Abstract
AmpC is a group I, class C ‐lactamase present in most Enterobacteriaceae and in Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli. The β‐lactam class of antibiotics is one of the most important structural classes of antibacterial compounds and act by inhibiting the bacterial D ,D - transpeptidases that are responsible for the final step of peptidoglycan cross-linking. Our main aim in the study is to screen possible inhibitors against AmpC / β ‐ lactamase (an enzyme responsible for antimicrobial activity in Pseudomonas aeruginosa), through virtual screening of 1364 NCI (National Cancer Institute) diversity set II compounds. Homology Model of AmpC / β ‐ lactamase was constructed using MODELLER and the Model was validated using PROCHECK and Verify 3D programs to obtain a stable structure, which was further used for virtual screening of NCI (National Cancer Institute) diversity set II compounds through molecular Docking studies using Autodock. The amino acid sequence of the β ‐ lactamase was also subjected to ScanProsite web server to find any pattern present in the sequence. After the prediction of 3-dimensional model of AmpC/ β‐lactamase, the possible Active sites ofβ ‐ lactamase were determined using LIGSITEcsc and CastP web servers simultaneously. The Docked complexes were validated and Enumerated based on the Autodock Scoring function to pick out the best inhibitor based on Autodock energy score. Thus from the entire 1364 NCI diversity set II compounds which were Docked, the best four docking solutions were selected (ZINC12670903, ZINC17465965, ZINC11681166 and ZINC13099024). Further the Complexes were analyzed through LIGPLOT for their interaction for the 4 best docked NCI diversity set II compounds. Thus from the Complex scoring and binding ability it is deciphered that these NCI diversity set II compounds could be promising inhibitors for Pseudomonas aeruginosa using AmpC /β ‐ lactamase as Drug target yet pharmacological studies have to confirm it.
Keywords: AmpC, beta lactamase, Pseudomonas aeruginosa, virtual screening
Background
Pseudomonas aeruginosa is an opportunistic pathogen, meaning that it exploits some break in the host defenses to initiate an infection. In fact, Pseudomonas aeruginosa is the epitome of an opportunistic pathogen of humans. The bacterium almost never infects uncompromised tissues, yet there is hardly any tissue that it cannot infect if the tissue defenses are compromised in some manner. It causes urinary tract infections, respiratory system infections, dermatitis, soft tissue infections, bacteremia, bone and joint infections, gastrointestinal infections and a variety of systemic infections, particularly in patients with severe burns and in cancer and AIDS patients who are immunosuppressed. Pseudomonas aeruginosa infection is a serious problem in patients hospitalized with cancer, cystic fibrosis, and burns. The case fatality rate in these patients is near 50 percent. [1,2,3]. Pseudomonas aeruginosa is intrinsically resistant to many antibiotics, or can develop resistance during treatment with consequent high mortality, and is, increasingly, a cause of infection in immunocompromised patients. The most relevant mechanism for the development of resistance to the antipseudomonal penicillins (such as ticarcillin or piperacillin) and cephalosporins (such as ceftazidime) is the selection of mutations leading to the hyperproduction of the chromosomal cephalosporinase AmpC [4,5,6]. AmpC is a group I, class C ‐lactamase present in most Enterobacteriaceae and in P. aeruginosa and other nonfermenting gramnegative bacilli [7,8]. The β‐lactam class of antibiotics is one of the most important structural classes of antibacterial compounds and act by inhibiting the bacterial D ,D ‐transpeptidases that are responsible for the final step of peptidoglycan cross-linking. The resistance mechanism in bacteria to β ‐ lactams is the production of β‐lactamases that catalyze the hydrolysis of the β ‐lactam ring, preventing their interaction with the D,D-transpeptidases. During treatment with lactams, resistant mutants showing constitutive high levels of AmpC production are frequently selected, leading to therapeutic failure [9]. Thus due to emergence of multidrug resistant and extremely drug resistant strains of Pseudomonas aeruginosa makes searching for drugs that are effective against these strains imperative.
Our main aim in the study is to screen possible inhibitors against AmpC / β ‐ lactamase (an enzyme responsible for antimicrobial activity in Pseudomonas aeruginosa), through virtual screening of 1364 NCI (National Cancer Institute) diversity set II compounds. NCI Diversity Set II is a collection of 1,364 compounds chosen (from 140,000 open compounds in the NCI chemical repository with more than or equal to 250 mg inventory) to cover a large, diverse range of molecular scaffolds and pharmacophore features, while also being relatively rigid. All compounds in the Diversity Set II have five or fewer rotatable bonds, facilitating pharmacophore development and conformational sampling [10].
Methodology
Comparative modeling of AmpC / β ‐ lactamase
As the tertiary structure of AmpC/ β‐lactamase is not available in the structure databases the structure was predicted by comparative modelling approach. The comparative modelling of P. aeruginosa β ‐ lactamase was done using MODELLER 9v6 [11] and five models were generated. The peptide sequence of P. aeruginosa β‐lactamase was retrieved from UniProt Knowledge Base, http://www.uniprot.org, (UniProt acc. No. P24735), ranging from 27 to 397 residues. ClustalW was used to produce alignment between the P. aeruginosa β ‐ lactamase sequence and the sequence of the templates (PDB: 2QZ6 and 1ZKJ) chosen from PDB BLAST hit. The predicted 3-D structures were evaluated using the PROCHECK [12] and Verify 3D programs [13].
Pattern detection in the sequence
The amino acid sequence of the β ‐ lactamase was subjected to ScanProsite web server [14] to find any pattern present in the sequence.
Active Site Determination
After the prediction of 3-dimensional model of AmpC/ β‐lactamase, the possible Active sites of β ‐ lactamase were determined using LIGSITEcsc.[15] and CastP [16] web servers simultaneously. LIGSITEcsc is based on the notion of surface-solvent-surface events and the degree of conservation of the involved surface residues where as CastP server uses the weighted Delaunay triangulation and the alpha complex for shape measurements. It provides identification and measurements of surface accessible pockets as well as interior inaccessible cavities, for proteins and other molecules.
Virtual Screening of NCI Diversity Set II against β ‐ lactamase using molecular docking
The ligand molecules of NCI Diversity Set II were obtained from ZINC database, a free database of commercially-available compounds for virtual screening in mol2 format, provided by the Shoichet Laboratory in the Department of Pharmaceutical Chemistry at the University of California, San Francisco (UCSF) [17]. Autodock4 program was used for molecular docking along with the help of the python scripts provided in the AutodockTools package for the preparation of the ligand, receptor, grid and dock parameter files. AutoDock4 uses Monte Carlo (MC) simulated annealing and Lamarckian genetic algorithm (LGA) to create a set of possible conformations, i.e. each string (coding for an individual in the population) passes on some of the characteristics it has acquired during its lifetime. LGA is used as a global optimizer and energy minimization as a local search method. Possible orientations are evaluated with AMBER force field model in conjunction with free energy scoring functions and a large set of protein-ligand complexes with known protein-ligand constants [18]. The active site center obtained by active site determination programs was used as the grid center in the grid parameter file, rest all the parameters were set to default values. The entire process of file preparation and docking was automated using an in house Perl script.
Molecular interactions
The LIGPLOT tool was used to generate molecular level interactions in the docked complexes [19].
Visualization
All the visualization of the structure files were done using PyMol molecular graphics system [20].
Discussion
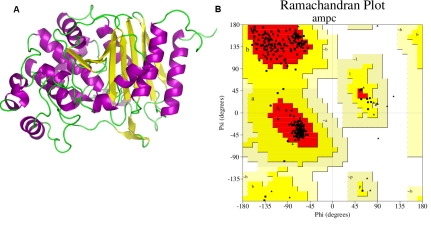
The hypothetical protein models generated in the modeling procedure were analyzed online by submitting to NIH MBI Laboratory for Structural Genomics and Proteomics' SAVES server. Validity reports generated by PROCHECK and Verfiy_3D judged accuracy of the protein models. A comparison of the results obtained from the abovementioned validation tools, showed that one of the models generated by Modeller is more acceptable in comparison to the others and was selected for further studies. For the final selected model Ramachandran plot generated by PROCHECK showed 95.1% residues in the most favored region (Figure 1). The compatibility of the atomic model (3D) with its own amino acid sequence (1D) computed by Verfy_3D reported 100.00% of the residues having an average 3D-1D score > 0.2 .
Figure 1.
(A) The final model of AmpC / β ‐ lactamase; (B) Validation of the model using Ramachandran plot computed with the PROCHECK program with 95.1 % of the residues in the most favored regions.
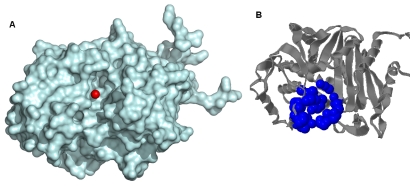
After the validation, the sequence of the β ‐ lactamase was scanned for any pattern present in the sequence using ScanProsite. The regular expression generated by the scan ([FY] ‐ E ‐ [LIVM] ‐ G ‐ S ‐ [LIVMG] ‐ [SA] ‐ K ) suggested β ‐ lactamase class-C active site region with first Serine residue as the catalytic residue. Active sites in the modeled protein structure were detected using LIGSITEcsc and CastP webserver. The LIGSITEcsc and CastP programs detected all the possible pockets in the structure which were compared with active site location from the homologous structures complexed with the substrates available in the PDB. Residues within the 5 A radius of the coordinates of the center of the active site detected by the LIGSITEcsc was also studied for the catalytic Serine residue as well as other residues found conserved in the ScanProsite. The various pockets obtained by the CastP program were also looked for the conserved residues and the spatial location of the pockets found by the program, the 2nd pocket detected was found better in all respects hence it was chosen as the most biologically favorable site for Docking study and other Identified Active sites were neglected (Figure 2).
Figure 2.
Active sites are shown. (A) The red sphere shows the coordinates of the center of the active site predicted by Ligsite program. (B) Blue spheres show the location and the area covered by the most appropriate active site as predicted by CastP program.
A set of 1364 compounds available in NCI Diversity Set II were obtained from ZINC database in mol2 format, the individual mol2 files converted into pdbqt file supported by Autodock4 using the python script prepare_ligand4.py available in Autodock Tools package. The receptor molecule was also converted into pdbqt format using prepare_receptor4.py script of Autodock Tools. The coordinates generated by the Ligsite in the grid parameter file, the grid was generated using Autogrid4 available in the Autodock4 suite. Docking of 1364 compounds was performed using Autodock4, the entire process of format conversion of files, preparation of grid and dock parameter files were automated using an in house Perl script. The algorithm exhaustively searches the entire rotational and translational space of the Ligand with respect to the receptor. The various solutions evaluated by a score giving Estimated Free Energy of Binding which is a sum total of Final Intermolecular Energy (vdW + Hbond + desolv Energy, Electrostatic Energy), Final Total Internal Energy, Torsional Free Energy and Unbound System's Energy of the Ligand in the protein environment. The best four docking solutions based on the energy scores were selected ( Table 1).
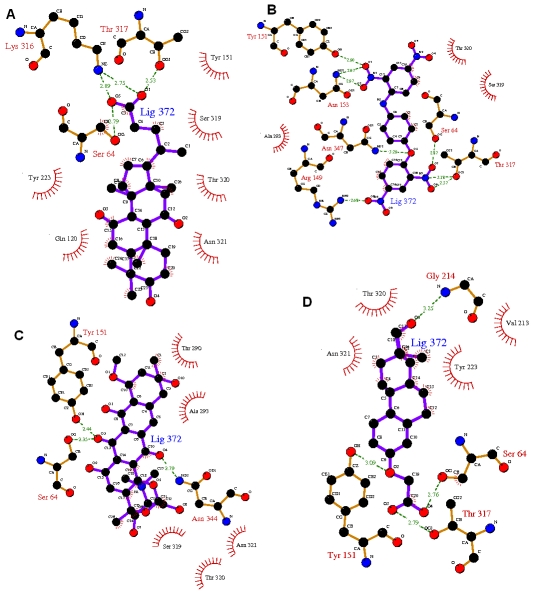
To understand the interaction between β ‐ lactamase and the four compounds from the NCI Diversity Set II, complexes were generated using AutodockTools for outputs of Autodock solutions. LIGPLOT were run for all the four complexes (Figure 3). It is evident from the analysis of the Docked complex that the Ligands are located in the center of the Active site, and is stabilized by hydrogen bonding interactions. The interaction analysis shows that Ser 64, Tyr 151 and Thr 317 are important anchoring residues for β ‐ lactamase and the main contributors towards compound interaction. Though the interaction energy does not include the contribution from the water of the extended compound structure, this preliminary data along with the list of hydrogen bond interactions between the compounds and the Active site residues clearly supports that Ser 64, Tyr 151 and Thr 317 are more preferred residues in binding.
Figure 3.
Receptor-ligand interaction is shown. (A) ZINC12670903 hydrogen bonded with Ser 64 OH, Lys 316 NH and Thr 317 OH at a distance of 2.79 Å, 2.75 Å and 2.53 Å, respectively whereas Gln 120, Tyr 151, Tyr 223, Ser 319, Thr 320 and Asn 321 are non-ligand residues involved in hydrophobic contact(s). (B) ZINC17465965 hydrogen bonded with Ser 64 OH, Arg 149 NH, Tyr 151 OH, Asn 153 NH, Thr 317 OH and Asn 347 NH at a distance of 2.87 Å, 2.68 Å, 2.90 Å, 2.97 Å, 2.57 Å and 3.29 Å respectively whereas Ala 293, Ser 319 and Thr 320 are non-ligand residues involved in hydrophobic contact(s). (C) ZINC11681166 hydrogen bonded with Ser 64 OH, Tyr 151 OH and Asn 344 NH at a distance of 2.35 Å, 2.44 Å and 2.79 Å respectively whereas Thr 290, Ala 293, Ser 319, Thr 320 and Asn 321 are non-ligand residues involved in hydrophobic contact(s). (D) ZINC13099024 hydrogen bonded with Ser 64 OH, Tyr 151 OH, Gly 214 NH and Thr 317 OH at distance of 2.76 Å, 3.09 Å, 3.25 Å and 2.79 Å respectively whereas Val 213, Tyr 223, Thr 320 and Asn 321 are non-ligand residues involved in hydrophobic contact(s).
Conclusion
AmpC/β ‐ lactamase is shown to be most potent cause for antimicrobial resistance in Pseudomonas aeruginosa. In this work, predicted 3‐ dimensional Model of AmpC / β ‐ lactamase was used for Virtual Screening of NIC Diversity Set II compounds. Docking results indicate that out of 1364 compounds, there were four inhibitory compounds for AmpC/β‐lactamase as target for antimicrobial resistance in Pseudomonas aeruginosa. Hydrogen bonding proved to play an important role for the structure and function of biological molecules, especially for inhibition in a complex. Thus our study confirms that ligand molecules having the following ZINC Ids ZINC12670903, ZINC17465965, ZINC11681166 and ZINC13099024 are potential inhibitor for AmpC / β ‐ lactamase as target for antimicrobial resistance in Pseudomonas aeruginosa. Yet to confirm it to be promising, Pharmacological studies need to be performed.
Supplementary material
Acknowledgments
The authors would like to acknowledge the support and facilities provided by the Department of Computational Biology and Bioinformatics, Sam Higginbottom Institute of Agricultural, Technology and Sciences, Allahabad, U.P., India.
Footnotes
Citation:Farmer et al, Bioinformation 4(7): 290-294 (2010)
References
- 1.Govan JRW, Deretic V. Microbiol Rev. 1996;60:539. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyczak JB, et al. Clin Microbiol Rev. 2002;15:194. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagaki M, et al. Chest. 2001;102:1464. doi: 10.1378/chest.102.5.1464. [DOI] [PubMed] [Google Scholar]
- 4.Giwercman B, et al. J Antimicrob Chemother. 1990;26:247. doi: 10.1093/jac/26.2.247. [DOI] [PubMed] [Google Scholar]
- 5.Livermore DM. Eur J Clin Microbiol. 1987;6:439. doi: 10.1007/BF02013107. [DOI] [PubMed] [Google Scholar]
- 6.Livermore DM. Clin Infect Dis. 2002;34:634. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 7.Bush K. Antimicrob Agents Chemother. 1995;39:1211. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore DM. Clin Microbiol Rev. 1995;8:557. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu PJ, Livermore DM. J Antimcrob Chemother. 1990;25:891. doi: 10.1093/jac/25.6.891. [DOI] [PubMed] [Google Scholar]
- 10. http://dtp.nci.nih.gov/branches/dscb/div2_explanation.html.
- 11.Sali A, Blundell TL. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 12.Laskowski RA, et al. J Appl Cryst. 1993;26:283. [Google Scholar]
- 13.Bowie JU, et al. Scienc. 1991;253:164. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 14. http://www.expasy.ch/tools/scanprosite/
- 15.Huang B, Schroeder M. BMC structural Biology. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dundas J, et al. Nucl Acids Res. 2006;34:116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irwin, Shoichet J Chem Inf Model. 2005;45:177. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris GM, et al. J of Comp Chem. 1998;19:1639. [Google Scholar]
- 19. http://www.pymol.org.
- 20.Wallace AC, et al. Prot Eng. 1995;8:127. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.