Abstract
Background
Although the clinical manifestations of alcoholic liver disease are well described, little is known about the molecular basis for liver injury. Recent studies have indicated that chronic alcohol consumption leads to the lysine-hyperacetylation of several hepatic proteins, and this list is growing quickly.
Methods
To identify other hyperacetylated proteins in ethanol-fed livers, we chose a proteomics approach. Cytosolic and membrane proteins (excluding nuclei) were separated on 2D gels, transferred to PVDF and immunoblotted with antibodies specific for acetylated lysine residues. Hyperacetylated proteins were selected for trypsin digestion and mass spectrometric analysis.
Results
In all, 40 proteins were identified, 11 of which are known acetylated proteins. Remarkably, the vast majority of hyperacetylated membrane proteins were mitochondrial residents. Hyperacetylated cytosolic proteins ranged in function from metabolism to cytoskeletal support. Notably, 3 key anti-oxidant proteins were identified whose activities are impaired in ethanol-treated cells. We confirmed that the anti-oxidant enzyme, glutathione peroxidase 1, actin and cortactin are hyperacetylated in ethanol-treated livers.
Conclusions
Alcohol-induced hyperacetylation of multiple proteins may contribute to the development of liver injury. The abundance of acetylated mitochondrial proteins further suggests that this modification is important in regulating liver metabolism and when perturbed, may contribute to the progression of a variety of metabolic diseases.
Keywords: Lysine Acetylation, Ethanol, Liver Injury, Oxidative Stress, Cytoskeleton
The liver is the major site of ethanol metabolism, thus it is highly susceptible to alcohol-induced injury. In the hepatocyte, ethanol is converted to acetaldehyde by alcohol dehydrogenase (ADH). This highly reactive intermediate is further metabolized in the mitochondria to acetate by acetaldehyde dehydrogenase (ALDH). Alcohol is also metabolized by cytochrome P450 2E1 (CYP2E1). CYP2E1-mediated metabolism not only leads to the formation of acetaldehyde, but also to the formation of oxygen and hydroxyethyl radicals that in turn promote the formation of other reactive intermediates (Tuma and Casey, 2003). Many of these metabolites can readily and covalently modify proteins, DNA, and lipids (Brooks, 1997; Fraenkel-Conrat and Singer, 1988; Kenney, 1982, 1984; Ristow and Obe, 1978; Wehr et al., 1993). More recently, alcohol exposure has been shown to induce posttranslational protein modifications that are part of the normal repertoire including methylation, phosphorylation, and acetylation (Kannarkat et al., 2006; Lee and Shukla, 2007; Lieber et al., 2008; Pal-Bhadra et al., 2007; Park et al., 2003; Picklo, 2008; Shepard and Tuma, 2009; You et al., 2008). In particular, numerous proteins have been identified that are hyperacetylated upon ethanol exposure, and this list is expanding rapidly (Shepard and Tuma, 2009).
There are 2 forms of protein acetylation: the irreversible, co-translational N-terminal acetylation of α-amino groups and the reversible, post-translational modification that occurs on ε-amino groups of lysine residues (Kouzarides, 2000; Polevoda and Sherman, 2002). The latter modification is the result of the coordinated activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Kouzarides, 2000; Polevoda and Sherman, 2002). Currently, there are at least 17 known families of HATs and 4 families of HDACs. Although most known HAT and HDAC substrates reside in the nucleus, a recent proteomics survey identified a diverse set of almost 200 lysine-acetylated proteins, many of which are non-nuclear (Kim et al., 2006). Remarkably, this survey also revealed that more than 20% of mitochondrial proteins are lysine-acetylated (Kim et al., 2006).
To date, the lysine-acetylation of a handful of hepatic proteins has been reported to be induced by ethanol exposure (Shepard and Tuma, 2009). So far this list includes histone H3, tubulin, sterol response element binding protein-1c, p53, peroxisome proliferator-activated receptor γ coactivator α and acetyl CoA synthetase 2 (AceCS2) (Shepard and Tuma, 2009). A recent study also determined that numerous mitochondrial proteins (not yet identified) are hyperacetylated after ethanol exposure, and that the acetylation remained long after ethanol withdrawal (Picklo, 2008). With the growing number of known acetylated proteins and the large number of modifying enzymes, it is likely that numerous proteins are hyperacetylated in ethanol-exposed hepatocytes.
To identify other ethanol-induced hyperacetylated proteins, we immunoblotted liver samples from control and ethanol-fed rats with antibodies generated against acetylated lysine. In whole homogenates from ethanol-fed animals, a striking increase in lysine acetylation was detected. We also immunoblotted nuclear, cytosolic, and membrane fractions from control and ethanol-fed livers and determined that all fractions contained numerous hyperacetylated proteins. We further analyzed the cytosolic and membrane fractions by 2D electrophoresis and immunoblotting. Selected hyperacetylated proteins were identified by mass spectrometry. In all, 40 non-nuclear proteins were identified, half of which were from the cytosol and half from the membrane fraction. Remarkably, almost all of the hyperacetylated proteins in the latter fraction were from mitochondria and most were metabolic enzymes. Hyperacetylation of 2 identified proteins, glutathione peroxidase 1 (GPx-1) and actin, was confirmed by immunoprecipitations and ELISAs. Further analysis also revealed that alcohol induced the hyperacetylation of cortactin, a known acetylated actin binding protein. Thus, alcohol-induced hyperacetylation may be a key factor in the development of liver injury.
MATERIALS AND METHODS
Reagents and Antibodies
BSA, anti-tubulin, and anti-HRP-conjugated secondary antibodies were purchased from Sigma-Aldrich (St Louis, MO). Acetylated BSA was purchased from USB (Cleveland, OH) and the polyclonal acetylated lysine antibodies were from Cell Signaling Technology (Danvers, MA). The histone H3, acetylated histone H3 and GPx-1 antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The actin and cortactin antibodies were purchased from Abcam (Cambridge, MA) and Millipore (Billerica, MA), respectively. The CE9 antibodies were kindly provided by A. Hubbard (Johns Hopkins University School of Medicine, Baltimore, MD).
Ethanol Treatment
Male Wistar rats (Charles River Laboratories, Wilmington, MA) were pair-fed control and ethanol Lieber-DeCarli liquid diets for 5 weeks as described (Lieber and DeCarli, 1989). The nutritionally adequate Lieber-DeCarli control and ethanol liquid diets were purchased from Dyets, Inc (Bethlehem, PA). The ethanol-containing diet consisted of 18% protein, 35% fat, 11% carbohydrate, and 36% ethanol. In the control diet, ethanol was replaced isocalorically with carbohydrate such that both ethanol-fed and control rats ingested identical amounts of all nutrients except carbohydrates. At time of sacrifice, the livers were excised and frozen at −70°C.
Liver Fractionation
Liver was Dounce-homogenized in 20% (w/v) of 0.25 M sucrose containing 10 mM Tris and protease inhibitors (2 µg/ml each of leupeptin, antipain, PMSF, and benzamidine). Homogenates were centrifuged at 900 × g at 4°C for 5 minutes. The supernatant was centrifuged at 150,000 × g at 4°C for 60 minutes to prepare the cytosolic and total membrane fractions. The nuclear pellet was washed by resuspending to volume and centrifuged at 14,200 × g at 4°C for 10 minutes. Samples were mixed with 2X Laemmli sample buffer (Laemmli, 1970) and boiled for 3 minutes.
Western Blotting
Proteins were separated using SDS–PAGE, transferred to nitrocellulose and immunoblotted with antibodies specific to acetylated lysine (1:1000), acetylated histone H3 (1:1000), tubulin (1:7500), CE9 (1:10,000), GPx-1 (1:2000), actin (1:2500), or cortactin (1:2000). The acetylated lysine and acetylated histone H3 antibodies were diluted in PBS containing 1% (w/v) BSA and 0.1% (v/v) Tween 20 (PBS-BT) and incubated overnight at 4°C. Immunoreactivity was detected using enhanced chemiluminescence (PerkinElmer, Crofton, MD). The fold increase in acetylation of 10 selected immunoreactive species in whole homogenate samples or of acetylated histone H3 in nuclear fractions was determined by densitometric analysis. Histone H3 acetylation levels were normalized to total histone H3 levels. Eight sets of pair-fed animals were examined in the whole homogenate analysis.
The anti-tubulin, CE9, GPx-1, actin and cortactin antibodies were diluted in PBS containing 5% (w/v) milk and 0.1% (v/v) Tween 20 for 1 hour at RT and processed as described above. For the preabsorption assays, 0.4 µg of the anti-acetylated lysine antibodies were incubated with 1.6 mg acetylated BSA diluted in PBS-BT for 2 hours on ice as described (Kim and Shukla, 2006). The mixture was diluted in an additional 3 ml PBS-BT and incubated overnight at 4°C and processed for immunoblotting as described above.
2D Gel Electrophoresis
Protein concentrations were determined using BCA Reagent (Thermo Scientific, Rockford, IL). 2D electrophoresis was performed by Kendrick Labs, Inc. (Madison, WI) using the carrier ampholine method of isoelectric focusing (O’Farrell, 1975); 645 µg of cytosolic proteins or 360 µg of total membrane proteins were loaded on each gel. Isoelectric focusing was carried out in a glass tube of inner diameter 3.0 mm using 2.0% pH 3.5–10 ampholines (GE Healthcare, Piscataway, NJ) for 20,000 volt-hours. After equilibrium for 10 minutes in buffer “0” (10% glycerol, 50 mM dithiothreitol, 2.3% SDS and 62.5 mM Tris, pH 6.8), the tube gel was sealed to the top of a stacking gel overlaying a 10% acrylamide slab gel. SDS slab gel electrophoresis was performed and the gel was dried between sheets of cellophane paper. Duplicate gels were transferred onto PVDF and immunoblotted with the acetylated lysine antibodies (1:2000).
To determine the fold increase in acetylation of proteins in samples from ethanol-fed animals, the density of individual spots on both the gels and immunoblots were determined. Because not all proteins were resolved in 2 dimensions into discrete spots, there were many smeared regions on both the gels and immunoblots that were excluded from our analysis. Thus, the numbers represent only the resolved spots. The level of each of the selected immunoreactive spots was normalized to the relative protein level of its corresponding spot in the gel. Fold-increase in acetylation was calculated by comparing the control ratios to those from ethanol-treated samples.
MALDI-MS Analysis
Mass spectrometry was performed by the Protein Chemistry Core Facility at Columbia University (New York, NY). In general, gel spots were prepared for digestion by washing twice with 50 mM Tris, pH 8.5/30% acetonitrile. Gel pieces were subsequently dried in a Speed-Vac concentrator and digested with trypsin (Roche Molecular Biochemicals, Indianapolis, IN) in 25 mM Tris, pH 8.5. Tubes were placed in a heating block at 32°C and left overnight. Peptides were extracted with 50% acetonitrile/2% trifluoroacetic acid (TFA) and suspended in a matrix solution containing 10 mg/ml 4-hydroxy-α-cyanocinnamic acid and 50% acetonitrile/0.1% TFA. The dried sample was analyzed by MALDI-MS analysis (Applied Biosystems Voyager DE Pro Mass spectrometer in linear mode). The MALDI spectra were manually searched against the NCBI database for protein matches. Parameters used in the search were Database: NBCI, taxonomy: rattus, enzyme: trypsin. MOWSE scores were generated from the MS-Fit program of Protein Prospector, v 5.3.0 (USCF Mass Spectrometry Facility). Mascot scores (probability based MOWSE scores) and expect values were generated from the Mascot search program (http://www.matrixscience.com). Up to one missed tryptic cleavage was allowed and cysteine propionamidation and methionine oxidation were considered. The peptide mass tolerance was 0.5 Da.
Immunoprecipitations
Cytosolic liver fractions (30 µl) were diluted to 500 µl with RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris, pH 8.0) containing protease inhibitors (2 µg/ml each of leupeptin, antipain, PMSF, and benzamidine). Anti-GPx-1 antibodies (0.5 µg) were added and samples incubated overnight at 4°C on a rotating shaker. Prewashed protein G-agarose (30 to 60 µl of a 50% slurry) (Thermo Scientific) was added and incubated for an additional 2 to 4 hours at 4°C on a rotating shaker. Agarose was collected by centrifugation. Unbound fractions were made into gel samples by addition of 5X Laemmli sample buffer. The bound fractions were resuspendend in 10 µl of 1X Laemmli sample buffer. In general, 10 µl of each unbound fraction and the entire bound sample were loaded on the gels.
Two-Antibody Sandwich ELISA
Twenty micrograms of the indicated antibodies were diluted in PBS and added to ELISA microplate strip wells (Bio-Rad, Hercules, CA). The antibodies were allowed to adhere for 2 hours in a humidified chamber, and nonlabeled sites were blocked with 1% BSA for 1 hour. The wells were washed with PBS and incubated with ~100 µg liver lysate for 2 hours. The bound protein was probed with the anti-acetylated lysine antibodies diluted 1:500 in 1% BSA for 2 hours and detected with anti-rabbit HRP-conjugated secondary antibodies. HRP levels were detected with the 1-step ABTS HRP detection solution (Thermo Scientific) and absorbance was measured at 405 nm.
RESULTS
Ethanol Induces Global Hepatic Protein Hyperacetylation
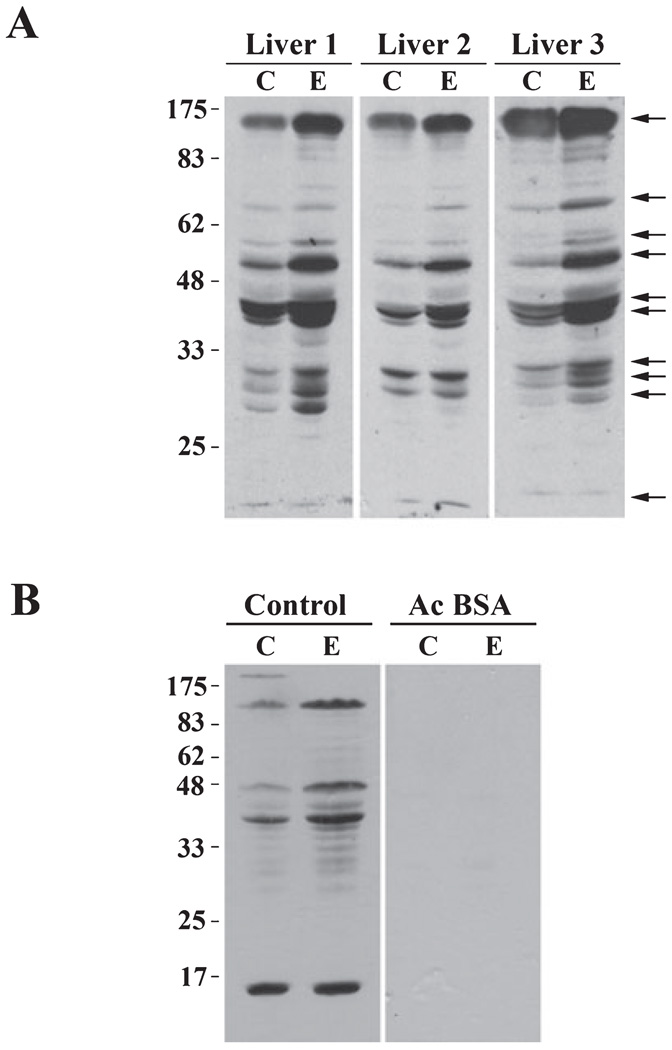
To determine whether other hepatic proteins are hyperacetylated by ethanol exposure, we began by immunoblotting whole homogenate samples from control or ethanol-fed livers with antibodies specific for acetylated lysine residues. Analysis was performed on 8 pair-fed liver sets from several different studies. In Fig. 1A, 3 representative pairs are shown. A striking increase in acetylation is apparent in the samples from ethanol-fed rats (Fig. 1A). In general, the same 10 proteins ranging in molecular weight from 17 to 175 kDa were more highly acetylated in ethanol-treated homogenates (Table 1). Among all samples examined, a cluster of 5 bands ranging from 30 to 50 kDa and a protein of 17 kDa were consistently hyperacetylated. For most proteins, acetylation was enhanced 2-to 3-fold, but in some cases, acetylation was increased to as much as ~14-fold (Table 1). To confirm the specificity of the anti-acetylated lysine antibodies, we preabsorbed them with 1% BSA in the absence or presence of 0.04% acetylated BSA (Fig. 1B). In the absence of the acetylated BSA, a similar pattern of hyperacetylation was observed in the ethanol-exposed samples (Fig. 1B). In contrast, addition of the acetylated BSA virtually abolished immunoreactivity (Fig. 1B).
Fig. 1.
Chronic ethanol treatment induces global hepatic protein hyperacetylation. (A) Livers from control (C) and ethanol (E) pair-fed rats were immunoblotted with the anti-acetylated lysine antibodies. Molecular weight standards are indicated on the left and arrows on the right indicate proteins with a 2-fold or greater increase in acetylation. Three representative pairs are shown. (B) The anti-acetylated lysine antibodies were preabsorbed in 1% BSA in the absence or presence of 0.04% acetylated BSA (Ac BSA) prior to immunoblotting the liver whole homogenate samples.
Table 1.
Increased Immunoreactivity Is Observed in a Conserved Set of Proteins in Livers From Ethanol-Fed Rats
| kDa | Fold increase | Subcellular Location |
|---|---|---|
| 175 | 2.9 ± 1.3 | N, C, M |
| 85 | 13.1 ± 11.1 | M |
| 62 | 5.5 ± 3.2 | N, M |
| 55 | 2.8 ± 1.0 | C |
| 50 | 2.0 ± 0.5 | N, C, M |
| 45 | 1.8 ± 0.3 | N, C, M |
| 40 | 2.7 ± 1.1 | N, C, M |
| 32 | 2.3 ± 0.8 | C |
| 30 | 1.6 ± 0.4 | M |
| 17 | 14.3 ± 9.9 | N |
The relative levels of 10 selected hyperacetylated proteins were determined by densitometric analysis of immunoreactive species of the indicated molecular weights. Values are averages ± SEM from 8 independent sets of pair-fed rats. The subcellular distribution of each of the species is also indicated.
N, nucleus; C, cytosol; M, membranes (without nuclei).
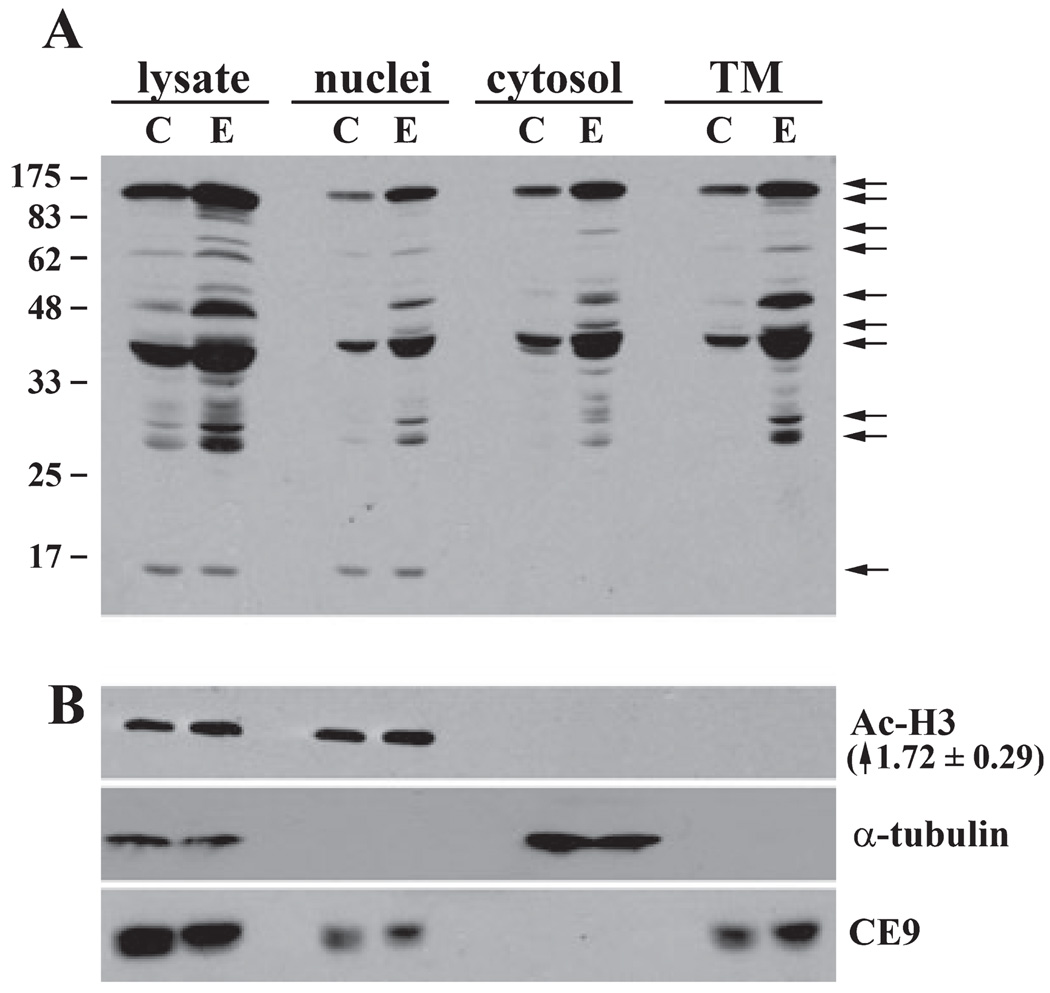
To further characterize the hyperacetylated proteins in the ethanol-fed rat livers, we prepared nuclear, cytosolic, and membrane fractions (excluding nuclei) by differential centrifugation (Fig. 2). As for the whole homogenates, multiple proteins were hyperacetylated in the various fractions. Although the 175 kDa protein equally distributed among all fractions, other proteins fractionated into distinct fractions. For example, 48, 62, and 85 kDa proteins were detected only in the total membrane fraction whereas a 30 to 35 kDa protein cluster distributedmainly to the cytosolic fraction (Fig. 2A).
Fig. 2.
Chronic ethanol treatment induces acetylation of nuclear, cytosolic, and membrane proteins. Liver homogenates from control (C) and ethanol (E) pair-fed rats were separated by differential centrifugation to prepare nuclei, cytosol, or total membranes (TM). (A) Fractions were immunoblotted with anti-acetylated lysine antibodies to detect hyperacetylated proteins (marked by arrows). Molecular weight standards are indicated on the left. (B) Fractions were immunoblotted for acetylated histone H3 (Ac-H3; a nuclear marker protein), tubulin (a cytosolic marker protein), and the basolateral resident protein, CE9 (a membrane marker protein) as indicated. The ethanol-induced increase in histone H3 acetylation is indicated in parentheses. The value is the average ± SEM from 3 independent sets of pair-fed animals.
To assess the purity of the 3 fractions, we immunoblotted them with acetylated histone H3 (a nuclear marker protein), tubulin (a cytosolic marker protein), and the basolateral resident protein, CE9 (a membrane marker protein). As shown in Fig. 2B, the marker proteins distributed to their corresponding fractions indicating their purity. Because the CE9 antibodies are extremely sensitive, we believe that the amount detected in the nuclear fraction likely reflects incomplete cell homogenization and sedimentation of intact cells at low speed. However, importantly, the cytosolic and membrane fractions are free of nuclei thereby allowing further analysis to identify non-nuclear hyperacetylated proteins in livers from ethanol-fed rats.
A low molecular weight protein was found exclusively in the nuclear fraction that was hyperacetylated in ethanol-treated samples. This low molecular weight and nuclear distribution suggested that it might be histone H3, a protein known to be hyperacetylated by ethanol exposure (Bardag-Gorce et al., 2007; Choudhury and Shukla, 2008; Kim and Shukla, 2005, 2006; Park et al., 2003, 2005). The acetylated histone H3 immunoblots confirmed this possibility (Fig. 2B) revealing an 1.72 ± 0.29-fold increase in acetylation in ethanol-fed samples.
Ethanol Induces Non-Nuclear Protein Acetylation
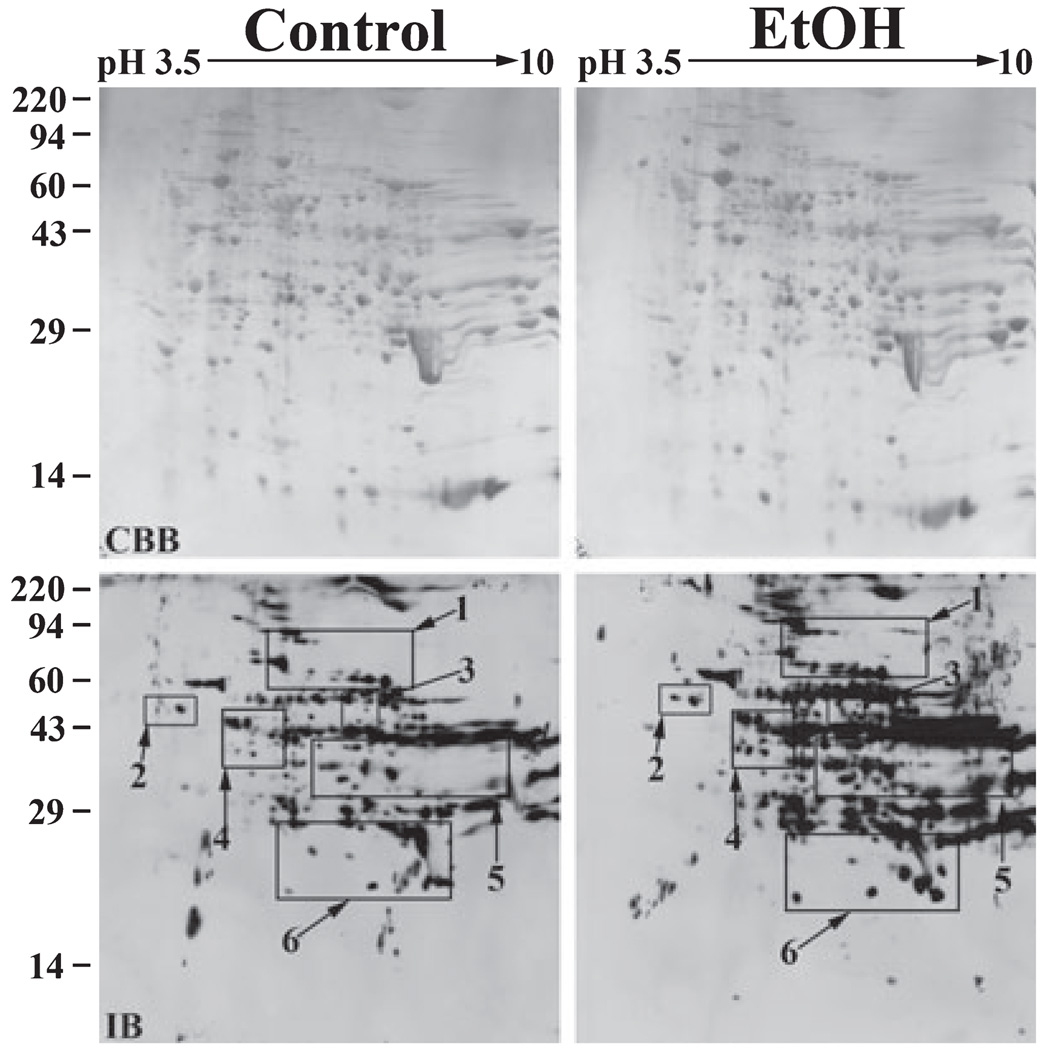
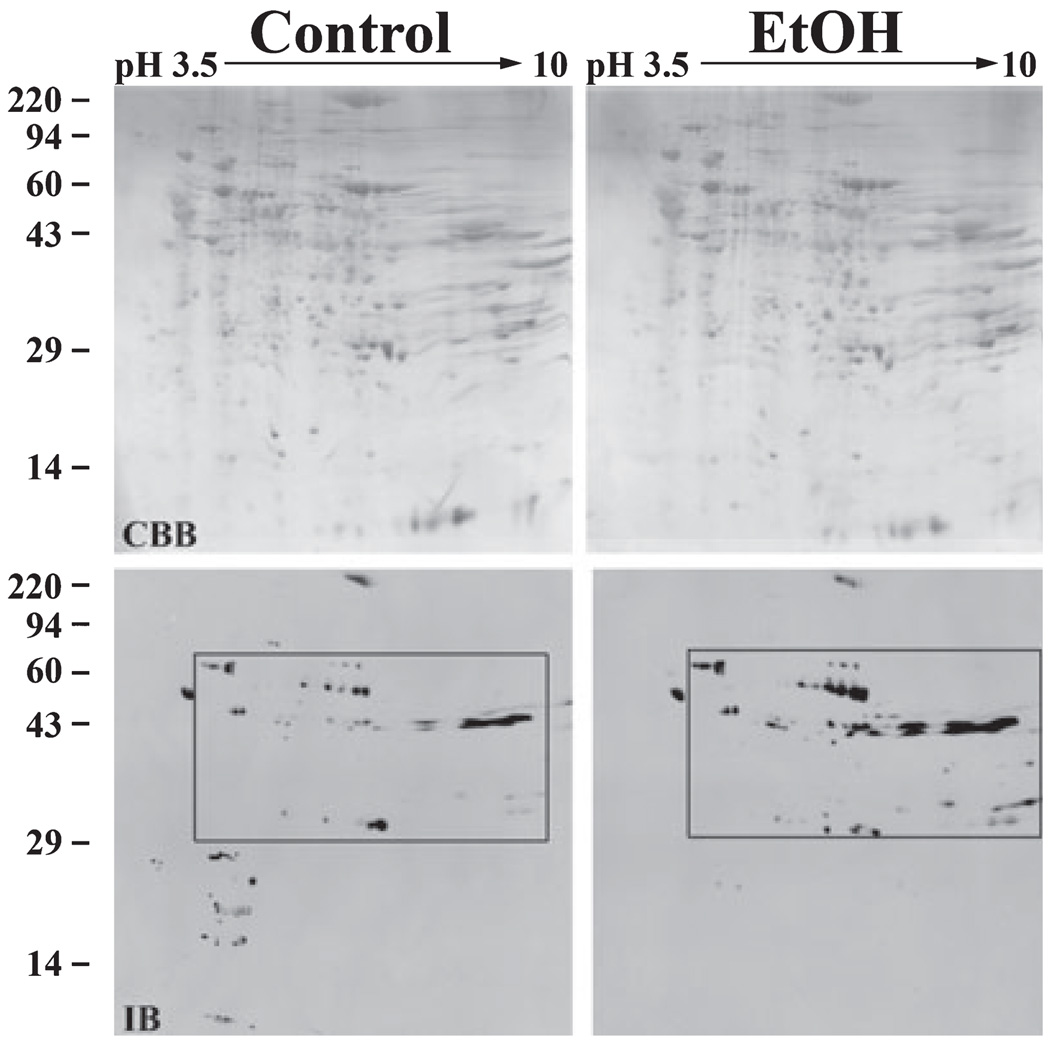
Because most known acetylated proteins are nuclear, we chose to further analyze cytosolic and total membrane fractions (excluding nuclei) to increase the likelihood of identifying novel acetylated proteins. We began by analyzing the cytosolic samples; 645 µg of total cytosolic protein from control or ethanol-treated livers was resolved on 2D gels and immunoblotted with the anti-acetylated lysine antibodies. Coomassie blue-stained gels revealed that the gels were equally loaded and displayed similar staining patterns (Fig. 3). In both the control and ethanol-treated gels, 426 discrete spots were resolved (Fig. 3). In the control gel, 191 acetylated spots were detected whereas 325 acetylated spots were detected in the gel loaded with the ethanol-treated sample indicating robust hyperacetylation (see Table 3).
Fig. 3.
Numerous cytosolic proteins are hyperacetylated in livers from ethanol-fed rats. Liver cytosolic extracts from control and ethanol pair-fed rats were prepared by differential centrifugation; 645 µg of total protein from each sample were subjected to 2D electrophoresis and immunoblotted with the anti-acetylated lysine antibodies. The pH gradient of the first dimension is indicated across the top and the molecular weight standards are indicated on the left. The Coomassie blue stained gels (CBB) are shown in the upper panels and the corresponding immunoblots (IB) are shown below. Regions of hyperacetylation are boxed and numbered. These boxes correspond with the regions of the blot enlarged in Fig. 4.
Table 3.
Most Alcohol-Induced Hyperacetylated Proteins Regulate Liver Metabolism
| Protein | Spot | Fold increase | Subcellular location | Function | Acetylated |
|---|---|---|---|---|---|
| δ-Aminolevulinic acid dehydratase | k | 14.1 | Cyto. | Porphyrin metabolism | |
| Adenosine kinase | i | 3.7 | Cyto. | Nucleotide metabolism | |
| β-Ureidopropionase | gg | 15.2 | Cyto. | Nucleotide metabolism | |
| 4-trimethylamino-butyraldehyde dehydrogenase | f | 23.1 | Cyto. | AA metabolism (NAD+) | |
| Argininosuccinate synthase | dd | 2.7 | Cyto. | AA metabolism | |
| 4-hydroxyphenyl pyruvate dioxygenase | aa | 6.5 | Cyto. | AA metabolism | |
| Glutamine synthetase | gg | 15.2 | Cyto. | AA metabolism | |
| 1-pyrroline-5-carboxylate dehydrogenase | b | 9.8 | Cyto. | AA metabolism (NAD+) | |
| Dihydropicolinate synthase | n | 8.8 | Cyto. | AA metabolism | |
| Cystathionine γ-lyase | ee | 4.5 | Cyto. | AA metabolism | |
| Glutathione peroxidase 1 | q | 8.1 | Cyto. | Oxidative stress | + |
| Glutathione S-transferase μ2 | r | 8.3 | Cyto. | Oxidative stress | |
| Superoxide dismutase | s | 2.9 | Cyto. | Oxidative stress | + |
| Fructose-1,6-bisphosphatase 1 | j | 28.6 | Cyto. | Gluconeogenesis | |
| Fructose-bisphosphate aldolase B | o | 5.6 | Cyto. | Glycolysis | + |
| p | 23.1 | ||||
| Transaldolase | k | 14.1 | Cyto. | Pentose phosphate shunt | |
| m | 6.2 | ||||
| Aconitate hydratase | d | 99.6 | Cyto. | TCA cycle | |
| β-Actin | i | 3.7 | Cyto. | Cytoskeleton | + (γ actin) |
| 26S protease regulatory Subunit S10 B | ff | 22.6 | Cyto. | Proteosomal degradation | |
| 60 kDa heat shock protein | t | 1.4 | Mito. | Chaperone | |
| u | 1.8 | ||||
| Peroxisomal δ3, δ2-enoyl CoA isomerase | p | 23.1 | Mito. | Lipid metabolism | + |
| Trans-2-enoyl-CoA reductase | o | 5.6 | Mito. | Lipid metabolism | |
| Acyl-CoA thioesterase 2 | aa | 3.6 | Mito. | Lipid metabolism | |
| bb | 4.5 | ||||
| cc | 6.5 | ||||
| Ethanolamine-phosphate cytidylyltransferase | h | 1.8 | Mito. | Lipid metabolism | |
| Medium-chain specific acyl-CoA | ee | 4.5 | Mito. | Lipid metabolism | |
| dehydrogenase | gg | 15.2 | |||
| Butyryl CoA synthetase 1 | a | 44.2 | Mito. | Lipid metabolism | |
| 3-oxo-5- β-steroid-4-dehydrogenase | m | 6.2 | Mito. | Lipid metabolism | |
| α-methylacyl-CoA racemase | l | 18.1 | Mito. | Lipid metabolism | |
| Enoyl-CoA hydratase | hh | 4.5 | Mito. | Lipid metabolism | |
| ii | 18.3 | ||||
| Hydroxyacyl-CoA dehydrogenase | jj | 6.2 | Mito. | Lipid metabolism (NAD+) | + |
| Hydroxymethylglutaryl-CoA lyase | jj | 6.2 | Mito. | Lipid metabolism (NAD+) | |
| 2,4-dienoyl-CoA reductase | kk | 13.8 | Mito. | Lipid metabolism | + |
| 3-ketoacyl-CoA thiolase | cc | 4.5 | Mito. | Lipid metabolism | |
| dd | 2.7 | ||||
| Acetyl-CoA acetyltransferase | ee | 4.5 | Mito. | Lipid metabolism AA metabolism | + |
| Ornithine transcarbamylase | m | 6.2 | Mito. | AA metabolism | + |
| Succinate-semialdehyde dehydrogenase | g | 7.2 | Mito. | AA metabolism (NAD+) | |
| Isovaleryl-CoA dehydrogenase | v | 2.2 | Mito. | AA metabolism | + |
| Glutamate dehydrogenase 1 | w | 7.0 | Mito. | AA metabolism (NAD+) | + |
| x | 27.0 | ||||
| y | 2.6 | ||||
| z | 2.5 | ||||
| Electron transfer flavoprotein subunit β | ii | 18.3 | Mito. | Electron transport chain | |
| ATP synthase β subunit | e | 4.6 | Mito. | ATP synthesis |
The identified hyperacetylated proteins listed in Table 2 were grouped according to function. To determine the fold increase in acetylation, the density of the individual spots on both the gels and immunoblots were determined. The level of each immunoreactive spot was normalized to the relative protein level of its corresponding spot in the gel. Fold-increase in acetylation was calculated by comparing the control ratios to those from ethanol-treated samples. The subcellular location of each protein is indicated. Proteins that are known to be acetylated at steady state are indicated with a “+”sign.
Cyto., cytosol; mito., mitochondria.
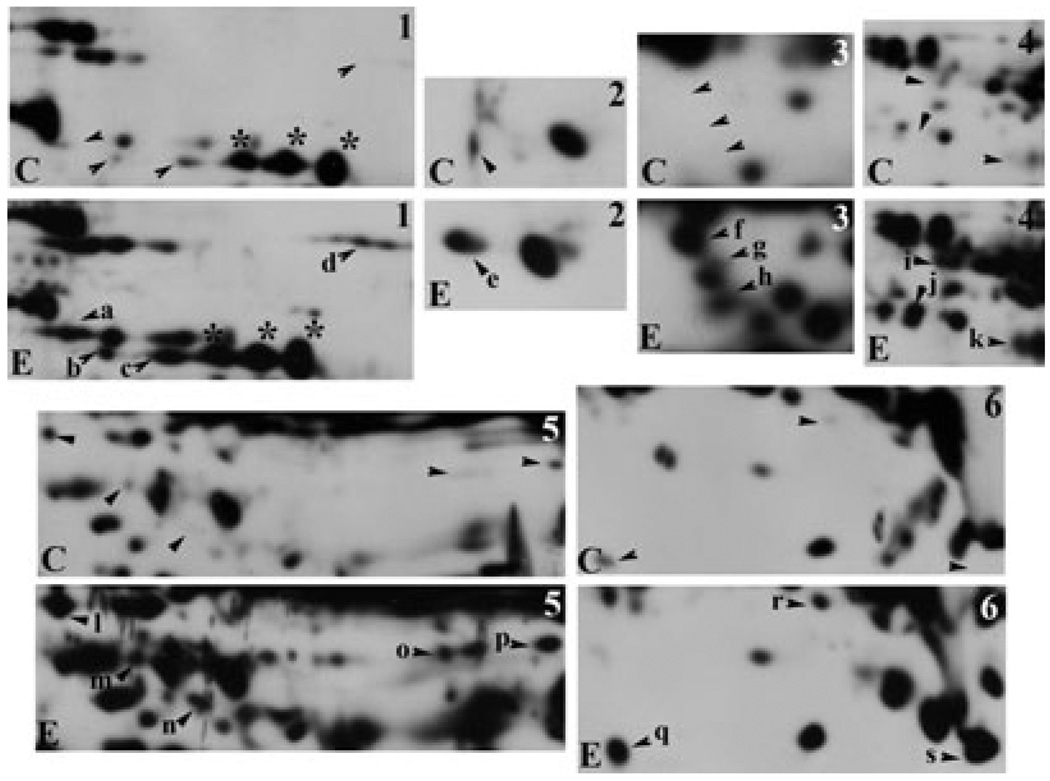
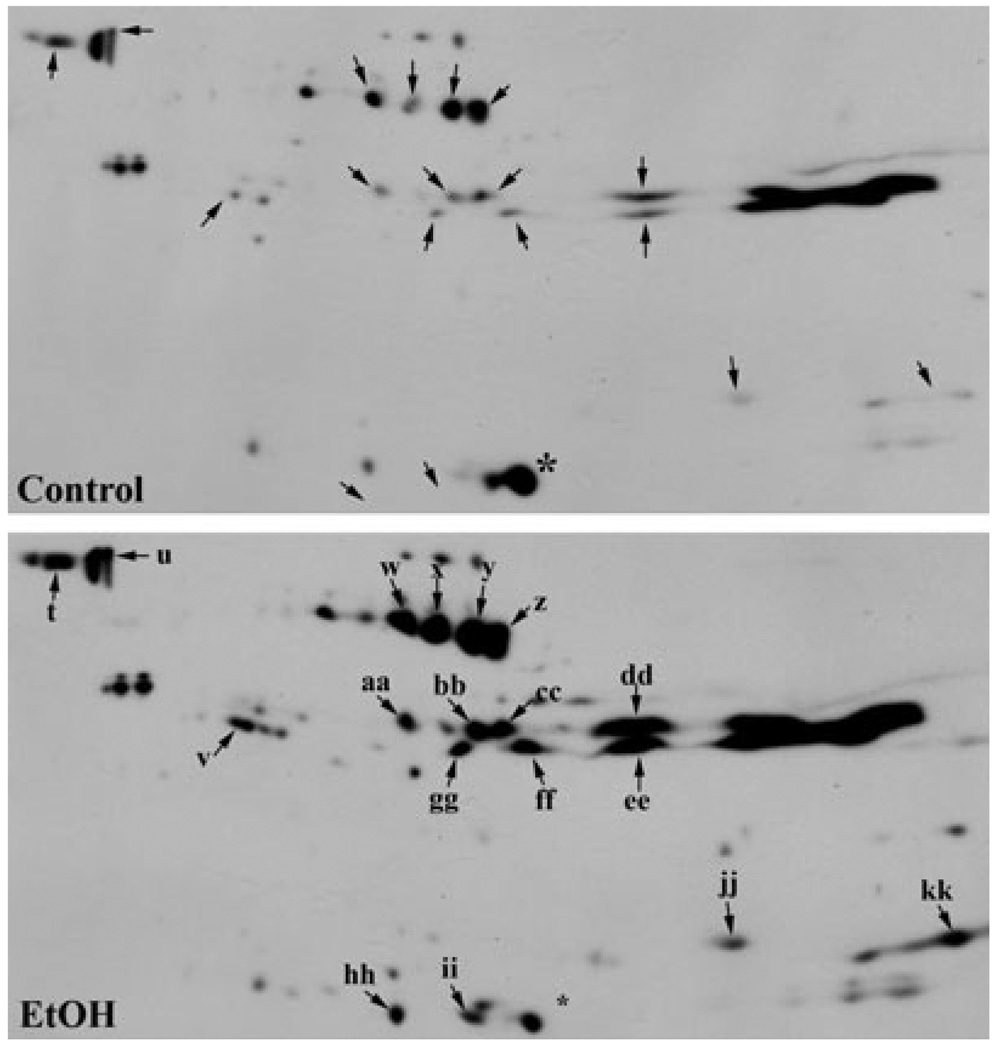
In general, cytosolic proteins ranging from 40 to 45 kDa with a pI of 7 to 10 exhibited substantial hyperacetylation (Fig. 3). A few distinct bands were also detected around 25 kDa. To better visualize individual hyperacetylated proteins, we enlarged portions of both the control and ethanol immunoblots (Fig. 4). In some cases, ethanol induced acetylation of proteins that were not detected in control blots (e.g., see spot a in Box 1). Many other proteins exhibited baseline acetylation levels that were either significantly increased in the presence of ethanol (e.g., see spot c in Box 1) or did not change (e.g., see the large immunoreactive species marked with asterisks in Box 1). The proteins exhibiting substantial hyperacetylation that were selected for MALDI-MS are indicated with arrowheads and are labeled (see Table 2). The corresponding spots in the control immunoblots are indicated with arrowheads.
Fig. 4.
Ethanol induces cytosolic protein hyperacetylation. Regions of hyperacetylation seen in the cytosolic 2D gels in Fig. 3 were boxed and enlarged. Control (C) and ethanol (E) samples were compared and hyperacetylated proteins were selected (marked with arrows) for mass spectrometric analysis. The labels on the blots from ethanol-treated samples correspond to the entries in Table 2. Asterisks are marking examples of acetylated proteins that were not changed by ethanol treatment.
Table 2.
Non-Nuclear Hyperacetylated Proteins Identified in Livers From Ethanol-Fed Rats
| Spot | Protein | NCBI accession | App MW | Calc MW | Calc PI | Peptides matched |
Sequence coverage (%) |
MOWSE score |
Mascot score |
Expect value |
|---|---|---|---|---|---|---|---|---|---|---|
| a | Butyryl CoA synthetase 1 | 65 | 65 | 7.6 | 16 | 44 | 1.32 × 1011 | 123 | 3.4 × 10−8 | |
| GI:197245828 | ||||||||||
| b | 1-pyrroline-5-carboxylate dehydrogenase | GI:149024431 | 67 | 62 | 8.3 | 12 | 32.6 | 1.05 × 107 | 70 | 6.4 × 10−3 |
| c | Keratin contamination | |||||||||
| d | Aconitate hydratase | GI:40538860 | 93 | 85 | 7.9 | 27 | 40 | 6.21 × 1014 | 170 | 6.9 × 10−13 |
| e | ATP synthase β subunit | GI:54792127 | 55 | 56 | 5.1 | 37 | 69.6 | 1.72 × 1018 | 267 | 1.1 × 10−22 |
| f | 4-trimethylamino-butyraldehyde dehydrogenase | GI:149058126 | 58 | 56 | 6.9 | 19 | 46.3 | 6.87 × 109 | 147 | 1.4 × 10−10 |
| g | Succinate-semialdehyde dehydrogenase | GI:182676407 | 53 | 56 | 8.4 | 19 | 46.1 | 3.06 × 1010 | 121 | 5.4 × 10−8 |
| h | Ethanolamine-phosphate cytidylyltransferase | GI:50925459 | 50 | 43 | 6.4 | 17 | 45.9 | 9.01 ×109 | 117 | 1.4 ×10−7 |
| i | Adenosine kinase | GI:149031258 | 43 | 38 | 5.8 | 15 | 43.8 | 7.18 × 106 | 101 | 5.4 × 10−6 |
| β-Actin | GI:4501885 | 43 | 42 | 5.3 | 9 | 28.8 | 4.07 × 104 | 43 | 3.4 | |
| j | Fructose-1,6-bisphosphatase 1 | GI:51036635 | 38 | 40 | 5.9 | 14 | 47.9 | 5.01 × 108 | 145 | 2.2 × 10−10 |
| k | δ-Aminolevulinic acid dehydratase | GI:6978483 | 36 | 36 | 6.3 | 17 | 54.5 | 5.80 × 108 | 126 | 1.7 × 10−8 |
| Transaldolase | GI:149061610 | 36 | 36 | 8.2 | 11 | 25.3 | 2.57 × 104 | 74 | 3.1 × 10−3 | |
| l | α-methylacyl-CoA racemase | GI:6981184 | 41 | 40 | 6.2 | 11 | 39.3 | 1.39 × 106 | 89 | 9.0 × 10−5 |
| m | 3-oxo-5-β-steroid-4-dehydrogenase | GI:20302063 | 36 | 37 | 6.2 | 14 | 45.4 | 4.65 × 109 | 62 | 3.9 × 10−2 |
| Transaldolase | GI:149061610 | 36 | 36 | 8.2 | 15 | 29.4 | 1.12 × 106 | 55 | 2.1 × 10−1 | |
| Ornithine transcarbamylase | GI:6981312 | 36 | 36 | 8.2 | 10 | 45.2 | 1.38 × 106 | 45 | 2.1 | |
| n | Dihydropicolinate synthase | GI:157822207 | 32 | 34 | 8.5 | 12 | 55.5 | 3.48 × 105 | 66 | 1.7 × 10−2 |
| o | Trans-2-enoyl-CoA reductase | GI:8393848 | 36 | 40 | 8.9 | 7 | 23.3 | 5.31 × 103 | ||
| Fructose-bisphosphate aldolase B | GI:158081751 | 36 | 40 | 8.7 | 15 | 50.8 | 1.03 × 107 | 92 | 3.9 × 10−5 | |
| p | Fructose-bisphosphate aldolase B | GI:158081751 | 38 | 40 | 8.7 | 8 | 30.1 | 1.92 × 104 | 72 | 4.9 × 10−3 |
| Peroxisomal δ 3, δ 2-enoyl CoA isomerase | GI:55741520 | 38 | 43 | 9.1 | 11 | 31.7 | 2.05 × 107 | 75 | 2.1 × 10−3 | |
| Peroxisomal δ3, δ 2-enoyl CoA isomerase | GI:55741520 | 38 | 43 | 9.1 | 11 | 31.7 | 2.05 × 107 | 75 | 2.1 × 10−3 | |
| q | Glutathione peroxidase 1 | GI:2654236 | 20 | 22 | 7.7 | 9 | 63.2 | 3.29 × 105 | 125 | 2.2 × 10−8 |
| r | Glutathione S-transferase μ2 | GI:62653546 | 27 | 26 | 6.9 | 16 | 55.0 | 1.32 × 109 | 143 | 3.4 × 10−10 |
| s | Superoxide dismutase | GI:8394331 | 24 | 25 | 9.0 | 5 | 33.3 | 9.07 × 103 | 58 | 1.1 × 10−1 |
| t | 60 kDa heat shock protein | GI:1334284 | 63 | 58 | 5.3 | 30 | 57.8 | 5.60 × 1017 | 220 | 6.9 × 10−18 |
| u | 60 kDa heat shock protein | GI:1334284 | 63 | 58 | 5.3 | 31 | 60 | 1.90 × 1018 | 219 | 8.6 × 10−18 |
| v | Isovaleryl-CoA dehydrogenase | GI:6981112 | 42 | 43 | 8.0 | 17 | 43.9 | 7.81 × 1010 | 105 | 2.2 × 10−6 |
| w | Glutamate dehydrogenase 1 | GI:6980956 | 55 | 61 | 8.1 | 17 | 39.6 | 2.49 × 108 | 119 | 8.6 × 10−8 |
| x | Glutamate dehydrogenase 1 | GI:6980956 | 55 | 61 | 8.1 | 25 | 50.0 | 3.78 × 1012 | 186 | 1.7 × 10−14 |
| y | Glutamate dehydrogenase 1 | GI:6980956 | 55 | 61 | 8.1 | 22 | 48.9 | 2.41 × 1012 | 170 | 6.9 × 10−13 |
| z | Glutamate dehydrogenase 1 | GI:6980956 | 55 | 61 | 8.1 | 27 | 51.1 | 1.75 × 1015 | 205 | 2.2 × 10−16 |
| aa | 4-hydroxyphenyl pyruvate dioxygenase | GI:8393557 | 42 | 45 | 6.3 | 13 | 42.2 | 5.52 × 108 | 94 | 2.7 × 10−5 |
| Acyl-CoA thioesterase 2 | GI:48675862 | 42 | 50 | 8.2 | 7 | 31.8 | 2.62 × 105 | 45 | 2.0 | |
| bb | Acyl-CoA thioesterase 2 | GI:48675862 | 42 | 50 | 8.2 | 13 | 39.5 | 2.76 × 105 | 86 | 1.6 × 10−4 |
| cc | Acyl-CoA thioesterase 2 | GI:48675862 | 42 | 50 | 8.2 | 9 | 33.8 | 2.18 × 104 | 60 | 6.2 × 10−2 |
| 3-ketoacyl-CoA thiolase | GI:149027156 | 42 | 50 | 8.2 | 13 | 46.3 | 8.50 × 108 | 77 | 1.3 × 10−3 | |
| dd | 3-ketoacyl-CoA thiolase | GI:149027156 | 42 | 50 | 8.2 | 9 | 35.6 | 1.47 × 107 | 60 | 6.2 × 10−2 |
| Argininosuccinate synthase | GI:25453414 | 42 | 46 | 7.6 | 13 | 41.5 | 2.67 × 109 | 75 | 2.4 × 10−3 | |
| ee | Cystathionine γ-lyase | GI:13699175 | 41 | 54 | 7.5 | 13 | 51.3 | 6.29 × 107 | 82 | 4.5 × 10−4 |
| Acetyl-CoA acetyltransferase | GI:135757 | 41 | 42 | 8.4 | 11 | 30.7 | 2.06 × 107 | 40 | 6.2 | |
| Medium-chain specific acyl-CoA dehydrogenase | GI:8392833 | 41 | 47 | 8.6 | 11 | 28.0 | 2.94 × 106 | 64 | 3.1 × 10−2 | |
| ff | 26S protease regulatory Subunit S10 B | GI:81294202 | 41 | 43 | 7.2 | 22 | 56.7 | 5.65 × 1012 | 113 | 3.4 × 10−7 |
| gg | β-Ureidopropionase | GI:16758704 | 41 | 44 | 6.5 | 9 | 33.8 | 5.16 × 105 | 45 | 2.3 |
| Glutamine synthetase | GI:142349612 | 41 | 42 | 6.6 | 11 | 33.5 | 2.29 × 107 | 49 | 8.4 × 10−1 | |
| Medium-chain specific acyl-CoA dehydrogenase | GI:8392833 | 41 | 47 | 8.6 | 13 | 39.9 | 3.75 × 106 | 75 | 2.0 × 10−3 | |
| hh | Enoyl-CoA hydratase | GI:17530977 | 30 | 32 | 8.4 | 15 | 50.0 | 4.52 × 106 | 120 | 6.9 × 10−8 |
| ii | Enoyl-CoA hydratase | GI:17530977 | 30 | 32 | 8.4 | 12 | 44.1 | 3.76 × 104 | 88 | 1.1 × 10−4 |
| Electron transfer flavoprotein subunit β | GI:51948412 | 30 | 28 | 7.6 | 6 | 25.5 | 2.84 × 104 | 28 | 1.1 × 102 | |
| jj | Hydroxyacyl-CoA dehydrogenase | GI:17105336 | 32 | 34 | 8.8 | 15 | 53.2 | 3.07 × 104 | 88 | 9.9 × 10−5 |
| Hydroxymethylglutaryl-CoA lyase | GI:13242293 | 32 | 34 | 8.7 | 11 | 39.4 | 1.60 × 107 | 58 | 1.1 × 10−1 | |
| kk | 2,4-dienoyl-CoA reductase | GI:67476443 | 32 | 36 | 9.1 | 15 | 38.5 | 2.47 × 1010 | 104 | 2.7 × 10−6 |
Although much less membrane protein was loaded (360 µg) than for the cytosolic samples, similar numbers of individual spots were resolved on both the control and ethanol gels (397 and 395 spots, respectively) (Fig. 5). Interestingly, the membrane samples displayed much less acetylation than the cytosolic samples, both in control (60 immunoreactive species) and in ethanol-treated (83 immunoreactive species) membranes. Nonetheless, robust hyperacetylation was observed in the ethanol-treated samples (see Table 3). In general, the hyperacetylated proteins were clustered in the middle of the gel ranging from 30 to 60 kDa with a pI of 5 to 9 (Fig. 5). When this region was enlarged, 18 hyperacetylated proteins were detected in the presence of ethanol and are marked with arrows (Fig. 6). Some proteins were significantly hyperacetylated (see spots w, x, y, and z) while others were not additionally modified (e.g., see the streak of proteins on the far right). Interestingly, ethanol also decreased acetylation of 2 proteins at 25 and 30 kDa. Only one was successfully identified as carbonic anhydrase 3 (Fig. 6, marked with an asterisk). The labeled spots were selected for MALDI-MS analysis (see Table 2). The corresponding spots in the control immunoblots are indicated with arrowheads.
Fig. 5.
Numerous membrane proteins are hyperacetylated in livers from ethanol-fed rats. Liver total membrane fractions from control and ethanol pair-fed rats were prepared by differential centrifugation; 360 µg of total protein from each sample was subjected to 2D electrophoresis and immunoblotted with the anti-acetylated lysine antibodies. The pH gradient of the first dimension is indicated across the top and the molecular weight standards are indicated on the left. The Coomassie blue stained gels (CBB) are shown in the upper panels and the corresponding immunoblots (IB) are shown below. The middle region of hyperacetylation is boxed and corresponds with the region enlarged in Fig. 6.
Fig. 6.
Ethanol induces total membrane protein hyperacetylation. The region of significant hyperacetylation found in Fig. 5 was boxed and enlarged. Control and ethanol samples were compared and hyperacetylated proteins were selected (marked with arrows) for mass spectrometric analysis. The labels on the blots from ethanol-treated samples correspond to the entries in Table 2. The asterisk indicates a hypoacetylated protein in ethanol-treated samples.
From the spots selected for mass spectrometric analysis, 40 proteins were positively identified. Table 2 provides a complete list of the selected proteins labeled in Figs. 4 and 6. Table 3 groups the identified proteins by subcellular location and function. Although our MALDI-MS approach used in these studies could not detect individual acetylated residues, 11 of these proteins were previously identified as acetylated proteins in a recent proteomic survey for acetylated residues (Kim et al., 2006) (Table 3, indicated with a “+”). Many of the cytosolic proteins were metabolic enzymes participating in amino acid metabolism, glycolysis, or gluconeogenesis. Of particular interest was the finding that GPx-1, glutathione S-transferase μ2 (GST μ2) and superoxide dismutase 1 (SOD1) were all hyperacetylated in ethanol-treated samples (see Discussion). We also identified β-actin, whose highly related γ-actin isoform is known to be acetylated (Kim et al., 2006). Remarkably, almost all of the hyperacetylated proteins in the membrane fraction were from mitochondria (Table 3). In general, these proteins fell into broad categories of lipid metabolism, amino acid metabolism and ATP synthesis indicating that these hyperacetylated proteins may be playing a role in the overall state of ethanol-induced mitochondrial dysfunction (see Discussion).
GPx-1 and Actin Hyperacetylation Are Confirmed
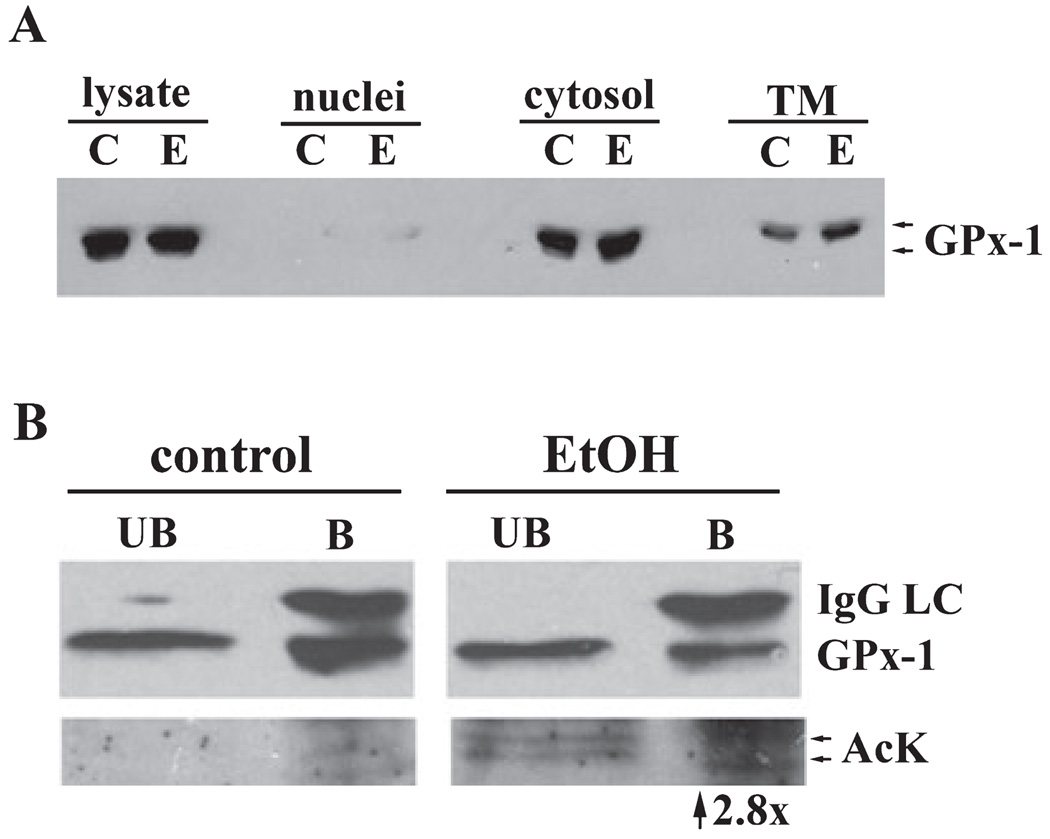
Considering that ethanol induces large changes in the redox state of the cell leading to oxidative stress and reactive oxygen species production, our identification of 3 hyperacetylated antioxidant proteins is intriguing. Since GPx-1 has previously been shown to be acetylated at steady state and the effects of ethanol on its activity are well characterized, we began our confirmation studies here. We first examined GPx-1 protein levels by immunoblotting control and ethanol liver fractions. A doublet at 23 kDa was observed for both the control and ethanol samples indicating that chronic ethanol consumption does not alter total or cytosolic GPx-1 levels, consistent with the literature (Bailey et al., 2001) (Fig. 7A). Importantly, the majority of GPx-1 was cytosolic further confirming the accuracy of our fractionation method. A small amount of GPx-1 was also detected in the total membrane population, which may reflect a population that is associated with the mitochondrial outer membrane (Bailey et al., 2001).
Fig. 7.
Alcohol-induced hyperacetylation of glutathione peroxidase 1. (A) Nuclear, cytosolic, and total membrane fractions were prepared from control (C) and ethanol-fed (E) rat livers. Fractions were immunoblotted for GPx-1. (B) Cytosolic fractions were immunoprecipitated for GPx-1 and both the unbound (UB) and bound (B) samples were blotted for GPx-1 (top) or acetylated lysine (AcK) (bottom). The IgG light chain (IgG LC) is detected in the bound fractions. Immunoreactive acetylated bands are detected in both the UB and B fractions from the ethanol-treated samples, but not in control.
To confirm that GPx-1 was hyperacetylated, we immunoprecipitated it from control and liver cytosols. The unbound and bound fractions were immunoblotted for GPx-1 (Fig. 7B, top panels) or acetylated lysine residues (Fig. 7B, bottom panels). In both the control and ethanol samples, we detected GPx-1 in the unbound and bound fractions indicating partial immunoprecipitation. The corresponding IgG light chain was only evident in the bound lanes indicating complete antibody recovery. In control immunoprecipitations, no immunoreactivity was detected in either the unbound or bound samples probed with anti-acetylated lysine antibodies. In contrast, a 23 kDa doublet was detected in both the unbound and bound ethanol-treated samples indicating that the enzyme was hyperacetylated.
We also confirmed ethanol-induced acetylation of GPx-1 using a two-antibody sandwich ELISA. Anti-GPx-1 antibodies were adhered to the wells of ELISA strip plates and then incubated with either control or ethanol cytosolic fractions. After incubation and subsequent washes, only the captured GPx-1 remained in the wells. The captured GPx-1 was then incubated with anti-acetylated lysine antibodies. HRP-conjugated secondary antibodies were added and binding was detected colorimectrically. From these assays we determined that GPx-1 from the ethanol-fed animals was hyperacetylated 2.8-fold more than from control samples (Fig. 7B).
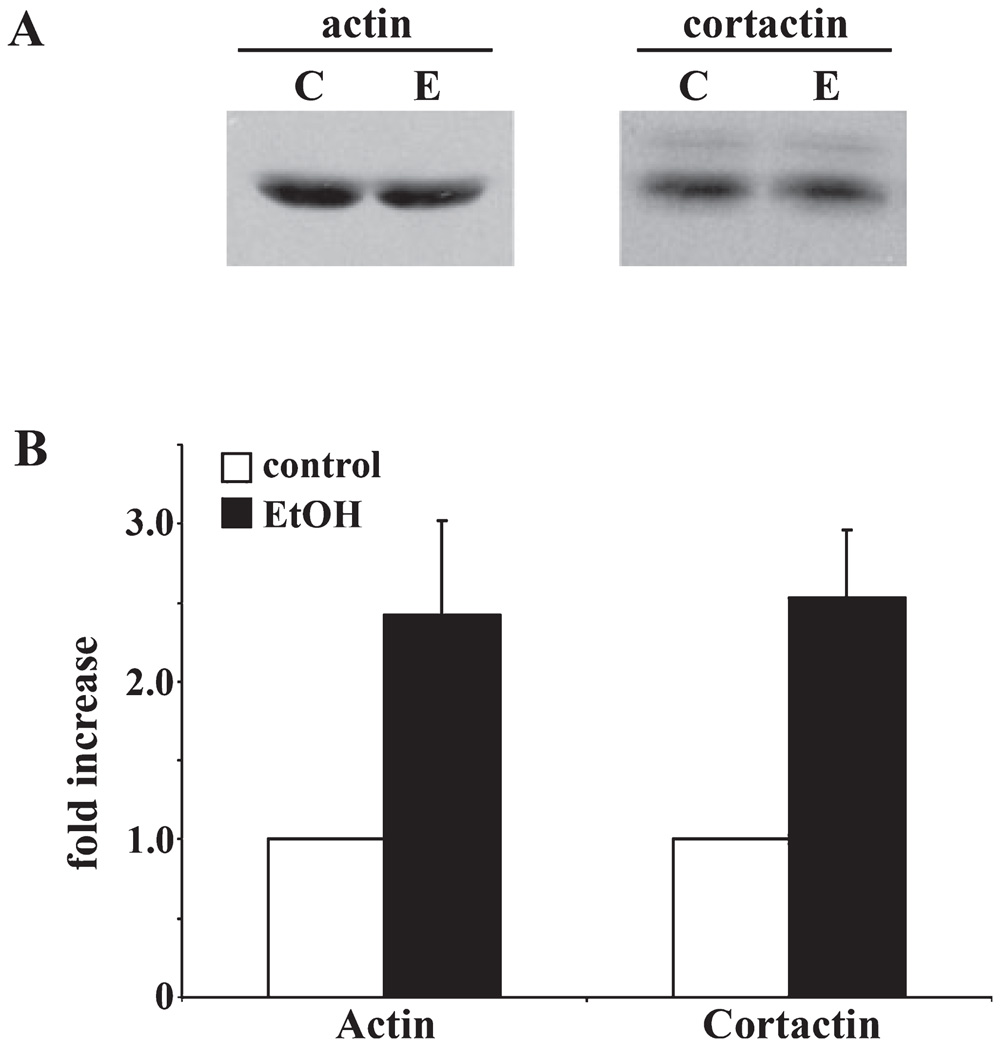
Previous work from our lab and others has found that ethanol impairs clathrin-mediated endocytosis, secretion and delivery of newly synthesized membrane proteins to the basolateral membrane (McVicker and Casey, 1999; Tuma and Sorrell, 1988; Tuma et al., 1990). In addition, studies using trichostatin A (a histone deacetylase inhibitor) have linked these impairments to increased protein acetylation (Joseph et al., 2008). Since both actin and its binding partner, cortactin, are likely required for clathrin-vesicle formation at the plasma membrane and TGN (Cao et al., 2003, 2005), an intriguing possibility is that actin hyperacetylation may contribute to the observed alcohol-induced defects in protein trafficking. Thus, we chose to confirm the acetylation of both actin and cortactin. We first immunoblotted liver whole homogenates for actin and cortactin protein expression levels. As observed for GPx-1, no changes were observed (Fig. 8A) indicating that hyperacetylation is not due to increased protein levels. To confirm hyperacetylation, we performed two-antibody sandwich ELISAs. Interestingly, acetylation of both actin and cortactin was increased to a similar extent as GPx-1. Actin acetylation was increased by 2.42 ± 0.60 whereas cortactin acetylation was enhanced 2.53 ± 0.43-fold in ethanol-treated samples (Fig. 8B).
Fig. 8.
Actin and cortactin are hyperacetylated in ethanol-treated liver cytosols. (A) Control (C) and ethanol (E) liver homogenates were immunoblotted for cortactin or actin as indicated. (B) Two-antibody sandwich ELISAs were performed to measure cortactin and actin acetylation in control or ethanol-treated samples. Twenty micrograms of each antibody was absorbed to wells, blocked, and lysates added. The captured antigen was further incubated with anti-acetylated lysine antibodies and detected with HRP-conjugated secondary antibodies. Absorbance was measured and hyperacetylation is plotted as a fold-increase over control. Values are expressed as the mean ± SEM from 3-independent experiments performed in duplicate.
DISCUSSION
A proteomics approach was used to identify cytosolic and membrane proteins that are hyperacetylated after chronic ethanol consumption. In all, we identified 40 non-nuclear proteins, half of which were from the cytosolic fraction and half from non-nuclear membranes. Remarkably, almost all of the hyperacetylated proteins in the latter fraction were from mitochondria and most were metabolic enzymes (Table 3). Similarly, cytosolic fractions were highly hyperacetylated after ethanol exposure and the proteins identified varied widely in function ranging from metabolic enzymes to proteins regulating oxidative stress to molecular chaperones. In order to confirm our proteomic results, we examined hyperacetylation of GPx-1 and actin directly. GPx-1 was found to be hyperacetylated by both immunoprecipitations and a two-antibody sandwich ELISA (2.8-fold). We also performed ELISAs to confirm actin hyperacetylation (2.42 ± 0.6-fold increase) and to establish ethanol’s impact on cortactin acetylation (2.53 ± 0.43-fold increase).
While our results provide compelling evidence for ethanol’s role in global hepatic acetylation, there are some limitations to the mass spectrometry method used. Because MALDI-MS is not optimized to detect post-translational modifications, acetylated residues could not be identified. However, 11 of these proteins were confirmed in a related study partially confirming our results (Kim et al., 2006). Clearly, the identification of the specific modified lysines will be required to not only confirm, but to determine the impact of such global hepatic protein acetylation.
Lysine Acetylation May Be a Regulator of Hepatic Protein Function
The reversibility of lysine acetylation and its presence on numerous proteins have led some to postulate that it might rival phosphorylation in its ability to regulate cellular processes (Kouzarides, 2000). In general, the added acetyl group likely neutralizes the lysine positive charge while increasing the size and hydrophobicity of the side chain. Such changes may result in protein conformational changes that alter function. Also, lysine acetylation sites have been identified that overlap with nuclear localization signals (Kim et al., 2006) such that the modification may induce altered protein subcellular distributions. In general, lysine acetylation has been shown to regulate protein stability, protein–protein interactions, protein-DNA interactions, and protein localization. Not only can lysines be acetylated, they can also be methylated, sumoylated, and ubiquitinylated such that ethanol-induced hyperacetylation may displace other modifications further altering protein function. In fact, p300 acetylation has been shown to prevent its sumoylation thereby repressing its activity (Bouras et al., 2005).
Not only is lysine acetylation is an emerging field, the understanding of ethanol’s role in protein acetylation is also in its infancy. Thus, the extent to which these modifications directly result from ethanol metabolism is not known. So far, the consequences of ethanol-induced protein acetylation have only been explored for a handful of proteins (Shepard and Tuma, 2009). From these studies, we can predict that ethanol-induced protein hyperacetylation greatly alters liver function ranging from metabolic processes to protein trafficking, some of which are more carefully considered below. Despite our predictions, mechanistic studies are clearly required to not only understand the functional consequences of acetylation in the normal liver, but also how alcohol-induced hyperacetylation alters hepatic function in the alcoholic liver.
Possible Mechanisms for Global Protein Acetylation
Currently, little is known about the mechanisms by which ethanol induces global non-nuclear hyperacetylation. Because acetylation is mediated by both HATs and HDACs, it is likely that these modifying enzymes are themselves altered leading to the observed hyperacetylation (Shepard and Tuma, 2009). Since most of these enzymes reside in the nucleus, attention is being turned to those that are exclusively cytosolic (HDAC6 and SirT2) and mitochondrial (SirT3-5) or shuttle between the nucleus and cytoplasm (PCAF, TIP60, and HDACs 5, 7, 9, and 10) (Shepard and Tuma, 2009). For mitochondrial proteins, only protein levels of SirT3 and 5 have been examined. Although SirT3 is considered the predominant mitochondrial deacetylase, its expression levels were not changed in rat livers from ethanol-fed rats (Picklo, 2008). In contrast, SirT5 protein levels were significantly decreased, but the specific sirtuin activity has not yet been addressed (Picklo, 2008).
We recently determined that the cytosolic HDAC activity in hepatocytes is exclusively HDAC6 (Shepard et al., 2008). While its activity was not changed in the presence of ethanol in hepatic WIF-B cells, we observed that both its protein levels and microtubule-association were significantly decreased (Shepard et al., 2008). Therefore, an exciting possibility is that HDCAC6 (or other cytosolic enzymes) exhibit altered binding to the identified proteins leading to hyperacetylation. Not only will studies on our identified proteins likely provide a better understanding of alcohol-induced protein acetylation and the progression of hepatotoxicity, they will undoubtedly identify new substrates for these well-established modifying enzymes.
Acetylation and Mitochondrial Dysfunction
The results presented here and results from others have shown that numerous mitochondrial proteins are hyperacetylated in the presence of ethanol (Picklo, 2008). However, little is known about the functional consequences of this global mitochondrial protein acetylation. To date, the effects of acetylation on mitochondrial activity have been examined only on glutamate dehydrogenase and AceCS2. In both cases, increased acetylation correlated with decreased activity (Schwer et al., 2006). Thus, the simple prediction is that acetylation functions as an on/off switch for these and other mitochondrial metabolic enzymes such that alcohol-induced changes in this modification alter hepatic metabolism.
Alternatively, it has been recently hypothesized that mitochondrial protein acetylation functions as a sensor for the overall energy status of the cell (Kim et al., 2006). According to this hypothesis, acetyl-CoA and NAD+ levels are the key indicators of energy status. This hypothesis stems from 2 observations. First, is that over 44% of mitochondrial dehydrogenases that require NAD+ for activity are known to be acetylated (Kim et al., 2006). Second, is that acetyl-CoA and NAD+ are cofactors for HATs and a subset of HDACs, respectively. Thus, one possibility is that lysine acetylation serves as a feedback mechanism for the regulation of dehydrogenases. For example, when cellular energy status is high (reflected in low NAD+ levels), the subset of HDACs are less active resulting in higher protein acetylation and dehydrogenase activities. In contrast, when acetyl-CoA levels are limiting (the energy status is low), HATs are inactivated leading to decreased protein acetylation and increased dehydrogenase activities. Thus, the simple prediction in the alcoholic liver where NAD+ is depleting, is that increased acetylation leads to impaired dehydrogenase activity and by extension, impaired mitochondrial function. Interestingly, our proteomics survey identified 4 mitochondrial enzymes that require NAD+ as a cofactor (Table 3). Although NAD+ levels may recover after prolonged ethanol exposure, the finding that hyperacetylation remains long after chronic ethanol withdrawal (Picklo, 2008) suggests that this mechanism may have physiologic relevance. Clearly, this exciting hypothesis needs to be rigorously tested.
Acetylation and Oxidative Stress
It is well-established that chronic alcohol consumption leads to increased hepatic oxidative stress (reviewed in Wu and Cederbaum, 2003). Not only are reactive oxygen species produced by CYP2E1-mediated ethanol metabolism, they are also produced as a result of alcohol-induced mitochondria dysfunction. Many of these and other reactive species can form covalent modifications with cellular proteins, lipids and DNA that can in turn, lead to hepatic dysfunction and disease (Tuma and Casey, 2003; Wu and Cederbaum, 2003). Alcohol consumption also induces overexpression of CYP2E1 while inhibiting the expression or activities of protective, antioxidant enzymes thereby reinforcing the vicious cycle. Thus, our findings that 3 key antioxidant enzymes (GPx-1, GSTμ2, and SOD1) are hyperacetylated in livers from ethanol-fed rats are particularly interesting.
The activities of many antioxidant enzymes are known to be impaired in ethanol-treated cells, including GPx-1and superoxide dismutaste (Bailey et al., 2001; Farbiszewski et al., 1991; Mari and Cederbaum, 2000; Oh et al., 1998). Overexpression of CYP2E1 has also been shown to impair GPx-1 activity suggesting ethanol metabolism may be required for this impairment (Mari and Cederbaum, 2000). Another intriguing possibility is that the alcohol-induced hyperacetylation of GPx-1 and other antioxidant enzymes may also be regulating enzymatic activity. If acetylation/deacetylation is functioning as a simple on/off switch as described above for the mitochondrial enzymes, the simple prediction is that alcohol-induced hyperacetylation leads to impaired antioxidant activities of GPx-1, GSTμ2, SOD1 and other antioxidant proteins thereby enhancing oxidative stress.
Actin Hyperacetylation and Altered Protein Trafficking
Defining the alcohol-induced defects in protein trafficking is an active area of research in understanding hepatotoxicity. To date, numerous proteins are known to have alcohol-induced alterations in their dynamics (Joseph et al., 2008; McVicker and Casey, 1999; Tuma and Sorrell, 1988; Tuma et al., 1990). In general, 2 transport pathways appear to be affected: transport of newly synthesized secretory or membrane proteins from the Golgi to the basolateral membrane and clathrin-mediated endocytosis from the basoalteral surface. Both impaired secretion and internalization require ethanol metabolism and are likely mediated by acetaldehyde (Clemens et al., 1998; Joseph et al., 2008).
We have determined that microtubule hyperacetylation induced by ethanol or by addition of a deacetylase inhibitor correlated with impaired clathrin-mediated endocytosis and secretion (Joseph et al., 2008). However, another interesting possibility is that actin and cortactin hyperacetylation may also lead to impaired endocytosis and secretion. Both of these proteins are known regulators of late stages of clathrin coated vesicle budding from the plasma membrane and the TGN (Cao et al., 2003, 2005). In general, cortactin is thought to promote actin polymerization at sites of vesicle formation and recruit dynamin (a GTPase required for vesicle fission) to the necks of budding vesicles (Cao et al., 2003, 2005). At present, the exact mechanism by which cortactin, actin, and dynamin function to promote vesicle release is not yet completely elucidated. However, acetylation of cortactin is known to prevent its association with actin and alters its subcellular localization (Zhang et al., 2007). From these results, we propose that alcohol-induced hyperacetylation leads to decreased interactions between actin and cortactin such that cortactin is no longer recruited to sites of clathrin-vesicle formation thereby inhibiting dynamin recruitment and subsequent vesicle fission. We are currently testing this exciting possibility.
Acetylation and Liver Disease
This study has revealed that chronic alcohol consumption leads to the hyperacetylation of numerous non-nuclear proteins. Although we have discussed some possible mechanisms by which hyperacetylation may contribute to alcohol-induced hepatotoxicity, future work is clearly needed to test these hypotheses. Furthermore, new therapeutic strategies for treating patients with chronic liver disease may be aimed at reducing protein acetylation. Currently, specific deacetylase activators (e.g., resveratrol and SRT-501) have been shown to be well-tolerated in humans and are in clinical trials for treatment of various metabolic diseases including type 2 diabetes (Elliott and Jirousek, 2008). Furthermore, resveratrol has been shown to attenuate fatty liver in alcohol-exposed mice (Ajmo et al., 2008). An exciting possibility is that this drug, other specific deacetylase activators or acetyltransferase inhibitors will be useful in treating alcoholic liver disease and other liver metabolic diseases.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Alcohol Abuse and Alcoholism (R21 AA015683) awarded to P.L.T.
REFERENCES
- Ajmo JM, Liang X, Rogers CQ, Pennock B, You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Patel VB, Young TA, Asayama K, Cunningham CC. Chronic ethanol consumption alters the glutathione/glutathione peroxidase-1 system and protein oxidation status in rat liver. Alcohol Clin Exp Res. 2001;25:726–733. [PubMed] [Google Scholar]
- Bardag-Gorce F, French BA, Joyce M, Baires M, Montgomery RO, Li J, French S. Histone acetyltransferase p300 modulates gene expression in an epigenetic manner at high blood alcohol levels. Exp Mol Pathol. 2007;82:197–202. doi: 10.1016/j.yexmp.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- Brooks PJ. DNA damage, DNA repair, and alcohol toxicity – a review. Alcohol Clin Exp Res. 1997;21:1073–1082. [PubMed] [Google Scholar]
- Cao H, Orth JD, Chen J, Weller SG, Heuser JE, McNiven MA. Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol Cell Biol. 2003;23:2162–2170. doi: 10.1128/MCB.23.6.2162-2170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- Choudhury M, Shukla SD. Surrogate alcohols and their metabolites modify histone H3 acetylation: involvement of histone acetyl transferase and histone deacetylase. Alcohol Clin Exp Res. 2008;32:829–839. doi: 10.1111/j.1530-0277.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Casey CA, Sorrell MF, Tuma DJ. Ethanol oxidation mediates impaired hepatic receptor-mediated enocytosis. Alcohol Clin Exp Res. 1998;22:778–779. doi: 10.1111/j.1530-0277.1998.tb03866.x. [DOI] [PubMed] [Google Scholar]
- Elliott PJ, Jirousek M. Sirtuins: novel targets for metabolic disease. Curr Opin Investig Drugs. 2008;9:371–378. [PubMed] [Google Scholar]
- Farbiszewski R, Chwiecko M, Holownia A, Pawlowska D. The decrease of superoxide dismutase activity and depletion of sulfhydryl compounds in ethanol-induced liver injury. Drug Alcohol Depend. 1991;28:291–294. doi: 10.1016/0376-8716(91)90063-5. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat H, Singer B. Nucleoside adducts are formed by cooperative reaction of acetaldehyde and alcohols: possible mechanism for the role of ethanol in carcinogenesis. Proc Natl Acad Sci USA. 1988;85:3758–3761. doi: 10.1073/pnas.85.11.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RA, Shepard BD, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Microtubule acetylation and stability may explain alcohol-induced alterations in hepatic protein trafficking. Hepatology. 2008;47:1745–1753. doi: 10.1002/hep.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannarkat GT, Tuma DJ, Tuma PL. Microtubules are more stable and more highly acetylated in ethanol-treated hepatic cells. J Hepatol. 2006;44:963–970. doi: 10.1016/j.jhep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Kenney WC. Acetaldehyde adducts of phospholipids. Alcohol Clin Exp Res. 1982;6:412–416. doi: 10.1111/j.1530-0277.1982.tb05000.x. [DOI] [PubMed] [Google Scholar]
- Kenney WC. Formation of Schiff base adduct between acetaldehyde and rat liver microsomal phosphatidylethanolamine. Alcohol Clin Exp Res. 1984;8:551–555. doi: 10.1111/j.1530-0277.1984.tb05728.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Shukla SD. Histone h3 modifications in rat hepatic stellate cells by ethanol. Alcohol Alcohol. 2005;40:367–372. doi: 10.1093/alcalc/agh170. [DOI] [PubMed] [Google Scholar]
- Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Shukla SD. Histone H3 phosphorylation at serine 10 and serine 28 is mediated by p38 MAPK in rat hepatocytes exposed to ethanol and acetaldehyde. Eur J Pharmacol. 2007;573:29–38. doi: 10.1016/j.ejphar.2007.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber CS, DeCarli LM. Liquid diet technique of ethanol administration: 1989 update. Alcohol Alcohol. 1989;24:197–211. [PubMed] [Google Scholar]
- Lieber CS, Leo MA, Wang X, Decarli LM. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–48. doi: 10.1016/j.bbrc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Mari M, Cederbaum AI. CYP2E1 overexpression in HepG2 cells induces glutathione synthesis by transcriptional activation of gamma-glutamylcysteine synthetase. J Biol Chem. 2000;275:15563–15571. doi: 10.1074/jbc.M907022199. [DOI] [PubMed] [Google Scholar]
- McVicker BL, Casey CA. Effects of ethanol on receptor-mediated endocytosis in the liver. Alcohol. 1999;19:255–260. doi: 10.1016/s0741-8329(99)00043-9. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Oh SI, Kim CI, Chun HJ, Park SC. Chronic ethanol consumption affects glutathione status in rat liver. J Nutr. 1998;128:758–763. doi: 10.1093/jn/128.4.758. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81:979–987. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PH, Lim RW, Shukla SD. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: potential mechanism for gene expression. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1124–G1136. doi: 10.1152/ajpgi.00091.2005. [DOI] [PubMed] [Google Scholar]
- Park PH, Miller R, Shukla D. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophys Res Commun. 2003;306:501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376:615–619. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3:6.1–6.6. doi: 10.1186/gb-2002-3-5-reviews0006. reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow H, Obe G. Acetaldehyde induces cross-links in DNA and causes sister-chromatid exchanges in human cells. Mutat Res. 1978;58:115–119. doi: 10.1016/0165-1218(78)90103-9. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard BD, Joseph RA, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Alcohol-induced alterations in hepatic microtubule dynamics can be explained by impaired histone deacetylase 6 function. Hepatology. 2008;48:1671–1679. doi: 10.1002/hep.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard BD, Tuma PL. Alcohol-induced protein hyperacetylation: mechanisms and consequences. World J Gastroenterol. 2009;15:1219–1230. doi: 10.3748/wjg.15.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma DJ, Casey CA. Dangerous byproducts of alcohol breakdown – focus on adducts. Alcohol Res Health. 2003;27:285–290. [PMC free article] [PubMed] [Google Scholar]
- Tuma DJ, Casey CA, Sorrell MF. Effects of ethanol on hepatic protein trafficking: impairment of receptor-mediated endocytosis. Alcohol Alcohol. 1990;25:117–125. doi: 10.1093/oxfordjournals.alcalc.a044986. [DOI] [PubMed] [Google Scholar]
- Tuma DJ, Sorrell MF. Effects of ethanol on protein trafficking in the liver. Semin Liver Dis. 1988;8:69–80. doi: 10.1055/s-2008-1040529. [DOI] [PubMed] [Google Scholar]
- Wehr H, Rodo M, Lieber CS, Baraona E. Acetaldehyde adducts and autoantibodies against VLDL and LDL in alcoholics. J Lipid Res. 1993;34:1237–1244. [PubMed] [Google Scholar]
- Wu D, Cederbaum AI. Alcohol, oxidative stress, and free radical damage. Alcohol Res Health. 2003;27:277–284. [PMC free article] [PubMed] [Google Scholar]
- You M, Liang X, Ajmo JM, Ness GC. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, Yao TP, Lane WS, Seto E. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]