineralization was detected as a sheet over the entire culture dishes (Figure 5C & E and data not shown). The extent of mineralization in 3.6-GFP+ cultures after 21 days was lower than primary cultures but similar to the secondary cultures (Figure 5G). The extent of mineralization in secondary cultures was about 66% lower than in primary cultures showing the effects of replating on the extent of mineralization (Figure 5G).
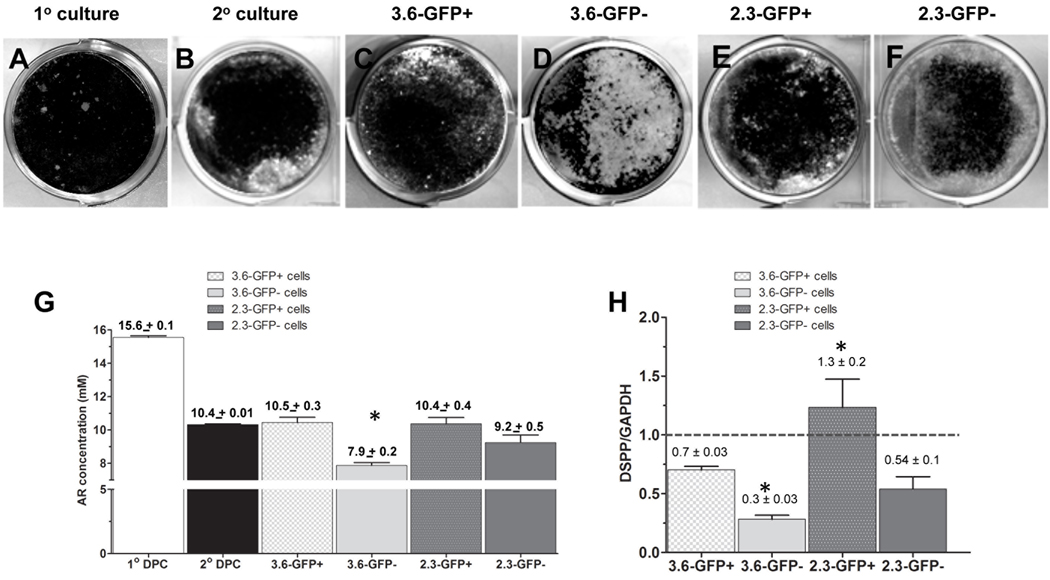
Figure 5. Differentiation potentials of cultures from Col1a1-GFP+ and Col1a1-GFP−populations.
A–F represent images of von Kossa staining of mineralized tissue in cultures established from unsorted cells without re-plating (primary cultures) (A), cultures from unsorted cells that were re-plated after 7 days (secondary culture) (B), 3.6-GFP+ (C), 3.6-GFP− (D), 2.3-GFP+ (E) and 2.3-GFP− (F) cells after 21 days.
(G) Histogram showing the amounts of extracted Alizarin Red staining in different cultures after 21 days. Note the decrease in the mineralization in secondary pulp cultures as compared to primary pulp cultures. The amounts of extracted Alizarin Red staining were similar between secondary unsorted cells, 3.6-GFP+, 2.3-GFP+ and 2.3-GFP− populations. There was mineralization in 3.6-GFP− cultures. Values represent the concentration of the extracted Alizarin Red calculated from the mean absorbance ± S.E. for at least three independent experiments with multiple samples in each experiment (*p<0.05).
(H) Histogram showing the relative levels of DSPP after 21 days in various cultures. Note the increased levels of DSPP in cultures from 2.3-GFP+ as compared to other cultures. Also note that the relative levels of DSPP were similar in 2.3-GFP− and 3.6-GFP+ cultures. The relative levels of DSPP in 3.6-GFP cultures were lower than all other cultures. The dash line represents the DSPP/GAPDH in secondary cultures that was arbitrary set to 1. Values represent the mean ± S.E. of DSPP/GAPDH for at least three independent experiments (*p<0.05).
The behaviour of 2.3-GFP+ cells in culture was similar to 3.6-GFP+ cells (Figures 3C & 4C). The intensity of the GFP in cultures from 2.3-GFP+ and 3.6-GFP+ populations exceeded those in unsorted and Col1a1- GFP+ cultures (Supplemental Figure 3).
The extent of mineralization in 2.3-GFP+ cultures was similar to 3.6-GFP+ cultures (Figure 5G). However, the relative levels of DSPP in 2.3-GFP+ cultures were significantly higher than in 3.6-GFP+ cultures (Figure 5H) suggesting that 2.3-GFP+ population contains either increased number of odonto-progenitors (pre-odontoblasts and/or polarizing odontoblasts) and/or odonto-progenitors at more advanced stage of differentiation.
Mineralization and dentinogenic potentials of Col1a1-GFP− populations
Analyses of 3.6-GFP− and 2.3-GFP− cultures after 21 days showed distinct differences in the extent of mineralization and relative levels of DSPP expression (Figure 5). The extent of mineralization and relative levels of DSPP expression in 3.6-GFP− cultures were significantly lower than in 2.3-GFP− cultures (Figures 5G & 5H). Interestingly, the extent of mineralization and relative levels of DSPP in 2.3-GFP− cultures was similar to 3.6-GFP+ cultures (Figures 5G & 5H).
The differences in the dentinogenic potentials of various subpopulations can be due to differences in the number of odonto-progenitors. To test this possibility, the distribution of CD90 and Sca-1, markers shown to be expressed by progenitors in dental pulp [13] in different populations were examined and compared. As shown in Supplemental Figure 4, Sca1+/CD45− and CD90+/CD45−cells were present in all four sub-populations. The percentage of Sca1+/CD45− cells was highest in 2.3-GFP− populations. (Supplemental Figure 4). Our interpretation of these results is that the differences in the dentinogenic potentials of different populations are not related to the number of odonto-progenitors and are related to differences in the stage of differentiation of odonto-progenitors in different populations.
DISCUSSION
The goal of our studies was to use pOBCol3.6GFP and pOBCol2.3GFP transgenic mice to distinguish and identify populations of cells at early stages of odontoblast differentiation. In our previous studies we characterized the expression of 3.6-GFP and 2.3-GFP transgenes in the developing teeth in vivo and showed similarities in the temporal and spatial expression of these transgenes during odontoblast differentiation [12, 33, 34].
In the present study we examined the expression of these transgenes during the mineralization and odontoblast differentiation in primary cultures derived from the coronal portions of dental pulp from these transgenic animals. Our studies showed the expression of these transgenes in scattered cells at day 7 before initiation of mineralization and expression of DSPP. These observations appear to be different from our previous in vivo studies that showed the expression of both transgene at high intensity in functional and in fully differentiated odontoblasts expressing high levels of DSPP [12, 33, 34]. However, in our in vivo studies, we reported the expression of low but detectable levels of both transgenes in differentiating odontoblasts at the tip of the mesio-lingual cusp of the first mandibular molar at the late bell stage (E18) of tooth development [12, 33, 34]. These observations together with previous tissue distribution studies showing that Col1a1 is first expressed in polarizing odontoblasts and its expression is maintained in secretory and terminally differentiated odontoblasts [26–28] indicated that during odontoblasts differentiation 2.3-GFP and 3.6-GFP are activated in polarizing odontoblasts.
The activation of these transgenes in polarizing odontoblasts is further supported by our studies in Col1a1-GFP+ populations. Our results showed that 2.3-GFP+ and 3.6-GFP+ populations contain highly proliferative cells giving rise to sheets of mineralized tissue with very little if any Col1a1-GFP− cells indicating their enrichment in progenitors committed to mineralization. Our further studies showed the formation of odontoblasts expressing DSPP in both populations. However, the levels of DSPP in 2.3-GFP+ cultures were significantly higher than in 3.6-GFP+ cultures. This difference most likely is related to increased number of polarizing odontoblasts in 2.3-GFP+ population as compared to 3.6-GFP+ population.
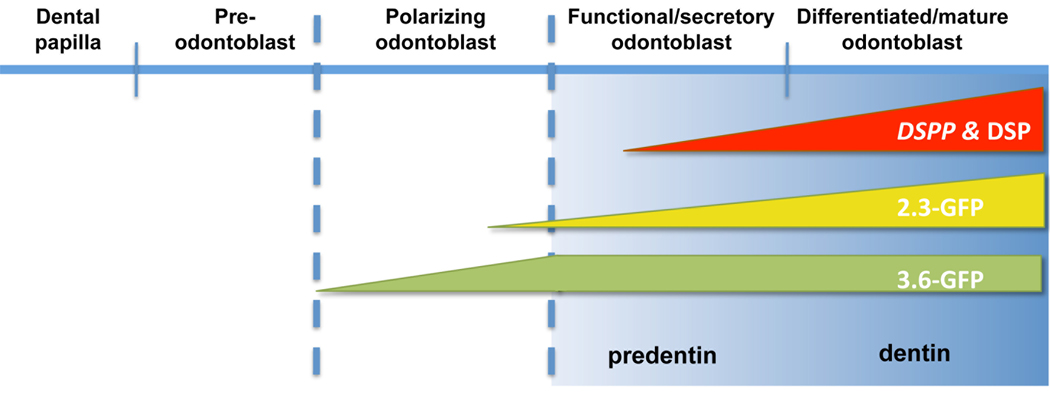
Furthermore, our results showed similarity in the levels of DSPP in 2.3-GFP− and 3.6-GFP+ cultures. This together with the lack of significant differences in the distributions of CD90+ and Sca1+ cells in these populations suggested that similarities in the levels of DSPP were related to similarties in the stage of differentiation of odonto-progenitors in these populations. These observations suggest differences in the stage of activation of the transgenes during odontoblast differentiation in that the 3.6-GFP transgene is activated at an earlier developmental stage (i.e; early stage of polarization) than the 2.3-GFP transgene (i.e; later stage of polarization just before transition to secretory odontoblasts) (see Figure 6). The differences in the stage of activation of these transgenes during odontoblast differentiation is also supported by the differences in the morphology of the 3.6-GFP+ and 2.3-GFP+ cells in early time points and before the mineralization in pulp cultures. These differences were not appreciated in the developing teeth in vivo because of the close proximity of cells in the early and late stage of polarization. Further experiments are in progress to examine the expression of these two transgene in dual GFP reporter mice in which different color GFPs (topaz and cyan) are driven by 3.6 and 2.3 kb of the Col1a1 promoter, respectively.
Figure 6.
Schematic representation of proposed stages of activation of 3.6-GFP and 2.3-GFP transgenes during odontoblast differentiation. DSPP was used as a marker of early and later stages of mineralization.
The patterns of expression of these transgenes in developing teeth and in dental pulp cultures share both similarities and differences with previous observations in the developing bones and calvaria that indicated that Col1a1-GFP transgenes can identify distinct sub-populations of cells during osteoblast differentiation [24, 38, 41, 42]. Histological analyses of calvaria and long bones showed that 3.6-GFP was expressed in the fibroblastic layer of the periosteum whereas 2.3-GFP was expressed in the osteoblastic layer of periosteum [41, 42]. Furthermore, in vitro studies revealed distinct differences in the time of activation of 3.6-GFP and 2.3-GFP transgenes during osteogenesis [38, 41]. In primary cultures derived from neonatal calvaria and bone marrow stromal cells, expression of 3.6-GFP was detected early prior to the appearance of mineralized nodules and was correlated with the expression of markers of pre-osteoblasts (i.e. mRNAs for alkaline phosphatase and Col1a1). On the other hand, expression of 2.3-GFP was not detected at early stages of differentiation and was detected at the time of mineralization. The expression of 2.3-GFP was correlated with the expression of markers of minerlization (i.e. mRNAs for bone sialoprotein and osteocalcin) [38, 41]. These observations together led to the conclusion that during osteogenesis activation of these transgenes is associated with different stages of osteoblast differentiation [38, 41]. 3.6-GFP is activated in pre-osteoblasts whereas 2.3-GFP is activated in osteoblasts [38, 41].
The dentinogenic and osteogenic potentials of dental pulp cells in vitro [12, 13, 43] and the expression of 2.3-GFP and 3.6-GFP in both osteoblasts and odontoblasts in dental pulp cultures makes the distinction between activation of these transgenes in odontoblasts vs. osteoblasts difficult. However, it is imporatnt to note that our studies showed that in pulp cultures both 3.6-GFP and 2.3-GFP are expressed prior to mineralization. The early expression of 3.6-GFP in pulp cultures is similar to the expression of this transgene in primary cultures derived from neonatal calvaria and bone marrow stromal cells suggesting that the expression of 3.6-GFP at day 7 in pulp cultures reflects the activity of this transgene in pre-osteoblasts and polarizing odontoblasts. On the other hand, the early expression of 2.3-GFP in pulp cultures is different from the expression of this transgene in primary cultures derived from neonatal calvaria and bone marrow stromal cells. This suggest that the expression of 2.3-GFP at day 7 in pulp cultures reflects the activation of this transgene in polarizing odontoblasts and not in the cells in the osteogenic lineage.
In summary, our studies showed that 2.3-GFP and 3.6-GFP transgenes mark cells first at polarizing stage of odontoblast differentiation and later in secretory and terminally differentiated odontoblasts. Furthermore, the differences in the extent of mineralization and levels of DSPP expression in sorted population also suggests that these transgene mark cells at different stages of polarization. The transition from pre-odontoblasts to polarizing odontoblasts and secretory odontoblasts involves many important changes, including changes in cell shape, reorganization of the cytoplasmic organelles, polarization and changes in cell-cell interactions. Recent studies showed expression of several transcription factors including Klf4 [44], Dlx3 [45], Wnt10a and Runx2 [46] in polarizing odontoblasts. Abnormalities in dentinogenesis in Ofd1 mutant mice that mimic the X-linked orofacial digital type I syndrome also have reveal the roles of cilium components in polarizing odontoblasts [47]. Identification of a marker for polarizing odontoblasts and two different Col1a1 promoter fragments with stage-specific expression in cells in odontoblast lineage would allow characterization of their gene expression profiles and facilitate many experimental approaches for studying odontoblasts in vivo and in vitro.
Previous studies showed that during odontoblast differentiation, expression of Col1a1 is followed by osteocalcin and osteopontin, which are expressed in late polarizing odontoblasts and secretory odontoblasts [27, 28, 48]. DSPP and DMP1 expression are first detected in secretory odontoblasts and increased in terminally differentiated odontoblasts [27, 28, 48]. Based on these reports, further experiments are in progress to examine the expression of OC-GFP, DMP1-GFP and DSPP-GFP during odontoblast differentiation in vivo and in vitro to identify and isolate cells at additional intermediate stages during odontoblast differentiation.
Supplementary Material
Fourteen day old primary cultures from dental pulp (A–H) and calvaria osteoblasts (I–L), established from 5–7 day old transgenic mice were processed for immunocytochemistry. Cultures were incubated with anti-DSP antibody (1:200), secondary antibody (Alexa Fluor 568 goat anti-rabbit antibody) (1:800) and TO-PRO3 (1:500). Cultures were examined under confocal microscope for the expression of 3.6-GFP (A, E, I), DSP (B, F, J) and TO-PRO3 (C, G, K). D, H, and L represent overlay of the three images generated by AxioVision software. Note that DSP expression was present in dental pulp (A–H) and not in calvaria (I–L) cultures. B, F, J represent higher magnification images of dental pulp cultures (E–H) showing DSP expression in the cytoplasm and in the extracellular matrix, but not in the nuclei (stained blue) as indicated by the overlay image (H). Scale bars =50µm.
Primary pulp cultures from control (non-GFP, CD1) (first row), pOBCol3.6GFP (second row) and pOBCol2.3GFP mice (third row) animals. Histograms show flow cytometry analysis of the expression of 2.3-GFP and 3.6-GFP transgenes at different time points. The X-axis represents the GFP intensity and the y-axis represents the PI staining used to gate only on viable cells. Numbers on the histograms represent the mean ± SD of GFP+ and GFP− in three independent analysis. Arrows indicate the separation point at 102 between higher and lower intensity of GFP that was set manually.
Dental pulp cultures were established from unsorted and sorted cells isolated from pOBCol3.6GFP (upper row) and pOBCol2.3GFP (bottom row) transgenic animals as previously described. GFP expression was examined by Zeiss scanning microscope. Each panel is a large image concentrated from 9 (3×3) small images captured by imaging workstation at 4× magnification. Note the differences in number of cells expressing GFP and the intensity of GFP expression in different sub-populations. Scale bars =1mm.
Histogram showing the percentage of CD90+/CD45− and Sca1+/CD45−cells in Col1a1-GFP+ and Col1a1-GFP− populations isolated from dental pulps from pOBCol13.6GFP pOBCol12.3GFP mice at day 7 in cultures. Values represent mean +S.E of at least three independent experiments.
Acknowledgements
We thank all the individuals who provided reagents, valuable input and technical assistance in various aspects of these studies including Drs. David Rowe, Ivo Kalajzic, Victor P. Francone, Mrs. Katie Lamothe, Miss. Diane Gran, and Mrs. Rodgers. We also thank Drs. Barbara Kream and William Upholt for critical review of this manuscript. This work was supported by a grant from National Institute of Health (NIDCR) to MM (DE016689).
Grant sponsor: National Institute of Health
Grant numbers: DE016689
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Thesleff I, Keranen S, Jernvall J. Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv Dent Res. 2001;15:14–18. doi: 10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- 2.Arana-Chavez VE, Massa LF. Odontoblasts: the cells forming and maintaining dentine. Int J Biochem Cell Biol. 2004;36:1367–1373. doi: 10.1016/j.biocel.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Sloan AJ, Smith AJ. Stem cells and the dental pulp: potential roles in dentine regeneration and repair. Oral Dis. 2007;13:151–157. doi: 10.1111/j.1601-0825.2006.01346.x. [DOI] [PubMed] [Google Scholar]
- 4.Lisi S, Peterkova R, Peterka M, Vonesch JL, Ruch JV, Lesot H. Tooth morphogenesis and pattern of odontoblast differentiation. Connect Tissue Res. 2003;44 Suppl 1:167–170. [PubMed] [Google Scholar]
- 5.Goldberg M, Smith AJ. Cells and Extracellular Matrices of Dentin and Pulp: A Biological Basis for Repair and Tissue Engineering. Crit Rev Oral Biol Med. 2004;15:13–27. doi: 10.1177/154411130401500103. [DOI] [PubMed] [Google Scholar]
- 6.Qin C, D'Souza R, Feng JQ. Dentin matrix protein 1 (DMP1): new and important roles for biomineralization and phosphate homeostasis. J Dent Res. 2007;86:1134–1141. doi: 10.1177/154405910708601202. [DOI] [PubMed] [Google Scholar]
- 7.Qin C, Baba O, Butler WT. Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med. 2004;15:126–136. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- 8.Steinfort J, van de Stadt R, Beertsen W. Identification of new rat dentin proteoglycans utilizing C18 chromatography. J Biol Chem. 1994;269:22397–22404. [PubMed] [Google Scholar]
- 9.Reichert T, Storkel S, Becker K, Fisher LW. The role of osteonectin in human tooth development: an immunohistological study. Calcif Tissue Int. 1992;50:468–472. doi: 10.1007/BF00296779. [DOI] [PubMed] [Google Scholar]
- 10.Bronckers AL, Gay S, Finkelman RD, Butler WT. Developmental appearance of Gla proteins (osteocalcin) and alkaline phosphatase in tooth germs and bones of the rat. Bone Miner. 1987;2:361–373. [PubMed] [Google Scholar]
- 11.Liu H, Gronthos S, Shi S. Dental pulp stem cells. Methods Enzymol. 2006;419:99–113. doi: 10.1016/S0076-6879(06)19005-9. [DOI] [PubMed] [Google Scholar]
- 12.Braut A, Kollar EJ, Mina M. Analysis of the odontogenic and osteogenic potentials of dental pulp in vivo using a Col1a1-2.3-GFP transgene. Int J Dev Biol. 2003;47:281–292. [PubMed] [Google Scholar]
- 13.Balic A, Aguila HL, Caimano MJ, Francone VP, Mina M. Characterization of stem and progenitor cells in dental pulps of the erupted and unerupted murine molars. Bone. 2010;46:1639–1651. doi: 10.1016/j.bone.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesot H, Lisi S, Peterkova R, Peterka M, Mitolo V, Ruch JV. Epigenetic signals during odontoblast differentiation. Adv Dent Res. 2001;15:8–13. doi: 10.1177/08959374010150012001. [DOI] [PubMed] [Google Scholar]
- 15.Ruch JV, Lesot H, Begue-Kirn C. Odontoblast differentiation. Int J Dev Biol. 1995;39:51–68. [PubMed] [Google Scholar]
- 16.Sloan AJ, Waddington RJ. Dental pulp stem cells: what, where, how? Int J Paediatr Dent. 2009;19:61–70. doi: 10.1111/j.1365-263X.2008.00964.x. [DOI] [PubMed] [Google Scholar]
- 17.Yu YA, Szalay AA, Wang G, Oberg K. Visualization of molecular and cellular events with green fluorescent proteins in developing embryos: a review. Luminescence. 2003;18:1–18. doi: 10.1002/bio.701. [DOI] [PubMed] [Google Scholar]
- 18.Hadjantonakis AK, Dickinson ME, Fraser SE, Papaioannou VE. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nat Rev Genet. 2003;4:613–625. doi: 10.1038/nrg1126. [DOI] [PubMed] [Google Scholar]
- 19.Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadjantonakis AK, Nagy A. FACS for the isolation of individual cells from transgenic mice harboring a fluorescent protein reporter. Genesis. 2000;27:95–98. doi: 10.1002/1526-968x(200007)27:3<95::aid-gene10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 21.Hadjantonakis AK, Nagy A. The color of mice: in the light of GFP− variant reporters. Histochem Cell Biol. 2001;115:49–58. doi: 10.1007/s004180000233. [DOI] [PubMed] [Google Scholar]
- 22.Misteli T, Spector DL. Applications of the green fluorescent protein in cell biology and biotechnology. Nat Biotechnol. 1997;15:961–964. doi: 10.1038/nbt1097-961. [DOI] [PubMed] [Google Scholar]
- 23.Bilic-Curcic I, Kronenberg M, Jiang X, Bellizzi J, Mina M, Marijanovic I, Gardiner EM, Rowe DW. Visualizing levels of osteoblast differentiation by a two-color promoter-GFP strategy: Type I collagen-GFPcyan and osteocalcin-GFPtpz. Genesis. 2005;43:87–98. doi: 10.1002/gene.20156. [DOI] [PubMed] [Google Scholar]
- 24.Kalajzic Z, Liu P, Kalajzic I, Du Z, Braut A, Mina M, Canalis E, Rowe DW. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–660. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 25.Kalajzic I, Braut A, Guo D, Jiang X, Kronenberg MS, Mina M, Harris MA, Harris SE, Rowe DW. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP−transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Andujar MB, Couble P, Couble ML, Magloire H. Differential expression of type I and type III collagen genes during tooth development. Development. 1991;111:691–698. doi: 10.1242/dev.111.3.691. [DOI] [PubMed] [Google Scholar]
- 27.Bleicher F, Couble ML, Farges JC, Couble P, Magloire H. Sequential expression of matrix protein genes in developing rat teeth. Matrix Biol. 1999;18:133–143. doi: 10.1016/s0945-053x(99)00007-4. [DOI] [PubMed] [Google Scholar]
- 28.Bronckers AL, D'Souza RN, Butler WT, Lyaruu DM, van Dijk S, Gay S, Woltgens JH. Dentin sialoprotein: biosynthesis and developmental appearance in rat tooth germs in comparison with amelogenins, osteocalcin and collagen type-I. Cell Tissue Res. 1993;272:237–247. doi: 10.1007/BF00302729. [DOI] [PubMed] [Google Scholar]
- 29.Pavlin D, Lichtler AC, Bedalov A, Kream BE, Harrison JR, Thomas HF, Gronowicz GA, Clark SH, Woody CO, Rowe DW. Differential utilization of regulatory domains within the alpha 1(I) collagen promoter in osseous and fibroblastic cells. J Cell Biol. 1992;116:227–236. doi: 10.1083/jcb.116.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebsbach PH, Harrison JR, Lichtler AC, Woody CO, Rowe DW, Kream BE. Transgenic expression of COL1A1-chloramphenicol acetyltransferase fusion genes in bone: differential utilization of promoter elements in vivo and in cultured cells. Mol Cell Biol. 1993;13:5168–5174. doi: 10.1128/mcb.13.9.5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogdanovic Z, Bedalov A, Krebsbach PH, Pavlin D, Woody CO, Clark SH, Thomas HF, Rowe DW, Kream BE, Lichtler AC. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J Bone Miner Res. 1994;9:285–292. doi: 10.1002/jbmr.5650090218. [DOI] [PubMed] [Google Scholar]
- 32.Thomas HF, Feldman JA, Bedalov A, Woody CO, Clark SH, Mack K, Lichtler AC. Identification of regulatory elements necessary for the expression of the COL1A1 promoter in murine odontoblasts. Connect Tissue Res. 1995;33:81–85. doi: 10.3109/03008209509016986. [DOI] [PubMed] [Google Scholar]
- 33.Braut A, Kalajzic I, Kalajzic Z, Rowe DW, Kollar EJ, Mina M. Col1a1-GFP transgene expression in developing incisors. Connect Tissue Res. 2002;43:216–219. doi: 10.1080/03008200290001078. [DOI] [PubMed] [Google Scholar]
- 34.Mina M, Braut A. New insight into progenitor/stem cells in dental pulp using Col1a1- GFP transgenes. Cells Tissues Organs. 2004;176:120–133. doi: 10.1159/000075033. [DOI] [PubMed] [Google Scholar]
- 35.Balic A, Mina M. Analysis of developmental potentials of dental pulp in vitro using GFP transgenes. Orthod Craniofac Res. 2005;8:252–258. doi: 10.1111/j.1601-6343.2005.00347.x. [DOI] [PubMed] [Google Scholar]
- 36.Balic A, Rodgers B, Mina M. Mineralization and expression of Col1a1-3.6GFP transgene in primary dental pulp culture. Cells Tissues Organs. 2009;189:163–168. doi: 10.1159/000154813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YH, Liu Y, Maye P, Rowe DW. Examination of mineralized nodule formation in living osteoblastic cultures using fluorescent dyes. Biotechnol Prog. 2006;22:1697–1701. doi: 10.1021/bp060274b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang YH, Liu Y, Buhl K, Rowe DW. Comparison of the action of transient and continuous PTH on primary osteoblast cultures expressing differentiation stage-specific GFP. J Bone Miner Res. 2005;20:5–14. doi: 10.1359/JBMR.041016. [DOI] [PubMed] [Google Scholar]
- 39.Boban I, Jacquin C, Prior K, Barisic-Dujmovic T, Maye P, Clark SH, Aguila HL. The 3.6 kb DNA fragment from the rat Col1a1 gene promoter drives the expression of genes in both osteoblast and osteoclast lineage cells. Bone. 2006;39:1302–1312. doi: 10.1016/j.bone.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 40.San Miguel SM, Fatahi MR, Li H, Igwe JC, Aguila HL, Kalajzic I. Defining a visual marker of osteoprogenitor cells within the periodontium. J Periodontal Res. 2010;45:60–70. doi: 10.1111/j.1600-0765.2009.01201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 42.Jiang X, Kalajzic Z, Maye P, Braut A, Bellizzi J, Mina M, Rowe DW. Histological analysis of GFP expression in murine bone. J Histochem Cytochem. 2005;53:593–602. doi: 10.1369/jhc.4A6401.2005. [DOI] [PubMed] [Google Scholar]
- 43.Ma D, Ma Z, Zhang X, Wang W, Yang Z, Zhang M, Wu G, Lu W, Deng Z, Jin Y. Effect of age and extrinsic microenvironment on the proliferation and osteogenic differentiation of rat dental pulp stem cells in vitro. J Endod. 2009;35:1546–1553. doi: 10.1016/j.joen.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Couble ML, Mouterfi N, Magloire H, Bleicher F. Spatial and temporal expression of KLF4 and KLF5 during murine tooth development. Arch Oral Biol. 2009;54:403–411. doi: 10.1016/j.archoralbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Choi SJ, Song IS, Feng JQ, Gao T, Haruyama N, Gautam P, Robey PG, Hart TC. Mutant DLX 3 disrupts odontoblast polarization and dentin formation. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.05.499. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashiro T, Zheng L, Shitaku Y, Saito M, Tsubakimoto T, Takada K, Takano-Yamamoto T, Thesleff I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;75:452–462. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 47.Thivichon-Prince B, Couble ML, Giamarchi A, Delmas P, Franco B, Romio L, Struys T, Lambrichts I, Ressnikoff D, Magloire H, Bleicher F. Primary cilia of odontoblasts: possible role in molar morphogenesis. J Dent Res. 2009;88:910–915. doi: 10.1177/0022034509345822. [DOI] [PubMed] [Google Scholar]
- 48.Bronckers AL, Lyaruu DM, Woltgens JH. Immunohistochemistry of extracellular matrix proteins during various stages of dentinogenesis. Connect Tissue Res. 1989;22:65–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fourteen day old primary cultures from dental pulp (A–H) and calvaria osteoblasts (I–L), established from 5–7 day old transgenic mice were processed for immunocytochemistry. Cultures were incubated with anti-DSP antibody (1:200), secondary antibody (Alexa Fluor 568 goat anti-rabbit antibody) (1:800) and TO-PRO3 (1:500). Cultures were examined under confocal microscope for the expression of 3.6-GFP (A, E, I), DSP (B, F, J) and TO-PRO3 (C, G, K). D, H, and L represent overlay of the three images generated by AxioVision software. Note that DSP expression was present in dental pulp (A–H) and not in calvaria (I–L) cultures. B, F, J represent higher magnification images of dental pulp cultures (E–H) showing DSP expression in the cytoplasm and in the extracellular matrix, but not in the nuclei (stained blue) as indicated by the overlay image (H). Scale bars =50µm.
Primary pulp cultures from control (non-GFP, CD1) (first row), pOBCol3.6GFP (second row) and pOBCol2.3GFP mice (third row) animals. Histograms show flow cytometry analysis of the expression of 2.3-GFP and 3.6-GFP transgenes at different time points. The X-axis represents the GFP intensity and the y-axis represents the PI staining used to gate only on viable cells. Numbers on the histograms represent the mean ± SD of GFP+ and GFP− in three independent analysis. Arrows indicate the separation point at 102 between higher and lower intensity of GFP that was set manually.
Dental pulp cultures were established from unsorted and sorted cells isolated from pOBCol3.6GFP (upper row) and pOBCol2.3GFP (bottom row) transgenic animals as previously described. GFP expression was examined by Zeiss scanning microscope. Each panel is a large image concentrated from 9 (3×3) small images captured by imaging workstation at 4× magnification. Note the differences in number of cells expressing GFP and the intensity of GFP expression in different sub-populations. Scale bars =1mm.
Histogram showing the percentage of CD90+/CD45− and Sca1+/CD45−cells in Col1a1-GFP+ and Col1a1-GFP− populations isolated from dental pulps from pOBCol13.6GFP pOBCol12.3GFP mice at day 7 in cultures. Values represent mean +S.E of at least three independent experiments.