Abstract
Genetic interactions represent the degree to which the presence of one mutation modulates the phenotype of a second mutation. In recent years, approaches for measuring genetic interactions systematically and quantitatively have proven to be effective tools for unbiased characterization of gene function and have provided valuable data for analyses of evolution. Here, we present protocols for systematic measurement of genetic interactions with respect to organismal growth rate for two yeast species.
1. Introduction
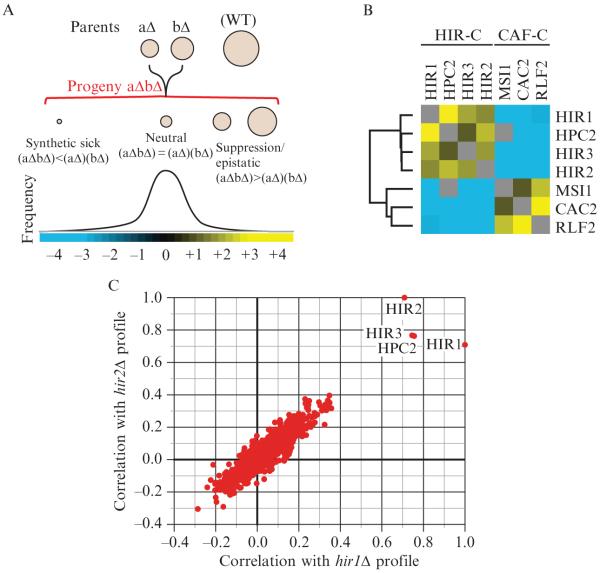
Genetic interactions, which represent the modulation of the phenotype of one mutation by the presence of a second mutation, have long been used as a tool to dissect the functional relationships among sets of genes (Guarente, 1993; Kaiser and Schekman, 1990). Classically, researchers have looked for strong qualitative differences between observed phenotypes of double mutants and the phenotypes of the two related single mutants. For example, a relationship referred to as synthetic lethality is observed when two mutations are not lethal when present individually but, when combined, result in an inviable organism. Synthetic sick/lethal, or negative, interactions have been used as evidence that two genes act in independent but complementary pathways. For example, strong negative genetic interactions exist between two key machines working in parallel pathways involved in chromatin assembly, the HIR complex (Hir1, Hir2, Hir3, and Hpc2) and the CAF complex (Msi1, Cac2, and Rlf2) (Collins et al., 2007a; Loyola and Almouzni, 2004) (Fig. 9.1B). On the other hand, if a normally deleterious mutation has no effect in the context of a second mutation, this is referred to as a positive genetic interaction, and often it identifies genes that act in the same pathway. Indeed, positive genetic interactions exist between members of the HIR complex as well as between the components that comprise the CAF complex (Collins et al., 2007a) (Fig. 9.1B). These two classes of interaction have been extremely useful for deciphering the organization of molecular pathways in model organisms, but in fact they represent only two special cases within a much larger spectrum of interactions (Fig. 9.1A).
Figure 9.1.
Epistatic interactions within and between chromatin assembly complexes. (A) The entire spectrum of genetic interactions. Quantitative genetic analysis can identify negative ((aΔbΔ) < (aΔ)(bΔ)), positive ((aΔbΔ) > (aΔ)(bΔ)), and neutral ((aΔbΔ) = (aΔ)(bΔ)) genetic interactions. (B) Genetic interactions between and within the HIR and CAF chromatin assembly complexes. Using the E-MAP approach (Collins et al., 2007a), strong negative interactions were detected between components of the HIR-C (HIR1, HPC2, HIR3, and HIR2) and the CAF-C (MSI1, CAC2, and RLF2), which are known to function in parallel pathways to ensure efficient chromatin assembly. Conversely, positive genetic interactions were observed between components within each complex. Blue and yellow interactions correspond to negative and positive genetic interactions, respectively. (C) Plot of correlation coefficients generated from comparison of the genetic profiles from hir1Δ and hir2Δ to all other ~750 profiles from the chromosome biology E-MAP (Collins et al., 2007a). Note the high pairwise correlations with HIR1, HIR2, HIR3, and HPC2.
Recent advances in technology now make it possible to measure large numbers of genetic interactions systematically and in parallel in yeast (Pan et al., 2004; Roguev et al., 2007; Tong et al., 2001), and it is possible to make these measurements quantitatively (Decourty et al., 2008; Roguev et al., 2008; Schuldiner et al., 2005). Synthetic growth defects of varying magnitudes as well as a broad range of suppressive and masking interactions can be detected. Importantly, the ability to make these measurements in a high-throughput fashion also provides a novel and valuable context for understanding quantitative genetic interactions. Work has demonstrated that the pattern of interactions (represented as a mathematical vector) for a given mutation can be used as a multidimensional phenotype. These patterns can be compared, and sets of genes producing similar patterns (and hence sets that are functionally closely related) can be accurately identified (Schuldiner et al., 2005; Tong et al., 2004; Ye et al., 2005). For example, the patterns of interactions for HIR complex gene deletions are more strongly correlated to each other than to the patterns for other genes (Collins et al., 2007a) (Fig. 9.1C).
We designed the epistatic miniarray profile (E-MAP) approach focusing on two key strategies to maximize the value of high-throughput genetic interaction measurements. In this approach, quantitative genetic interactions are measured (using a simple growth phenotype) systematically between all pairwise combinations of 400–800 rationally chosen mutations. The first strategy is measuring interactions quantitatively, which allows the detection and analysis of the complete spectrum of interaction strengths. We have found that this both improves our ability to use patterns of genetic interactions to identify sets of genes acting in a common pathway (Collins et al., 2006), and that positive interactions (i.e., where a double mutant is fitter than expected) are particularly useful for making conjectures about gene function. Second, we aim to measure all pairwise interactions among a rationally chosen set of genes. This approach has several advantages. It increases the signal-to-noise ratio because the frequency of genetic interactions is higher between genes acting in related pathways (thus signal increases while noise is constant). It also provides a richer set of patterns for analysis. For example, having data for the components of all known DNA damage repair genes in an organism allows a researcher not only to classify a newly identified gene as a damage repair component, but also to assess which (if any) of the known repair pathways the new gene is most likely closely involved in. Importantly, with such a strategy, expansion is easy. New mutations of interest can rapidly be screened, and the results can be readily compared to and merged with the existing dataset.
Genetic interactions are measured not only in terms of growth or viability, but also can be (and have been) derived from other phenotypes (Jonikas et al., 2009). In general, we can define a genetic interaction (εAB) between mutations A and B in terms of any quantitative phenotype P as the difference between the observed phenotype of the double mutant (PAB,observed) and the expected phenotype of the double mutant (PAB,expected) if no interaction exists between the two mutations:
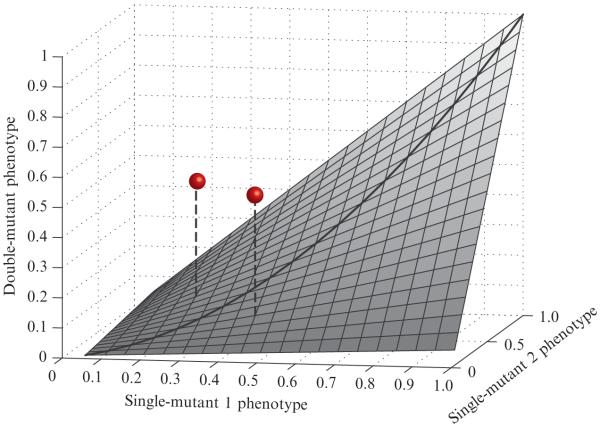
This formulation clearly depends on our ability to compute PAB,expected as a function of PA,observed and PB,observed. A theoretically derivable form for PAB,expected does not necessarily exist. However, a practically useful rule is that PAB,expected should account for the typical combined effect of two individual mutations with phenotypes PA,observed and PB,observed. Since strong genetic interactions are rare (Pan et al., 2004; Schuldiner et al., 2005; Tong et al., 2004), they manifest themselves as outliers that deviate from the surface that describes the broad trends in the majority of double-mutant data (Fig. 9.2). With this motivation, we define PAB,expected empirically to represent this typical combined effect. Additionally, if a simple functional form (e.g., the product PA,observed × PB,observed) captures accurately the empirical relationship, this can be an extremely useful simplification.
Figure 9.2.
Genetic interactions as deviations from expected double-mutant phenotypes. An idealized smooth surface is shown to represent the expected combined effects of independent mutations. The surface shown is not based on real data, but is intended to serve as an abstract example. The x- and y-axes represent single-mutant phenotypes, scaled between 0 and 1. The height of the surface (along the z-axis) represents the corresponding expected double-mutant phenotype. This surface should accurately describe the empirical typical double-mutant phenotypes, and it should be symmetric about the line y = x. The grey spheres represent observations for specific double mutants. The quantitative interaction is represented by the vertical distance from the point to the surface. As both of these points lie above the surface, they represent positive interactions.
In this chapter, we describe in detail a method for measuring quantitative genetic interactions based on the area of yeast colonies. We have used this strategy (with some technical differences) for both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Roguev et al., 2007; Schuldiner et al., 2006), and we provide here the details for each protocol. We also give a brief description of how analysis of the resulting data can be used to generate specific biological hypotheses.
2. Selection of Mutations for Genetic Analysis
Ultimately, comprehensive genetic interaction maps in both budding and fission yeasts may be generated where every possible pairwise double mutant is created and phenotypically assessed. Completion of such comprehensive genome-wide maps will represent a major accomplishment; however, it will still be some time until this is achieved. Additionally, our observations so far indicate that such maps will consist primarily of neutral genetic interactions. As mentioned above, the frequency of strong negative or positive genetic interactions between randomly chosen gene pairs has been estimated to be as little as 0.5%, but interactions are much more frequent between functionally related pairs of genes (Schuldiner et al., 2005; Tong et al., 2004). The E-MAP approach was devised to take advantage of this fact by specifically targeting genes that are likely to be functionally related. There are various ways by which sets of mutations can be selected for high-density, quantitative genetic interaction screening, and which is best strongly depends on the biological process that one wishes to interrogate, and the types of answers one would like to uncover. Genes can be chosen with the goal of identifying missing links within and between known pathways (e.g., what genes control the deposition of variant histones?), and they can also be chosen to address broader questions (to what extent are genetic interactions conserved during evolution?). We provide here an overview of strategies that have been used in the past.
The first E-MAP focused on approximately 400 genes likely to be involved in the early secretory pathway. In this case, gene selection was based largely on the localization of the corresponding proteins to either the Golgi apparatus or the endoplasmic reticulum (Schuldiner et al., 2005). The rationale was that proteins residing in a common subcellular compartment are more likely to be functionally related. Of course, gene selection solely based on localization would miss important factors acting in an indirect fashion. For example, signaling proteins such as kinases may mediate strong effects on spatially distant processes.
Another recently generated E-MAP focused on factors involved in various aspects of chromosome function, including transcription, DNA repair/replication, and chromosome segregation (Collins et al., 2007a). For this study, protein–protein interaction datasets were used as a primary source for gene selection. Using information from systematic affinity tag/purification mass spectrometry experiments (Collins et al., 2007b; Gavin et al., 2006; Krogan et al., 2006), the genes whose corresponding proteins were contained within one or more complexes involved in the chromatin-related functions were targeted. Once again, this method alone would miss factors indirectly impinging on the processes.
Sets of genes have also been targeted based on the presence of characterized domains (e.g., kinase domain, SET domain, etc.) or their likely molecular activity. For instance, an E-MAP was generated that comprised all kinases and phosphatases (both protein and nonprotein), regulatory subunits for these enzymes, phospho-binding proteins, and many factors known to be phosphorylated (Fiedler et al., 2009). The genes in this E-MAP impinge on all processes in the cell and therefore this approach allowed for a global view of the genetic architecture of the signaling apparatus instead of high-density information on one specific biological process. This work revealed an enrichment of positive genetic interactions between kinases, phosphatases, and their corresponding substrates (Fiedler et al., 2009) which would have been difficult to assess without a broad survey of these protein classes.
Finally, genes can be selected based on global, unbiased phenotypic screens. A recent study began with a genome-wide screen for mutations that affect the activity of the unfolded protein response (UPR) pathway (Jonikas et al., 2009). Then, approximately 400 mutations that modulate basal UPR activity were selected and further characterized using systematic double-mutant analysis. Of course, gene selection in this fashion requires single mutants to have a measurable effect on the process in question, whereas multiple mutations may be required to see an effect when compensatory pathways exist.
In general, a combination of the approaches described above is likely to be most effective. For example, in the final chromosome function E-MAP (Collins et al., 2007a), selection was not only based on composition of protein complexes but also on genome-wide genetic interaction screens using mutations of genes known to be integrally involved in processes of interest. In this way, a number of genes not associated with a known chromatin remodeling complex and not yet functionally annotated could be included in the analysis. Using this approach, we included the previously uncharacterized protein Rtt109 in the genetic analysis. Based on the genetic interaction profiling, we were able to link its function to the chromatin assembly protein Asf1 and identify it as the founding member of a new family of histone acetyltransferases responsible for K56 histone H3 acetylation (Collins et al., 2007a; Driscoll et al., 2007; Han et al., 2007).
Recently, this analysis was extended to another, evolutionarily distant organism (S. pombe). Selection of genes for analysis was guided by the same criteria. Additionally, mapping of direct orthologs between the two organisms made it possible to observe some of the general trends in genetic interaction network evolution (Roguev et al., 2007, 2008).
3. Generation and Measurement of Double Mutant Strains
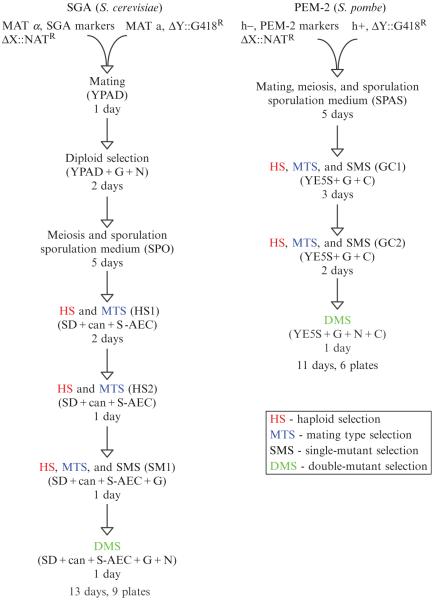
Here, we describe protocols for screening in both S. cerevisiae (SGA, synthetic genetic array) (Collins et al., 2006; Schuldiner et al., 2006; Tong and Boone, 2006) and S. pombe (PEM, pombe epistatic mapper) (Roguev et al., 2007). A flowchart comparing the two protocols is presented in Fig. 9.3. Both methodologies are, in essence, high-throughput procedures for random spore analysis where the growth of double mutant cell pools is monitored on agar using high-density cell arrays. The same logic is followed in both model organisms. In each screen, a query strain containing one NAT-marked mutation is crossed to an array of strains carrying G418-marked mutations. An array of diploid strains is generated by mating and a change of growth media is used to induce meiosis and sporulation. A series of selections are then used to select for haploid double-mutant cells of a particular mating type carrying both mutations. The growth phenotypes of the resulting double mutants are assessed by measuring colony area after a defined period of time.
Figure 9.3.
Overview of the experimental protocol. Flow charts outlining the series of selections used in S. cerevisiae E-MAP screens (left) and S. pombe PEM screens (right) are presented.
The two protocols differ in several important technical details due to biological differences between the two organisms. In S. cerevisiae, the mating step occurs on rich medium and nitrogen starvation is used to induce meiosis and sporulation. However, in S. pombe, the entire sequence (mating, meiosis, and sporulation) is induced by limited nitrogen, allowing the whole process to be carried out in a single step. Unlike in S. pombe, the diploid phase in S. cerevisiae is very stable, so an additional step enriching for diploids (diploid selection) is usually included in the SGA screen. In both systems, after the spores are germinated, the remaining diploid cells are killed (during haploid selection steps, HS), and only haploid cells of one mating type are allowed to grow (mating type selection, MTS). Importantly, one marker used in this selection comes from the parent strain of the opposite mating type, thus requiring that only haploid progeny from the initial mating can pass the selection. HS in both systems and MTS in S. pombe take advantage of recessive selectable markers. In S. cerevisiae, canavanine and S-AEC are used to select against the parent diploid cells, which are heterozygous for the CAN1 and LYP1 genes encoding transporters needed for import of these toxic compounds. A mating type-specific promoter driving the transcription of a conditionally essential metabolic gene (HIS3) is used for MTS. In S. pombe, a recessive allele providing resistance to cycloheximide has been engineered to perform both selections using a single marker (Roguev et al., 2007). In both the SGA and PEM protocols, two rounds of HS and MTS are carried out. In S. cerevisiae, an additional single mutant selection (SMS) step enriching for single and double mutant haploids is performed. In the PEM protocol, the same media is used for HS, MTS, and SMS. Finally, a double-mutant selection (DMS) is performed, the arrays are photographed and the images analyzed.
All selections before the final DMS step (HS, MTS, and SMS) let single and double mutants compete within the same cell mixture. These competition steps greatly improve the sensitivity and dynamic range of the method by allowing the detection of subtle synthetic growth defects. The final colony sizes measured after growth on the DMS plates reflect both the growth rate of the double mutant cells during this final stage, as well as the fraction of cells deposited on the final plate which has the right genotype (both NAT- and KAN-marked mutations). This latter quantity is largely determined by competitive growth during earlier steps.
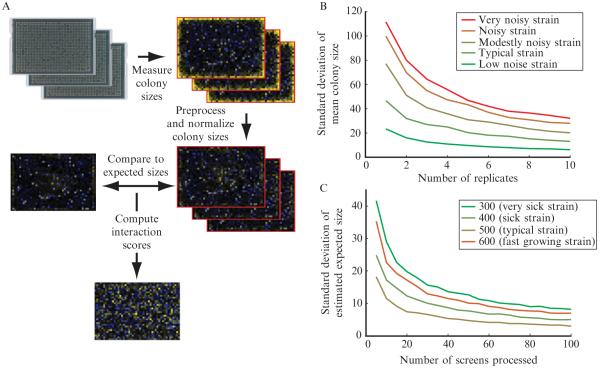
In planning a set of screens, one must decide how many replicate measurements will be sufficient to generate a high-quality dataset. By reanalyzing our earlier published data (Schuldiner et al., 2005), we find that the first three independent replicate measurements give substantial improvements in the precision of the measured average colony size (Fig. 9.4B). Additional replicates beyond three improve data quality, but it may be worth sacrificing these marginal gains in exchange for the ability to complete more screens at lower cost.
Figure 9.4.
Overview of the data processing procedure. (A) A flowchart describing the data processing procedure for a single screen is shown. The first images are digital photographs of arrays of yeast colonies. In the following images heatmaps of either measured colony sizes or genetic interaction scores are shown. In colony size heatmaps, blue represents small colonies, black represents average-sized colonies, and yellow represents large colonies. In the genetic interaction heatmap, blue represents negative interactions, black represents neutral interactions, yellow represents positive interactions, and gray represents missing data (or data filtered out during quality control). (B) The variability in measured growth phenotype (mean colony size over the replicate measurements) is shown as a function of number of experimental replicates. The curves shown were generated using data from 36 replicates of a control screen run while generating the early secretory pathway E-MAP (Schuldiner et al., 2005). On a given curve, the point corresponding to N replicates was generated by randomly drawing N of the 36 replicates for a particular strain and computing the mean normalized colony size. This process was repeated 1000 times, and the standard deviation over these 1000 repeats was plotted. Each curve represents data for a different strain. Five different representative strains with different levels of measurement variability were chosen for analysis. (C) The variability of the empirically determined expected double-mutant phenotype was estimated as a function of the number of screens analyzed in parallel. In this case, sets of screens of the indicated size were drawn at random from a set of 329 screens completed at approximately the same time from the early secretory pathway E-MAP (Schuldiner et al., 2005). For each point on each curve, 1000 random draws were completed, and each time expected colony size values were computed. The standard deviation of the expected colony sizes (over the 1000 random draws) was plotted for four representative strains with different single mutant phenotypes.
Detailed SGA and PEM protocols using a Singer RoToR pinning station are given below. The protocols below may also be executed with other types of pinning devices such as hand pinning tools or alternate robotic pinners (Schuldiner et al., 2006). If hand-pinning tools are used, a pinner with small diameter pins (e.g., VP384FP4 from V&P Scientific, San Diego, CA) should be used for the last step. The length of incubation times may need to be adjusted slightly for other pinning systems. We have used two days rather than one for the diploid selection and for the SMS steps for S. cerevisiae screens in 768 colony format with hand pinning tools. In our experience, results obtained using the Singer RoToR have better signal-to-noise than those obtained with hand-pinning devices. However, satisfactory results can be obtained with either method.
3.1. Basic SGA protocol
3.1.1. Genotypes
Query: MATα; his3Δ1; leu2Δ0; ura3Δ0; LYS2+; can1::STE2pr-SpHIS5 (SpHIS5 is the S. pombe HIS5 gene); lyp1Δ::STE3pr-LEU2; XXX::NatMX
Library: MATa; his3Δ1; leu2Δ0; ura3Δ0; met15Δ0; LYS2+; CAN1+; LYP1+; YYY::KanMX
3.1.2. Growth media (all recipes for 1 l of media)
YPAD (YEPD + adenine)—Mix 10 g yeast extract, 20 g peptone, 120 mg adenine, 20 g Difco Agar, and DDW up to a final volume of 1 l. Autoclave. Add 50 ml of sterile 40% glucose.
SPO (sporulation media: NGS Agar)—Mix 20 g Difco Agar and 820 ml DDW in one flask. Mix 0.5 g –ura-trp amino acid powder mix (Sunrise Science #1010-100), 2.5 ml 20 mM uracil, 2.5 ml 20 mM tryptophan, and 163 ml DDW in a second flask. Autoclave each flask separately. Mix the two flasks and add 20 ml of 500 mg/ml filter sterilized potassium acetate.
Note: do not autoclave the potassium acetate!
SD–HIS–LYS–ARG (for haploid selections)—Mix 20 g Difco Agar and 850 ml DDW in one flask. Mix 6.7 g yeast nitrogen base without amino acids, 2 g amino acid drop out mix (recipe below), and 100 ml DDW in a second flask. Autoclave both flasks. Mix the flasks and add 50 ml of 40% glucose.
SD(MSG)–HIS–LYS–ARG (for single- and double-mutant selections)—Mix 20 g Difco Agar and 850 ml DDW in one flask. Mix 1.7 g yeast nitrogen base without amino acids and without ammonium sulfate (Becton, Dickinson and Company #233520), 2 g amino acid drop out mix (recipe below), 1 g monosodium glutamatic acid, and 100 ml DDW in a second flask. Autoclave the first flask and filter sterilize the second. Mix the two flasks together and add 50 ml 40% glucose.
Amino acid drop out mix: Mix 3 g adenine, 10 g leucine, 0.2 g paraaminobenzoic acid, and 2 g each of alanine, asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, inositol, isoleucine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, uracil, and valine.
Note: Be sure to pour level plates, as this is very important for the effectiveness of the robotic pinning steps.
3.1.3. Drug concentrations
NAT (N)—100 mg/l; G418 (G)—100 mg/l; S-AEC (S)—50 mg/l; canavanine (c)—50 mg/l
Note: for adding labile compounds (NAT, G418, S-AEC, and canavanine), the media should be cooled first until is hot, but no longer painful to touch the container.
Note: previous protocols used 200 mg/l NAT and G418, but we have found 100 mg/l to be sufficient.
3.1.4. Plates nomenclature
YPAD = YPAD; SPO = SPO; Diploid = YPAD + N + G; NAT = YPAD + N; G418 = YPAD + G; HS = SD–HIS–LYS–ARG + S + c; SM = SD–HIS–LYS–ARG + S + c + G; DM = SD–HIS–LYS–ARG + S + c + G + N
HS1 and HS2 are HS plates used in two consecutive steps of the protocol.
3.1.5. Procedure
3.1.5.1. Preparation of query lawn
A lawn of cells of the query strain is prepared for use in the mating step.
Inoculate a liquid culture in YEPD from a single colony of the query strain (from a YEPD + NAT plate) and grow to saturation overnight.
Prepare a lawn of cells—Spread up to 500 μl of thick culture onto a NAT plate using glass beads and incubate at 30 °C for 2 days.
3.1.5.2. Preparation of library arrays (or “T-arrays” for “Target arrays”) in 1536 format
The library array is replicated for use in the mating step.
Program: Agar-Agar. Replicate. Replicate Many. 1536
Parameters: Source plate: G418; Target plate: G418; Source pressure: 100%; Target pressure: 100%; Offset: Manual; Number of replicas: 3; Economy: ON; Revisit source: ON
Note: More than three replicas/source at this density may not be consistent and slow growing mutants (e.g., small colonies) may be lost. Incubate at 30 °C for 1 day.
3.1.5.3. Mating
Query and array cells are pinned on top of each other on a fresh plate for mating. First pin the T-array twice and then pin the lawn twice on top of it.
Program: Agar-Agar. Replicate. 1536
Parameters: Source plate: T-array and lawn; Target plate: YPAD; Source pressure: 100%; Target pressure: 100%; Offset: Manual; Economy: OFF
Incubate at 30 °C for 1 day.
3.1.5.4. Diploid selection
Cells are pinned from the mating plate and diploids are selected by using NAT and G418.
Program: Agar-Agar. Replicate. 1536
Parameters: Source plate: mating; Target plate: Diploid; Source pressure: 100%; Target pressure: 100%; Offset: Manual; Economy: OFF
Incubate at 30 °C for 1 day.
3.1.5.5. Sporulation
Replicate the arrays from Diploid onto SPO plates using 384 pads.
Do 2 pins per array onto the same target GC plate. This is the most critical stage of the protocol and transferring enough cells on the target plate is very important.
Program: Agar-Agar. Replicate. Replicate One. 384
Parameters: Source plate: Diploid; Target plate: SPO; Source pressure: 100%; Target pressure: 100%; Offset: OFF; Economy: OFF
Incubate for 5 days at room temperature in a humid environment.
Note: When loading the pads click “Modify” to change to 384 pads.
Note: The plates should be packed in plastic bags and kept in a humid environment to prevent drying.
3.1.5.6. HS1
Replicate the arrays from the SPO plates onto HS plates using 384 pads. Do 2 pins per array. The cells do not divide on the SPO media, so maximizing cell transfer at this step and at the previous step is critical.
Program: Agar-Agar. Replicate. Replicate One. 384
Parameters: Source plate: SPO; Target plate: HS (HS1); Source pressure: 100%; Target pressure: 100%; Offset: Automatic
Note: Incubate for 2 days at 30 °C.
3.1.5.7. HS2
Replicate the arrays from the HS1 plates onto HS2 plates using 1536 pads. Do 1 pin per array.
Program: Agar-Agar. Replicate. Replicate One. 1536
Parameters: Source plate: HS1; Target plate: HS (HS2); Source pressure: 100%; Target pressure: 100%; Offset: Automatic
Note: Incubate for 1 day at 30 °C.
3.1.5.8. SM
Replicate the arrays from the HS2 plates onto SM plates using 1536 pads. Do 1 pin per array.
Program: Agar-Agar. Replicate. Replicate One. 1536
Parameters: Source plate: HS1; Target plate: HS (HS2); Source pressure: 100%; Target pressure: 100%; Offset: Automatic
Note: Incubate for 1 day at 30 °C.
3.1.5.9. DM
Replicate the arrays from the SM plates onto DM plates using 1536 pads. Do 1 pin per array.
Program: Agar-Agar. Replicate One. 1536
Parameters: Source plate: SM; Target plate: DM; Source pressure: 100%; Target pressure: 100%; Offset: Automatic
Incubate at 30 °C.
Take pictures of the DM after 48 h.
Store the final plates in coldroom.
3.2. Basic PEM protocol
3.2.1. Genotypes
Query: h−; ade6-M210; leu1-32; ura4-D18; mat1_m-cyhS, smt0; rpl42::cyhR (sP56Q); XXX::NatMX6
Library: h+; ade-M210 (or M216); ura4-D18; leu1-32; YYY::KanMX6
3.2.2. Growth media
YE5S (general purpose rich media)—5 g/l yeast extract; 30 g/l glucose; 225 mg/l of each adenine, leucine, histidine, uracil, and lysine; 20 g/l Difco Agar
SPAS (mating and sporulation media)—10 g/l glucose;1 g/l KH2PO4 45 mg/l of each adenine, histidine, leucine, uracil, and lysine hydrochloride; 1 ml 1000× vitamin stock (1 g/l pantotenic acid; 10 g/l nicotinic acid; 10 g/l inositol; 10 mg/l biotin)
3.2.3. Drug concentrations
NAT (N)—100 mg/l; G418 (G)—100 mg/l; CYH (C)—100 mg/l
3.2.4. Plates nomenclature
YE5S = YE5S; SPAS = SPAS; NAT = YE5S + N; G418 = YE5S + G; GC = YE5S + G + C; GNC = YE5S + G + N + C
GC1 and GC2 are GC plates used in two consecutive steps of the protocol.
3.2.5. Procedure
3.2.5.1. Preparation of Query arrays (Q-arrays) in 1536 format
Prepare a lawn of cells—Spread up to 500 μl of thick culture onto a NAT plate using glass beads and incubate at 30 °C for 2–3 days.
Program: Agar-Agar. Replicate. Replicate One. 1536
Parameters: Source plate: NAT; Target plate: NAT
Source pressure: 100%; Target pressure: 100%; Offset: Manual; Offset radius: 1 mm
Note: Do 2 pins per plate picking cells from different parts of the lawn plate. Incubate at 30 °C for 2–3 days.
3.2.5.2. Preparation of library arrays (T-arrays) in 1536 format
Program: Agar-Agar. Replicate. Replicate Many. 1536
Parameters: Source plate: G418; Target plate: G418; Source pressure: 100%; Target pressure: 100%; Offset: Manual; Number of replicas: 3; Economy: ON; Revisit source: ON
Note: More than three replicas/source at this density may not be consistent and slow growing mutants (e.g., small colonies) may be lost. Incubate at 30 °C for 2–3 days.
3.2.5.3. Mating
Combine the T-array and the Q-array onto a SPAS plate generating a 1536 density mating array. First pin the T-array twice (2×) and then pin the Q-array twice on top of it with agar mixing.
Program: Agar-Agar. Replicate. 1536
Parameters: Source plate: T-array and Q-array; Target plate: SPAS; Source pressure: 100%; Target pressure: 100%; Offset: Manual; Economy: OFF
Note: Incubate for 5–6 days at room temperature packing the plates in plastic bags to prevent drying.
3.2.5.4. SPAS-GC1
Replicate the mating arrays from SPAS onto GC plates using 384 pads.
Do 2 pins per array onto the same target GC plate. You will need eight (Schuldiner et al., 2005) 384 pads per array. This is the most critical stage of the protocol and transferring enough cells on the target plate is very important.
Program: Agar-Agar. Replicate. Replicate One.1536
Parameters: Source plate: SPAS; Target plate: GC; Source pressure: 100%; Target pressure: 100%; Offset: OFF; Economy: OFF
Note: When loading the pads click “Modify” to change to 384 pads. Incubate for 3 days at 30 °C.
3.2.5.5. GC1-GC2
Replicate the arrays from the GC1 plates onto GC2 plates using 1536 pads. Do 1 pin per array.
Program: Agar-Agar. Replicate. Replicate One.1536
Parameters: Source plate: GC (GC1); Target plate: GC (GC2); Source pressure: 100%; Target pressure: 100%; Offset: Automatic
Note: Incubate for 2 days at 30 °C.
3.2.5.6. GC2-GNC
Replicate the arrays from the GC2 plates onto GNC plates using 1536 pads. Do 1 pin per array.
Program: Agar-Agar. Replicate One.1536
Parameters: Source plate: GC (GC2); Target plate: GNC; Source pressure: 100%; Target pressure: 100%; Offset: Automatic
Note: Incubate at 30 °C. Take pictures of the GNC at 24, 36, and 48 h. Store the final plates in coldroom.
3.3. Digital photography
We take color digital photographs of the final plates using a Canon PowerShot S3 IS camera (6.0 megapixels) at a resolution of 180 dpi, focal length = 18.2 and F-number = 8. Images are taken at a distance of 60 cm (24 in.) by mounting the camera on a KAISER camera stand (Germany). The position of the plate is fixed using a custom-made metal plate holder permanently bolted to the camera stand. The base of the camera stand is covered with black velvet to create a uniform black background for the images. Illumination is provided by two fixed luminescent lamps (25–30 W) outside of a 20 × 20 in. nylon soft light tent which serves to even the illumination and prevent reflections.
4. Data Processing and Computation of Scores
The data from the screens is collected as digital photographs of arrays of yeast colonies. These images are converted into measures of interactions between mutations through a multistep computational process (Collins et al., 2006). Colony areas are measured digitally using the HT Colony Grid Analyzer Java program (Collins et al., 2006). The resulting sizes are then processed using a software toolbox we have developed for use with MATLAB. Both pieces of software are freely available for download (http://sourceforge.net/projects/ht-col-measurer/ and http://sourceforge.net/projects/emap-toolbox/). The computational steps for converting colony area measurements into genetic interaction scores is depicted in Fig. 9.4A and outlined in the following steps:
4.1. Preprocessing and normalization
A preprocessing and normalization step is used to correct for systematic artifacts (uneven image lighting, artifacts due to physical curvature of the agar surface on which the colonies grew, differences in growth time, etc.), and also to account for the growth phenotype of the query strain.
Several types of systematic artifacts can arise in the data collection process that give rise to spatial patterning of measured colony sizes which is independent of the growth properties of the yeast strains in the experiment. For instance, uneven lighting can result in apparently larger colonies in areas with brighter light. Additionally, an uneven agar surface can result in deposition of a larger number of cells in vertically elevated areas of the agar surface, and deposition of fewer cells in lower areas. In our experience, this agar curvature artifact is much more pronounced using the Singer robot plastic pad-based cell deposition method rather than a floating pin-based method.
We correct for these artifacts using a spatial flattening of the colony sizes. Specifically, the colony size measurement at each position on one agar plate is compared to the median size at that position over all plates in the dataset to compute a log-ratio indicating whether that colony is larger or smaller than is typical. The resulting set of log-ratios is fit using robust linear regression (using MATLAB’s robustfit function) to a second order surface (i.e., one of the form: z = Ax2 + By2 + Cxy + Dx + Ey + F). The resulting surface is then subtracted from the log-ratios to remove spatial artifacts, and the corrected colony sizes are recovered by exponentiating the result and multiplying by the original median size at the corresponding position. This correction requires only six parameters and is calculated using 384, 768, or 1536 measurements, depending on the number of colonies on the plate, so it is unlikely to be strongly affected by real genetic interactions. Additionally, robust regression, rather than standard linear regression, is used to minimize the impact of individual real interactions on the calculated correction. Colony sizes of zero and the colonies on the edges of the plate are excluded from the correction calculation. In our experience, this correction effectively removes spatial patterning, without compromising detection of interactions. The MATLAB toolbox contains a graphical user interface which displays heatmaps (similar to those seen in Fig. 9.4A) before and after the spatial flattening so that the success of this step can be assessed.
A separate correction is applied for the colonies on the edges of the plate. For each edge row or column, the colony sizes are scaled such that the median size in that row or column is equal to the median size in the interior of the plate. This correction is applied because we have found that edge rows and columns can be systematically larger or smaller (usually larger) than other colonies on the plate due to proximity or distance from the physical edge of the agar.
Finally, colony sizes are normalized to account for differences in the growth phenotype of the query mutation which is present in all double-mutant colonies on the same plate. We apply this normalization in addition to the above-described spatial flattening to account for the possibility that the query mutation may have more synthetic interactions (where double mutants grow more slowly than expected) than positive interactions (where double mutants grow faster), or vice versa. We do assume that most mutations in the array will have little or no growth defect, as is the case for gene deletions in yeast (Breslow et al., 2008; Giaever et al., 2002), and that most mutation pairs will be noninteracting. We, therefore, normalize the colony sizes according to the peak of the histogram of colony sizes on a given plate (this is the “Parzen” setting in the toolbox menu). Heatmap images showing the spatial pattern of colony size measurements at different stages of the normalization process can be seen in Fig. 9.4A. All of these preprocessing and normalization steps are implemented in the MATLAB toolbox.
4.2. Computing expected colony sizes
After normalization, the growth phenotype of the query mutation has been accounted for, and we assume that differences in normalized colony size now result from either the growth phenotype of the array mutation or a genetic interaction. For a given array mutation (which is always present in the exact same spatial position within the array), we then empirically estimate the expected normalized colony size as the typical normalized colony size over a set of screens. If the number of screens is large (generally 50 or more), we prefer to use the peak of the histogram of normalized sizes (the “Parzen” setting in the toolbox), similar to our normalization procedure. However, if the number of screens analyzed is smaller we have found that the median normalized size is a more robust estimate.
We sometimes observe batch-to-batch variability, where the typical colony size estimated for one group of screens completed at approximately the same time using the same preparation of media differ from the values estimated for another group of screens completed at a different time (perhaps weeks or months apart). If such batch-to-batch variability is apparent, it is preferable to compute the expected colony sizes independently for each batch. This can be done easily with options included in the MATLAB toolbox (web address provided above).
A natural question is then how many different screens need to be included in a batch, such that the estimated expected colony size values will be reliable? We have investigated this question empirically, using measurements from the early secretory pathway E-MAP (Schuldiner et al., 2005). As increasingly many screens are completed in the same batch, the error in estimates of expected colony sizes decreases (Fig. 9.4C). There can be substantial error if a batch includes fewer than 20 screens. On the other hand, each additional screen beyond about the 40th gives only marginal improvement.
4.3. Computing genetic interaction scores
We compute genetic interaction scores as we have previously described (Collins et al., 2006) as S-scores, which are closely related to t-values and account for both the magnitude of the interaction effect, as well as our confidence that the measurement constitutes a real genetic interaction. The S-score differs from a standard t-value in several ways. Rather than comparing experimental observations explicitly to control screens, we use the expected colony size estimates described above. Additionally, a standard t-value can be very sensitive to the standard deviation of a small number of experimental replicate measurements. In particular, if the replicates are unusually similar, resulting in an unusually small standard deviation, this can result in a large score even if the magnitude of the interaction is small. We have found that the reproducibility of S-scores is substantially improved by implementing a lower bound on the standard deviation measurement (i.e., if the measured standard deviation is below the lower bound, we use the lower bound instead) (Collins et al., 2006). This lower bound is an estimate of the typical standard deviation for double-mutant measurements with similar parent query and array strain phenotypes.
4.4. Quality control
Careful quality control is an essential part of the screening process. In our experience, it is not uncommon for ~10% of all strains (both array and query) to be incorrect. These incorrect strains need to be systematically identified, and removed from the analysis.
The most effective tool for identifying incorrect strains is analysis of the apparent interaction scores between mutations at chromosomally linked loci (Collins et al., 2006; Roguev et al., 2008). The absence of apparent negative interactions between a mutation and mutations at loci within ~100 kb in S. cerevisiae or ~200 kb in S. pombe is strong evidence that a strain is incorrect. The MATLAB toolbox contains a graphical user interface for browsing the linkage data which facilitates the identification of incorrect strains. Additionally, cases where the data for an array strain is completely uncorrelated to data for a corresponding query strain should be identified systematically, and if found, the corresponding strains should be checked by PCR.
5. Extraction of Biological Hypotheses
Completion of a genetic interaction map creates a huge quantity of data. These data have proven useful in numerous instances for generating new hypotheses and helping guide ongoing work (Collins et al., 2007a; Fiedler et al., 2009; Keogh et al., 2005; Kornmann et al., 2009; Kress et al., 2008; Laribee et al., 2007; Morrison et al., 2007; Nagai et al., 2008; Schuldiner et al., 2005; Wilmes et al., 2008). However, determining the best way to navigate these maps and generate hypotheses from them remain areas of active work. We expect that we and others will continue to find new and better ways to use the data, but we also describe here several general approaches that have proven useful in the past.
5.1. Identifying genes acting in the same pathway using patterns of interactions
One of the simplest and most useful applications of high-throughput genetic interaction data is the identification of sets of genes whose products work together very closely in a common biochemical pathway. For each pair of mutations in a quantitative genetic interaction map, two distinct measures of their relationship can be recognized. First, the genetic interaction score (S-score) represents the degree of synergizing or mitigating effects of the two mutations in combination; this can be neutral (e.g., no interaction), positive (e.g., suppression), or negative (e.g., synthetic lethality). Second, the similarity (typically measured as a correlation) of their genetic interaction profiles represents the congruency of the phenotypes of the two mutations across a wide variety of genetic backgrounds. One would logically expect both measures to be indicative of whether two genes act in the same pathway. Indeed, pairs of genes exhibiting positive genetic interactions and highly correlated genetic interaction profiles very frequently encode proteins that are physically associated (Collins et al., 2007a). Furthermore, in cases where the proteins do not physically associate, they tend to act coherently in a biochemical pathway. This latter case is particularly informative as these are very close functional relationships which could be difficult to detect by other methods. The combined signature of a positive genetic interaction and highly correlated interaction patterns can be formalized into a score (the COP score (Collins et al., 2006, 2007a; Schuldiner et al., 2005), and the sets of genes with this signature are also often readily apparent after hierarchical clustering of genetic interaction profiles.
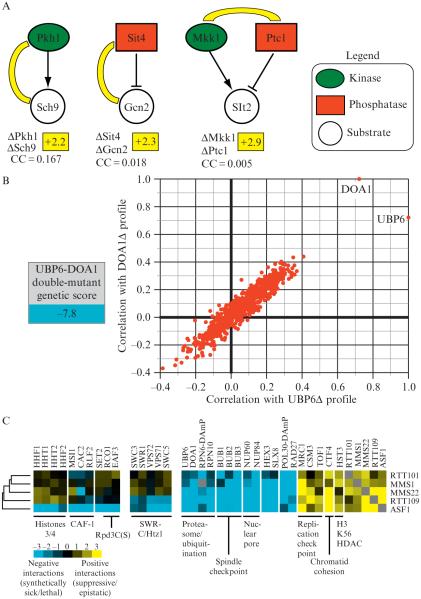
In general, the similarity of interaction profiles is much stronger evidence for membership in the same pathway than an individual positive interaction. However, the direct interaction score sometimes provides the critical distinction. For example, deletions of DOA1 and UBP6 give very similar patterns of genetic interactions (Fig. 9.5B), which likely reflects the fact that each deletion leads to depletion of ubiquitin (Collins et al., 2007a). However, the double deletion results in a strong negative genetic interaction, presumably because they largely function independently of each other to maintain ubiquitin levels.
Figure 9.5.
Quantitative genetic data reveals insight into functional pathways. (A) Individual genetic interactions identify enzyme–substrate relationships. An E-MAP focused on the regulation of phosphorylation in budding yeast (Fiedler et al., 2009) revealed positive genetic interactions between the kinase Pkh1 and it substrate, Sch9 (+2.2), between the phosphatase Sit4 and its substrate, Gcn2 (+2.3), and between a kinase (Mkk1) and a phosphatase (Ptc1) (+2.9) that acts on Slt2. The correlation of genetic interaction profiles between these pairs of genes is below the individual genetic interactions and was derived from the kinase E-MAP (Fiedler et al., 2009). (B) Functional connection between the deubiquitinase enzyme, Ubp6 and the ubiquitin chaperone, Doa1. A strain containing deletions in both UBP6 and DOA1 results in a strong negative genetic interaction (−7.8) and the genetic profiles generated from these deletions are highly correlated. (C) Using genetic interaction profiles to identify a pathway involved in genome integrity. Subsets of interactions (both negative and positive) for rtt101Δ, mms1Δ, mms22Δ, rtt109Δ, and asf1Δ are displayed. Some interactions are observed for all five deletions, interactions with the SWR complex are seen with only asf1Δ and rtt109Δ whereas only deletion of ASF1 result in negative interactions with histones H3/H4, the CAF complex, and factors involved in the Rpd3C(S) pathway (Set2, Eaf3, and Rco1).
It should be noted that interpretation of genetic interaction data derived from hypomorphic alleles of essential genes may differ from the data obtained from deletions of nonessential genes. For example, two hypomorphic alleles may, in fact, give rise to a negative genetic interaction, if each mutation partially cripples the same pathway. However, a positive genetic interaction may still be observed if the two encoded proteins form a tight heterodimer, where the minimum of the two concentrations determines the cellular phenotype.
5.2. Using individual interactions to predict enzyme–substrate relationships
In addition to identifying the core components of coherent pathways, one would like to have efficient strategies to suggest points of integration or cross-talk between pathways. One important class of such connections between pathways includes protein-modifying enzymes and their cognate substrates. We have found that kinase–substrate and phosphatase–substrate pairs are enriched for positive genetic interactions (Fiedler et al., 2009). Pkh1-Sch9 and Sit4-Gcn2 correspond to kinase–substrate and phosphatase–substrate relationships, respectively, and in both cases, the double deletions result in positive genetic interactions (Fig. 9.5A) (Fiedler et al., 2009). However, in neither case is the correlation coefficient between the pairs notable. Thus, when looking for a critical substrate of a kinase, phosphatase, or other protein-targeting enzyme, the genes with the highest scoring positive interactions represent excellent candidates, even if the correlation between the interaction profiles is weak. It should be noted though, that kinases and phosphatases may have multiple substrates. In these cases, only enzyme–substrate pairs corresponding to modifications that significantly affect the phenotype being measured are likely to be detectable.
5.3. Using individual interactions to predict opposing enzyme relationships
Similarly, we have found that pairs of opposing kinases and phosphatases which share a common substrate are highly enriched for positive genetic interactions. Since these enzymes have opposing effects, they will also tend not to have correlated interaction patterns. An example is the mitogen-activated protein kinase kinase Mkk1 and the phosphatase Ptc1. These enzymes have opposing roles regulating the activity of the down-stream kinase Slt2. The two deletion mutations have a strong positive genetic interaction, but the interaction profiles are not correlated (Fiedler et al., 2009) (Fig. 9.5A). Such positive interactions can be key clues for deciphering the role of uncharacterized genes and pathways. Indeed, strong positive interactions with the H3 K56 histone deacetylase Hst3 was an important piece of data suggesting that Rtt109 was the opposing acetylase (see below for further discussion of this pathway) (Collins et al., 2007a).
5.4. Dissecting multiple roles of a single gene by detailed comparison of interaction patterns
A major challenge in interpreting genetic interaction data is that many genes encode multifunctional proteins acting in multiple pathways. While it would be useful to be able to extract pathway-specific information, genetic interactions for these genes may arise from a role in one pathway or another, or the interactions may represent a mixture of effects from more than one pathway. We have found that in some cases, comparison of the interaction profile for a multifunctional gene with the profiles for other related genes can give important pathway-specific insights. For example, we identified a pathway involving the chromatin assembly protein Asf1, a then-uncharacterized protein Rtt109, and a putative ubiquitin ligase complex containing Rtt101, Mms1, and Mms22 (Collins et al., 2007a). Asf1 was the best characterized member of the group, but it had also been implicated in multiple cellular roles (Loyola and Almouzni, 2004). Comparison of the genetic interaction profiles for this group of genes suggested that several of these roles were Asf1-specific and that all of these factors work together in a pathway intimately related to histone H3 K56 acetylation and maintenance of genomic integrity during DNA replication. Further experiments identified Rtt109 as the acetylase (Collins et al., 2007a; Driscoll et al., 2007; Han et al., 2007).
In the above case, a critical observation was that a subset of asf1Δ’s interactions was unique to asf1Δ, and another large subset is shared uniformly with the rest of the group. All members of this pathway display positive genetic interactions with one another, positive interactions with replication checkpoint genes (MRC1, TOF1, and CSM3), and negative genetic interactions with genes involved in DNA replication (POL30, ELG1, RAD27), the spindle checkpoint (BUB1, BUB2, and BUB3), DNA Repair (NUP60, NUP84, HEX3, SLX8) and ubiquitin regulation (UBP6, DOA1, RPN6, RPN10). Asf1 was also known to be required for histone H3 K56 acetylation, and the shared positive interaction between all members of this pathway and the gene encoding a K56 deacetylase (HST3) suggested that this role of Asf1 was likely to be central to the pathway (Celic et al., 2006; Maas et al., 2006).
On the other hand, only asf1Δ displays negative interactions with factors involved in general chromatin assembly: the histone genes HHF1, HHF2, HHT1, HHT2 and components of the CAF complex (MSI1, CAC2, and RLF2), arguing that Asf1’s general role in chromatin assembly is independent of its function with Rtt109. Similarly, only deletion of ASF1 results in negative interactions when combined with deletions of the Rpd3C(S) histone deacteylation complex (EAF3 and RCO1) and SET2, which codes for a histone methyltransferase enzyme. Eaf3, Rco1, and Set2 function together in a histone deacetylation/methylation pathway that is required for maintaining chromatin integrity during transcription and suppressing cryptic initiation by RNAPII (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005), again suggesting that Asf1 alone impinges on this process. Indeed, deletion of ASF1 and not the other factors functioning in the K56 acetylation pathway results in spurious transcription rising from the inability to suppress cryptic initiation (Schwabish and Struhl, 2006).
6. Perspective
The term “epistatic” was originally used in 1907 by Bateson to describe a masking effect whereby a variant or allele at one locus prevents the variant at another locus from manifesting its effect (Bateson, 1907). Over the past one hundred years, geneticists have uncovered important biological insight from “epistatic” interactions, first qualitatively and then much more efficiently through quantitative analysis. The vast majority of genetic interaction data has been collected from simpler systems like yeast and bacteria using basic read-outs like colony size or growth rates. We are now in a position to collect this type of data in multicellular systems and the phenotypic read-outs, which are expanding exponentially, can occur at the organismal level, providing invaluable information about not only functional pathways that comprise key biological processes but also about evolution and behavior.
REFERENCES
- Bateson W. Facts limiting the theory of heredity. Lipid-modifying therapy and attainment of cholesterol goals in Hungary: the return on expenditure achieved for lipid therapy (REALITY) study. Science. 1907;26:647–660. doi: 10.2165/00044011-200727090-00006. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007a;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2007b;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, Kemmeren P, Holstege FC, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Guarente L. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. USA. 2007;104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, Weissman JS, Delrow J, et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol. Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat. Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, Hoe KL, Kim DU, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, Ideker T, Guthrie C, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Peyser BD, Pan X, Boeke JD, Spencer FA, Bader JS. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100034. 2005.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]