Abstract
A genomics approach was used to identify nutritionally regulated genes involved in growth of fast skeletal muscle in Atlantic salmon (Salmo salar L.). Forward and reverse subtractive cDNA libraries were prepared comparing fish with zero growth rates to fish growing rapidly. We produced 7,420 ESTs and assembled them into nonredundant clusters prior to annotation. Contigs representing 40 potentially unrecognized nutritionally responsive candidate genes were identified. Twenty-three of the subtractive library candidates were also differentially regulated by nutritional state in an independent fasting-refeeding experiment and their expression placed in the context of 26 genes with established roles in muscle growth regulation. The expression of these genes was also determined during the maturation of a primary myocyte culture, identifying 13 candidates from the subtractive cDNA libraries with putative roles in the myogenic program. During early stages of refeeding DNAJA4, HSPA1B, HSP90A, and CHAC1 expression increased, indicating activation of unfolded protein response pathways. Four genes were considered inhibitory to myogenesis based on their in vivo and in vitro expression profiles (CEBPD, ASB2, HSP30, novel transcript GE623928). Other genes showed increased expression with feeding and highest in vitro expression during the proliferative phase of the culture (FOXD1, DRG1) or as cells differentiated (SMYD1, RTN1, MID1IP1, HSP90A, novel transcript GE617747). The genes identified were associated with chromatin modification (SMYD1, RTN1), microtubule stabilization (MID1IP1), cell cycle regulation (FOXD1, CEBPD, DRG1), and negative regulation of signaling (ASB2) and may play a role in the stimulation of myogenesis during the transition from a catabolic to anabolic state in skeletal muscle.
Keywords: myogenesis, nutrition, refeeding, fasting, heat shock proteins, myogenic cells
atlantic salmon (Salmo salar L.) is an important food species and one of a small number of teleost model organisms with its genome currently being sequenced to draft level (http://web.uvic.ca/grasp/). Teleost fish such as Atlantic salmon represent excellent model species for investigating postembryonic myogenesis and the regulation of muscle mass in vertebrates because of a number of features facilitating experimental analysis. These include the fact that myotube production continues in juvenile and adult stages (39, 85), the anatomical separation of different fiber types into discrete layers (40), and the natural cycles of muscle wasting and concomitant mobilization of amino acids for energy metabolism and gonadogenesis during sexual maturation and/or migration (51).
The underlying molecular mechanisms regulating myogenesis in postembryonic stages are common to all vertebrates. Quiescent myogenic progenitor cells (MPCs) are present in adult skeletal muscle and require expression of the transcription factor Pax7 for their maintenance (70). Once activated by appropriate physiological stimuli, which in mammals include hormonal signals, stretch, overload, and exercise (31, 59, 21) MPCs proliferate before entering a differentiation program orchestrated by a redundant network of four myogenic regulatory factors (MRFs): MyoD (25, 75), Myf5 (16, 24), myogenin (26, 87), and Mrf4/Myf6 (15, 49, 62). MRFs contain highly conserved basic and helix-loop-helix (HLH) domains (53). The HLH domains allow the formation of heterodimers with bHLH containing E-proteins, which together with accessory proteins, bind via the basic domains to a conserved CIS-acting site (CANNTG) found in the promoter sequences of hundreds of muscle-specific genes (45). MRFs act synergistically with members of the MEF2 gene family to initiate and stabilize the differentiation program (50). Other genes that are involved in the specification (SVEP1) (73), proliferation (Megf10) (36), and differentiation (TAK1) (6) of myogenic progenitor cells have been described recently, and many more undoubtedly remain to be discovered (83).
Fish exhibit indeterminate growth and, compared with mammals, show a much greater increase in body size during ontogeny, which explains the requirement to continue producing myotubes in juvenile and adult stages to support growth (38). By the early juvenile stage a resident population(s) of MPCs can be observed scattered throughout the myotome (64). The progeny of these MPCs have one of two fates depending on ontogenetic stage. In fish less than ∼40% of their ultimate body length some of the myoblasts fuse to form myotubes on the surface of muscle fibers giving rise to a mosaic of fiber diameters (mosaic hyperplasia) (64). The majority of the MPCs are absorbed into muscle fibers as they expand in length and diameter, and this process of nuclear accretion continues until some limiting diameter is reached and occurs throughout the life cycle (39). In Atlantic salmon, the recruitment of fast muscle fibers continues until the fish reach 1.5–2 kg body mass during the first spring to summer of seawater life (38, 39).
Muscle fiber growth represents the net balance between protein anabolism and catabolism. Several of the genetic pathways regulating muscle fiber hypertrophy and/or nuclear accretion have been identified including calcineurin signaling (54), interleukin 6 signaling (71), insulin-like growth factor signaling (63), and the myostatin (47) and PCG-1α pathways (68). IGF-signaling regulates muscle atrophy and hypertrophy through the PI3K/AKT/mTOR pathway (79). During feeding, activation of this pathway by IGF-I activates a phosphorylation cascade that ultimately results in increased translation and protein synthesis (63) and inhibition of protein degradation pathways via phosphorylation of FOXO transcription factors (8). The phosphorylation of FOXO results in the downregulation of key regulators of skeletal muscle atrophy, the muscle-specific E3 ubiquitin ligases MAFbx and MuRF1 (8). Conversely, during periods of nutrient restriction, dephosphorylation of FOXO results in increased expression of MAFbx and MuRF1, which are involved in the targeting of proteins, such as myosin light chain 2, for degradation via the ubiquitin pathway.
Satiation feeding of fasted fish results in increased growth rates relative to continuously fed control groups. This phenomenon, referred to as compensatory or catch-up growth, is primarily achieved by increased feeding intensity in Atlantic salmon (56). Fasting-refeeding protocols have become one of the main manipulative tools used to investigate the molecular and genetic mechanisms regulating muscle growth in teleost fish (13, 22, 29, 61). The aim of the present study was to use suppression subtraction hybridization (SSH) in combination with a fasting-refeeding protocol designed to perturb muscle growth to identify nutritionally regulated and growth-related genes in Atlantic salmon. By combining normalization and subtraction, rare transcripts are enriched, increasing the probability of discovering novel or rare genes, which would otherwise be missed from standard cDNA libraries (41). Single-pass sequencing of cDNA to produce expressed sequence tags (ESTs) enables gene sequences to be identified by homology searches and functional motifs within these sequences to be classified. The subsequent assignment of gene ontology identifiers allows for the placement of these genes within a biochemical framework. The identified candidates were further investigated by determining their expression in a completely independent fasting-feeding experiment in the context of the expression of known growth regulatory genes. The genes regulated by nutrition in both experiments were screened for a role in myogenesis by examining their expression patterns during the differentiation of a primary muscle cell culture.
MATERIALS AND METHODS
Ethical Approval
This study was conducted on Atlantic salmon (Salmo salar L.) of aquaculture origin. Fish were humanely killed by a blow to the head following Schedule 1 of the Animals (Scientific Procedures) Act 1986 (Home Office Code of Practice. HMSO: London January 1997), and all husbandry procedures and experimental protocols were approved by University of St Andrews Animal Ethics and Welfare Committee.
Fasting-feeding Experiments
Two independent experiments were performed to manipulate growth.
First growth manipulation experiment.
The first experiment was used to obtain samples for preparation of the subtractive libraries and has been described previously (13). A strain of Norwegian farmed salmon (Salmo Breed) was used comprising 110 fish, 840 ± 152 g, mean ± SD. Fish were reared in replicate tanks under continuous light at an average temperature of 7.8°C, 13.96 mg/l oxygen, and average salinity 28.9 ppm at Ewos Innovation (Dirdal, Norway). The salmon were individually passive integrated transponder tagged (Fish Eagle, Lechlade, Gloucestershire, UK) to monitor growth and fed a maintenance diet (25% normal ration) to achieve a zero or slightly negative growth rate over a period of 22 days. Growth was measured as the thermal growth coefficient (TGC) = [(M20.333 − M0.333)(degree days)−1 *1,000], where M1 and M2 were start and final body weights, respectively. Degree days values are the sum of the °C values for each day of the experiment. At the start of the experiment [0 day (d)], fish were fed to satiation with a commercial feed (EWOS Innovation) to stimulate fast growth. Three fish were sampled at 0 d (fasted, 897 ± 72.8 g, n = 3), and at 3 d (855 ± 99 g, n = 3) and 14 d (842 ± 56 g, n = 3) following satiation feeding. Fast muscle was dissected from the dorsal epaxial myotome between 0.6 and 0.7 standard length (tip of snout to last vertebrae). Tissues were snap-frozen in liquid nitrogen and stored at −80°C until analyzed.
Second growth manipulation experiment.
A second independent experiment involving a change in feeding/growth status was carried out to validate the discovery of muscle growth-related genes from the first experiment with high stringency. The expression of candidate growth related genes was investigated in the context of 26 other myogenic genes with well established roles in growth regulation (13, 14, 29). By using a different population, size class, and rearing conditions of fish for validation of expression patterns we hoped to identify genes that changed with feeding and growth in a variety of contexts, i.e., were not specifically regulated at one particular life stage or set of environmental conditions. For this purpose we used a population of farmed salmon parr sourced from Scotland reared in duplicate tanks at the University of St Andrews comprising 150 fish, 59.8 ± 7.9 g, mean ± SD. The average temperature was 10.6°C, and fish were reared in aerated freshwater under a photoperiodic regime of 12 h light:12 h dark. Fish were fed a maintenance diet (25% normal ration) for 21 days, fasted for 7 d, and fed to satiation (starting at 0 d) with a commercial feed (EWOS Innovation). Sampling of fish occurred at 0, 1, 3, 5, 8, 15, and 21 d with three fish sampled from both replicate tanks at each time point. Individual mass and fork lengths were measured. Dissection and preparation of fast skeletal muscle samples were exactly as described for the first experiment. In addition, slow muscle, heart, liver, brain, kidney, gill, skin, gut, and eye tissues were dissected from fasted fish at 0 d and from fish fed to satiation for 8 and 21 d. RNA was extracted from these tissues from three fish at each time point, pooled and cDNA synthesized as described below.
Myogenic Cell Culture
Primary cell cultures from fast skeletal muscle were performed as previously described (10) using the same population of fish and rearing conditions as described for the second growth manipulation experiment above.
RNA Extraction
Total RNA was extracted by addition of 100 mg of muscle to Lysing matrix D (Qbiogene, Irvine, CA) with 1 ml Tri Reagent (Sigma, Gillingham, Dorset, UK) as per manufacturer's recommendations and homogenized using a Fast Prep instrument (Qbiogene). Total RNA was quantified based on absorbance at 260 nm. Genomic DNA contamination was removed by treatment with Turbo DNA-free (Ambion, Austin, TX), and the integrity of purified RNA confirmed by agarose gel electrophoresis. For myogenic cell culture, RNA was immediately extracted from duplicate wells of three separate cell cultures. RNA extraction and genomic DNA removal was performed using a RNeasy plus kit (Qiagen, Chatsworth, CA) as per manufacturer's recommendations.
Subtractive Library Production
RNA was extracted and pooled from the fast skeletal muscle of three fish from each of the 0 (fasted), 3, or 14 d (satiation feeding) samples from the first growth manipulation experiment. The SMART cDNA synthesis kit (Clontech Laboratories, Mountain View, CA) was used to synthesize cDNA. SMART cDNA was amplified for 17 cycles and digested with RsaI (Clontech Laboratories), and used for SSH using the PCR-select cDNA subtraction kit (Clontech Laboratories). Four subtractions were performed in total, two forward (where cDNA from day 3 or day 14 was used as tester and cDNA from day 0 was used as driver), and two reverse (cDNA from day 0 was used as tester and cDNA from days 3 or 14 used as driver). The subtracted PCR products were ligated into pCR4-TOPO vector (Invitrogen, Carlsbad, CA) and transformed into TOP10 chemically competent Escherichia coli cells (Invitrogen). Individual colonies were picked and grown overnight in 200 μl of Luria broth containing 100 μg/ml ampicillin. Four subtractive libraries were produced consisting of 3,072 clones in the day 3 forward (3F); 3,264 clones in the day 14 forward (14F); 1,536 clones in the day 3 reverse (3R); and 1,920 clones in the day 14 reverse (14R).
EST Sequencing
Inserts were amplified by transferring 1 μl of culture to a PCR reaction. PCR was performed in a 20 μl reaction volume using BioTaq PCR kit (Bioline, London, UK) under limiting conditions of primers and dNTPs, using short T3 (5′-ATTAACCCTCACTAAAG-3′) and short T7 (5′-AATACGACTCACTATAG-3′) primers. PCR was performed with an initial denaturation at 96°C for 2 min, followed by 35 cycles of amplification (96°C for 20 s, 49°C for 20 s, and 72°C for 45 s), and final extension at 72°C for 5 min.
PCR products were diluted two fold and directly sequenced using a T3 primer (5′-AATTAACCCTCACTAAAGGG-3′) and Big Dye terminator sequencing mix (Applied Biosystems). The sequencing reaction cycle (95°C for 20 s and 60°C for 140 s) was repeated 25 times. DNA sequences were read using an Applied Biosystems 3700 capillary sequencer (Geneservice, Oxford, UK).
Bioinformatic Analysis
Sequence trace files were first analyzed by trace2dbEST (58) with removal of low-quality sequence, vector sequence, and contaminating E. coli sequences and polyA tails. The processed sequences were subjected to BLAST similarity searches using the BLASTX algorithm against the National Center for Biotechnology Information (NCBI) nonredundant protein sequence database. Good-quality sequences and their preliminary annotation were submitted to dbEST (9). Clustering of sequences was performed using the Phrap algorithm within the Codon Code program (Codon Code, Dedham, MA).
Annotation of ESTs was performed with Blast2GO (23) using the default parameter settings. Annotations were assigned to three ontologies defined by the Gene Ontology (GO) consortium and were further summarized by assigning GO slim terms.
Quantitative Real-time PCR
The following procedures were performed to comply with the Minimum Information for Publication of Quantitative Real-Time PCR experiments MIQE guidelines (18). RNA was concentrated by ethanol precipitation and quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Only RNA with an A260/280 ratio between 1.8 and 2.1 and an A260/230 >1.9 was used for cDNA synthesis. Residual genomic DNA was removed using the genomic DNA wipeout buffer included in the Quantitect reverse transcription kit (Qiagen). We reverse transcribed 1 μg and 800 ng of RNA from the second growth manipulation experiment and myogenic culture, respectively, into cDNA for 30 min at 42°C using a Quantitect reverse transcription kit (Qiagen) as per manufacturer's recommendations.
qPCR was performed using a Stratagene MX3005P QPCR system (Stratagene, La Jolla, CA) with Brilliant II SYBR (Stratagene). cDNA used in qPCR was first diluted 80-fold with nuclease-free H20. Each qPCR reaction mixture contained 7.5 μl 2× Brilliant II SYBR green master mix (Surestart Taq DNA polymerase, 2.5 mM MgCl2), 6 μl cDNA (80-fold dilution), 500 nM each primer and RNase-free water to a final volume of 15 μl. Amplification was performed in duplicate in 96 well plates (Stratagene) with the following thermal cycling conditions: initial activation 95°C for 10 min, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Control reactions included a no template control and no reverse transcription control. Dissociation analysis of the PCR products was performed by running a gradient from 60 to 95°C to confirm the presence of a single PCR product. Products were also sequenced to confirm identity. The PCR amplification efficiency of each primer pair was calculated with LingregPCR 2009 using the average efficiency of 92 PCR reactions for each gene (67).
Primer Design
Primers for reference genes (10) and for 25 other genes known to be involved in myogenesis and growth (11–14) have been previously published as have primers for CTSL1 (88) and FXYD8 (82). Primers were designed using Primer 3 (65) to have Tm of 60°C, and where possible, were designed to cross an exon-exon junction to avoid amplification of contaminating genomic DNA. To determine exon-intron junction sites, genomic sequences for orthologous genes from Danio rario, Gasterosteus aculeatus, Oryzias latipes, Takifugu rubripes, and Tetraodon nigroviridis were retrieved from Ensembl (http://ensembl.org/) and compared with the Salmo salar cDNA sequences using the Spidey software tool (http://www.ncbi.nlm.nih.gov/spidey/). Primers were designed across conserved exon-intron junctions and used at a final concentration of 500 nM. The primers used for qPCR are listed in Supplementary Table S1.1
qPCR Data Analysis
GeNorm (84) was used to analyze the stability of transcription of several reference genes including hypoxanthine phosphoribosyltransferase 1 (HPRT1), β-actin, RNA polymerase II, elongation factor 1 alpha (ef1α), ribosomal protein s29 (rps29), and ribosomal protein L13 (rpl13). GeNorm Analysis revealed rps29 and rpl13 to be the most stable genes in the refeeding experiment (geNorm stability value M = 0.19) and also across the different tissues. Normalization of gene expression was performed using the geometric average of rpl13 and rps29, and values are shown as arbitrary units. Statistical analysis was performed using minitab (Minitab). Normalization of gene expression in myogenic cell culture was performed using the geometric average of ef1α, PPIA, and HPRT1 as described by Bower et al. (10). Significant differences in expression between time points were calculated by ANOVA using Fisher's individual error rate post hoc tests. Correlations in gene expression were calculated using Spearman rank correlation. Hierarchical clustering analysis of z-score normalized gene expression data was performed with Permutmatrix (20).
RESULTS
First Growth Manipulation Experiment
cDNA library characterization.
To identify genes that were upregulated in fish that showed no net growth or slightly negative growth and that responded early and late following satiation feeding to stimulate fast growth, a total of four subtractive libraries were produced, with forward and reverse subtractions performed to compare 0 d to 3 d, and 0 d to 14 d. Fish at 0 d had a zero or slightly negative growth rate (TGC = −0.26 ± 0.4, mean ± SD, n = 10), while after 14 d satiation feeding growth rate was positive (TGC = 1.4 ± 0.3, mean ± SD, n = 10). Forward and reverse subtractions were performed to compare 0 d to 3 d, and 0 d to 14 d groups. Forward subtracted libraries (3F and 14F) were enriched for transcripts that increased in response to feeding, while the reverse libraries (3R and 14R) were enriched for transcripts that decreased in response to feeding. A total of 10,000 clones were single-pass sequenced giving a total of 7,320 good quality ESTs (2,330 from 3F, 2,421 from 14F, 1,173 from 3R, and 1,396 from 14R). These ESTs, with an average read length of 567 bases, were submitted to dbEST under the accession numbers GE617488–GE624934. Sequence assembly using Phrap identified 222 contigs and 490 singletons in the 3F library, 242 contigs and 530 singletons in the 14F library, 133 contigs and 200 singletons in the 3R library, and 162 contigs and 206 singletons in the 14R library. The largest clusters present in the libraries comprised muscle structural proteins such as troponin I and actin, and metabolic genes such as glycogen phosphorylase, triose phosphate isomerase and glyceraldehyde 3 phosphate dehydrogenase; however, in the 3F library, two large clusters were present comprising 99 and 100 ESTs that corresponded to HSP90A, and a third cluster comprising 31 ESTs corresponding to HSPA1B.
To identify sequences that were specific to a particular library, the nonredundant sequences (i.e., consensus sequences from contigs and singletons) from each library were aligned. Within the reverse libraries (3R + 14R), there were 82 nonredundant sequences that formed contigs and 523 singletons (data not shown). A total of 238 nonredundant sequences formed contigs from the 3F and 14F libraries, 173 formed contigs from the 3F and reverse libraries, 171 formed contigs from the 14F and reverse libraries, and 196 formed contigs from 3F, 14F and 3R + 14R libraries (Supplementary Fig. S1). The contigs mainly comprised muscle structural proteins and metabolic enzymes.
Many of the contigs from each library did not have significant hits against the NCBI nonredundant protein database (51% of 3F, 48% of 14F, 42% of 3F, 48% of 14R) and are therefore novel sequences or untranslated regions. The nonredundant collection of singletons and contigs that did have significant hits were annotated and assigned to three ontologies defined by the GO consortium and further summarized by assigning GO slim terms. There were 72, 75, 38, and 39 contigs from the 3F, 14F, 3R, and 14R libraries, respectively, that did not have an associated GO term and are therefore unclassified. A summary of the GO classifications is shown in Supplementary Fig. S2.
To identify processes and genes that were differentially regulated between the libraries, individual ESTs were annotated using Blast2GO (23), and the relative abundance of the GO terms in the forward and reverse libraries compared using the GOSSIP software package within Blast2GO (23) using a false discovery rate P-value cutoff of P < 0.01 (7). Based on the GO-term distribution, in the 3F library, genes associated with vesicle formation, nucleotide binding, developmental processes, cytoskeleton organization, response to stimulus, and cell differentiation were enriched compared with the 3R library. In the 3R library, genes with GO terms associated with cell death and transferase activity were enriched compared with the 3F library (Supplementary Fig. S3). Comparing the GO term distribution for the 14F and 14R libraries revealed genes associated with ion channel activity, ion binding, vesicle formation, and transporter activity were enriched in the 14F library, while those associated with macromolecular complex and cellular process were enriched in the 14R library (Supplementary Fig. S4). The GO-term distribution suggests that there are more genes differentially expressed at day 3 than at day 14 of feeding. Many of the GO terms that were differentially distributed between forward and reverse libraries corresponded to heat shock proteins and in particular HSP90A. The accession numbers, top blastx hit, and GO terms for the 3F, 14F, 3R, and 14R libraries are listed in Supplementary Tables S2, S3, S4, and S5, respectively.
Selection of Candidate Genes
Transcripts putatively identified as being nutritionally regulated and growth related are referred to as candidate genes throughout the rest of the paper. The results obtained from the interlibrary assembly of nonredundant sequences and the GO term enrichment analysis were used to make an initial selection of candidate genes based on the following criteria: 1) Contigs that contained ESTs present in the forward library and absent from the reverse (or vice versa) and 2) contigs significantly enriched in the forward or reverse library based on the relative abundance of GO terms. These selection criteria identified differentially expressed genes of known function as well as genes with no associated GO term and allowed for the selection of a subset of 40 genes for detailed investigation including abundant transcripts, transcription factors, and signaling molecules. We selected 30 genes putatively upregulated with feeding from the forward libraries along with 10 genes putatively downregulated with feeding from reverse libraries.
For ease of interpretation, throughout results and discussion, when homology between the candidate gene and top blastx hit were high (blastx e-value <1e-3) we will refer to the candidate genes using the gene symbol from their corresponding top blastx hit (Table 1). When there is poor or no homology between candidate gene and known genes (blastx e-value >1e-3), we will refer to the candidate gene by their contig number (Table 1).
Table 1.
List of genes that were differentially expressed in an independent refeeding experiment (second growth manipulation experiment)
| Contig | GenBank Accession No. | # ESTs | Gene Symbol | BlastX Hit (GenBank Accession No.) |
|---|---|---|---|---|
| FCon1 | GE618547 | 6 | SMYD1 | NP_991103 SET and MYND domain containing 1 (Danio rerio) |
| FCon2 | GE617873 | 4 | RTN1 | ACQ58869 Reticulon-1-A (Anoplopoma fimbria) |
| FCon3 | GE621733 | 6 | MID1IP1 | ACM09805 Mid1-interacting protein 1 (Salmo salar) |
| FCon4 | GE623473 | 100 | HSP90A | BAF92789 cytosolic heat shock protein 90 alpha (Solea senegalensis) |
| FCon5 | GE617747 | 6 | no significant hit | |
| FCon6 | GE621845 | 3 | FOXD1 | NP_571346 forkhead box D1 (D. rerio) |
| FCon7 | GE618332 | 5 | DRG1 | NP_001134621 Developmentally regulated GTP-binding protein 1 (S. salar) |
| FCon8 | GE623079 | 5 | no significant hit | |
| FCon9 | GE623156 | 6 | DNAJA4 | ABA54277 DnaJ-like subfamily A member 4 (Paralichthys olivaceus) |
| FCon10 | GE622980 | 14 | HSPA1B | ACI34374 Heat shock 70 kDa protein (S. salar) |
| FCon11 | GE617931 | 31 | CHAC1 | NP_001133807 Cation transport regulator-like protein 1 (S. salar) |
| FCon12 | GE622026 | 2 | NHP2L1 | ACI69124 NHP2-like protein 1 (S. salar) |
| FCon13 | GE617673 | 3 | DDX21 | ACH85363 DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 (S. salar) |
| FCon14 | GE617505 | 14 | no significant hit | |
| FCon15 | GE622033 | 15 | HSP30 | NP_001134440 Heat shock protein 30 (S. salar) |
| RCon1 | GE624208 | 6 | CEBPD | ACF94990 CCAAT/enhancer binding protein delta (S. salar) |
| RCon2 | GE621159 | 5 | ASB2 | CAI21253 novel protein similar to vertebrate ankyrin repeat and SOCS box-containing 2. (D. rerio) |
| RCon3 | GE623928 | 6 | no significant hit | |
| RCon4 | GE624303 | 3 | CTSL1 | NP_001140018 Cathepsin L1 (S. salar) |
| RCon5 | GE624833 | 7 | GBP | ACI67421 GSK-3-binding protein (S. salar) |
| RCon6 | GE624221 | 4 | FXYD8 | NP_001117203 FXYD domain containing ion transport regulator 8 (S. salar) |
| RCon7 | GE620453 | 4 | CCT8 | NP_001117705 chaperonin containing TCP1 subunit 8 (Oncorhynchus mykiss) |
| RCon8 | GE620675 | 6 | no significant hit |
EST, expressed sequence tag.
Gene Expression Profiling of Candidate Genes
Second growth manipulation experiment.
The aim of this experiment was to determine which of the selected gene candidates were nutritionally regulated and associated with a change in growth status over a broad range of physiological contexts. To this end we used a different population of Atlantic salmon from the first experiment, investigated a juvenile freshwater rather than adult seawater stage, and maintained the fish under very different environmental conditions. To obtain an insight into the function of the candidates, their expression following fasting and refeeding was clustered in relation to the expression of 26 other genes with well-characterized roles in myogenesis and growth regulation (Fig. 1). These 26 genes were examined in the same cDNA, and the data processed identically to that of the candidate genes.
Fig. 1.
Heat map summary and hierarchical clustering for candidate genes (red) with other genes involved in muscle growth regulation during refeeding of fasted Atlantic salmon [0 days (d)] and fish fed to satiation for 1, 3, 5, 8, 15, and 21 d. Rows are standardized to have mean 0 and standard deviation (SD) 1 so that yellow indicates high and blue indicates low values.
Table 1 lists the 23 genes from the 40 selected candidate genes that were significantly differentially regulated with feeding across both experiments. Expression of several other candidate genes was also modulated with feeding in the second experiment; however, the changes in expression were not statistically significant, and these were discarded from further analysis. Apart from the expression of HSP30 (which was expected to be upregulated but was actually downregulated), the candidate genes gave the expected expression profiles as predicted by the selection criteria.
Genes upregulated during feeding.
The upregulated genes showing the greatest fold change with feeding in the second growth manipulation experiment were RTN1 (21-fold), FCon14 (19-fold), HSPA1B (9-fold), and HSP90A (5-fold). There were distinct phases of gene expression observed during the feeding response. For example, genes which responded early to refeeding included HSPAIB (Fig. 2A, P < 0.05) at 1 d and CHAC1 (Fig. 2B, P < 0.05) and FOXD1 (Fig. 2C, P < 0.05) at 1 d–5 d. FOXD1 and HSPA1B expression clustered with myoD1b, myoD1a, and the IGFBP-2 paralogs, respectively (Fig. 1). FCon8 (Fig. 2D) and MID1IP1 (Fig. 2E) expression was correlated (R = 0.80, P < 0.001) and had highest levels at 3 and 5 d, which then remained elevated throughout the experiment (P < 0.05). This pattern was similar to that of SMYD1 (Fig. 2F) and DDX21 (Fig. 2G), and together the expression of these genes formed a cluster that included IGFBP-5.1, AdipoQ, Pax7, and Myf5 (Fig. 1).
Fig. 2.
Expression profiles for HSPA1B (A), CHAC1 (B), FOXD1(C), FCon8 (D), MID1IP1 (E), SMYD1 (F), and DDX21 (G) in fasted fish (0 d) and fish fed to satiation for 1, 3, 5, 8, 15, and 21 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
Another group of candidate genes displayed more sustained increase in expression after feeding. For example, DNAJA4 (Fig. 3A), DRG1 (Fig. 3B), HSP90A (Fig. 3C), and NHP2L1 (Fig. 3D) all showed peak expression within 1 d of refeeding (P < 0.05), which, with the exception of DRG1 (Fig. 3B) and NHP2L1 (Fig. 3D) at 21 d, remained above the levels observed in fasted fish (P < 0.05). RTN1 (Fig. 3E) and FCon14 (Fig. 3F) showed a monotonic increase in expression throughout refeeding, similar to that of FCon5 (Fig. 3G), and had peak expression at 21 d (P < 0.05). Together, these genes formed a cluster that included IGF-I and IGFBP-4 (Fig. 1), with expression of several genes correlated including DNAJA4 and HSP90A (R = 0.74, P < 0.001), RTN1 and FCon14 (R = 0.81, P < 0.001), RTN1 and IGF-I (R = 0.82, P < 0.001) and FCon14 and IGF-I (R = 0.75, P < 0.001).
Fig. 3.
Expression profiles for DNAJA4 (A), DRG1 (B), HSP90A (C), NHP2L1 (D), RTN1 (E), FCon14 (F), and FCon5 (G) in fasted fish (0 d) and fish fed to satiation for 1, 3, 5, 8, 15, and 21 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
Genes downregulated during feeding.
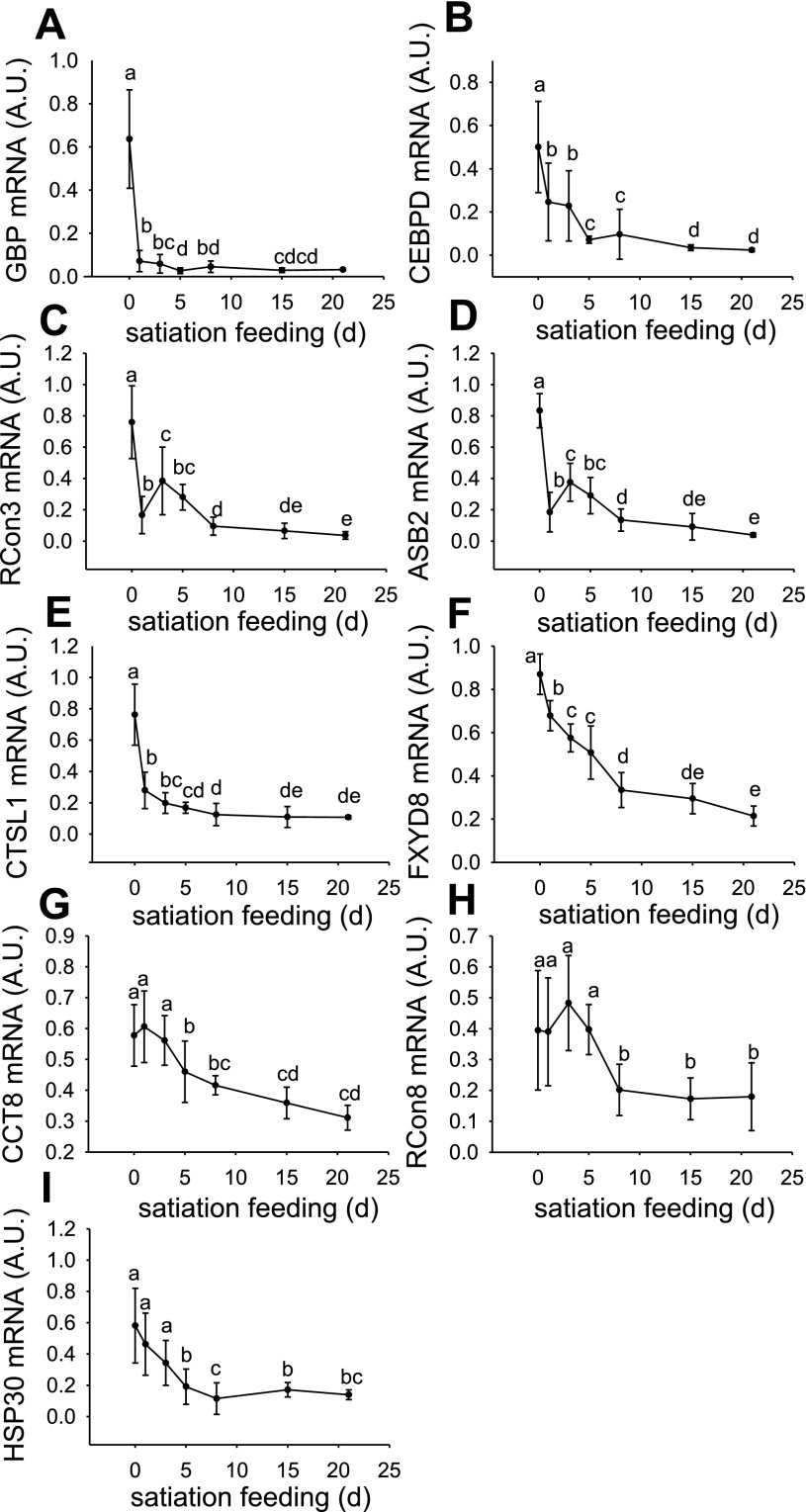
Of the downregulated genes, those showing the greatest fold change were GBP (22-fold, Fig. 4A), CEBPD (21-fold, Fig. 4B), RCon3 (21-fold, Fig. 4C), ASB2 (21-fold, Fig. 4D), CTSL1 (7-fold, Fig. 4E), and FXYD8 (4-fold, Fig. 4F). GBP, CEBPD, RCon3, ASB2, CTSL1, and FXYD8 (Figs. 4, A–F) were all strongly downregulated as early as 1 d following refeeding (P < 0.05) and remained at this depressed level of expression throughout the experiment. The expression of these genes clustered and was highly correlated with MAFbx and MuRF1 genes known to be involved in protein degradation (Fig. 1). For example, GBP and CEBPD were highly correlated with MAFbx (R = 0.93, P < 0.001 and R = 0.76, P < 0.001, respectively; Fig. 1), while CTSL1 had high correlation with MAFbx (R = 0.88, P < 0.001) and MuRF1 (R = 0.95, P < 0.001). ASB2 and RCon3 expression was highly correlated with each other (R = 0.91, P < 0.001) and also with that of IGF-II (R = 0.81, P < 0.001 and R = 0.81, P < 0.001, respectively; Fig. 1). Expression of CCT8 (Fig. 4G), RCon8 (Fig. H), and HSP30 (Fig. 4I) showed a slower response, being significantly downregulated from 8 and 5 d (P < 0.05).
Fig. 4.
Expression profiles for GBP (A), CEBPD (B), RCon3 (C), ASB2 (D), CTSL1 (E), FXYD8 (F), CCT8 (G), RCon8 (H), and HSP30 (I) in fasted fish (0 d) and fish fed to satiation for 1, 3, 5, 8, 15, and 21 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
Tissue Distribution of Candidate Genes
To further characterize the candidate genes and to identify genes with high expression in muscle, we examined their expression across several tissues in fasted fish and fish fed to satiation for 8 and 21 d. The relative abundance of transcripts and their response to refeeding in various tissues were examined using QPCR and are shown in Table 2 and Supplementary Figs. S5–S7. Twelve of the candidate genes had high expression in skeletal muscle compared with other tissues and included SMYD1, RTN1, MID1IP1, HSP90A, FCon5, FOXD1, FCon8, DNAJA4, HSP30, CEBPD, ASB2, RCon3, and FXYD8 (Table 2). Many of these genes gave the same response to refeeding in slow muscle as in fast muscle (Supplementary Figs. S5–S7). The remaining genes were expressed at similar levels across the tissues, although RTN1 had high expression in the brain and RCon8 had high expression in gut (Table 2).
Table 2.
Expression patterns of candidate genes brain, eye, gill, gut, heart, kidney, liver, slow muscle, skin, and fast muscle of Atlantic salmon normalized to the geometric average of Rps29 and Rpl13
| Tissue |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Contig | Gene Symbol | Brain | Eye | Gill | Gut | Heart | K | Liver | Skin | SM | FM |
| FCon1 | SMYD1 | + | + | + | + | ++ | + | + | + | ++ | +++ |
| FCon2 | RTN1 | +++ | + | + | + | + | + | + | + | + | ++ |
| FCon3 | MID1IP1 | + | + | + | + | + | + | + | + | +++ | ++ |
| FCon4 | HSP90A | − | + | − | + | ++ | − | − | + | ++ | +++ |
| FCon5 | − | − | − | − | + | − | − | + | + | +++ | |
| FCon6 | FOXD1 | + | + | + | ++ | + | + | + | + | + | +++ |
| FCon7 | DRG1 | + | ++ | + | + | + | + | + | + | + | +++ |
| FCon8 | + | ++ | + | + | − | + | + | + | ++ | +++ | |
| FCon9 | DNAJA4 | + | + | + | + | + | + | + | + | + | +++ |
| FCon10 | HSPA1B | +++ | ++ | + | + | ++ | + | + | + | + | + |
| FCon11 | CHAC1 | +++ | +++ | + | + | + | + | + | ++ | + | ++ |
| FCon12 | NHP2L1 | + | + | + | + | + | + | + | + | + | +++ |
| FCon13 | DDX21 | ++ | +++ | ++ | +++ | ++ | ++ | ++ | ++ | ++ | +++ |
| FCon14 | + | + | + | ++ | + | + | + | + | + | ++ | |
| FCon15 | HSP30 | − | + | + | + | ++ | − | − | ++ | + | + |
| RCon1 | CEBPD | + | + | + | + | + | + | + | ++ | +++ | +++ |
| RCon2 | ASB2 | − | − | − | − | − | − | − | − | ++ | +++ |
| RCon3 | − | − | − | − | − | − | − | − | ++ | +++ | |
| RCon4 | + | + | + | ++ | + | + | +++ | + | +++ | +++ | |
| RCon5 | GBP | + | + | + | + | + | + | + | + | +++ | +++ |
| RCon6 | FXYD8 | + | + | + | + | + | + | + | + | +++ | ++ |
| RCon7 | CCT8 | +++ | ++ | + | + | ++ | ++ | + | ++ | +++ | +++ |
| RCon8 | + | ++ | + | ++ | + | + | + | + | + | + | |
Expression is shown relative to the highest tissue expression, so that (−) represents no expression, (+) 1–33% maximal expression, (++) 34–66% maximal expression, and (+++) 67–100% maximal expression. Genes from the forward libraries are expressed relative to the highest fed expression level, and genes from the reverse libraries are expressed relative to the highest fasted expression level. Graphs depicting the tabulated information are supplied as Supplementary Figs. S5–S7. K, kidney; SM, slow muscle; FM, fast muscle.
Expression of Candidate Genes During Myogenesis in Cell Culture
To further characterize the candidate genes and investigate their possible role in myogenesis, we examined their expression profile in vitro during the maturation of a primary cell culture. Primary cell cultures derived from fast muscle of Atlantic salmon showed distinct phases of maturation. Mononuclear cells and proliferating cells predominated at 2–5 d, after which short myotubes became evident. Myotube elongation subsequently occurred resulting in continuous sheets of multinucleated myotubes at 20 d (Fig. 5A). The expression of MRFs and components of the IGF-signaling pathway and cell proliferation rates (Fig. 5, B and C) have been previously examined using the same culture used to characterize the candidates from the subtractive library (11, 12). These data were produced from the same cDNA samples and analyzed identically to the subtractive library candidates. To place the expression of candidate genes within the myogenic program, we combined and compared the subtractive library candidates with this data set (Fig. 5C).
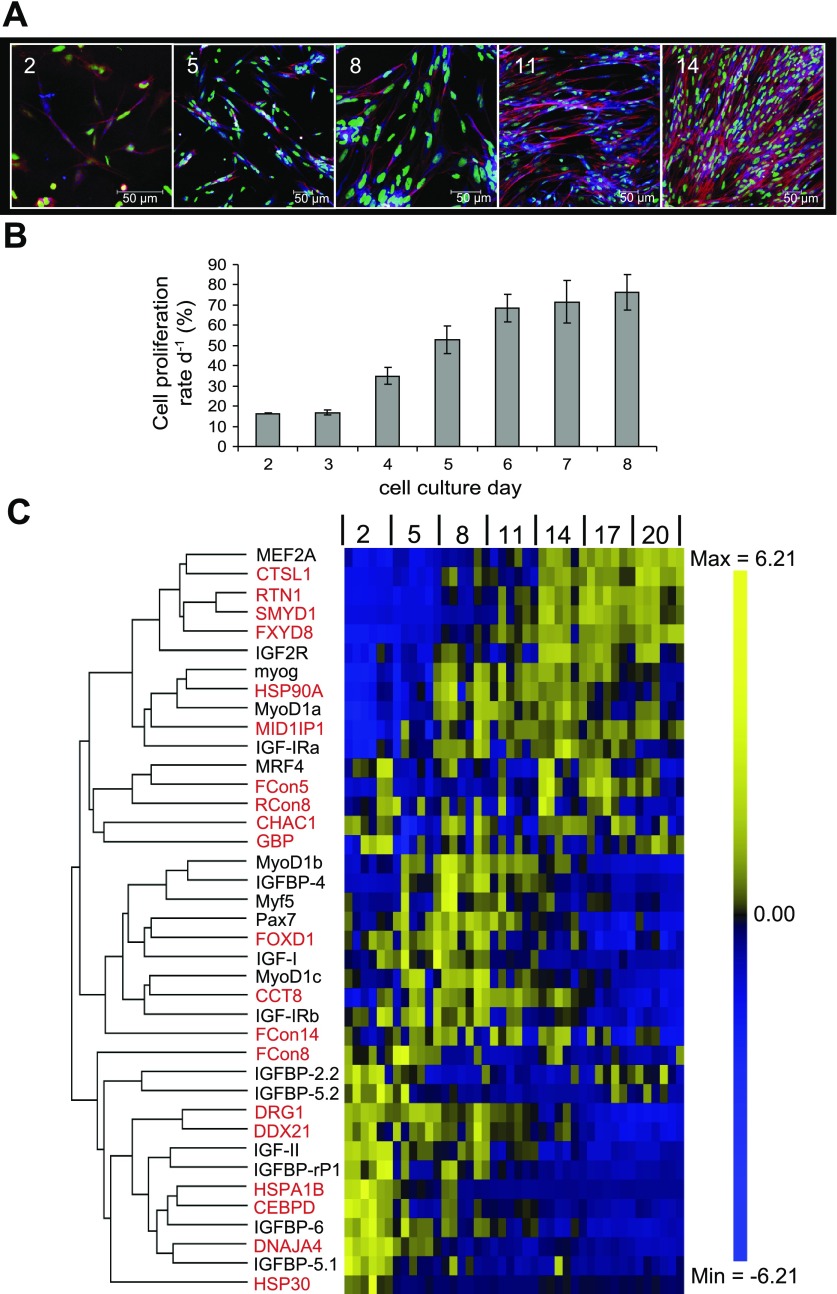
Fig. 5.
Phenotype of the cell culture at 2, 5, 8, 11, and 14 d (A). Cell proliferation rates of myogenic cells from 2 d to 8 d (B). Heat map summary and hierarchical clustering for candidate genes (red) with MRFs, MEF2A, pax7, and components of the IGF signaling pathway during Atlantic salmon myogenic cell culture at 2, 5, 8, 11, 14, 17, and 20 d (C). Rows are standardized to have mean 0 and SD 1 so that yellow indicates high and blue indicates low values (C).
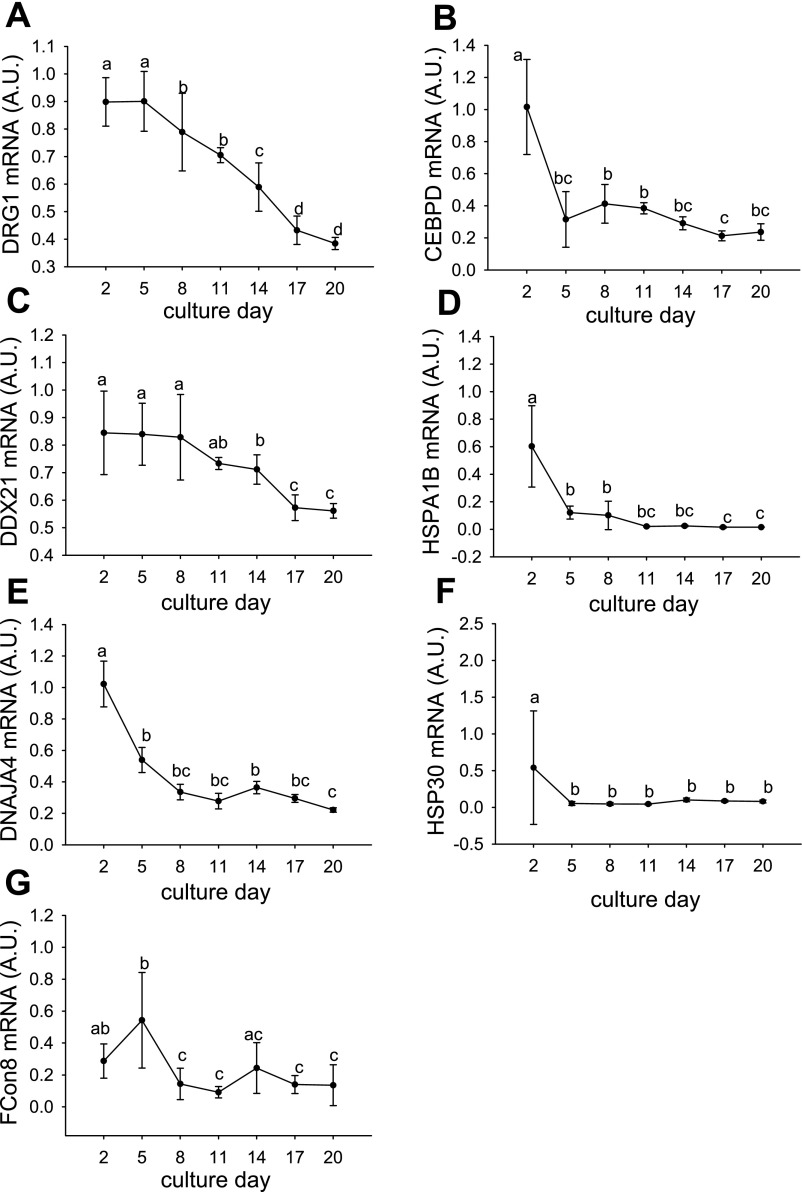
There was a cluster of genes that were highly expressed in day 2 of the culture corresponding to a phase when mostly mononuclear cells were present (Fig. 5A) and proliferation rates were low (Fig. 5B). These genes decreased in expression to varying extents at day 5 and day 8 before being downregulated during the phases of myotube elongation and differentiation (Fig. 5, A and C). This cluster included IGF-II, IGFBP-rP1, IGFBP-6, IGFBP-5.1 (Fig. 5C), as well as the candidate genes DRG1 (Fig. 6A), CEBPD (Fig. 6B), DDX21 (Fig. 6C), HSPA1B (Fig. 6D), DNAJA4 (Fig. 6E), and HSP30 (Fig. 6F). External to, but associated with, this cluster was FCon8 (Figs. 5C, 6G). A second major cluster of transcripts were weakly expressed at day 2, showed high expression at days 5 and 8 as proliferation of myoblasts became active (Fig. 5B) and short myotubes formed (Fig. 5A), but were then downregulated to low levels at later stages of differentiation when the cultures of myotubes became confluent. This second cluster included Pax7, Myf5, myoD1b, myoD1c, IGF-I, IGFBP-4 (Fig. 5C), as well as the candidates FOXD1 and CCT8 (Fig. 7, A and B). The third major cluster of transcripts comprised IGF2R, IGF-Ira, MEF2A, myog, and myoD1a (Fig. 5C) together with the candidates CTSL1, RTN1, SMYD1, FXYD8, HSP90A, and MID1IP1 (Fig. 8, A, B, C, D, E, and F). These transcripts were expressed at low levels at day 2 and day 5 of the culture but were strongly upregulated with differentiation and the formation of confluent multinucleated myotubes (Fig. 5, A and C). Associated with the third cluster was MRF4, which showed a biphasic expression at early and late stages of the culture (Fig. 5C). MRF4 formed a subcluster with the candidates CHAC1, GBP, and the novel transcripts FCon5 and RCon8 (Fig. 9, A–D). There was no significant change in expression of FCon14 (data not shown), very low expression of ASB2 and RCon3 was observed in some samples at 2 d (data not shown), and NHP2L1 expression was below the limits of detection (data not shown). Of the genes that we had previously identified as upregulated with feeding, several were also upregulated during myogenesis in vitro and included FCon8, HSP90A, MID1IP1, RTN1, FOXD1, FCon5, and SMYD1. These genes are therefore excellent candidates as promyogenic factors. CEBPD and HSP30 were downregulated during refeeding and from 2 d in the myogenic culture and are thus excellent candidates as myogenic inhibitory genes, as their levels decreased as cells began to grow and proliferate. Several genes showed correlated expression patterns during myogenesis. Expression of SMYD1 and RTN1 was highly correlated (R = 0.96, P < 0.001), and also with that of MEF2A (R = 0.84, P < 0.001, R = 0.84, P < 0.001, respectively) and IGF2R (R = 0.78, P < 0.001, R = 0.78, P < 0.001, respectively). HSP90A and MID1IP1 were correlated with myoD1a (R = 0.84, P < 0.001, and R = 0.67, P < 0.001, respectively) and myog (R = 0.88, P < 0.001 and R = 0.76, P < 0.001, respectively). CTSL1 correlated with IGF2R (R = 0.79, P < 0.001) and MEF2A (R = 0.87, P < 0.001), and FXYD8 was also correlated with MEF2A (R = 0.87, P < 0.001). CEBPD clustered with IGF-II (R = 0.75, P < 0.001) and IGFBP-6 (R = 0.81, P < 0.001), HSPA1B with IGFBP-6 (R = 0.76, P < 0.001), FCon5 with MRF4 (R = 0.68, P < 0.001) and DNAJA4 with IGFBP-5.1 (R = 0.81, P < 0.001).
Fig. 6.
Expression of candidates in primary myogenic cultures with peak expression at 2–5 d. Expression profiles for DRG1 (A), CEBPD (B), DDX21 (C), HSPA1B (D), DNAJA4 (E), HSP30 (F), and FCon8 (G) in Atlantic salmon myogenic cell culture at 2, 5, 8, 11, 14, 17, and 20 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
Fig. 7.
Expression of candidates in primary myogenic cultures with peak expression at 5 and 8 d. Expression profiles are shown for FOXD1 (A) and CCT8 (B) in Atlantic salmon myogenic cell culture at 2, 5, 8, 11, 14, 17, and 20 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
Fig. 8.
Expression of candidates in primary myogenic cultures with peak expression coincident with the formation of confluent multinucleated myotubes during late stages of differentiation. CTSL1 (A), RTN1 (B), SMYD1 (C), FXYD8 (D), HSP90A (E), and MID1IP1 (F). Data points are from cell culture at 2, 5, 8, 11, 14, 17, and 20 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
Fig. 9.
Expression profiles for genes that formed a subcluster with MRF4. Expression profiles are shown for CHAC1 (A), GBP (B), FCon5 (C), and RCon8 (D) in Atlantic salmon myogenic cell culture at 2, , 8, 11, 14, 17, and 20 d. Values represent means ± SD, 6 samples per point. Significant differences between means are indicated by different letters.
DISCUSSION
We have identified 23 previously unrecognized genes in fast myotomal muscle of Atlantic salmon that were differentially regulated with growth stimulation in two feeding experiments that involved a distinct population, lifecycle stage, and set of environmental conditions. Thirteen of these genes were also transcriptionally regulated during the maturation of primary myogenic cell culture and are thus excellent candidates for playing a role in myogenesis. To characterize further the identified genes we examined their expression in the context of other genes with well-characterized roles in myogenesis and growth regulation.
Genes Associated With Muscle Fiber Differentiation and Positive Growth
The transition from a proliferative to differentiation state requires the coordinated expression of prodifferentiation genes and silencing of genes involved in proliferation, a process that requires alterations in chromatin structure. Two of the genes we identified, SMYD1 and RTN1, may play opposing roles in chromatin modification. Within the tissues examined, SMYD1 expression was largely restricted to cardiac, slow and fast muscle (Table 2). During myotube maturation, SMYD1 expression increased >30-fold from 2 d to 14 d (Fig. 8C) and remained at elevated levels for 17–20 d when myotubes predominated (Fig. 5, A and C). Its expression was highly correlated with that of MEF2A (R = 0.84, P < 0.001) and IGF2R (R = 0.78, P < 0.001) (Fig. 5C), giving strong evidence for a role in myogenic differentiation. The SMYD1 protein contains evolutionary conserved SET and MYND domains with histone methyltransferase and deacetylation activity, respectively (60, 80), indicative of a role in chromatin modification (74) leading to transcriptional suppression. Indeed, acetylation of histones results in decreased levels of transcription, and Gottlieb et al. (30) demonstrated that SMYD1 can function as a transcriptional repressor in mouse skeletal muscle. SMYD1 is expressed in the developing skeletal muscle cells of the somite as well as differentiated muscle fibers in mice (30, 37) and is required for normal skeletal muscle development in zebrafish (81).
RTN1, a member of the poorly understood reticulon gene family, increased in expression 21-fold during refeeding (Fig. 3E). Its expression clustered with IGFBP-4 and was correlated with IGF-I (R = 0.82, P < 0.001) in vivo (Fig. 1) and with MEF2A (R = 0.84, P < 0.001) and IGF2R (R = 0.78, P < 0.001) in myogenic culture (Fig. 5C). Correlations with such genes are indicative of a role in myogenic differentiation. Through interaction with SNARE proteins RTN1 is involved in vesicle trafficking and exocytosis, including growth hormone secretion (78). RTN1 is also potentially a nuclease and an inhibitor of histone deacetylases (48, 55), a function that leads to the dissociation of histones from DNA resulting in increased transcription. Thus, whereas SMYD1 may function to repress the expression of genes that inhibit myogenic differentiation, RTN1 may promote the expression of genes required for differentiation. Interestingly, SMYD1 and RTN1 expression during myotube maturation was highly correlated (R = 0.97, P < 0.0001, Fig. 5C), which suggests that these genes share similar regulatory elements or are present on the same signaling pathway.
Microtubules play important roles in many myogenic processes including cell morphology and sarcomere assembly and are reorganized to form stable microtubules during differentiation (32). Proteins that stabilize microtubule formation have been shown to be essential for skeletal muscle cell differentiation (77). Mid1 interacting protein (MID1IP1), also known as MIG12, stabilizes microtubule formation through its interaction with Mid1 and is implicated in processes requiring microtubule stabilization (5). We observed that expression of MID1IP1 increased during myotube maturation (Fig. 8F), clustered with genes involved in differentiation and was correlated with myog (R = 0.76, P < 0.001, Fig. 5C). This, together with its high expression level in slow and fast muscle relative to other tissues (Table 2), suggests an involvement in the differentiation of myogenic cells.
HSP90A is required for the folding and activation of proteins such as myoD (72) and many proteins involved in signal transduction (90). Inhibition of HSP90 activity disrupted zebrafish somite development (42), decreased myogenin expression, inhibited myotube formation, and depleted levels of three protein kinases in C2C12 cells (90), including AKT. During refeeding, we observed a biphasic response in HSP90A expression (Fig. 3C), with increased expression at 1 d, which then decreased to 0 d values by 5 d and then increased again for 8 d to 21 d. This response likely represents the diverse functions of this protein during the unfolded protein response (UPR) (1 d–3 d, discussed below). At later time points (8 d–21 d), the observed increase in expression is likely associated with differentiation processes, as during myogenic cell culture we observed correlated gene expression with myoD1a (R = 0.83, P < 0.001, Fig. 5C) and myog (R = 0.88, P < 0.001, Fig. 5C).
Genes Associated With Cell Cycle Control and Myoblast Proliferation
Growth and regeneration of muscle requires the proliferation of myogenic progenitor cells from a pool of metabolically quiescent cells (46) in a process that is activated by exercise, muscle damage, and degenerative muscle diseases (21, 31, 59). There is evidence that fasting checks myogenic progenitors at the G1/S stage of the cell cycle in teleosts (17). Fasting and refeeding stimulates proliferation of fish myogenic cells in vivo (17) and in vitro (28). Two of the candidate genes had expression profiles characteristic of genes playing a role in the proliferation of myoblasts, or controlling entry into the cell cycle. Transcripts of FOXD1, a poorly characterized transcription factor, increased significantly 1 d after feeding, decreased to 0 d values by 8 d (Fig. 2C) and clustered with myoD1b (Fig. 1). Interestingly, during myogenic culture its expression was elevated at early time points as cells started to proliferate (Fig. 5B) but then decreased as cells differentiated (Figs. 5A, 7A). Expression during myogenesis clustered with myoD1b, myoD1c (Fig. 5C) and was highly correlated with Pax7 (R = 0.68, P < 0.001, Fig. 5C) and IGF-I (R = 0.67, P < 0.001, Fig. 5C), all of which are expressed in proliferating myoblasts (11, 27, 91). This, combined with its high expression largely restricted to fast muscle (Table 2), suggests a possible role for this transcription factor in myoblast proliferation. Interestingly, in T-cells, FOXD1 regulates FOXJ1, to suppress the NF-κ β-pathway (43), which is implicated in muscle wasting (19).
DRG1 expression was also increased at early time points during refeeding, with levels returning to 0 d values by 21 d (Fig. 3B); however, in myogenic culture, expression started to decrease after 5 d (Fig. 6A) and was correlated with IGF-II (R = 0.72, P < 0.001) (Fig. 5C). This expression pattern suggests that DRG1 is also expressed in proliferating cells, which increase in number during refeeding (17) and decrease in number as the cell culture progressed and differentiated cells predominated (Fig. 5A). DRG1 has been implicated in regulation of the cell cycle and may be important in growth and differentiation of cells (76).
As well as genes expressed during the proliferative and differentiation phases of myogenesis, we identified the leucine zipper family member CEBPD, which appears to be specifically expressed in quiescent cells. CEBPD expression is highly upregulated during G0 phase in growth-arrested mammary epithelial cells (92) and in human limbal stem cells, where it regulates the cell cycle and promotes self renewal of holo-clone forming cells (2). Although CEBPD is strongly upregulated during the differentiation of adipocytes in mammals (89), in salmon it was 21-fold downregulated with feeding (Fig. 4B), whereas adipoQ, a marker of adipocyte differentiation was upregulated (Fig. 1). CEBPD was also downregulated concomitant with myoblast proliferation in vitro (Figs. 5B, 6B). It is therefore likely that CEBPD is expressed in quiescent myogenic progenitor cells in teleosts. CEBPD is regulated by CREB through binding to CREB elements in the promoter of CEBPD (4), and we found a strong correlation between CREBA and CEBPD expression (R = 0.73, P < 0.001, Fig. 1).
Genes Associated With Protein Degradation Pathways
A microarray study involving rainbow trout (Oncorhynchus mykiss) revealed 1,000 transcripts that were overexpressed in fast muscle of fish fasted for 30 d and then downregulated after 4 d of feeding, with a preponderance of transcripts involved in ubiquitin-proteasomal and lysosomal protein degradation pathways and fatty acid oxidation (61). Similar changes were found in a comparison of gravid and sterile rainbow trout (69), and refeeding of fasted salmon also results in the downregulation of genes associated with proteasomal protein degradation pathways such as the E3 ubiquitin ligases MuRF1 and MAFbx (14). The present study identified genes that were strongly downregulated in refed fish and were highly correlated with the expression of MuRF1 and MAFbx. CTSL1, a cysteine protease involved in the lysosomal degradation of proteins, was strongly downregulated as soon as 1 d of feeding (Fig. 4E) and expression highly correlated with the E3 ubiquitin ligases MuRF1 and MAFbx (R = 0.89, P < 0.001 and R = 0.93, P < 0.001, respectively; Fig. 1). This expression suggests that during fasting, protein degradation occurs via both proteasomal and lysosomal systems.
RCon2 has homology to a novel protein with similarities to the vertebrate ankyrin repeat-containing protein with a suppressor of cytokine signaling box-2 (ASB2) gene. ASB2 interacts with Elongin BC complex and assembles with Cullin 5/Rbx1, forming an E3 ubiquitin complex (35). In myeloid leukemia cells, ASB2 responds to retinoic acid, inhibits growth, and promotes differentiation (35). ASB2 targets filamin B for proteasomal degradation, and during differentiation of C2C12 myoblasts its knockdown delays myoblast fusion and the expression of muscle contractile proteins (3). We observed that the high expression of ASB2 in fasted fish was strongly downregulated in response to feeding (Fig. 4D) and was restricted to skeletal muscle (Table 2). However, expression during myogenesis was below the limit of detection by qPCR, which, considering the high expression we observed in fish muscle (Cq values 22–28 cycles), suggests that this protein is expressed specifically in quiescent cells or expressed in another cell type other than myoblasts, similar to the restricted expression of ASB6 in adipocytes (86). The expression pattern we observe suggests that S. salar ASB2 is not involved in muscle differentiation but may play a role in growth inhibition, as has been demonstrated for other ASB family members (86, 66). Interestingly, RCon3 expression was highly correlated with ASB2 (R = 0.91, P < 0.001) (Fig. 5C), was also restricted to skeletal muscle (Table 2), and was expressed below the limits of detection during myogenesis. BLAST searches using this contig found no homology to known genes; however, further in silico analysis of this sequence revealed significant homology (91% identity) with ASB2, which, combined with its similar expression pattern, suggests that this sequence may be a paralog of ASB2 or a related family member.
Protein Chaperones Are Activated in Response to Feeding
During refeeding of fasted fish we observe increased expression of genes with homology to several heat shock proteins including DNAJA4, HSPA1B, and HSP90A (Figs. 3A, 2A, 3C) and a gene with homology to CHAC1 (Fig. 2B). The contigs corresponding to these genes were some of the largest in the EST collection with 31 and 100 ESTs corresponding to CHAC1 and HSP90A, respectively. The expression of these genes may be indicative of activation of the unfolded protein response (UPR) pathway, a stress response mechanism activated when endoplasmic reticulum (ER) stress occurs, due to the accumulation of unfolded proteins in the ER. Accumulation of unfolded proteins can occur when the amounts of newly synthesized proteins exceeds that of the protein folding capacity in the ER (57). This situation is likely to occur during the transition from a fasted state to that of satiation feeding where increased protein synthesis occurs through activation of the AKT/mTOR pathway. Stimulation of the UPR leads to the activation of several transcription factors, resulting in the expression of DNAJ, HSPA1B, HSP90, and CHAC1 (52). There are three different UPR response pathways, regulated by PERK, ATF-6, and IRE-1. Activation of the PERK pathway leads to phosphorylation of eIF2α, resulting in reduced translation (34). The ATF-6 pathway leads to the induction of ER chaperones involved in protein folding (52), and the IRE-1 pathway increases the production of ER-associated degradation factors involved in the degradation of unfolded proteins (33). Our observations suggest that two of these pathways are activated during refeeding of fasted fish, with the heat shock proteins characteristic of the ATF-6 pathway, and Chac1 indicative of PERK pathway activation. Thus, with refeeding, unfolded proteins are cleared from the ER through decreased rates of translation and increased folding capacity, ensuring the quality of the secreted proteins. This response appears to be short lived as levels of DNAJA4, HSPA1B, and CHAC1 were reduced by day 3 (Fig. 3A, 2A, 2B), presumably as levels of the ER chaperones have reached levels sufficient to cope with the increased flux through the ER. These data also highlight the increased production of secreted proteins that occurs in muscle during the refeeding response (61).
Genes of Unknown Identity
The identity of the majority of the contigs analyzed by qPCR could be assigned through homology based sequence searches; however, several contigs had no homology to known genes including FCon5, FCon8, FCon14, RCon3, and RCon8. By retrieving salmonid ESTs from dbEST (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html) (9) with homology to these unknown sequences, we were able to generate longer contigs giving far greater sequence coverage; however, the resulting contigs still had no homology to known genes and could represent novel genes, pseudogenes, or untranslated regions. Two of these contigs had expression patterns that make them interesting candidates as promyogenic factors. FCon5 expression was restricted to skeletal muscle and was highest in fast muscle (Table 2). During refeeding, expression was elevated at all time points (Fig. 3G), clustered with that of IGF-I and IGFBP-4 (Fig. 1), and was positively correlated with that of MRF4 in myogenic cell culture (R = 0.68, P < 0.001) (Fig. 5C). FCon8 had peak expression at 1–5 d that remained elevated at all time points following refeeding (Fig. 2D), was most abundant in fast muscle (Table 2), and was transiently increased at 5 d of myogenic culture (Fig. 6G). Bioinformatic analysis reveals that these contigs are restricted to salmonids and do not contain an open reading frame. Therefore, if they are part of a gene, they are likely to represent untranslated regions.
We have screened nutritionally regulated genes in vivo against their expression pattern in cell culture in an attempt to identify genes specifically involved in muscle growth. However, this approach has limitations since in vivo muscle is composed of many cell types including fibroblasts and adipocytes with known cross talk and involvement in muscle fiber growth (1). It is therefore not possible to exclude roles in muscle growth for the remaining genes without further investigation.
Conclusion
In summary, we have identified several genes that have expression patterns restricted to muscle, increased expression during refeeding, and, based on their expression in myotube maturation, may be involved in proliferation (FOXD1 and DRG1) and differentiation (SMYD1, RTN1, MID1IP1, HSP90A, and FCon5) phases of myogenesis and are thus excellent candidates as promyogenic genes. We have also identified genes that could be considered inhibitory to myogenesis, or expressed in quiescent cells (CEBPD, ASB2, RCon3, and HSP30). Most of these genes have human orthologs; however, we also identified genes that may be salmonid specific (FCon5 and FCon8) and could be important regulators of growth in Atlantic salmon, a species of indeterminate growth. Future analysis of these candidate genes using gene knockdown experiments in zebrafish could shed further light on their possible roles in myogenesis.
GRANTS
This work was supported by Biotechnology and Biological Research Council Grant BB/D015391/1.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
S. salar samples used for subtractive library production were kindly supplied by Dr. Richard Taylor (EWOS innovation, Dirdal, Norway). S. salar samples for the independent refeeding experiment and cell culture were provided by Landcatch (Lochgilphead, UK). The authors thank Dr. Daniel Macqueen for primers for Rpl13 and Rps29 and Luke Bower for help with fish maintenance.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Argiles JM, Lopez-Soriano J, Almendro V, Busquets S, Lopez-Soriano FJ. Cross-talk between skeletal muscle and adipose tissue: a link with obesity? Med Res Rev 25: 49–65, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Barbaro V, Testa A, Di Iorio E, Mavilio F, Pellegrini G, De Luca M. C/EBP delta regulates cell cycle and self-renewal of human limbal stem cells. J Cell Biol 177: 1037–1049, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bello NF, Lamsoul I, Heuze ML, Metais A, Moreaux G, Calderwood DA, Duprez D, Moog-Lutz C, Lutz PG. The E3 ubiquitin ligase specificity subunit ASB2 beta is a novel regulator of muscle differentiation that targets filamin B to proteasomal degradation. Cell Death Differ 16: 921–932, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmonte N, Phillips BW, Massiera F, Villageois P, Wdziekonski B, Saint-Marc P, Nichols J, Aubert J, Saeki K, Yuo A, Narumiya S, Ailhaud G, Dani C. Activation of extracellular signal-regulated kinases and CREB/ATF-1 mediate the expression of CCAAT/enhancer binding proteins beta and -delta in preadipocytes. Mol Endocrinol 15: 2037–2049, 2001. [DOI] [PubMed] [Google Scholar]
- 5.Berti C, Fontanella B, Ferrentino R, Meroni G. Mig12, a novel Opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co-operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol 5, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatnagar S, Kumar A, Makonchuk DY, Li H. Transforming growth factor-beta-activated kinase 1 Is an essential regulator of myogenic differentiation. J Biol Chem 285: 6401–6411, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluthgen N, Brand K, Cajavec B, Swat M, Herzel H, Beule D. Biological profiling of gene groups utilizing Gene Ontology. Genome Inform 16: 106–115, 2005. [PubMed] [Google Scholar]
- 8.Bodine SC, Latres E, Baumhueter S, Lai VKM, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na EQ, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001. [DOI] [PubMed] [Google Scholar]
- 9.Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST-Database for expressed sequence tags. Nat Genet 4: 332–333, 1993. [DOI] [PubMed] [Google Scholar]
- 10.Bower NI, Johnston IA. Selection of reference genes for expression studies with fish myogenic cell cultures. BMC Mol Biol 10: 80, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bower NI, Johnston IA. Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am J Physiol Regul Integr Comp Physiol 298: R1615–R1626, 2010. [DOI] [PubMed] [Google Scholar]
- 12.Bower NI, Johnston IA. Transcriptional regulation of the IGF signaling pathway by amino acids and insulin-like growth factors during myogenesis in Atlantic salmon. PLoS One 5: e11100, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bower NI, Li X, Taylor R, Johnston IA. Switching to fast growth: the insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J Exp Biol 211: 3859–3870, 2008. [DOI] [PubMed] [Google Scholar]
- 14.Bower NI, Taylor RG, Johnston IA. Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Front Zool 6: 18, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun T, Bober E, Winter B, Rosenthal N, Arnold HH. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J 9: 821–831, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun T, Buschhausendenker G, Bober E, Tannich E, Arnold HH. A novel human-muscle factor related to but distinct from Myod1 induces myogenic conversion in 10t1/2 Fibroblasts. EMBO J 8: 701–709, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodeur JC, Calvo J, Johnston IA. Proliferation of myogenic progenitor cells following feeding in the sub-antarctic notothenioid fish Harpagifer bispinis. J Exp Biol 206: 163–169, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622, 2009. [DOI] [PubMed] [Google Scholar]
- 19.Cai DS, Frantz JD, Tawa NE, Melendez PA, Oh BC, Lidov HGW, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKK beta/NF-kappa B activation causes severe muscle wasting in mice. Cell 119: 285–298, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 21: 1280–1281, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Charge SBP, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev 84: 209–238, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Chauvigne F, Gabillard JC, Weil C, Rescan PY. Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen Comp Endocrinol 132: 209–215, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676, 2005. [DOI] [PubMed] [Google Scholar]
- 24.Cornelison DDW, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol 191: 270–283, 1997. [DOI] [PubMed] [Google Scholar]
- 25.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000, 1987. [DOI] [PubMed] [Google Scholar]
- 26.Edmondson DG, Olson EN. A gene with homology to the Myc similarity region of Myod1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev 3: 628–640, 1989. [DOI] [PubMed] [Google Scholar]
- 27.Engert JC, Berglund EB, Rosenthal N. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J Cell Biol 135: 431–440, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fauconneau B, Paboeuf G. Effect of fasting and refeeding on in vitro muscle cell proliferation in rainbow trout (Oncorhynchus mykiss). Cell Tissue Res 301: 459–463, 2000. [DOI] [PubMed] [Google Scholar]
- 29.Gabillard JC, Kamangar BB, Montserrat N. Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss). J Endocrinol 191: 15–24, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31: 25–32, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Grounds MD, Yablonka-Reuveni Z. Molecular and cell biology of skeletal muscle regeneration. Mol Cell Biol Hum Dis Ser 3: 210–256, 1993. [DOI] [PubMed] [Google Scholar]
- 32.Gundersen GG, Khawaja S, Bulinski JC. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J Cell Biol 109: 2275–2288, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampton RY. ER stress response: getting the UPR hand on misfolded proteins. Curr Biol 10: R518–R521, 2000. [DOI] [PubMed] [Google Scholar]
- 34.Harding HP, Zhang YH, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274, 1999. [DOI] [PubMed] [Google Scholar]
- 35.Heuze ML, Guibal FC, Banks CA, Conaway JW, Conaway RC, Cayre YE, Benecke A, Lutz PG. ASB2 is an elongin BC-interacting protein that can assemble with cullin 5 and Rbx1 to reconstitute an E3 ubiquitin ligase complex. J Biol Chem 280: 5468–5474, 2005. [DOI] [PubMed] [Google Scholar]
- 36.Holterman CE, Le Grand F, Kuang S, Seale P, Rudnicki MA. Megf10 regulates the progression of the satellite cell myogenic program. J Cell Biol 179: 911–922, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang I, Gottlieb PD. The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J Immunol 158: 1165–1174, 1997. [PubMed] [Google Scholar]
- 38.Johnston IA. Environment and plasticity of myogenesis in teleost fish. J Exp Biol 209: 2249–2264, 2006. [DOI] [PubMed] [Google Scholar]
- 39.Johnston IA, Manthri S, Alderson R, Smart A, Campbell P, Nickell D, Robertson B, Paxton CGM, Burt ML. Freshwater environment affects growth rate and muscle fibre recruitment in seawater stages of Atlantic salmon (Salmo salar L.). J Exp Biol 206: 1337–1351, 2003. [DOI] [PubMed] [Google Scholar]
- 40.Johnston IA, Ward PS, Goldspink G. Studies on the swimming musculature of the rainbow trout I. Fibre types. J Fish Biol 7: 451–458, 1975. [Google Scholar]
- 41.Laffin JJS, Scheetz TE, Bonaldo MD, Reiter RS, Chang S, Eyestone M, Abdulkawy H, Brown B, Roberts C, Tack D, Kucaba T, Lin JJC, Sheffield VC, Casavant TL, Soares MB. A comprehensive nonredundant expressed sequence tag collection for the developing Rattus norvegicus heart. Physiol Genomics 17: 245–252, 2004. [DOI] [PubMed] [Google Scholar]
- 42.Lele Z, Hartson SD, Martin CC, Whitesell L, Matts RL, Krone PH. Disruption of zebrafish somite development by pharmacologic inhibition of Hsp90. Dev Biol 210: 56–70, 1999. [DOI] [PubMed] [Google Scholar]
- 43.Lin L, Peng SL. Coordination of NF-kappa B and NFAT antagonism by the forkhead transcription factor Foxd1. J Immunol 176: 4793–4803, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG, Glass C, Seto E, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol 18: 7176–7184, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik S, Huang CF, Schmidt J. The role of the canntg promoter element (E-Box) and the myocyte-enhancer-binding-factor-2 (Mef-2) site in the transcriptional regulation of the chick myogenin gene. Eur J Biochem 230: 88–96, 1995. [DOI] [PubMed] [Google Scholar]
- 46.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9: 493–495, 1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94: 12457–12461, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melino S, Nepravishta R, Bellomaria A, Di Marco S, Paci M. Nucleic acid binding of the RTN1-C C-terminal region: toward the functional role of a reticulon protein. Biochemistry 48: 242–253, 2009. [DOI] [PubMed] [Google Scholar]
- 49.Miner JH, Wold B. Herculin, a 4th member of the Myod family of myogenic regulatory genes. Proc Natl Acad Sci USA 87: 1089–1093, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83: 1125–1136, 1995. [DOI] [PubMed] [Google Scholar]
- 51.Mommsen TP. Paradigms of growth in fish. Comp Biochem Physiol B Biochem Mol Biol 129: 207–219, 2001. [DOI] [PubMed] [Google Scholar]
- 52.Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182: 466–476, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murre C, McCaw PS, Baltimore D. A new DNA-binding and dimerization motif in immunoglobulin enhancer binding, daughterless, Myod, and Myc proteins. Cell 56: 777–783, 1989. [DOI] [PubMed] [Google Scholar]
- 54.Musaro A, McCullagh KJA, Naya FJ, Olson EN, Rosenthal N. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400: 581–585, 1999. [DOI] [PubMed] [Google Scholar]
- 55.Nepravishta R, Bellomaria A, Polizio F, Paci M, Melino S. Reticulon RTN1-C-CT peptide: a potential nuclease and inhibitor of histone deacetylase enzymes. Biochemistry 49: 252–258. [DOI] [PubMed] [Google Scholar]
- 56.Nicieza AG, Metcalfe NB. Growth compensation in juvenile Atlantic salmon: Responses to depressed temperature and food availability. Ecology 78: 2385–2400, 1997. [Google Scholar]
- 57.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J 366: 585–594, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parkinson J, Anthony A, Wasmuth J, Schmid R, Hedley A, Blaxter M. PartiGene - constructing partial genomes. Bioinformatics 20: 1398–1404, 2004. [DOI] [PubMed] [Google Scholar]
- 59.Pavlath GK, Horsley V. Cell fusion in skeletal muscle–central role of NFATC2 in regulating muscle cell size. Cell Cycle 2: 420–423, 2003. [PubMed] [Google Scholar]
- 60.Rea S, Eisenhaber F, O'Carroll N, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599, 2000. [DOI] [PubMed] [Google Scholar]
- 61.Rescan PY, Montfort J, Ralliere C, Le Cam A, Esquerre D, Hugot K. Dynamic gene expression in fish muscle during recovery growth induced by a fasting-refeeding schedule. BMC Genomics 8:438, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhodes SJ, Konieczny SF. Identification of Mrf4 - a new member of the muscle regulatory factor gene family. Genes Dev 3: 2050–2061, 1989. [DOI] [PubMed] [Google Scholar]
- 63.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009–1013, 2001. [DOI] [PubMed] [Google Scholar]
- 64.Rowlerson A, Veggetti A. Cellular mechanisms of post-embryonic muscle growth in aquaculture species. In: Muscle Development and Growth. Fish Physiology Series, vol. 18, edited by Johnston IA. San Diego, CA: Academic Press, 2001. [Google Scholar]
- 65.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386, 2000. [DOI] [PubMed] [Google Scholar]
- 66.Rui LY, Yuan MS, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem 277: 42394–42398, 2002. [DOI] [PubMed] [Google Scholar]
- 67.Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sainz N, Rodriguez A, Catalan V, Becerril S, Ramirez B, Gomez-Ambrosi J, Fruhbeck G. Leptin administration favors muscle mass accretion by decreasing FoxO3a and increasing PGC-1 alpha in ob/ob Mice. PLoS One 4, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salem M, Kenney PB, Rexroad CE, 3rd, Yao J. Microarray gene expression analysis in atrophying rainbow trout muscle: a unique nonmammalian muscle degradation model. Physiol Genomics 28: 33–45, 2006. [DOI] [PubMed] [Google Scholar]
- 70.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell 102: 777–786, 2000. [DOI] [PubMed] [Google Scholar]
- 71.Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, Munoz-Cinoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab 7: 33–44, 2008. [DOI] [PubMed] [Google Scholar]
- 72.Shaknovich R, Shue GL, Kohtz DS. Conformational activation of a basic helix-loop-helix protein (Myod1) by the C-terminal region of murine Hsp90 (Hsp84). Mol Cell Biol 12: 5059–5068, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shefer G, Benayahu D. SVEP1 is a novel marker of activated pre-determined skeletal muscle satellite cells. Stem Cell Rev 6: 42–49, 2010. [DOI] [PubMed] [Google Scholar]
- 74.Sims RJ, Weihe EK, Zhu L, O'Malley S, Harriss JV, Gottlieb PD. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem 277: 26524–26529, 2002. [DOI] [PubMed] [Google Scholar]
- 75.Smith CKI, Janney MJ, Allen RE. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J Cell Physiol 159: 379–385, 1994. [DOI] [PubMed] [Google Scholar]
- 76.Song H, Kim S, Ko MS, Kim HJ, Heo JC, Lee HJ, Lee HS, Han IS, Kwack KB, Park JW. Overexpression of DRG2 increases G(2)/M phase cells and decreases sensitivity to nocodazole-induced apoptosis. J Biochem (Tokyo) 135: 331–335, 2004. [DOI] [PubMed] [Google Scholar]
- 77.Spencer JA, Eliazer S, Ilaria RL, Richardson JA, Olson EN. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J Cell Biol 150: 771–784, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steiner P, Kulangara K, Sarria JCF, Glauser L, Regazzi R, Hirling H. Reticulon 1-C/neuroendocrine-specific protein-C interacts with SNARE proteins. J Neurochem 89: 569–580, 2004. [DOI] [PubMed] [Google Scholar]
- 79.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents short article expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14: 395–403, 2004. [DOI] [PubMed] [Google Scholar]
- 80.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. SET domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276: 25309–25317, 2001. [DOI] [PubMed] [Google Scholar]
- 81.Tan XG, Rotllant J, Li HQ, DeDeyne P, Du SJ. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA 103: 2713–2718, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tipsmark CK. Identification of FXYD protein genes in a teleost: tissue-specific expression and response to salinity change. Am J Physiol Regul Integr Comp Physiol 294: R1367–R1378, 2008. [DOI] [PubMed] [Google Scholar]
- 83.Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J 17: 403–405, 2003. [DOI] [PubMed] [Google Scholar]
- 84.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weatherley AH, Gill HS, Lobo AF. Recruitment and maximal diameter of axial muscle-fibers in teleosts and their relationship to somatic growth and ultimate size. J Fish Biol 33: 851–859, 1988. [Google Scholar]
- 86.Wilcox A, Katsanakis KD, Bheda F, Pillay TS. Asb6, an adipocyte-specific ankyrin and SOCS box protein, interacts with APS to enable recruitment of elongins B and C to the insulin receptor signaling complex. J Biol Chem 279: 38881–38888, 2004. [DOI] [PubMed] [Google Scholar]
- 87.Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to Myod. Cell 56: 607–617, 1989. [DOI] [PubMed] [Google Scholar]
- 88.Wynne JW, O'Sullivan MG, Cook MT, Stone G, Nowak BF, Lovell DR, Elliott NG. Transcriptome analyses of amoebic gill disease-affected Atlantic salmon (Salmo salar) tissues reveal localized host gene suppression. Mar Biotechnol 10: 388–403, 2008. [DOI] [PubMed] [Google Scholar]
- 89.Yeh WC, Cao ZD, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by 3 members of the C/Ebp family of leucine-zipper proteins. Genes Dev 9: 168–181, 1995. [DOI] [PubMed] [Google Scholar]
- 90.Yun BG, Matts RL. Differential effects of Hsp90 inhibition on protein kinases regulating signal transduction pathways required for myoblast differentiation. Exp Cell Res 307: 212–223, 2005. [DOI] [PubMed] [Google Scholar]
- 91.Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119: 1824–1832, 2006. [DOI] [PubMed] [Google Scholar]
- 92.Zhang YJ, Sif S, DeWille J. The mouse C/EBP delta gene promoter is regulated by STAT3 and Sp1 transcriptional activators, chromatin remodeling and c-Myc repression. J Cell Biochem 102: 1256–1270, 2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.