Abstract
Active sorting at the endoplasmic reticulum (ER) drives efficient export of fully folded secretory proteins into coat protein complex II (COPII) vesicles, whereas ER-resident and misfolded proteins are retained and/or degraded. A number of secretory proteins depend upon polytopic cargo receptors for linkage to the COPII coat and ER export. However, the mechanism by which cargo receptors recognize transport-competent cargo is poorly understood. Here we examine the sorting determinants required for export of yeast alkaline phosphatase (ALP) by its cargo receptor Erv26p. Analyses of ALP chimeras and mutants indicated that Erv26p recognizes sorting information in the lumenal domain of ALP. This lumenal domain sorting signal must be positioned near the inner leaflet of the ER membrane for Erv26p-dependent export. Moreover, only assembled ALP dimers were efficiently recognized by Erv26p while an ALP mutant blocked in dimer assembly failed to exit the ER and was subjected to ER-associated degradation. These results further refine sorting information for ER export of ALP and show that recognition of folded cargo by export receptors contributes to strict ER quality control.
Keywords: ALP, cargo receptor, ER, sorting signal
Since the discovery that secretory proteins are transported in eukaryotic cells through a discontinuous set of membrane compartments (1), significant progress has been made in identifying molecular components and characterizing mechanisms in the secretory pathway. Coat protein complexes are known to select secretory cargo and bud transport vesicles from donor membranes. Budded intermediates are then targeted to and fuse with specific acceptor membrane compartments (2). While a conceptual framework for intracellular transport is emerging, fundamental questions remain regarding how the secretory pathway accommodates such a broad diversity of secretory cargo and how organelle identity is maintained as cargo proteins advance.
Current estimates indicate that 20–30% of translated proteins in the eukaryotic proteome enter the secretory pathway (3) and then must be folded and sorted at the endoplasmic reticulum (ER). Anterograde transport from the ER requires the coat protein complex II (COPII), which selects fully folded secretory proteins into COPII-formed vesicles that bud from ER exit sites (4). The COPII coat consists of the small GTPase Sar1p and the larger heteromeric Sec23/24 and Sec13/31 complexes. The Sec23/24 complex in conjunction with Sar1p recognizes sorting information displayed by many transmembrane cargo proteins (5,6), while the outer layer Sec13/31 complex forms a cage structure that deforms ER membranes and produces transport vesicles (7). The Sec24p subunit of the coat contains binding sites for linear sorting signals that have been identified in specific secretory cargo (8,9). However, not all COPII vesicle cargo contain apparent sorting signals and it is known that certain secretory proteins depend on cargo receptors for linkage to the COPII coat. In addition to positive sorting information for selective incorporation into COPII vesicles, misfolded or unassembled secretory cargo are monitored by an ER quality control system, inefficiently packaged by the COPII export machinery and may be subjected to endoplasmic reticulum-associated degradation (ERAD) (10,11). How the ER quality control process is coordinated with COPII recognition and export is not well understood.
To address these basic questions, we investigated a cargo/receptor pair to dissect sorting mechanisms in a model system. In this example, vacuolar alkaline phosphatase (ALP) in Saccharomyces cerevisiae depends on the polytopic membrane protein, Erv26p, for efficient packaging into COPII vesicles. Moreover, the ER form of ALP is detected in association with Erv26p when isolated from ER membranes. On the basis of these findings, Erv26p is proposed to act as a cargo receptor that cycles between the ER and Golgi compartments and selects fully folded secretory proteins into COPII vesicles (12). Both ALP and Erv26p are highly conserved in nature; therefore, an understanding of this sorting mechanism will likely provide insights on the biogenesis of mammalian ALPs, genetic diseases connected to missense mutations in the human ALPL gene (13) and on mechanisms underlying other receptor-dependent sorting events in the early secretory pathway.
Yeast ALP is a type II vacuolar glycoprotein synthesized in a pro-form and converted to the active form by C-terminal pro-sequence cleavage in the vacuole (14). Vacuolar processing of ALP has been used in previous studies as a reporter for clathrin-dependent trafficking signals through later secretory organelles (14,15).
However, sorting signals required for early trafficking steps of this model protein have not been examined. This study aims to refine the sorting information in pro-ALP required for Erv26p-dependent ER export. An in vivo assay to monitor ER to Golgi transport of pro-ALP allowed us to examine the fate of pro-ALP chimeras and mutants in distinct genetic backgrounds. On the basis of these findings, we conclude that the lumenal domain of pro-ALP, when positioned at an appropriate distance from the ER inner membrane leaflet, is necessary and sufficient for Erv26p-dependent ER export. Moreover, assembly of the native pro-ALP dimer in the ER is critical for its stability and recognition by Erv26p.
Results
Pro-ALP is a 566 amino acid type II vacuolar glycoprotein with a 33 residue N-terminal cytosolic tail segment, a single transmembrane domain (TMD) and a 50 residue C-terminal pro-sequence (14). To determine which domain(s) of the protein are important for Erv26p-dependent ER export, we engineered a series of deletion and chimeric constructs (see Figure 1) for analyses.
Figure 1.
ALP constructs used in this study. Diagram of pro-ALP protein with the TMD (black) and pro-sequence (dark grey) indicated. Putative N-glycosylation sites are shown by asterisks and scissors indicate the pro-sequence cleavage site. The Och1p TMD and cytosolic segments are shown as light grey boxes and the α-factor cleavable signal sequence as a black line. The triple HA epitope is represented as HA on the carboxyl terminus.
The rate of ER to Golgi transport of ALP is reduced in an erv26Δ background in vivo
To monitor the ER to Golgi transport of pro-ALP in vivo, we measured the rate of acquisition of an early Golgi-specific modification: the α1,6-mannose (α1,6-man) branching of N-glycans (16). Pro-ALP contains two potential N-glycosylation sites (Arg 268 and Arg 401), of which at least one is modified (17). In pulse-chase experiments, the level of Golgi-modified pro-ALP was measured in a secondary immunoprecipitation (IP) using α1,6-man-specific polyclonal antibodies. Under our experimental conditions, maximal levels of pro-ALP reached the Golgi apparatus and acquired α1,6-man modification after 5 min of chase in a wild-type background (Figure 2). After 20 min of chase, most pro-ALP had been cleaved to mature ALP, indicating delivery to the vacuole. Strikingly, in an erv26Δ background, less than 10% of ALP was recovered in the α1,6-man IP after a 20 min chase and no significant pro-sequence cleavage was observed at this time-point. In the absence of Erv26p, we had previously reported that after a 60 min chase only one third of labelled ALP was delivered to the vacuole as assessed by pro-sequence cleavage (12), suggesting a slow, `bulk-flow' type of transport. The maximum recovery of α1,6-man modified ALP was ~60% of total ALP in a wild-type background. This incomplete recovery could be due to our experimental method which heat denatures proteins between the first and the second IPs or because of heterogeneity in outer-chain mannose modification. A similar recovery of α1,6-man modified carboxypeptidase Y (CPY) was also observed (Figure 2A). Pro-CPY is similarly transported to and matured in the vacuole after outer-chain mannose additions in the Golgi complex. Importantly, the rate of α1,6-man modification of CPY was not influenced by erv26Δ, which indicates that the Golgi glycosylation machinery was functional in this deletion strain but that pro-ALP was very slowly transported to early Golgi compartments. These results are entirely consistent with the role of Erv26p in selective packaging of pro-ALP into COPII vesicles. Therefore, this in vivo pulse-chase approach allows us to identify sorting information contained in pro-ALP necessary for its Erv26p-dependent transport.
Figure 2.
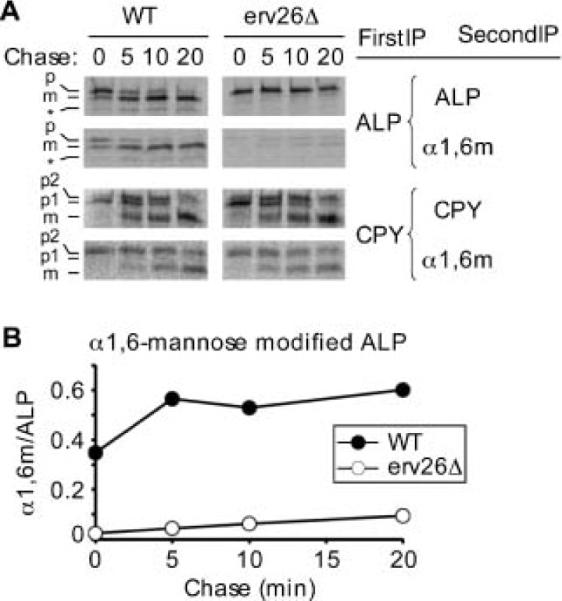
ALP is inefficiently transported to the Golgi complex in erv26Δ cells. A) Wild-type (CBY2151) and erv26Δ (CBY2240) strains expressing full-length pro-ALP (pFL8) were monitored in pulse-chase experiments as described under Materials and Methods. Primary IPs were performed with anti-ALP or anti-CPY serums followed by secondary IPs to measure α1,6-mannose modified carbohydrate. Positions of pro-ALP (p), mature ALP (m) and a soluble breakdown product (*) are indicated as are the ER (p1), Golgi (p2) and mature (m) forms of CPY. B) Graphical representation of the results in (A) showing α1,6-mannose modified target protein divided by total target protein at each time-point.
The cytosolic tail sequence of ALP is not required for Erv26p-dependent ER export
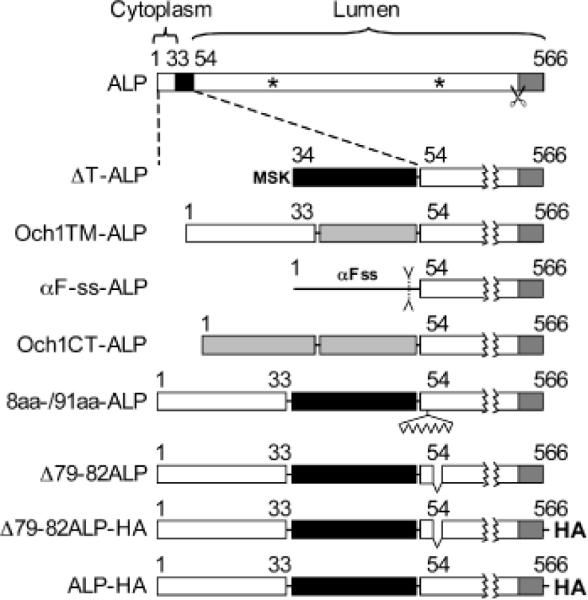
The N-terminal 33 amino acid cytoplasmic tail segment of ALP contains a defined vacuolar targeting signal for AP3-dependent transport from the late Golgi complex to the vacuole (15,18,19). Deletion of the first 31 amino acids produces stable pro-ALP that is slowly trafficked to the vacuole through the Vps pathway (18). To determine if the cytoplasmic tail sequence influences pro-ALP transport from the ER, we examined the same ΔT-ALP construct in wild-type and erv26Δ strains. Under steady-state conditions, most ΔT-ALP protein was processed to the mature form in wild-type whereas in an erv26Δ strain, about 30% of total ΔT-ALP was detected in the pro-form (Figure 3A). Moreover, a subcellular fractionation experiment showed that the pro-form of ΔT-ALP co-fractionated with ER membranes in the erv26Δ strain (Figure 3B). In pulse-chase experiments shown in Figure 3C,D, the ΔT-ALP protein acquired Golgi-specific α1,6-man modifications at near normal rates in the wild-type strain and was then slowly delivered to the vacuole. In contrast, the rate of ΔT-ALP transport to the Golgi was greatly reduced in the erv26Δ strain compared to wild-type. In both strains, a small fraction of ΔT-ALP was cleaved after 60 min chase indicating eventual transport to the vacuole (data not shown). These results indicate that the cytoplasmic tail sequence in pro-ALP is not required for Erv26p-dependent export from the ER and suggested that ER sorting information resides in the transmembrane or lumenal domain of pro-ALP.
Figure 3.
The cytosolic tail of ALP is dispensable for ER export. A) Wild-type (CBY2151) and erv26Δ (CBY2240) cells expressing full-length pro-ALP (pSN92) or the tail deletion construct (pNB4) were spheroplasted by lyticase treatment. Total cell extracts were resolved on 8% polyacrylamide gels for immunoblots. Erv41p, a protein localized to ER and Golgi membranes, was used as a loading control. The pro-ALP (p), mature ALP (m) and a soluble breakdown product (*) are indicated. B) Cell extracts as in (A) were fractionated on sucrose density gradients and fractions analysed by immunoblot as described under Materials and Methods. Och1p and Sec61p served as Golgi and ER membrane markers, with peak fractions for Och1p (G) and Sec61p (ER) indicated by arrowheads. C) Pulse-chase analysis of ΔT-ALP in the wild-type and erv26Δ cells expressing the tail deletion construct (pNB4). D) Graphical representation of the results in (C) showing α1,6-mannose modified ΔT-ALP protein divided by total ΔT-ALP at each time-point.
The TMD of ALP does not contain ER export information but the membrane-bound nature of the protein is crucial
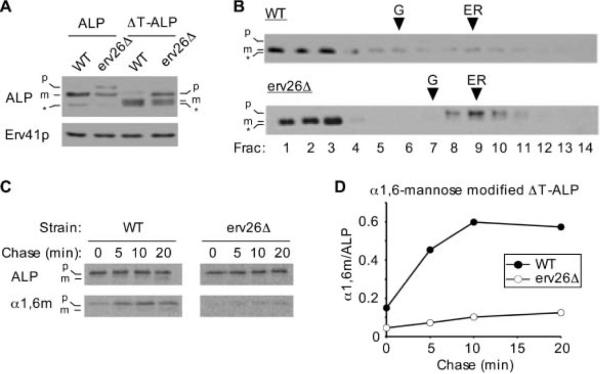
The TMD of ALP is predicted to be 20 amino acids in length (http://phobius.cgb.ki.se/, http://www.cbs.dtu.dk/services/TMHMM/) and contains four polar residues as well as a cysteine. To study if ALPs TMD is relevant for ER export, we sought to replace it with a signal-less TMD from Och1p, a Golgi-localized glycosyltransferase with type II membrane topology (20). Och1p and other Golgi glycosyltransferases rely on cytoplasmic tail signals for their Golgi localization (21). Although it is not known if sorting information also resides in the Och1p TMD or lumenal domain, we have found that transport of Och1p does not depend on Erv26p because the localization of Och1p was not influenced by erv26Δ(12) and the glycosylation activity of Och1p was normal in the erv26Δ strain (see Figure 2A). Therefore, the Och1p TMD appears to be a good candidate for a signal-less segment, leading us to construct the Och1TM-ALP chimera. We observed that Och1TM-ALP was transported to the vacuole in a wild-type background whereas the pro-form of Och1TM-ALP accumulated in the ER of the erv26Δ mutant (Figure 4A,B). Kinetic analyses indicated that Och1TM-ALP was efficiently transported to the Golgi complex and acquired α1,6-man modification (Figure 4C,D). Moreover, the overall transport rate of Och1TM-ALP to the vacuole was similar to wild-type ALP indicating the content of this TMD is not critical for vacuolar targeting. In contrast, Och1TM-ALP persisted in the ER of an erv26Δ strain and very slowly acquired α1,6-man modification as observed for other Erv26p-dependent versions of ALP. Because the Och1p TMD is quite different from the ALP TMD, in both composition and length, we conclude that ALPs TMD does not specify Erv26p-dependent export from the ER.
Figure 4.
The TMD of ALP does not specify Erv26p-dependent transport. A) Alignment of the ALP and Och1p TMD sequences. A two amino acid linker (underlined) was introduced for cloning purposes between the Och1p TMD and ALP lumenal domain in Och1TM-ALP. Wild-type (CBY2151) and erv26Δ (CBY2240) cells expressing full-length pro-ALP (pFL8) or the Och1TM-ALP construct (pAF13) were converted to spheroplasts and proteins detected by immunoblot. The pro-ALP (p), mature ALP (m) and a soluble breakdown product (*) are indicated. B) Cell extracts from strains expressing pAF13 were separated on sucrose density gradients and fractions analysed by immunoblot. C) Pulse-chase analysis of Och1TM-ALP in the wild-type and erv26Δ cells expressing pAF13. Graphical representation of the results in (C) showing α1,6-mannose modified Och1TM-ALP protein divided by total Och1TM-ALP at each time-point.
Altogether these results suggested that information needed for Erv26p-dependent export out of the ER resides in the lumenal domain of ALP. To explore this hypothesis, we engineered a chimera consisting of the lumenal domain of ALP linked to the signal sequence of pre-pro- α-factor, which is known to target this chimera for translocation into the ER and to efficiently produce a signal sequence cleaved lumenal protein (22). Figure 5A shows that at steady state, αF-ss-ALP was mainly secreted to the extracellular medium with a smaller intracellular pool consisting of full-length protein and a cleaved form, which presumably reflected a low level of delivery to the vacuole. Importantly, however, the pattern of αF-ss-ALP forms expressed in the wild type compared to erv26Δ was the same in contrast to other Erv26p-dependent versions of ALP.
Figure 5.
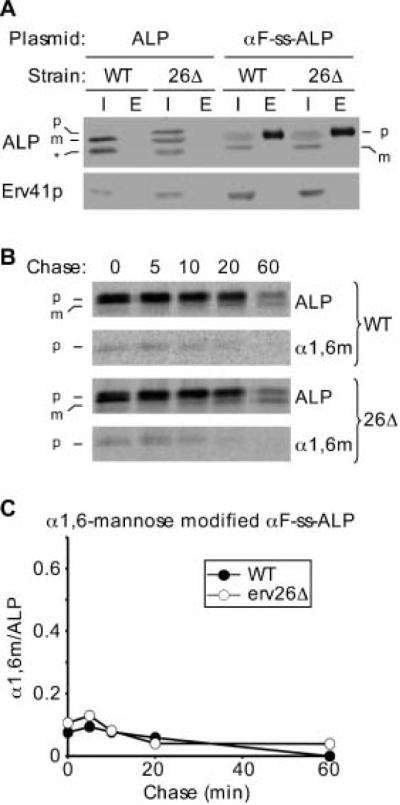
A soluble lumenal form of pro-ALP slowly exits the ER in an Erv26p-independent manner. A) Wild-type (CBY2151) and erv26Δ (CBY2240) cells expressing full-length pro-ALP (pFL8) or the αF-ss-ALP construct (pQC43) were lysed and equal amounts of intracellular (I) and extracellular (E) proteins detected by immunoblot as described under Materials and Methods. The pro-ALP (p), mature ALP (m) and a soluble breakdown product (*) are indicated. B) Pulse-chase analysis of αF-ss-ALP in the wild-type and erv26Δ cells expressing pQC43. C) Graphical depiction of the results in (C) showing α1,6-mannose modified F-ss-ALP protein divided by total F-ss-ALP at each time-point.
In pulse-chase analyses, the soluble αF-ss-ALP construct displayed very slow transport rates to the Golgi in both wild-type and erv26Δ strains (Figure 5B,C), which likely reflects a bulk flow rate of transport. The level of intracellular α1,6-man modified αF-ss-ALP reached a maximum at the 5 min time-point then decreased gradually. This decrease correlated with the appearance of αF-ss-ALP in the extracellular medium (data not shown). We interpret these results to indicate that αF-ss-ALP slowly traffics to the Golgi complex in an Erv26p-independent manner and then this soluble version of ALP predominantly follows an exocytic route to the plasma membrane. Importantly, αF-ss-ALP localization and trafficking was not influenced by erv26Δ. Therefore, we conclude that pro-ALP must be membrane bound for recognition and interaction with Erv26p.
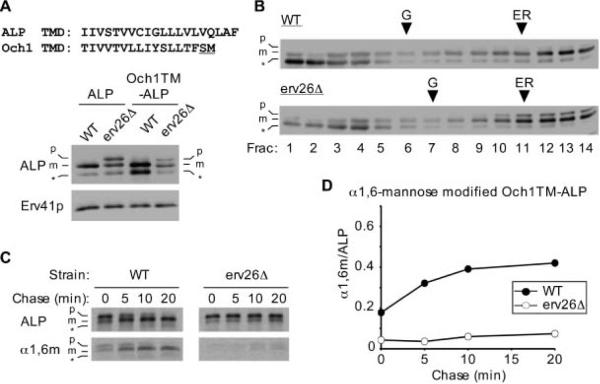
To test if the membrane-bound lumenal domain of pro-ALP was sufficient for Erv26p-dependent export, we constructed a fusion appending this domain to the cytosolic and TMD portions of Och1p. As observed for the Och1TM-ALP, the Och1CT-ALP fusion displayed Erv26p-dependent localization and transport properties (Figure 6). These findings indicate that the lumenal domain of ALP, once bound to the membrane, is necessary and sufficient for Erv26p-dependent ER export.
Figure 6.
The membrane-bound lumenal domain of ALP is sufficient to specify Erv26p-dependent export. A) Wild-type (CBY2151) and erv26Δ (CBY2240) cells expressing full-length pro-ALP (pFL8) or the Och1CT-ALP construct (pCIP2) were converted to spheroplasts and proteins detected by immunoblot. The pro-ALP (p), mature ALP (m) and a soluble breakdown product (*) are indicated. B) Extracts from the wild-type and erv26Δ cells expressing Och1CT-ALP were fractionated on sucrose gradients and proteins detected by immunoblot. C) Pulse-chase analysis of Och1CT-ALP in wild-type and erv26Δ cells expressing pCIP2. D) Graphical representation of the results in (C) plotting α1,6-mannose modified Och1CT-ALP protein divided by total Och1CT-ALP.
Membrane positioning of the lumenal ALP domain influences Erv26-dependent export
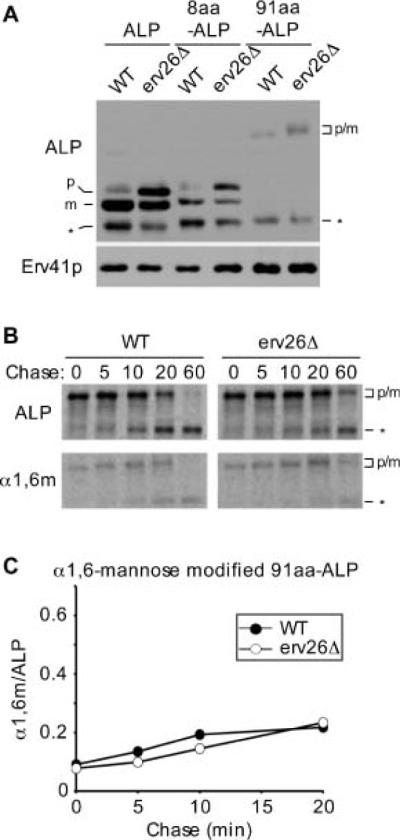
Our results indicated that a membrane-bound form of pro-ALP was required for Erv26p recognition. We next tested if inserting amino acid spacers to distance the lumenal domain of ALP away from the inner membrane bilayer influenced Erv26p-dependent transport. Two insertion constructs were generated in which 8 and 91 amino acid residue spacers were inserted into the lumenal domain of pro-ALP just after its TMD. The rational was that these insertions would extend the lumenal domain of ALP away from the inner leaflet of ER membranes by approximately 10 and 100 angstroms, respectively. We observed that the 8aa-ALP construct exhibited the same Erv26p-dependent transport properties as wild-type ALP (Figure 7A). In contrast, the 91aa-ALP construct was transported to the Golgi at a very slow rate that was independent of Erv26p (Figure 7B,C). We note that the steady-state expression level of 91aa-ALP was reduced compared to wild-type ALP (Figure 7A) and approximately half of the protein was degraded over this pulse-chase time–course (Figure 7B). However, a fraction of the 91aa-ALP does appear to fold, assembles into dimers (Figure S1) and was ultimately transported to the vacuole in an Erv26p-independent manner (Figure 7B, 60 min time-point). These results suggest that efficient recognition of pro-ALP by the Erv26p cargo receptor depends on an optimum spatial positioning of ALPs lumenal domain with respect to the ER membrane. However, we cannot exclude the possibility that this extension somehow obscures an ER export signal in pro-ALP.
Figure 7.
The position of pro-ALPs lumenal domain from the inner ER membrane influences Erv26p-dependent export. A) Wild-type (CBY2151) and erv26Δ (CBY2240) cells expressing either full-length pro-ALP (pFL8), the 8 amino acid insertion (pEC5) or the 91 amino acid insertion construct (pQC72) were converted to spheroplasts and proteins detected by immunoblot. The pro-ALP (p), mature ALP (m) and a soluble breakdown product (*) are indicated. B) Pulse-chase analysis of the 91aa-ALP protein in wild-type and erv26Δ strains expressing pQC72. C) Graphical depiction of the results in (B) plotting α1,6-mannose modified 91aa-ALP protein divided by total 91aa-ALP at the start of the time–course.
Dimerization of pro-ALP is required for stability, Erv26p recognition and ER export
Many questions remain regarding coordination between the ER quality control process and the ER export machinery. To explore this issue for export of pro-ALP from the ER, we followed the behaviour of a mutant in which the assembly of dimeric pro-ALP was disrupted. The lumenal catalytic domain of yeast ALP is highly conserved across prokaryotic and eukaryotic species. Molecular structures of ALP have been obtained for bacterial (PhoA), shrimp and human orthologs, all of which crystallize as homodimeric species (23–25). Several lines of evidence indicate that native forms of ALP are functionally dimeric and it has been shown that purified yeast ALP migrates as a homodimer on non-denaturing gels (17,26). Boulanger et al. (27) showed that a single amino acid substitution at position 81 in bacterial PhoA prevented dimerization. A primary sequence alignment of yeast ALP with several orthologs (Figure 8A) and a homology search against quaternary structures in the Protein DataBank (PDBLAST) (28) suggested that amino acids 79 to 82 of yeast ALP corresponded to a conserved α-helical region of the dimer interface. Therefore, we generated a construct to express Δ79-82ALP for studies on the role of ALP dimerization in ER export.
Figure 8.
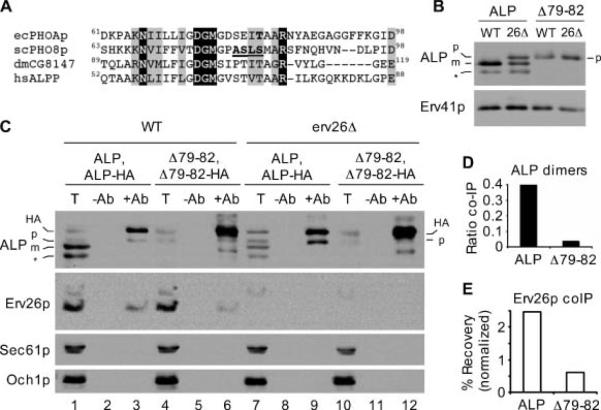
Dimerization of pro-ALP is required for efficient binding to Erv26p and ER export. A) Primary sequence alignments of Escherichia coli PhoA (GenBankTM accession BAE76164), Saccharomyces cerevisiae ALP (GenBankTM accession P11491), Drosophila melanogaster ALP (GenBankTM accession NP_649897) and human placental ALP (GenBankTM accession AAC97139). Invariant amino acids are shaded black, conserved amino acids in grey. Residues of interest are bold and underlined (see text). B) Wild-type (CBY2151) and erv26Δ (CBY2240) cells expressing full-length pro-ALP (pFL8) or the Δ79-82ALP construct (pQC30) were converted to spheroplasts and proteins detected by immunoblot. C) Immunoblots of anti-HA co-immunoprecipitation experiments as described in Materials and Methods. Totals (T) represent 5% of the solubilized input material, mock immunoprecipitations (−Ab) and anti-HA immunoprecipitations (+Ab) are labelled above each lane. Sec61p (ER) and Och1p (Golgi) serve as negative controls to demonstrate specificity of the immunoprecipitations. D) Graphical representation of the data in panel (C) showing ALP dimer formation as the ratio of HA-tagged and untagged pro-forms of ALP in lanes 9 and 12. E) The percentage of total Erv26p that co-immunoprecipitated with ALP-HA in lanes 3 and 6 normalized by the ALP-HA signals in these same lanes. The positions of pro-ALP (p), mature ALP (m), pro-ALP-HA (HA) and a soluble breakdown product (*) are indicated.
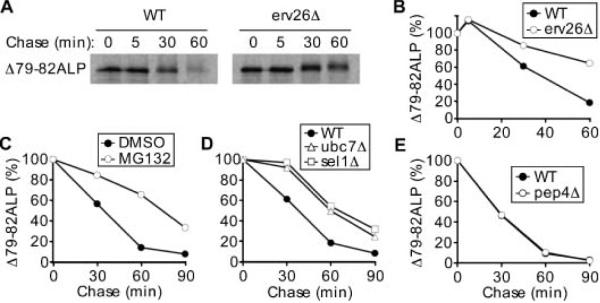
Figure 9A, the Δ79-82ALP mutant was stabilized in erv26Δ and displayed a t1/2 of ~ 35 min in wild-type and >60 min in the erv26Δ strain (Figure 9B). The lack of vacuolar cleavage even after 1 h of chase (and at steady state, Figure 8A) suggests that the degradation is prevacuolar. A decrease in the electrophoretic mobility of the Δ79-82ALP protein was observed at 30 and 60 min. We speculate that this dimerization mutant received additional post-translational modification with prolonged residence in the ER. The Δ79-82ALP protein did not efficiently receive Golgi-specific α1,6-man modifications in either background (data not shown).
The steady-state level of Δ79-82ALP expression was lower than wild-type ALP suggesting an increased turnover rate of this protein. Strikingly, no pro-sequence cleavage of the Δ79-82ALP protein was observed indicating that this mutant protein does not reach the vacuole in either wild-type or erv26Δ backgrounds (Figure 8B). To determine if this deletion mutation produced a dimerization defect, we devised an IP assay to monitor dimer assembly using Hemagglutinin-tagged (HA-tagged) and untagged copies of either wild-type ALP or Δ79-82ALP (Figure 1). First, we appended the 3X-HA tag to the C-terminus of wild-type ALP (ALP-HA) and of Δ79-82ALP (Δ79-82ALP-HA) allowing us to pull-down prevacuolar complexes because cleavage of the pro-sequence and HA tag occurs upon vacuolar delivery. For these experiments, the endogenous ALP gene (PHO8) was deleted and plasmids expressing either ALP and ALP-HA or Δ79-82ALP and Δ79-82ALP-HA were introduced into the wild-type and erv26Δ strains. When ALP-HA was expressed in a wild-type strain and protein immunoprecipitated with anti-HA, untagged pro-ALP was immunoprecipitated (Figure 8C, lane 3) indicating recovery of mixed pro-ALP-HA/pro-ALP dimers. In the erv26Δ background, more pro-ALP-HA/pro-ALP was recovered (Figure 8C, lane 9) because of an accumulation of assembled dimers in the ER. In both wild-type and erv26Δ strains, pro-ALP was recovered in pro-ALP-HA IPs at levels that approximated the ratio (1:3) of the pro-ALP-HA/pro-ALP mixed dimer compared to pro-ALP/pro-ALP and pro-ALP-HA/pro-ALP-HA dimers expected to assemble in the ER. These results indicate that wild-type pro-ALP assembles into homodimers that can be monitored by this IP method and that erv26Δ does not decrease homodimer assembly.
Next, we tested if the Δ79-82 deletion disrupted assembly of pro-ALP homodimers. In anti-HA IP experiments, Δ79-82ALP-HA was recovered (Figure 8C, lanes 6 and 12); however, untagged Δ79-82ALP was inefficiently co-immunoprecipitated under these conditions. The ratio of untagged to HA-tagged pro-ALP immunoprecipitated was 0.05 for the Δ79-82 mutant compared to 0.38 for wild-type pro-ALP. Moreover, expression of the Δ79-82ALP-HA protein in wild-type strains with endogenous ALP did not influence the transport rate of endogenous ALP (data not shown). These results indicate that the Δ79-82 deletion prevented stable assembly of pro-ALP dimers. The Δ79-82 deletion does not appear to cause major folding problems for pro-ALP monomers because no changes in disulfide bonding were detected upon analysis on non-reducing polyacrylamide gels (Figure S2).
Erv26p normally forms a complex with pro-ALP in the ER (12), therefore we measured the amount of this export complex under conditions in which dimer assembly was blocked by the Δ79-82 deletion. As seen in Figure 8C, IP of HA-tagged pro-ALP recovered ~ 2.5% of total Erv26p (lane 3), whereas the amount of Erv26p recovered in IP of the Δ79-82ALP-HA mutant was reduced to ~ 1.0%. However, it should be noted that there was a much higher level of the Δ79-82ALP-HA mutant in the ER compared to wild-type ALP-HA and therefore substantially more Δ79-82ALP-HA was immunoprecipitated than wild-type protein (compare HA-tagged protein recovered in lanes 3 and 6). Therefore, if Erv26p recovery is normalized relative to the amount of HA-tagged protein precipitated, we calculate that the amount of mutant Δ79-82ALP-HA bound to Erv26p was at least fivefold less than wild-type ALP-HA. Together, these experiments demonstrate that the Δ79-82ALP mutant does not assemble into stable dimers and does not efficiently bind to Erv26p.
The Δ79-82ALP dimerization mutant is stabilized by erv26Δ
We had observed that the steady-state expression level of Δ79-82ALP was reduced compared to wild- type ALP and that this dimerization mutant was partially stabilized in the erv26Δ strain (Figure 8B). To investigate the fate of Δ79-82ALP, we performed a pulse-chase experiment in both wild-type and erv26Δ strains. As shown in The ER plays a major role in the degradation of terminally misfolded secretory proteins. To investigate whether the degradation of Δ79-82ALP was because of the proteasome-dependent ERAD pathway (29), we investigated the degradation rates of Δ79-82ALP in cells treated with MG132, a well-known proteasome inhibitor (30). Wild-type yeast strains are impermeant to MG132, therefore this experiment was conducted in an ise1Δ background, which renders cells more permeable and sensitive to the drug (30). In this background, we detected a strong stabilization of the Δ79-82ALP mutant protein in cells treated with MG132 (Figure 9C), indicating that the degradation of Δ79-82-ALP was at least partially dependent on proteasome activity. To confirm that the effect of MG132 on Δ79-82ALP was because of involvement of ERAD, we expressed this dimerization mutant in cells deleted for UBC7, which encodes a ubiquitin-conjugating enzyme involved in the targeting of ERAD substrates (31), or SEL1/UBX2, which encodes a protein that coordinates Cdc48p-dependent stages in ERAD (32). Figure 9D shows that Δ79-82ALP was stabilized in both ubc7Δ and sel1Δ backgrounds, confirming that turnover of the dimerization mutant was mediated by the ERAD pathway. Finally, expression of Δ79-82ALP in a pep4Δ background, in which vacuolar proteases are inactivated (33), did not alter the turnover kinetics of the mutant indicating that vacuolar proteolysis does not contribute to degradation of the Δ79-82ALP protein (Figure 9E). Based on these results, we conclude that when pro-ALP fails to assemble into stable dimers the protein does not exit the ER and is subjected to ERAD. Interestingly, the erv26Δ deletion stabilizes Δ79-82ALP protein. These findings indicate an intriguing connection between the ER quality control and the ER export machineries.
Figure 9.
Δ79-82ALP is degraded by a proteasome-dependent ERAD pathway. A) Pulse-chase analysis of Δ79-82ALP in wild-type (CBY2151) and erv26Δ (CBY2240) strains. B) Graphical representation of the degradation rates in panel (A) showing the level of Δ79-82ALP as a percentage of initial pulsed (t = 0) Δ79-82ALP protein. C) Degradation rates of Δ79-82ALP-HA assessed in ise1Δ cells (CBY2771) pretreated either with 100 μm MG132 (in DMSO) or 0.5% DMSO for 45 min prior to pulse. D) Degradation rates of Δ79-82ALP-HA in ubc7Δ (CBY2773) and sel1Δ (CBY1323) strains compared to an isogenic wild-type strain. E) Plots showing the degradation rates of Δ79-82ALP-HA protein in a pep4Δ strain (CBY173) compared to an isogenic wild-type strain (CBY80).
Discussion
In this report, we characterize sorting determinants within secretory ALP that are required for Erv26p-dependent export from the ER. Our studies show that the ER export information resides within the lumenal domain of this type II transmembrane protein and represent the first example in which lumenal domain export signals have been identified in a transmembrane secretory protein. In addition, we find that only fully assembled ALP dimers are efficiently recognized by Erv26p, supporting a role for this export factor after cargo are transport competent. We propose that Erv26p-dependent selection of fully folded cargo contributes to the overall ER quality control process. Finally, we document that Erv26p expression specifically influences the degradation rate of unassembled ALP monomers indicating a connection between the ER quality control and the ER export machineries.
Characterized ER export signals within transmembrane secretory proteins typically reside in cytoplasmic regions and bind to subunits of the COPII coat (2). For example, short linear diacidic and hydrophobic export signals have been identified in the cytoplasmic C-terminal tails of type I membrane proteins VSV-G, Sys1p and ERGIC53 (34–36). Cytoplasmic export signals have also been defined in type II membrane proteins including Sed5p and GalT2 (9,37). Therefore, our finding that the cytoplasmic tail sequence of pro-ALP was dispensable for COPII export was unexpected. In the case of pro-ALP, however, ER export had been reported to depend on the adaptor protein Erv26p (12). Here we demonstrate that Erv26p-dependent export of pro-ALP from the ER can be specified solely through sorting information contained within the lumenal domain of pro-ALP. We note that both Erv26p and ALP are highly conserved in nature with both transmembrane (14) and glycosylphosphatidylinositol (GPI)-anchored (38) homologs of ALP found in different intracellular compartments and at the cell surface. We speculate that lumenal domain interaction between yeast pro-ALP and Erv26p represents a conserved interaction module that is necessary for efficient transport of eukaryotic pro-ALP proteins through the early secretory pathway. Homologous ALP proteins appear to present additional sorting determinants to specify transport routes in later branches of the secretory pathway. For example, in yeast a dileucine-like signal in the cytoplasmic tail sequence specifies AP3-dependent transport to the vacuole (18). In contrast, the four human ALP homologs are GPI-anchored proteins generally targeted to the outer leaflet of the plasma membrane in a variety of cell types (38). To date, however, all eukaryotic ALPs are synthesized as membrane-bound proteins consistent with our observation that the lumenal domain of yeast pro-ALP must be proximal to the inner ER membrane for efficient Erv26p-dependent export.
There are likely to be other instances of lumenal domain recognition of transmembrane secretory proteins by the ER export machinery. For example, Erv14p in yeast and the homologous cornichon protein in flies are required for ER export of integral membrane secretory proteins (39,40). In the case of cornichon and its cargo ligand, the transforming growth factor α(TGF α)-like cargo Gurken, lumenal domain interactions may be required for transport and loss of cornichon function can be overcome by appending a COPII-binding signal to the cytoplasmic tail of Gurken (41). In addition, we proposed that the Golgi glycosylation enzyme Ktr3p, which accumulates in the ER of erv26Δ mutants (42), depends on Erv26p for efficient export from the ER. Recent in vitro budding experiments on Ktr3p indicate that Erv26p is required for Ktr3p packaging into COPII vesicles (C. Bue and C. Barlowe, unpublished). Therefore, Ktr3p may contain lumenal domain sorting information. At this point we have not succeeded in further defining sequences or surface residues in yeast pro-ALP or Ktr3p that are recognized by Erv26p. Such recognition may rely on formation of tertiary or quaternary sorting elements in the lumenal domain of these secretory proteins. Structural studies combined with mutant analyses will be necessary to more fully define binding surfaces between pro-ALP and Erv26p.
Across many species, ALPs are homodimeric enzymes with very few exceptions (38). We observed that deletion of specific residues (Δ79-82) at the dimer interface in yeast pro-ALP generated a relatively stable protein that failed to dimerize, was inefficiently recognized and exported by Erv26p, and was ultimately subjected to ERAD. There are several examples where unassembled homo- and hetero-oligomeric secretory proteins fail to exit the ER and are degraded by the ERAD machinery (43–47). However, our results appear to be the first example where an export receptor was shown to discriminate between assembled and unassembled oligomers. Even with an accumulation of monomeric pro-ALP in the ER, we estimate at least a fivefold lower level of Erv26p binding than for wild-type pro-ALP. We hypothesize that the dimeric nature of Erv26p (42) allows for higher binding affinity to homodimeric pro-ALP than the monomeric species. Further experimentation with purified Erv26p and various species of pro-ALP will be needed to directly test this hypothesis. Regardless, our observation that Erv26p fails to efficiently recognize monomeric pro-ALP provides strong evidence for the idea that cargo packaging into COPII vesicles is tightly linked to ER quality control (4).
Regarding the mechanisms by which monomeric pro-ALP proteins are recognized as ERAD substrates and degraded in the ER, we found that general ERAD components Ubc7p, Sel1p and proteasome activity were required for turnover. Several lines of evidence indicate that multiple pathways operate upstream of these components to efficiently remove diverse ERAD substrates depending on if the misfolded determinant resides in the lumen (ERAD-L), membrane (ERAD-M) or cytoplasmic (ERAD-C) region of the substrate (11). We are currently determining which ERAD components and pathway(s) are responsible for turnover of monomeric Δ79-82ALP.
Interestingly, we observed that erv26Δ has a stabilizing influence on the Δ79-82ALP mutant. A second ERAD substrate, CPY*, was not stabilized in erv26Δ (Figure S3) suggesting that the effect on Δ79-82ALP was specific. A similar relationship has been reported for the cargo receptor Erv29p in which erv29Δ stabilizes its misfolded cargo ligand CPY*(48). It was also demonstrated that a fraction of CPY* continues to bind to Erv29p and can exit the ER in a wild-type background (48,49). Based on these and other results, it has been proposed that ER exit and retrieval from post-ER compartments makes CPY* more susceptible to ERAD (49). Given these findings, it seems possible that weak binding of the pro-form of Δ79-82ALP to Erv26p could yield a slow rate of export from the ER followed by its retrieval to facilitate ERAD. However, a review of the literature indicates that distinct pathways can operate on soluble and membrane-bound ERAD substrates (11) raising the possibility that unassembled membrane proteins may not need to exit the ER for their turnover. As an alternative explanation, we can envisage that a transient interaction of monomeric pro-ALP with Erv26p in the ER could temporarily draw unassembled pro-ALP monomers away from the folding machinery and in a position more susceptible to ERAD. It is also possible that an accumulation of Erv26p-dependent cargo in the ER of erv26Δ cells indirectly interferes with this class of ERAD substrates and slows Δ79-82ALP turnover.
Finally, we note that several genetically defined cases of hypophosphatasia, a human disorder characterized by defective bone and teeth mineralization, arise from missense mutations in the ALPL gene (13). ALPL encodes a dimeric GPI-anchored ALP protein that is present on the surface of cells in the liver, bone and kidney (50). More than 190 mutations in ALPL have been characterized and estimates indicate that severe forms of the disorder occur at a frequency of 1/100 000. Most of the missense mutations found in severe hypophosphatasia produce mutant ALP that accumulates in the early secretory pathway and fails to reach the cell surface (13). While ER export of GPI-anchored proteins may be distinct (51), if Erv26p functions in human cells as in yeast, it may be possible to manipulate the level of Erv26 expression and/or activity to influence ALPL expression. Future studies can now be directed at exploring the role of mammalian Erv26 in the biogenesis of homologous ALP proteins.
Materials and Methods
Bacterial and S. cerevisiae strains were propagated and transformed using standard procedures (52,53). Yeast semi-intact cell preparations and serum specific for α1,6-mannose carbohydrate have been described (54). Rabbit polyclonal antibodies directed against ALP, CPY, Sec61p, Erv41p and Och1p have been previously described (12). SDS-PAGE followed by immunoblotting on nitrocellulose membranes was performed (55). Immunoblots were developed using enhanced chemiluminescence (ECL) reagent (GE Healthcare); images were obtained using the UVP ChemiDoc Imaging System and formatted using Adobe Photoshop.
Strain and plasmids construction
Yeast strains and plasmids used in this study are listed in Table 1. Detailed methods used to generate these strains and plasmids are provided in the Supporting Information.
Table 1.
Strains and plasmids used in this study.
| Name | Description | Reference |
|---|---|---|
| Saccharomyces cerevisiae strains | ||
| CBY 1480 | MAT α his3 leu2 lys2 ura3 erv26∷KAN | (56) |
| CBY 2151 | MAT α his3 leu2 lys2 ura3 pho8∷KAN | (56) |
| CBY 2240 | MAT α his3 leu2 lys2 ura3 erv26∷KAN pho8 ∷LEU2 | This study |
| CBY 2771 | MAT α his3 leu2 lys2 ura3 ise1∷KAN | (56) |
| CBY 2773 | MAT α his3 leu2 lys2 ura3 ubc7∷KAN | (56) |
| CBY 1323 | MAT α his3 leu2 lys2 ura3 sel1∷KAN | (56) |
| CBY 80 | MAT α his3 leu2 lys2 ura3 trp1 | Barlowe lab |
| CBY 173 | MAT α his3 leu2 lys2 ura3 trp1 pep4 ∷URA3 | Barlowe lab |
| Plasmids | ||
| pFL8 | pRS313 (CEN HIS3) with EcoRI/BamHI full-length PHO8 | This study |
| pSN92 | pRS316 (CEN URA3) with full-length PHO8 | (18) |
| pNB4 | pRS316 (CEN URA3) with ΔT-ALP (Δ2 – 31ALP) | (18) |
| pAF13 | pRS313 (CEN HIS3) with Och1TM-ALP | This study |
| pQC43 | pRS313 (CEN HIS3) with αF-ss-ALP | This study |
| pCIP2 | pRS313 (CEN HIS3) with Och1CT-ALP | This study |
| pEC5 | pRS313 (CEN HIS3) with 8aa-ALP | This study |
| pQC72 | pRS313 (CEN HIS3) with 91aa-ALP | This study |
| pQC30 | pRS313 (CEN HIS3) with Δ79-82ALP | This study |
| pAE4 | pRS313 (CEN HIS3) with ALP-HA | This study |
| pCE1 | pRS316 (CEN URA3) with ALP | This study |
| pCF3 | pRS316 (CEN URA3) with Δ79-82ALP | This study |
| pCC1 | pRS313 (CEN HIS3) with Δ79-82ALP-HA | This study |
| pFA1 | pRS313 (CEN HIS3) with 91aa-ALP-HA | This study |
| pFB2 | pRS316 (CEN URA3) with 91aa-ALP | This study |
Pulse chase and immunoprecipitation
IP of ALP, CPY and HA-tagged proteins from [35S]-labelled cells was performed as previously described (57) with minor modifications. Briefly, cells were grown overnight at 30^C to stationary phase in minimal medium supplemented with appropriate amino acids. Cells were then back diluted to 0.1 OD600/mL in sulphate-reduced medium, grown for two cell divisions at 30^C and washed with sulphate-depleted medium. Twelve OD600 units of washed cells (3 OD600/mL) were incubated at 27^C (unless otherwise noted) in sulphate-depleted medium for 5 min before pulse with 25μCi/OD600 of [35S] Promix (GE Healthcare) for 7 min. Chase was initiated by adding unlabelled methionine and cysteine (2 mm each). Labelled cells (1 mL) were collected at indicated times and processed as described (58).
To measure α1,6-mannose modification of secretory proteins, primary IPs were performed with anti-ALP (or anti-CPY) antibodies from extract as described above. After washing, immunoprecipitates were denatured in 0.1 mL of 1% SDS, 50 mm dithiotreitol, 10% β-mercaptoethanol at 95^C for 5 min, centrifuged, and supernatants recovered. For secondary IPs, supernatants were divided into two parts, each diluted to 1 mL and immunoprecipitated with anti-ALP, anti-CPY or anti-α1,6-manose antibody. Immunoprecipitates were resolved on 8% gels and labelled species visualized by fluorography. Phosphor images were acquired with the STORM imager system and quantified using ImageQuant software.
Analysis of steady-state protein expression
To monitor protein expression levels, semi-intact yeast cells were prepared (54) and equivalent amounts of cellular protein resolved on polyacrylamide gels followed by immunoblotting. Protein concentrations were estimated from A280 reading in 2% SDS. In the case of αF-ss-ALP expression, cells were grown at 30^C to stationary phase in minimal medium supplemented with appropriate amino acids. Cultures were then back diluted to 0.1 OD600/mL in rich medium and grown at 30^C to mid-logarithmic phase. Five OD600 units of cells were collected by centrifugation and resuspended in 0.3 mL of buffer containing 20 mm Hepes pH 7.4, 100 mm sorbitol, 50 mm KOAc, 2 mm ethylenediaminetetraacetic acid (EDTA) and 1 mm phenylmethanesulphonyl fluoride (PMSF). Cell lysates were prepared by agitation with an equal volume of 0.5 mm acid-washed glass beads and spun at 1000×g for 3 min to pellet unbroken cells. Secreted protein was assessed from 1 mL of culture supernatants after trichloroacetic acid precipitation and washes with ice-cold 95% ethanol (57).
Cell fractionation on sucrose gradients
Cellular organelles were resolved on sucrose density gradients as previously described (12) with minor modification. Briefly, 200 OD600 units of mid-logarithmic cells were converted to spheroplasts, resuspended in buffer containing 10 mm Hepes pH 7.5, 1 mm EDTA, 12.5% sucrose, 0.5 mm PMSF, and lysed using a motorized Dounce homogenizer. Homogenates were spun at 1000×g for 5 min and the supernatant fluid loaded onto a 12-step sucrose gradient from 18 to 60% sucrose in 10 mm Hepes pH 7.5, 1 mm MgCl2. Gradients were centrifuged at 165,000×g for 3 h in an SW-40 rotor and divided into 15 or 16 fractions starting from the top of the gradient. Aliquots of each fraction were assessed by immunoblot.
Native immunoprecipitation (IP)
Methods to detect co-immunoprecipitating proteins by native IP have been described (12). To measure ALP dimerization, semi-intact cell membranes expressing HA-tagged and untagged versions of ALP were solubilized in B88-8 buffer [20 mm Hepes (pH 8), 250 mm sorbitol, 150 mm KOAc, 5 mm Mg(OAc)2] containing 5 mm EDTA, 10 mm PMSF and 1% digitonin. After a clearing spin, the solubilized material was subjected to IP in B88-8 buffer containing 0.05% digitonin, protein A-Sepharose beads plus or minus 0.5 μg of monoclonal anti-HA antibody (Covance) at 4^C for 1.5 h. Immune complexes bound to beads were washed four times with B88-8/0.05% digitonin and precipitated proteins detected by immunoblot.
Supplementary Material
Acknowledgments
We thank the Wickner laboratory for reagents and advice. This work was funded by the National Institutes of Health.
References
- 1.Jamieson JD, Palade GE. Synthesis, intracellular transport, and discharge of secretory proteins in stimulated pancreatic exocrine cells. J Cell Biol. 1971;50:135–158. doi: 10.1083/jcb.50.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 3.Kanapin A, Batalov S, Davis MJ, Gough J, Grimmond S, Kawaji H, Magrane M, Matsuda H, Schönbach C, Teasdale RD, Yuan Z. Mouse proteome analysis. Genome Res. 2003;13:1335–1344. doi: 10.1101/gr.978703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 5.Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 6.Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stagg SM, Gürkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 8.Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 9.Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 10.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 11.Sayeed A, Ng DT. Search and destroy: ER quality control and ER-associated protein degradation. Crit Rev Biochem Mol Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- 12.Bue CA, Bentivoglio CM, Barlowe C. Erv26p directs pro-alkaline phosphatase into endoplasmic reticulum-derived coat protein complex II transport vesicles. Mol Biol Cell. 2006;17:4780–4789. doi: 10.1091/mbc.E06-05-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mornet E. Hypophosphatasia. Orphanet J Rare Dis. 2007;2:40. doi: 10.1186/1750-1172-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 16.Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onishi HR, Tkacz JS, Lampen JO. Glycoprotein nature of yeast alkaline phosphatase. Formation of active enzyme in the presence of tunicamycin. J Biol Chem. 1979;254:11943–11952. [PubMed] [Google Scholar]
- 18.Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vowels JJ, Payne GS. A role for the lumenal domain in Golgi localization of the Saccharomyces cerevisiae guanosine diphosphatase. Mol Biol Cell. 1998;9:1351–1365. doi: 10.1091/mbc.9.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama K, Nagasu T, Shimma Y, Kuromitsu J, Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters MG, Evans EA, Blobel G. Prepro-alpha-factor has a cleavable signal sequence. J Biol Chem. 1988;263:6209–6214. [PubMed] [Google Scholar]
- 23.Sowadski JM, Handschumacher MD, Murthy HM, Foster BA, Wyckoff HW. Refined structure of alkaline phosphatase from Escherichia coli at 2.8 A resolution. J Mol Biol. 1985;186:417–433. doi: 10.1016/0022-2836(85)90115-9. [DOI] [PubMed] [Google Scholar]
- 24.De Backer M, McSweeney S, Rasmussen HB, Riise BW, Lindley P, Hough E. The 1.9 A crystal structure of heat-labile shrimp alkaline phosphatase. J Mol Biol. 2002;318:1265–1274. doi: 10.1016/s0022-2836(02)00035-9. [DOI] [PubMed] [Google Scholar]
- 25.Llinas P, Stura EA, Ménez A, Kiss Z, Stigbrand T, Millán JL, Le Du MH. Structural studies of human placental alkaline phosphatase in complex with functional ligands. J Mol Biol. 2005;350:441–451. doi: 10.1016/j.jmb.2005.04.068. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama Y, Ito K. Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J Biol Chem. 1993;268:8146–8150. [PubMed] [Google Scholar]
- 27.Boulanger RR, Kantrowitz ER. Characterization of a monomeric Escherichia coli alkaline phosphatase formed upon a single amino acid substitution. J Biol Chem. 2003;278:23497–23501. doi: 10.1074/jbc.M301105200. [DOI] [PubMed] [Google Scholar]
- 28.Rychlewski L, Jaroszewski L, Li W, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 30.Lee DH, Goldberg AL. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:30–38. doi: 10.1128/mcb.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 32.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 33.Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura N, Balch WE. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 35.Votsmeier C, Gallwitz D. An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 2001;20:6742–6750. doi: 10.1093/emboj/20.23.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kappeler F, Klopfenstein DRC, Foguet M, Paccaud JP, Hauri HP. The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J Biol Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- 37.Giraudo CG, Maccioni HJ. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol Biol Cell. 2003;14:3753–3766. doi: 10.1091/mbc.E03-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millan JL. Alkaline phosphatases structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signal. 2006;2:335–341. doi: 10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers J, Barlowe C. Transport of Axl2p depends on Erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Queenan AM, Barcelo G, Van Buskirk C, Schüpbach T. The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech Dev. 1999;89:35–42. doi: 10.1016/s0925-4773(99)00196-3. [DOI] [PubMed] [Google Scholar]
- 41.Bökel C, Dass S, Wilsch-Br äuninger M, Roth S. Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development. 2006;133:459–470. doi: 10.1242/dev.02219. [DOI] [PubMed] [Google Scholar]
- 42.Inadome H, Noda Y, Adachi H, Yoda K. Immunoisolaton of the yeast Golgi subcompartments and characterization of a novel membrane protein, Svp26, discovered in the Sed5-containing compartments. Mol Cell Biol. 2005;25:7696–7710. doi: 10.1128/MCB.25.17.7696-7710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Silva AM, Balch WE, Helenius A. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J Cell Biol. 1990;111:857–866. doi: 10.1083/jcb.111.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shenkman M, Ehrlich M, Lederkremer GZ. Masking of an endoplasmic reticulum retention signal by its presence in the two subunits of the asialoglycoprotein receptor. J Biol Chem. 2000;275:2845–2851. doi: 10.1074/jbc.275.4.2845. [DOI] [PubMed] [Google Scholar]
- 45.Hill K, Cooper AA. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belbeoc'h S, Massoulié J, Bon S. The C-terminal T peptide of acetylcholinesterase enhances degradation of unassembled active subunits through the ERAD pathway. EMBO J. 2003;22:3536–3545. doi: 10.1093/emboj/cdg360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang Z, Veeraprame H, Bayan N, Li G. The C-terminus of prenylin is important in forming a dimer conformation necessary for endoplasmic-reticulum-to-Golgi transport. Biochem J. 2004;380:43–49. doi: 10.1042/BJ20031788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- 49.Kincaid MM, Cooper AA. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol Biol Cell. 2007;18:455–463. doi: 10.1091/mbc.E06-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jemmerson R, Low MG. Phosphatidylinositol anchor of HeLa cell alkaline phosphatase. Biochemistry. 1987;26:5703–5709. doi: 10.1021/bi00392a019. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe R, Riezman H. Differential ER exit in yeast and mammalian cells. Curr Opin Cell Biol. 2004;16:350–355. doi: 10.1016/j.ceb.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Ausubel RM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York Greene Publishing Associates; Wiley-Interscience; 1988. pp. 3.0.1–3.14.3.. [Google Scholar]
- 53.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 54.Baker D, Hick L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- 55.Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 56.Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 57.Belden WJ, Barlowe C. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J Biol Chem. 1996;271:26939–42696. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- 58.Belden WJ, Barlowe C. Deletion of p24 genes activates the unfolded protein response. Mol Biol Cell. 2001;12:957–969. doi: 10.1091/mbc.12.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.