Abstract
A new method is described for automatic detection of subtle morphological phenotypes in mouse embryos. Based on high-resolution magnetic resonance imaging scanning and nonlinear image alignment, this method is demonstrated by comparing the morphology of two inbred strains, C57BL/6J and 129Sv/S1ImJ, at 15.5 days postconception. Mouse embryo morphology was found to be highly amenable to this kind of analysis with very low levels (on average 110 μm) of residual anatomical variation within strains after linear differences in pose and scale are removed. Mapping of local size differences showed that C57BL/6J embryos were larger than 129Sv/S1ImJ embryos, although these differences were not uniformly distributed across the anatomy. Expressed in terms of organ volumes, heart and lung were larger in C57BL/6J embryos, while brain and liver were comparable in volume between strains. The positive relationship between organ size and embryo size was consistent for the two strains but differed by organ, with the brain and liver being the least variable. Together these findings suggest the power of this technique for detecting subtle phenotypic differences arising from mutated genes.
Keywords: mouse embryo, morphometry, magnetic resonance imaging, phenotype comparison
mice are the major model system for studying the genetic basis of mammalian development and disease processes.They have a short gestation and relatively low maintenance costs; moreover, a wealth of genetic techniques is available to produce genetically modified mice. These advantages, along with the high degree of homology between the mouse and human genomes (14), have motivated an international effort to annotate the completed genetic sequences with functional information (6). Projects such as the KnockOut Mouse Project (KOMP), the EUropean Conditional Mouse Mutagenesis Program (EUCOMM), North American Conditional Mouse Mutagenesis Project (NorCOMM), and the Collaborative Cross (7) are generating large numbers of mutated strains so as to establish gene function. However, for the value of these genetic resources to be fully realized, a comparable systematic approach is needed to assay the phenotype of each mutant. Arguments based on economy, the role of development in disease, and the prevalence of embryonic lethal mutations all point to embryonic phenotypes as the place to start such analysis.
Three-dimensional (3D) imaging of mouse embryos is one approach to systematic phenotypic characterization that has the potential to provide complete coverage with high sensitivity. The likelihood for any given mutation to produce a complex phenotype, the difficulty of predicting which body systems will be affected, and the prevalence of peripheral or unanticipated phenotypes among known genes all point to a whole organism survey as the appropriate starting point for assessing a novel mutation. Anatomy can be a sensitive indicator of dysfunction in underlying molecular processes that perturb the normal developmental trajectory (17). The interplay between molecular programming and physical constraints encodes much of the developmental history in the structure of an organism. For this reason, we sought to establish whether a computational assessment of whole embryo anatomy based on 3D magnetic resonance imaging (MRI) would allow identification of subtle phenotypic differences.
Gross phenotypes where organs are radically realigned, transposed, or absent are readily detected on MRI without computational techniques (20) and are not the focus this work. Subtle phenotypes are defined here as perturbations on the normal anatomy that are not readily detected by visual inspection. The computational approach that we applied is based on anatomical homology between different phenotypes such that the correspondence of every anatomical feature, whether displaced, scaled, or rotated, is identified in all specimens. This was implemented in an automated manner such that homologous correspondence between anatomical features was established by warping one 3D image to match another. Applied to a group of embryo images, this method summarizes the anatomical variation within the population and provides a quantitative method for comparisons of volumes, lengths, and other features of shapes such as curvature. In practice, anatomic homology is only an approximation. Variation between individuals can be thought of as some mix of true anatomical features, postural differences that could be mistaken as anatomical features, and meso- and microscopic-scale structural features with stochastic qualities. One of the goals of this work was to establish whether enough separation exists between these three concepts of variation as to allow for meaningful anatomical comparisons among embryos.
Here we present a comparison of embryo morphology from two common inbred mouse lines (C57BL/6J and 129S1/SvImJ) using group-wise image alignment (image registration) to quantitatively assess anatomical similarities and differences.
MATERIALS AND METHODS
Animals
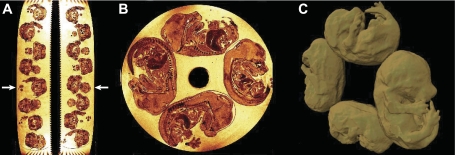
Sample preparation and image acquisition were carried out at the University of Oxford as previously described in Ref. 21. Animal procedures were approved by the Oxford University Local Ethical Review Panel and performed under UK Home Office Project Licenses PPL 30/2126 and PPL 30/2639. In brief, C57BL/6J and 129S1/SvImJ pregnant female mice were killed by cervical dislocation at 15.5 days postcoitum (dpc). After being harvested, the embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline at 4°C for ∼1 wk. The embryos were then embedded in 1% agarose gel containing 2 mM gadolinium-diethylenetriamine pentaacetic anhydride. A 28 mm MR tube was used to stack 32 embryos in eight layers of four embryos each (Fig. 1, A and B). To facilitate identification of orientations, the left forelimb was removed from all embryos before they were embedded in the MR tube. In addition, other limbs and/or tails were removed in a unique combination to allow for identification of each embryo in a given layer. All the embryos used in this study were male.
Fig. 1.
High-resolution magnetic resonance (MR) images of 32 mouse embryos. Embryos were stacked into 8 layers (A) with 4 embryos in each layer (B). The result of segmenting 4 embryos in 1 layer is shown in C as a surface rendering.
MRI
Briefly, a high spatial resolution MRI technique was employed to image the 32 embryos simultaneously (21). An 11.7 T (500 MHz) vertical magnet (Magnex Scientific, Oxon, UK) with a 3D spoiled gradient echo sequence (TE = 10 ms, flip angle = 90°) with a short repetition time (TR = 30 ms) was used to obtain strong T1 contrast. The total scan time was ∼12 h, and the voxel size after twofold zero filling of the dataset was 25.4 × 25.4 × 24.4 μm3 (acquisition matrix size: 512 × 512 × 1,024; field of view: 26 × 26 × 50 mm3).
Image Processing
Sample selection.
Images of 40 embryos, 20 of each strain, were selected arbitrarily for analysis and together comprised 10 central layers of four embryos each from two different sample tubes.
Preprocessing.
The intensity of the agar gel that was used as an embedding medium for the embryos had a bright contrast in the MR images that needed to be suppressed for proper function of the image registration algorithm. Prior to registration, the individual embryos within each tube were digitally outlined using semiautomatic purpose-written software. This delineation of the embryos in 3D was reviewed and edited where necessary with the software package Amira 4.1 (Visage Imaging, San Diego, CA) as shown in Fig. 1C. An automated version of this embryo segmentation has been reported elsewhere (1). Individual embryo images were corrected for intensity nonuniformity artifacts using the N3 algorithm (23).
Image registration.
Image registration finds a smooth spatial transformation that best aligns one image to another image such that corresponding anatomical features over the whole 3D images are superimposed. The deformation (i.e., local expansions, contractions, rotations, and translations) that bring the two into alignment becomes a summary of how the two embryo specimens differ spatially. We used an automated intensity-based group-wise registration approach (12) to align all specimens within each strain group of genetically identical embryos and produce a mean anatomy. The algorithm proceeds in a hierarchical manner (5) beginning with overall posture and finishing with the fine deformations to bring small anatomical features into alignment. This processing yields an unbiased average image and a set of deformations that relate the individual images to this average. Combining the individual embryo images as a simple arithmetic average after removing all of the identified anatomical differences is one means of representing the mean anatomy for the group. Total computation time for 20 embryos was 18 h on a cluster of AMD Opteron-based computers with one embryo allocated to each processor.
Evaluation of the registration accuracy.
Although the image registration algorithm has been previously used successfully to analyze adult mouse brains (2, 16, 24), we needed to verify its accuracy for MR images of mouse embryos and to provide a metric for tuning application-specific parameters of the algorithm. Heart volumes (excluding chambers) were manually outlined on six embryos from the 129S1/SvImJ strain. The quantitative fractional overlap between these volumes following alignment was assessed by the Jaccard index (22) as a measure of alignment accuracy.
Morphometric Analysis
The deformation fields obtained by image registration encode a detailed description of the anatomical differences between each embryo and the group average, including the displacements, pose, and relative scale of every anatomical feature. Here we summarize that information as the variation in location of anatomical features for each strain, the relative size of anatomical features in the two strains, and the volumes of specific organs.
Within-strain variation.
The positional variation of anatomical features was computed as a 3D map for each strain using the root mean square of the deformation magnitudes (RMSDM) (11). Expressed in micrometers, this scalar quantity is a measure of the average variation in the position of anatomical features with respect to the strain average and reflects the residual differences between individuals after overall linear differences such as embryo size have been removed. In addition to the 3D map, a mean variation was computed for each strain by averaging the RMSDM across all locations within the strain average embryo.
The mean variation of the two strains was compared, and the statistical significance of the difference assessed based on a nonparametric bootstrap analysis (3). The latter used 1,000 resamples and established a 95% confidence interval for each estimate of strain variation based on the percentile method (8).
Local size differences between strains.
The Jacobian determinant of the total linear and nonlinear transformation was computed for each embryo as a measure of whether the local anatomy was expanded or compressed with respect to the overall average anatomy (4). Jacobian values >1 correspond to local expansion and values <1 correspond to local compression. In practice this means reducing the deformation fields to a map of expansion/contraction factors that show for every voxel element of the average embryo the volume that element would occupy in the original image of the individual embryo. Mean differences in strain anatomy were assessed by subtracting the mean log Jacobian maps for the two strains after smoothing the deformation fields with a 500 μm Gaussian kernel. Statistical significance was assessed by a Student's t-test at every point with the threshold for significance adjusted for multiple comparisons by the false discovery rate (FDR) method (9). The smoothing improves the sensitivity for detecting local volume differences at the expense of not detecting features <500 μm.
Automated organ volume estimation.
The volumes of specific organs were computed for each embryo in both strains. The organs of interest were first manually outlined on the average embryo image for each strain using the Amira software package. Heart myocardium (excluding chambers), brain, liver, and lungs were outlined according to the Kaufman's atlas of mouse development (10), which is based on serially sectioned (C57BL × CBA) F1 hybrid mouse embryos. All of the labeled structures had clearly visible boundaries on both the MR images and the histological atlas. The volume of each organ with reference to the average was then computed automatically for each embryo by summing the Jacobian values within the organ volume defined on the average image (13).
An additional metric, crown-to-rump length (CRL), defined as the maximum distance from the cephalic pole to the caudal end (15), was measured manually from the images for each embryo.
RESULTS
Sample Yield
All 20 of the 129S1/SvImJ embryos and 18 of the 20 C57BL/6J embryos could be unambiguously distinguished from the mounting gel by the semiautomatic segmentation procedure. The remaining two C57BL/6J embryos had insufficient contrast at the boundary with the gel to make this distinction. Of the remaining 38 embryos, assessed visually, 37 were accurately aligned by the automated registration algorithm. The C57BL/6J embryo for which registration failed was uncharacteristically small, with a CRL of 12.4 mm (3 SDs below the mean) and did not satisfy the perturbational assumptions of the algorithm.
Qualitative Features of the Average Image
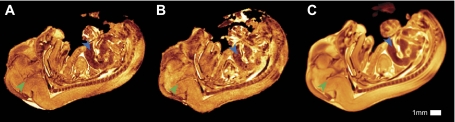
Two representative individual images of 129S1/SvImJ embryos are shown in Fig. 2 along with the average of 20 embryos. The averaging process tends to sharpen features in the image that are present in all specimens while blurring textural and stochastic features that vary between specimens (12, 18). The average images obtained from both strains demonstrate enhanced structural and anatomical definition compared with individual images. Compare for instance the 129S1/SvImJ embryos in Fig. 2, A and B, to the average in 2C. Following alignment, high-contrast features such as somites corresponded exactly for all specimens. Structures such as intestine, which have a high degree of variability among the embryos, show a reduction in the level of definition and were blurred in the average images.
Fig. 2.
Examples of 2 different individual embryos of the 129S1/SvImJ strain after rigid registration (A, B); final nonlinear group-wise average of 20 embryos (C). Consistently appearing features such as in the brain (green arrows) sharpen in the average image, whereas stochastic textural features such as in the liver (blue arrows) are blurred in the average.
Registration Accuracy
The overlap measure of the manually segmented myocardia of the six 129S1/SvImJ embryos was calculated at each stage of the hierarchical registration procedure. Overlap increased monotonically from 0.4 following linear alignment to 0.8 at the final (finest scale) nonlinear stage. Visually, the residual discrepancies corresponded to the rater's uncertainty of assigning particular volume elements on the organ boundary as being either inside or outside the organ.
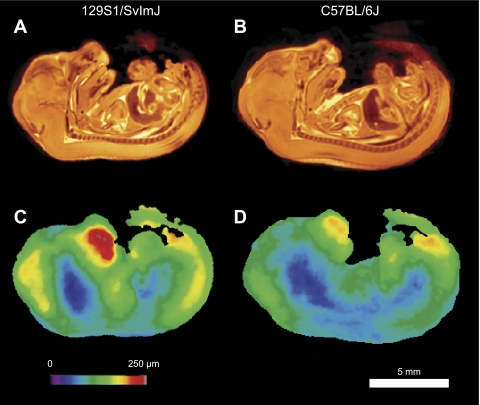
Within-Strain Variation
The within-strain variation maps are shown for the two strains in Fig. 3, C and D. Comparing these positional variation maps to the average images (Fig. 3, A and B) highlights anatomical regions with greater positional variability such as the nose and the somites adjacent to the tail. It can be observed that the regions of highest variability (shown in red) are those most subject to postural differences such as the nose. Internal regions of the embryos and in particular the spine show the lowest variability (shown in blue). The mean RMSDM (± SD) was 118 ± 33 μm in the 129S1/SvImJ strain and 100 ± 27 μm in the C57BL/6J strain. These values are remarkably small compared with the 13 mm CRL typical of a 15.5 dpc embryo. Bootstrap analysis did not provide evidence that variability differed between the two strains.
Fig. 3.
Strain average images for 129S1/SvImJ (A) and C57BL/6J (B). Root mean square of the deformation magnitudes (RMSDM) images for 129S1/SvImJ (C) and C57BL/6J (D).
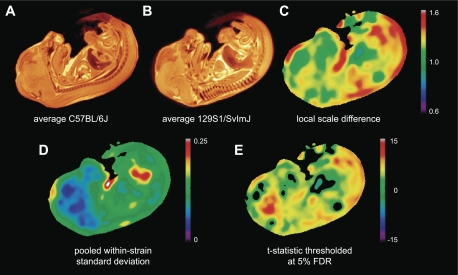
Local Size Differences Between Strains
Local size differences between the mean anatomy of the two strains are demonstrated by the Jacobian map shown in Fig. 4C. Values >1 (warm colors) indicate areas where the C57BL/6J strain is larger than the 129S1/SvImJ strains, whereas values <1 (cool colors) indicate the reverse. A Student's t-test at every voxel allowing for a 5% FDR showed significant local volume differences across ∼90% of the embryo (Fig. 4E). C57BL/6J embryos were in general larger than their 129S1/SvImJ counterparts although the relative increase in size of the C57BL/6J embryos compared with the 129S1/SvImJ embryos varied significantly from region to region. For example, greater expansion is seen in and around the heart and lungs than the brain and liver. Within-strain variation among specimens in the Jacobian maps as shown in Fig. 4D was lowest in the brain and largest around the intestines. The CRL of the C57BL/6J strain (13.6 ± 0.4 mm) was larger than that of the 129S1/SvImJ (12.5 ± 0.7 mm) (P < 0.001).
Fig. 4.
Final nonlinear average images of the C57BL/6J strain (A) and 129S1/SvImJ strain (B). C: local size difference expressed as relative volume between the 2 strains. Values >1 (warm colors) indicate regions where C57BL/6J embryos were larger than 129S1/SvImJ embryos, whereas values <1 (cool colors) indicate the reverse. D: within-strain SD of local volume differences among specimens pooled for the 2 strains. E: t-statistic map showing regions where the C57BL/6J strain was significantly larger (warm colors) or smaller (cool colors) than the 129S1/SvImJ strain. The threshold |t| > 2.06 corresponds to a 5% false discovery rate (FDR).
Comparison of Organ Volumes
Organ volumes (mean ± SD) for the two strains are reported in Table 1. The volumes of the heart and lungs were significantly larger (P < 0.001) in the C57BL/6J strain compared with the 129S1/SvImJ, whereas the volumes the brain and liver were comparable between strains. Covarying for CRL when comparing organ volumes reversed this pattern of significance but did not eliminate the differences between strains. The coefficient of variation defined as the ratio of the SD to the mean is provided as a normalized volumes variation in Table 1. The lungs were the most variable in both strains (16% in 129S1/SvImJ and 12% in C57BL/6J), whereas the brain was the least variable organ (7% in 129S1/SvImJ and 6% in C57BL/6J) and differed significantly (P < 0.001) in variability with respect to the lung.
Table 1.
Average volume and SD (in mm3) of selected organs for the two strains
| 129S1/SvImJ |
C57BL/6J |
|||
|---|---|---|---|---|
| Organ | Mean ± SD, mm3 | Coeff. Var., % | Mean ± SD, mm3 | Coeff. Var., % |
| Brain | 42.2 ± 3.0 | 7 | 41.8 ± 2.4 | 6 |
| Liver | 26.5 ± 3.5 | 13 | 28.6 ± 2.3 | 8 |
| Lungs | 6.4 ± 1.1 | 17 | 8.3 ± 1.0* | 13 |
| Myocardium | 2.1 ± 0.24 | 11 | 2.4 ± 0.20* | 8 |
Coefficient of variation (Coeff. Var.) is ratio of SD over mean.
Significant difference (P < 0.001) between the mean organ volumes of the 2 strains.
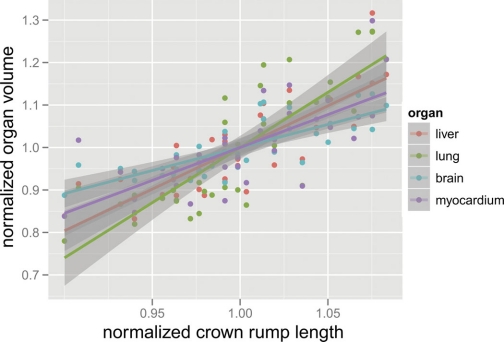
The relationship between organ volumes and embryo size is presented in Fig. 5. Computing an organ volume and a CRL normalized by the mean for the given strain allowed the relationship between organ volumes and embryo size to be compared for the different organs. As the regression slopes did not significantly differ between strains, all 37 embryos were pooled for this analysis following normalization by the mean organ volumes for each strain. Organ volumes were found to be strongly correlated with CRL (R2 > 0.52 for all organs, P < 0.0001). The regression slopes were 2.6 (lung), 2.0 (liver), 1.5 (heart), and 1.1 (brain), respectively. An analysis of variance in the regression slopes revealed significant differences (P = 0.002) between organs.
Fig. 5.
Scatter plot of normalized organ volume vs. normalized crown-to-rump length (n = 37) for each organ. Best-fit linear regression lines with 95% confidence intervals shaded are also plotted.
DISCUSSION
Here we describe the first use of a computational anatomy approach to detecting phenotypic differences between mouse embryos based on 3D MRI data. A group-wise nonlinear registration algorithm was employed to create an average embryo anatomy for two inbred strains (129S1/SvImJ and C57BL/6J), and the differences between the two were expressed quantitatively in terms of a number of summary statistics. The average anatomy for each strain was represented by averaging the individual embryo images after alignment. As can been seen from Fig. 3, A and B, the averaging process tended to enhance the definition of anatomical structures by cancelling image acquisition noise and suppressing textural differences in the images. These observations are consistent with an embryo anatomy that is highly conserved among individuals. Further evidence of this is provided by the maps of within-strain variation seen in Fig. 3, C and D. These maps, which show the residual interindividual variation after linear (overall scale) differences are removed, show remarkably small levels of variation, on average around a 110 μm displacement of a given anatomical landmark from the mean anatomy. Treating linear differences as nuisance parameters and nonlinear differences as anatomical variation is a useful distinction but not the only partitioning that one could choose. An alternate analysis might define a set of smooth nonlinear transformations accounting for postural differences as nuisance parameters as well and, thereby, remove the variation in the position of the nose seen in Fig. 3, C and D. Outside of the extremities, the low variation seen in the present study suggests that the benefit of this more complex approach would be modest. Within-strain variation was comparable for the two strains.
The pattern of increase from the middle outward of the variation maps seen in Fig. 3, C and D, likely reflects both geometry and anatomy. The removal of linear differences between embryos ensures alignment of their centers thereby reducing central variation; moreover, the greater range of postural variation afforded to the extremities likely accounts for the largest variation seen at the nose. Conversely, central location, strong image features, and the high contrast of the spine may explain the low variability in this region. However, the concordance between low anatomical variation and the central nervous system seen in these maps may also reflect the tightly controlled development of these structures. The brain showed low variation as measured by organ volume and by within-strain Jacobian variation maps (Fig. 4D).
The principal application of the described technology is detecting systematic differences between groups of embryos. The detection of subtle anatomical phenotypes is of interest both for identifying local effects of gene function and for the understanding of quantitative traits. The detection problem is somewhat different than that of computer-aided diagnosis in that the question is posed with respect to populations of embryos, and the requirement to avoid false negatives can be balanced against requirements for specificity. Here we have used local size differences expressed as a scale factor with respect to 1.0 as a measure of how the two strains differ. It is clear from the prevalence of warm colors on the map shown in Fig. 4 that C57BL/6J embryos are larger than their 129Sv/S1ImJ counterparts. However, not all regions were larger in the C57BL/6J strain and the increase in size varied between regions. For instance, part of the liver as seen in Fig. 4C is smaller in the C57BL/6J strain. An important feature of this analysis is that no a priori definition of anatomical structures or regions of interest is required. The whole embryo is examined, and the size and shape of the detected difference map are only constrained by the imposed smoothness, in this case a 500 μm smoothing filter, and not by the anatomical boundaries.
Absolute embryo size is one of the measures yielded by this analysis, reported here in terms of CRL. The measured CRL of C57BL/6J embryos of 13.6 ± 0.4 mm is within the bounds of 10.4 ± 1.6 mm at 14.5 dpc and 14.7 ± 1.4 mm at 16.5 dpc reported from in utero ultrasound measurements (25) and compares with ultrasound measurements for the CD-1 strain of 14.2 mm at 15.5 dpc (15). CRL for 129Sv/S1ImJ embryos in the present study at 15.5 dpc was 12.5 ± 0.7 mm. The agreement among these measurements suggests that overall size differences caused by fixation and sample preparation were modest if present at all.
Organ volumes are another measure obtained from this analysis. Here brain, heart, lung, and liver volumes were computed in an automated fashion using a single manually created template. Heart and lungs were larger in the C57BL/6J strain, whereas brain and liver volumes were comparable between strains. Noting the smaller size of the 129Sv/S1ImJ strain, it is interesting to note that mature 129Sv/S1ImJ mice have also been reported to have a larger brain/body weight ratio compared with the C57BL/6J strain (19). Relating organ size to embryo size also provides a means to explore variation within the population. We regressed organ volumes against CRL with both normalized with reference to strain means to examine organ scaling compared with the whole organism. While regression slopes were similar between strains, the slopes differed by organ. Only the lung approaches the 3-to-1 slope that would correspond to proportional scaling based on the dimensionality. The remaining organs had less volume variation than CRL variation would predict, and the brain had the least. We speculate that the differences in organ scaling could relate to organ growth rates, either because organ sizes are more variable when growing rapidly or because the half day uncertainty in the conception date of the embryo creates greater dispersion in the observed volumes of rapidly growing organs.
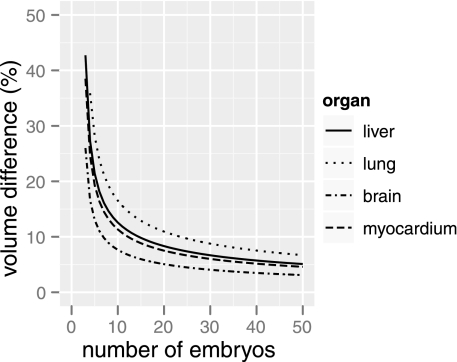
A power analysis based on the organ volumes suggests that relatively modest sample sizes could be used when comparing a test group of embryos to a large reference sample (see Fig. 6). For example, eight embryos would be sufficient to detect volume differences between 8% (in brain) and 20% (in lung). Small sample sizes such as this are of particular interest for screening applications where hundreds or thousands of phenotypes may need to be assessed.
Fig. 6.
Power analysis for detecting differences in organ volume (%) as a function of the number of 15.5 postcoitum embryos in the test sample. Curves are based on a 1-sample t-test with P < 0.05 and a study power of 0.9.
A limitation of the presented methodology is the assumption that anatomical differences represent a small perturbation. This is reflected in the algorithm's failure to align an unusually small embryo with the remaining specimens. This also has the implication that radical phenotypes such as transposition of the heart that can be readily detected visually (20) are not amenable to the present analysis. This is not of particular concern, however, because quantitative metrics are less meaningful when specimens are qualitatively different. In the present analysis both strains were at a comparable stage of development at 15.5 dpc and showed a high degree of anatomic homology. As a way to ensure consistency of specific features, one could consider other criteria besides gestational age to select embryos for comparison such as for example the number of somites or the Theiler stage (26). Embryos at other stages of development should also be amenable to this analysis technique provided that the differences between specimens are well modeled as an anatomical perturbation.
The main finding of the present work is that mouse embryos are readily amenable to a perturbational (deformation based) analysis. One of our goals was to assess the mix of true anatomical features, postural differences that could be mistaken as anatomical features, and meso- and microscopic-scale structural features with stochastic qualities. While in principle all three types of differences could be intractably overlapped in their extent and scale, the present analysis demonstrates that a window exists from which useful, interesting, and important anatomical differences can be drawn in an efficient and fully automated manner. This technical development has a wealth of applications for interrogating gene function including its use as a screening technology, in testing hypotheses on specific mutations, and as the basis of quantitative trait analysis. In all of these applications, the breadth and extent of quantitative anatomical information that can be obtained from a single assay (3D imaging) make the present approach attractive.
GRANTS
This work was funded by grants from Genome Canada, the Ontario Research Fund, the British Heart Foundation, and the Wellcome Trust.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank Angela Franklyn for technical help.
REFERENCES
- 1.Baghdadi L, Sled JG, Schneider JE, Henkelman RM, Lerch JP. Semi-automatic segmentation of mouse embryo MRIs. Int Soc Magn Reson Med: 1779, 2009 [Google Scholar]
- 2.Chen XJ, Kovacevic N, Lobaugh NJ, Sled JG, Henkelman RM, Henderson JT. Neuroanatomical differences between mouse strains as shown by high-resolution 3D MRI. Neuroimage 29: 99–105, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Chernick MR. Bootstrap Methods: A Guide for Practitioners and Researchers. Hoboken, NJ: Wiley-Interscience, 2008 [Google Scholar]
- 4.Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. Neuroimage 14: 595–606, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Collins DL, Evans AC. Animal: validation and applications of nonlinear registration-based segmentation. Int J Patt Recogn Artif Intell 11: 1271–1294, 1997 [Google Scholar]
- 6.Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell 128: 9–13, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Complex Trait Consortium The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36: 1133–1137, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman & Hall, 1993 [Google Scholar]
- 9.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15: 870–878, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Kaufman MH. The Atlas of Mouse Development. New York: Academic Press, 1992 [Google Scholar]
- 11.Kovacevic N, Chen J, Sled JG, Henderson J, Henkelman M. Deformation based representation of groupwise average and variability. Lecture Notes in Computer Science: 615–622, 2004 [Google Scholar]
- 12.Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, Chen XJ. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex 15: 639–645, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Maheswaran S, Barjat H, Bate ST, Aljabar P, Hill DL, Tilling L, Upton N, James MF, Hajnal JV, Rueckert D. Analysis of serial magnetic resonance images of mouse brains using image registration. Neuroimage 44: 692–700, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Mu J, Slevin JC, Qu D, McCormick S, Adamson SL. In vivo quantification of embryonic and placental growth during gestation in mice using micro-ultrasound. Reprod Biol Endocrinol 6: 34, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieman BJ, Flenniken AM, Adamson SL, Henkelman RM, Sled JG. Anatomical phenotyping in the brain and skull of a mutant mouse by magnetic resonance imaging and computed tomography. Physiol Genomics 24: 154–162, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Nieman BJ, Lerch JP, Bock NA, Chen XJ, Sled JG, Henkelman RM. Mouse behavioral mutants have neuroimaging abnormalities. Hum Brain Mapp 28: 567–575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbins S, Evans AC, Collins DL, Whitesides S. Tuning and comparing spatial normalization methods. Med Image Anal 8: 311–323, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Roderick TH, Wimer RE, Wimer CC, Schwartzkroin PA. Genetic and phenotypic variation in weight of brain and spinal cord between inbred strains of mice. Brain Res 64: 345–353, 1973 [DOI] [PubMed] [Google Scholar]
- 20.Schneider JE, Bhattacharya S. Making the mouse embryo transparent: identifying developmental malformations using magnetic resonance imaging. Birth Defects Res C Embryo Today 72: 241–249, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Schneider JE, Bose J, Bamforth SD, Gruber AD, Broadbent C, Clarke K, Neubauer S, Lengeling A, Bhattacharya S. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol 4: 16, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. Neuroimage 13: 856–876, 2001 [DOI] [PubMed] [Google Scholar]
- 23.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17: 87–97, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage 35: 1424–1433, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Spurney CF, Leatherbury L, Lo CW. High-frequency ultrasound database profiling growth, development, and cardiovascular function in C57BL/6J mouse fetuses. J Am Soc Echocardiogr 17: 893–900, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Theiler K. The House Mouse: Atlas of Embryonic Development. New York: Springer-Verlag, 1989 [Google Scholar]