Abstract
The major birch pollen allergen, Betv1 of Betula verrucosa is the main causative agent of birch pollen allergy in humans. Betv1 is capable of binding several physiological ligands including fatty acids, flavones, cytokinins and sterols. Until now, no structural information from crystallography or NMR is available regarding binding mode of any of these ligands into the binding pocket of Betv1. In the present study thirteen ligands have been successfully docked into the hydrophobic cavity of Betv1 and binding free energies of the complexes have been calculated using AutoDock 3.0.5. A linear relationship with correlation coefficient (R2) of 0.6 is obtained between ΔGbs values plotted against their corresponding IC50 values. The complex formed between Betv1 and the best docking pose for each ligand has been optimized by molecular dynamics simulation. Here, we describe the ligand binding of Betv1, which provides insight into the biological function of this protein. This knowledge is required for structural alteration or inhibition of some of these ligands in order to modify the allergenic properties of this protein.
Keywords: Betula verrucosa, Birch pollen allergy, Docking, Molecular dynamics simulation
Background
Betv1 an allergic protein with molecular wt of 17 kd is a constituent of the pollen of white birch tree Betula verrucosa. White birch pollen is one of the main causes of Type I allergic reactions (allergic rhinoconjunctivitis, allergic bronchial asthma) in Middle and Northern Europe, North America [1]. Type I allergy represents a major health problem in these countries, since 10-15% of the population suffer from these diseases. The structures of Betv1 were determined independently by two methods X-ray diffraction and NMR spectroscopy [2]. The main structural feature of Betv1 consists of a seven stranded anti-parallel β- sheet that wraps around a 25 residue-long C-terminal amphipathic α- helix. The β-sheet and the C-terminal part of the long helix are separated by two small consecutive helices [2]. The presence of a large, forked, hydrophobic, solvent-accessible cavity spanning through the entire protein is the most unique feature of the structure. Besides, the protein contains a Gly rich P-loop motif found in many nucleotide binding proteins. The biological function of Betv1 has not yet been elucidated. It is homologous to a group of pathogenesis-related proteins, the PR-10 proteins, which are expressed in disease and stress condition [3] such as during microorganism infection. Structural similarity of Betv1 with the START domain of MLN64 [4] suggests a similar function for Betv1 as a steroid binding protein. Previous workers identified a range of physiologically relevant ligands including fatty acids, flavonoids and cytokinin that are able to bind in the cavity of Betv1 with moderate to high affinity [5]. So far, no structural information about the binding modes of any of these ligands into the hydrophobic cavity of Bev1 is available. However these ligands i.e fatty acids, flavonoids and cytokinins, are involved in a myriad of cell-cell recognition and signaling events. Here, in this paper we employ molecular docking to study possible interactions of these ligands into the hydrophobic cavity of Bev1 using the program AutoDock 3.0.5 [6]. The availability of experimentally determined IC50 values of these ligands gives us a unique opportunity to correlate the same with the binding free energies (ΔGb) derived from AutoDock. The complex formed between the protein and the best docking pose for each ligand has been optimized by molecular dynamics simulation. An implication of these ligands for a possible biological function of Betv1 has also been discussed.
Methodology
Docking
The crystal structure of Betv1 (pdb code: 1BV1) [2] was obtained from PDB [7]. The structures of ANS, kinetin, flavone and naringenin were downloaded from Cambridge Structural Database (CSD) [8]. All other ligands were constructed using the Biopolymer utility of the software Insight II. Docking calculations were carried out using AutoDock 3.0.5 [6]. The grid maps representing the protein in the actual docking process were calculated with the aid of AutoGrid. The dimension of the grid was
40X40X40 points in each dimension for smaller ligands (ANS and lower chain length fatty acids) and
60X60X60 points in each dimension for rest with spacing of 0.375 Å between the grid points.
Gasteiger charges computed by ADT (AutoDock tools) were used on the atoms for each ligand. The AUTOTORS utility, included in the AutoDock software, was used to define all possible torsions of ligand molecules for the docking algorithm. Docking simulations were carried out using the Lamarckian Genetic Algorithm, with an initial population of 150 randomly placed individuals, a maximum number of 2,50,000 energy evaluations, a mutation rate of 0.02, a cross over rate of 0.08 and an elitism value of 1. For the local search, the pseudo-Solis and Wets algorithm was applied. Each job consisted of 100 independent runs. Finally, resulting docking orientations lying within 1.5 Å in the root-mean square deviation (rmsd) tolerance of each other were clustered together and represented by the result with the most favorable free energy of binding (ΔGb). The lowest free energy conformers are listed in Table 1.
MD simulation
Molecular dynamics simulations of all Betv1-ligand complexes as well as the crystal structure of unliganded Betv1 were performed using the GROMACS 3.3.1 package [9] and gmx (modified GROMOS 87) force field implemented on LINUX architecture. The ligand topology files were prepared using PRODRG2 server [10]. The protein-ligand complexes were solvated in a triclinic water box having a dimension of 7.22 nm containing approximately 8800 SPC water molecules. All protein atoms are at a distance equal or greater than 1.0 nm from the box edges. Appropriate number of NA+ ions was added to neutralize the charge of the system followed by energy minimization for 2000 steps by steepest descents. The minimized systems were equilibrated for 50 ps each at 300 K by position restrained molecular dynamics simulation in order to relax the solvent. The equilibrated systems were then subjected to molecular dynamics simulations for 10 ns each at 300 K. The LINCS algorithm [11] was used to constrain bond lengths using a time step of 2 fs for all calculations. Periodic boundary conditions combined with the minimum image convention were used under isothermal, isobaric conditions using Berendsen coupling algorithm [12] with relaxation times of 0.1 ps and 0.5 ps respectively. Electrostatic interactions were calculated using the Particle Mesh Ewald (PME) [13] summation scheme. van der Waals and Coulomb interactions were truncated at 0.9 nm. The non-bonded pair list was updated every 10 steps and conformations were stored every 2 ps. Secondary structure analysis was performed using the program DSSP [14]. Other analyses were performed using scripts included with the Gromacs [9] distribution. LPC software [15]was used to calculate the ligand-protein interactions.
Discussion
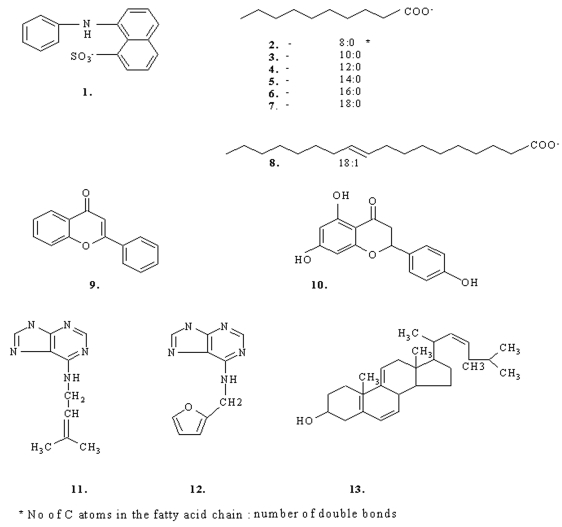
Molecules 1-13 (Figure 1) have been successfully docked into the hydrophobic cavity of Betv1. AutoDock estimates binding free energies (ΔGb) based on AMBER force field and empirical weighting factors. Table 1 shows the results of the docking experiments: calculated free energy of binding for the ligands and their corresponding IC50 values. We have plotted ΔGbs values against the corresponding IC50 and obtained a good linear relationship with correlation coefficient (R2) of 0.6. The complex formed between the protein and the best docking pose for each ligand has been optimized by molecular dynamics simulation.
Figure 1.
Overview of the structure of the ligands for Betv1.
Interaction with ANS
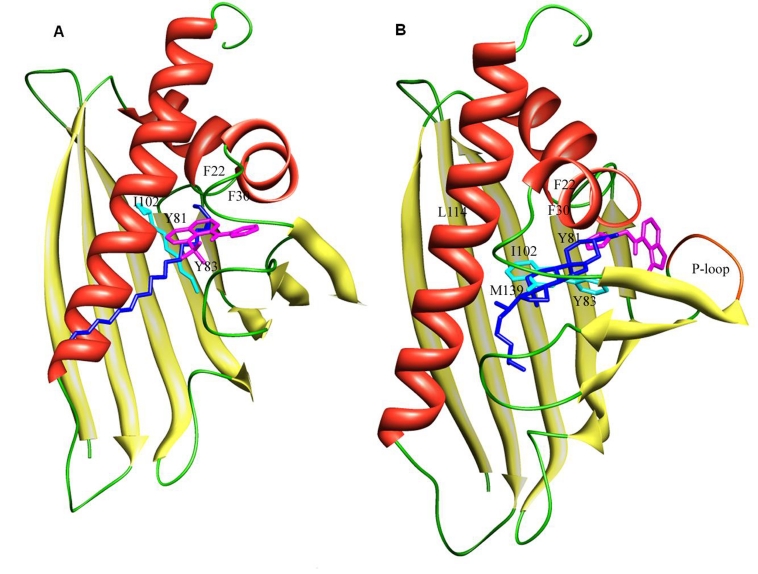
Previous NMR study indicates that fluorescent probe 8-anilino-1- naphthalenesulfonic acid (ANS) binds in the cavity of Betv1 but the authors did not pinpoint specific ANS binding site [5]. Our docking and subsequent molecular dynamics simulation result of ANS shows that it is involved in hydrophobic interaction with side-chains of residues Ile23, Phe30, Pro31, Ile38, Val41, Lys54, Ile56, Ile102, Ile116 and Leu143 of Betv1 (Figure 2A). The anilino and naphthalene-sulphonic acid moiety of ANS interact with the side-chain residues of Phe30, Phe58, Tyr81 and Phe22, Phe30, Phe58, Tyr81, Tyr83 respectively via π-π interaction. Our result corroborates well with the NMR experiment where the perturbed protons are shown to form a large patch along the β sheet and several distinct regions in α–helices [5]. Most of the interacting residues found in our study also belong to these two regions.
Figure 2.
(A) Graphical representation of docked orientation of ligands 1, 2 and 7 with Betv1. Compounds 1, 2 and 7 are shown in pink, cyan and blue respectively. (B): Graphical representation of docked orientation of ligands 9, 12 and 13 with Betv1. Compounds 9, 12 and 13 are shown in cyan, pink, and blue respectively. P-loop is shown in red.
Interaction with Fatty acids
It has been shown experimentally that fatty acids are important ligands for Betv1 [5]. Our result with lower chain fatty acids (caprylate, caprate and laurate) shows that the binding modes of these fatty acids into the cavity of Betv1 are quite similar. The C atoms of these fatty acids make hydrophobic interactions with side chains residues of Phe22, Ile23, Gly26, Phe30, Tyr81, Tyr83, Ile102, Ile116, Gly140 of Betv1 (Figure 2A). The binding affinity of fatty acids increases with the rise in the number of C atoms (Table 1). Betv1 displays highest affinities for medium length fatty acids (myristic, palmitic and stearic) (Table 1). These medium length fatty acids generate several new hydrophobic interactions with the adjacent apolar residues i.e Ala21, Val41, Ile56, Phe58, Phe64, Val85, Leu114, Met139 in addition to the above mentioned interactions occurring between small chain fatty acids and Betv1. These additional hydrophobic interactions stabilize medium chain fatty acids even more into the cavity of Betv1. Saturated and unsaturated fatty acids (stearic acid and oleic acid) exhibit quite similar binding affinity for Betv1 (Table 1). Our results are in agreement with the previous experimental results, which show that the binding affinity of fatty acids with Betv1 increases from lower to medium chain fatty acids and the unsaturation has no effect on binding [5].
Interaction with Flavonoids
Flavone and 4',5,7-trihydroxyflavone (naingenin) are two important plant pigments belonging to the flavonoid group. These compounds are shown to bind Betv1 with moderate affinity [5]. The exact binding site has not been suggested in that study. Our result shows that the binding modes of flavone and naringenin within the cavity of Betv1 are quite similar and both the compounds show similar binding affinities (Table 1). Flavone and naringenin adopt a position in a hydrophobic cage surrounded by residues Phe22, Phe23, Phe30, Ile38, Val41, Ile56, Phe58, Val71, Tyr81, Tyr83, Ile102, Leu114, Ile116, Met139 and Leu143 (Figure 2B). The benzopyran group of flavone has strong hydrophobic interaction with side chain of residues Ile23, Ile38, Ile56, Ile102, Leu114, Ile116 and Leu143, while the phenyl group interacts with Ile56, Val71, Lys54 and Lys46. In addition to these residues benzopyran and phenyl group of naringenin have additional hydrophobic interaction with Phe22, Phe30, Tyr83 and Ile23, respectively. The phenyl and benzopyran moieties of the flavones make a strong π-π interaction with residues Phe22, Phe30, Tyr81 and Try83.
Interaction with Cytokinins
Cytokinins are plant growth hormones that regulate differentiation and proliferation of plant cells [5]. N6-(Δ2-isopentenyl)adenine (IPA) and N6-furfuryladenine (kinetin) are two extremely potent cytokinins. Our result shows that the adenine ring of kinetin and IPA interacts hydrophobically with Gly46, Asn47 and Gly48, residues belonging to the P-loop motif (-G-X-G-G-X-G) of Betv1 (Figure 2B). In addition to these residues cytokinins interact with Ile23, Leu24, Phe30, Thr52, Val71, Val74, Tyr81 and Ile102 of Betv1 via hydrophobic contact. The adenine ring of kinetin forms two hydrogen bonds, one with ND atom of Asn47 and the other with main chain N atom of Gly49, while the adenine ring of IPA forms two hydrogen bonds, one with the side chain O atom of Asn28 and the other with main chain O atom of Leu24. Our result is in agreement with the previous experimental study, which shows that cytokinins bind near the P-loop motif of Betv1 [5].
Interaction with DHE
Dehydroergosterol (DHE), a naturally occurring sterol with similar properties to cholesterol is capable of binding within the hydrophobic cavity of Betv1. DHE interacts with Betv1 via extensive van der waals contacts between the hydrophobic side-chains lining the binding cavity (Phe22, Ile23, Phe30, Ile38, Val41, Ile56, Phe58, Leu62, Pro63, Phe64, Val67, Val85, Ile98, Ile102, Met139 and Leu143) and the polycyclic ring system of DHE (Figure 2B). The binding mode of DHE within the hydrophobic cavity of Betv1 is very similar to deoxycholate an important family of plant steroid [16]. Most of the residues involved in the interaction of both the ligands are highly conserved.
Implications of ligand binding on biological function of Betv1
Betv1 is an allergenic protein, which release allergenic proteins via their pollen or accumulate allergens in tissues used for the production of plant food [5]. The internal cavity of Betv1 plays a key role in the biological function of Betv1. The presence of large quantities of Betv1 in pollen grain suggests that Betv1 is involved in the processes leading to plant reproduction such as germination of pollen grain on stigma, directional pollen tube growth and fertilization [5]. Most of the ligands studied here such as cytokinins, fatty acids, flavones participate in such processes. Cytokinins are plant growth hormones that control differentiation and proliferation of plant cells [5]. It has been suggested that cytokinin binding proteins may act as storage compartments for cytokinins in seeds allowing rapid release of cytokinins upon germination [17]. Fatty acids are responsible for hydration of the pollen grain. They form a watertight seal between pollen and stigma facilitating rapid transport of water into the pollen through channels in the stigma and pollen membranes. Favonoids are required for pollen fertility [18]. The ability of Betv1 to bind long chain fatty acids and flavonoids suggests probable role for Betv1 in ensuring proper hydration and germination of pollen by transporting the lipids or flavonoids to the stigmatic surface and releasing them there [5]. Our study demonstrates that these naturally occurring ligands bind to Betv1 with moderate to high affinity. Binding modes of these ligands with Betv1 has also been studied. Since these ligand-binding activity is an important aspect of the biological function of Betv1 structural modification or inhibition of some of these ligands could modify the biological function of this protein.
Conclusion
In the present study thirteen ligands have been successfully docked into the hydrophobic cavity of Betv1 and binding free energies of the complexes have been calculated using AutoDock 3.0.5. A linear relationship with correlation coefficient (R2) of 0.6 is obtained between ΔGbs values plotted against their corresponding IC50 values. The complex formed between Betv1 and the best docking pose for each ligand has been optimized by molecular dynamics simulation. We described the ligand binding of Betv1, which provides insight into the biological function of this protein. This knowledge is required for structural alteration or inhibition of some of these ligands in order to modify the allergenic properties of this protein.
Supplementary material
Acknowledgments
This study was supported by grants from Department of Biotechnology Govt. of India.
Footnotes
Citation:Lim et al, Bioinformation 4(7): 326-330 (2010)
References
- 1.Breiteneder H, et al. EMBO J. 1989;8:1935. doi: 10.1002/j.1460-2075.1989.tb03597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gajhede M, et al. Nat Struct Biol. 1996;3:1040. doi: 10.1038/nsb1296-1040. [DOI] [PubMed] [Google Scholar]
- 3.van Loon LC, van Strien EA. Physiol Mol Plant Pathol. 1999;55:85. [Google Scholar]
- 4.Tsujishita Y , Hurley JH. Nat Struct Biol. 2000;7:408. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 5.Mogensen JE, et al. J Biol Chem. 2002;277:23684. doi: 10.1074/jbc.M202065200. [DOI] [PubMed] [Google Scholar]
- 6.Morris GM, et al. J Comput Chem. 1998;19:1639. [Google Scholar]
- 7.Berman HM, et al. Nucleic Acid Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen FH. Acta Crystallogr. 2002;B58:380. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 9.van der Spoel D, et al. J Comput Chem. 2005;26:1701. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 10.Schuettelkopf AW, van Aalten DMF. Acta Crystallographica. 2004;D60:1355. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 11.Hess B, et al. J Comp Chem. 1997;18:1463. [Google Scholar]
- 12.Berendsen HJC, et al. J Chem Phys. 1984;81:3684. [Google Scholar]
- 13.Essmann U, et al. J Chem Phys. 1995;103:8577. [Google Scholar]
- 14.Kabsch W, Sander C. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 15.Sobolev V, et al. Bioinformatics. 1999;15:327. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 16.Marković-Housley Z, et al. J Mol Biol. 2003;325:123. doi: 10.1016/s0022-2836(02)01197-x. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi K, et al. Plant Cell Physiol. 2000;41:148. doi: 10.1093/pcp/41.2.148. [DOI] [PubMed] [Google Scholar]
- 18.Mo Y, et al. Proc Natl Acad Sci. 1992;89:7213. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.