Abstract
The application of “omics” technologies to biological samples generates hundreds to thousands of biomarker candidates; however, a discouragingly small number make it through the pipeline to clinical use. This is in large part due to the incredible mismatch between the large numbers of biomarker candidates and the paucity of reliable assays and methods for validation studies. We desperately need a pipeline that relieves this bottleneck between biomarker discovery and validation. This paper reviews the requirements for technologies to adequately credential biomarker candidates for costly clinical validation and proposes methods and systems to verify biomarker candidates. Models involving pooling of clinical samples, where appropriate, are discussed. We conclude that current proteomic technologies are on the cusp of significantly affecting translation of molecular diagnostics into the clinic.
Keywords: Biomarker verification, Multiple reaction monitoring, Targeted proteomics
1 Introduction
Biomarkers are a cornerstone of medical care. In the acute care setting (e.g., emergency rooms), blood biomarker measurements are routinely used to differentiate causes of patient symptoms such as chest pain (troponins for heart attack) or abdominal pain (transaminases for hepatitis, alkaline phosphatase for biliary problems, and human chorionic gonadotropin (β-hCG) for pregnancy). Biomarkers also have a proven track record in other clinical applications such as risk stratifying patients for preventive interventions [1], screening populations for early disease detection [2], subtyping disease to facilitate molecularly tailored therapy [3], and monitoring response to treatment [4]. Additionally, biomarkers spur the development of new generations of therapeutics by providing accepted surrogates that reduce the cost of screening drugs in humans (e.g., LDL cholesterol for risk of stroke or heart attack, viral load for HIV).
Given the tremendous track record of biomarkers for impacting patient care and the medical community’s growing interest in personalized medicine, there is considerable activity toward the development of more and better biomarkers. With the recent application of genomics and proteomics technologies, hundreds-to-thousands of biomarker candidates are routinely identified in biomarker discovery experiments, spawning great hope that a new onslaught of clinically useful biomarkers is imminent. However, enigmatically, diminishingly few new protein biomarkers are achieving FDA approval [5], leading to a community that is disgruntled and questioning the value of proteomic technologies to biomarker discovery [6–10].
In this article, we will focus on the development of novel biomarkers that can be measured relatively noninvasively in plasma. We will review the current status of the biomarker development pipeline, with a focus on biomarker verification, the stage of the pipeline where our opportunities for improvement using emerging proteomic technologies are greatest. We will argue that clinical proteomics in the post-genomics era is in its infancy, and despite having produced no novel biomarkers to date, is poised to impact the clogged biomarker pipeline now more than at any other time in history. We will propose one possible path forward to apply emerging proteomic technologies in novel ways to improve flux through and overall success of the biomarker pipeline.
2 Current status of the biomarker development pipeline
2.1 Overview
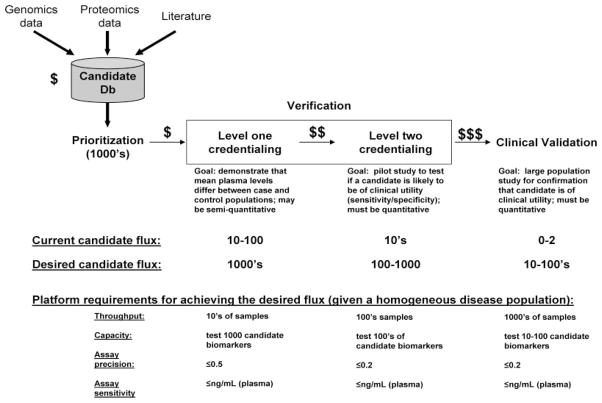
A schematic of the biomarker pipeline is shown in Fig. 1. The major stages in the pipeline are shown including biomarker candidate identification, prioritization, verification, and clinical validation. Additionally, typical numbers of candidate biomarkers making it through each stage are shown, highlighting the aforementioned enigma wherein despite our ability to generate long lists of candidates, a mere 0–2 per year are achieving FDA approval (across all diseases).
Figure 1.
Current and desired flux of candidates through the biomarker discovery pipeline. Note that despite our ability to generate long lists of candidate biomarkers using omics approaches, a mere 0–2 per year are achieving FDA approval (across all diseases). Our immediate goal is to improve this process 100-fold, which may be possible using a staged approach with sequential, cost-effective credentialing of candidate biomarkers. If conventional proteomics is to meet its promise for delivering molecular diagnostics, we must hone our ability to credential candidates in the most cost-effective manner. One approach is to stage candidate credentialing such that a small amount of resources are invested in early testing of a very large number of candidates, whereas greater resources are invested for assay development in only the subset of candidates showing greatest promise for clinical utility.
Low flux through the biomarker pipeline has been compared to that of the drug discovery pipeline, for which a high failure rate [11, 12] is responsible for the rising cost of bringing a new drug to market, which is now approaching $1 billion [13, 14]. Although the emergence of molecularly targeted therapies will likely change the situation, diagnostics have historically been perceived as being of lesser value than drugs [15]. Hence, diagnostics are reimbursed at significantly lower levels than therapeutics, and inadequate reimbursement is a profound impediment to development of new diagnostics [15]. Until this situation changes, we must minimize the cost of developing new diagnostics if we are to succeed in bringing new tests into clinical use.
In this article, we will discuss each stage of the protein biomarker pipeline, maintaining a focus on how each stage impacts the overall success rate of the pipeline. This will naturally lead us to a discussion of what can be done to improve our success rates (and reduce costs), especially in the verification stage, the “tar pit” of the pipeline wherein the largest bottleneck is encountered.
2.2 The high bar of clinical validation
Although it is the last step of the pipeline, we will begin our discussion with clinical validation since it sets the context for all of the upstream steps. Validation is expensive and time consuming, and the bar for success is daunting. It is not sufficient that a protein’s mean abundance differs between populations of cases and controls and that a clinical-grade assay is available; the successful protein biomarker must also perform responsibly and economically in a given clinical scenario.
Each clinical scenario will have different requirements for success. For example, a biomarker to be used for population screening for early cancer detection must meet very high standards; millions of healthy people are screened, and the overall disease incidence is low leading to a low pretest probability and high risk for false positives. Hence, to be beneficial, the test must have extraordinarily high specificity (or it must have a more specific follow-up test), and there must be a clinical intervention that will improve the quality of or prolong life.
Let’s take prostate-specific antigen (PSA) as an example. PSA is a protein secreted by normal and cancerous prostate epithelial cells. Currently PSA is the only serum- or plasma-based population screening tool we have for any cancer. Blood PSA levels are used to screen men over 50 years old for early diagnosis of prostate cancer. However, the specificity of this test is low; only 25–30% of men with a PSA elevated at 7.0 ng/mL will actually have prostate cancer on biopsy [16]. Hence, this low specificity screening marker (PSA) is coupled to a more specific follow-up test (biopsy) for definitive diagnosis. The end result is that the poor specificity of PSA leads to an annual cost of $750 million in unnecessary medical follow up [17]. There are other issues with PSA screening. For example, ideally we would diagnose only disease that will become clinically significant, otherwise intervention may cause more harm than good (this is called “overdiagnosis”). For example, since PSA screening has achieved widespread use, a man’s lifetime risk of being diagnosed with prostate cancer has increased to ~17%, yet his lifetime risk of dying from prostate cancer is only ~3% [18, 19]. So most men die “with” and not “of” their prostate cancer, and overdiagnosis in this population has been a major problem associated with significant treatment-related cost and morbidity.
In contrast to biomarkers for screening millions of mostly healthy individuals to detect asymptomatic disease, a biomarker intended to diagnose patients who have presented with a specific symptom must meet different minimum performance standards. For example, a patient with chest pain has an elevated prior probability that he is having a heart attack compared to the general population; because far fewer patients will be screened (only those with chest pain) and the prior probability of a heart attack is elevated over the general population, the number of false positive results requiring follow up will be much lower. These tailored clinical requirements, coupled to the need to validate markers in thousands of individuals with clinical follow-up information, create a situation wherein trials for validating biomarkers are lengthy, multimillion dollar endeavors. Hence, it is absolutely essential that we give priority to investing in clinical validation studies for only the most highly credentialed candidate biomarkers. In other words, each step in the biomarker pipeline must be designed with the clinical application in mind, rather than proceeding in a vacuum.
2.3 Biomarker candidate discovery
During the discovery of candidate biomarkers, a candidate database is populated via de novo discovery using genomic and/or proteomic technologies and/or through the curation of candidates from the scientific literature. Discovery efforts produce candidates (hypotheses), not biomarkers [20], and these efforts are inherently error-prone for multiple reasons. First, conventional technologies are not currently capable of globally interrogating biological proteomes, resulting in low sensitivity for directly detecting putative protein biomarkers. This is especially true for low concentration biomarkers or biomarkers resulting from disease-associated mutations, aberrant PTMs, or alternative splicing. Second, although genomics technologies are more comprehensive (and quantitative) than proteomics for discovering candidates, the correlation between DNA or mRNA copy number and protein abundance is imperfect [21–25], and thus many candidates discovered based on gene or mRNA copy number will not be elevated at the protein level. Third, proteomic-based discovery of biomarkers directly in plasma is challenging due to a small number of overwhelmingly high abundance proteins [5]. Thus, increasingly, discovery efforts are focusing first on tissues or proximal fluids for discovery, only moving to plasma once candidates have been identified. However, we have no good rules for predicting tissue proteins likely to be successful as plasma biomarkers, and the error rate is high. Fourth, discovery efforts are often poorly designed without clear understanding of the nuances of interpreting high dimensional datasets, often leading to biases [26] and high false discovery rates (FDR). Finally, discovery efforts rarely use pertinent clinical information to prioritize markers that will ultimately have the highest likelihood of success in the desired clinical setting. As a result, discovery efforts create large lists of candidate biomarkers, many of which may discriminate two classes of interest but are unlikely to meet the high bar of clinical validation, as discussed above.
Certainly, we can stack the deck in favor of success during the discovery process by using appropriate study designs that avoid bias, carefully estimating and controlling the FDR of our discovery technologies, and integrating clinical information early in the discovery or prioritization process. However, despite our earnest efforts to implement these measures, the success rate for translating biomarker candidates into biomarkers capable of meeting the high bar of clinical validation will still be disturbingly low. Our best hope for success is to relieve the bottleneck of candidate verification (Fig. 1) and allow the maximum number of candidate biomarkers to be tested, improving our odds of finding clinically useful markers. Strategies for achieving this goal are discussed in detail below.
2.4 Biomarker candidate prioritization
Because discovery efforts generate more candidates (100s–1000s) than there are available resources for follow up, an ill-defined prioritization step ensues. Often, candidates that show the most significant differences between cases and controls in discovery datasets are prioritized, without any information as to whether these may be the most useful analytes for clinical decision making. For example, many such candidates are a part of a generalized inflammatory response, and as individual clinical markers these are of very little diagnostic use due to their lack of specificity [27].
Proteins discovered in diseased tissues and predicted to be secreted or on the cell surface (based on the presence of a signal sequence, or N-linked glycosylation site) are often given priority based on the assumption that they might have greater access to plasma [28]. A better understanding of the biology of plasma biomarkers would help us develop meaningful criteria for prioritizing candidates. For example, what are the predominant modes by which cellular proteins from diseased tissues access the plasma? Should we be focused on proteins predicted to be secreted, based on the abundance of secreted proteins amongst the known plasma proteome [29], or does this rule not apply for proteins not in plasma by design, but rather leaked or shed from diseased tissue? Alternatively, an additional targeted proteomics step such as dynamic inclusion [30, 31] or multiple reaction monitoring (MRM) MS [32] can be used as an empirical prioritization step to test selected biomarker candidates to determine if they can be detected in plasma.
Other times prioritization is done based on a biological hypothesis. For example, proteins acting in cellular pathways known or hypothesized to be deregulated in the diseased state are targeted for testing. Although there are examples of success using this approach [33], our biological knowledge base is far too incomplete to rely entirely on this method for prioritizing candidates. In this sense it will likely be of use to use gene expression and protein interaction network analyses based on genomics data to refine candidate lists [34, 35] by selecting candidates from neighborhoods of interest within the network.
2.5 Biomarker candidate verification: The tar pit
To improve our success rate of moving candidate biomarkers successfully into clinical use, we must accommodate the harsh reality described above wherein even if our discovery efforts are honed to perfection, many candidates (perhaps the majority) will still not meet the high bar of clinical validation. In other words, despite our best efforts, it is highly likely that the majority of protein biomarker candidates will ultimately fail as useful clinical biomarkers. Hence, to succeed, we must develop a staged pipeline that incorporates a verification step that allows us to test (in pilot studies) the maximal number of candidates with highest possible throughput and lowest possible cost to ensure even a few successes [36].
Verification has a singular goal: to determine if there is sufficient evidence for potential clinical utility of a given candidate plasma biomarker to warrant further investment in that candidate for clinical validation studies. Because of the high cost (in terms of time, money, and consumption of clinical samples) of follow-up clinical validation studies, these “pilot” verification data are essential for credentialing a candidate to be moved forward.
In the current pipeline, the same assays are used for verification and clinical validation of biomarkers. In an ideal world this is advantageous since measurement of biomarkers can vary across different assays. One current assay often employed is the ELISA, which is understandable since a well-functioning ELISA can be relatively high throughput and has extraordinary sensitivity for quantifying the target analyte. Unfortunately, ELISA development is costly ($100 000–$2 million per biomarker candidate) and associated with a long development lead time (>1 year) and a high failure rate [37, 38], making it impractical to develop an ELISA for all putative biomarkers. As a result, even in the best-funded efforts, only a few percent of total candidate biomarkers for any given disease are actually tested (Fig. 1), not surprisingly leading to a high failure rate and an abysmal return on investment. Even if the immunoassay is to remain the gold standard for ultimate clinical application of validated biomarkers, we desperately need affordable bridging technologies to facilitate testing of a large number of potential candidates [37] if we are to identify the few that are likely to be of clinical use. To succeed, we must aim to increase our capacity in the verification stage by a 100-fold or more (Fig. 1).
The remainder of this article will focus on: (i) an exploration of the technological capabilities required for efficient verification of large numbers of candidates, (ii) a discussion of how current and emerging proteomic technologies measure up to these requirements, and (iii) a proposal for a staged approach to credentialing biomarkers in the most cost-effective manner.
3 Defining the platform requirements for verification studies
Let’s assume that a rigorously determined list of 1000 candidate plasma protein biomarkers for detecting prostate cancer has been produced, and that the discovery process was well orchestrated using a study design that avoided bias, used well-characterized technologies with low FDRs, and that information on the likelihood of clinical relevance was also included in the prioritization or discovery of these candidates (e.g., markers were generated that correlate with clinical outcome). We lack sufficient resources to build ELISAs for all of these 1000 candidates, yet we would like to perform verification studies for as many of the candidates as possible to maximize our chances to find the subset of the most clinically promising candidates for validation studies. So we desperately need a novel experimental approach, and potentially a new assay method, to measure 1000 candidates to accommodate our conundrum.
Let’s use known information about the PSA biomarker to define the boundaries of performance that will be required of our new approach to candidate verification. As discussed above, despite its widespread use, the performance of the PSA marker is marginal at best. Hence, we will use its performance characteristics to set the minimum standard that we will accept in our ongoing search for new markers for the detection of prostate cancer. In other words, we do not want to aim to discover markers that perform worse than PSA; we will only aim to discover markers that perform as well as or better than PSA.
Using the empirical distribution of PSA levels described from a previous population study [16], we will simulate different experimental scenarios for verification studies. We will consider two levels of candidate credentialing as part of the verification stage (Fig. 1):
Level one credentialing: demonstrating that the mean plasma levels of a given candidate are significantly different between a population of cases and a population of controls. Once the mean levels have been determined in the two populations, a statistical test (e.g., t-test) can be applied to assess the significance of any difference between cases and controls. (For the PSA example, the average level in the cancer group (7.63 ng/mL) is about 3.8-fold higher than the average level in the normal group (2.01 ng/mL; see Table 1 for statistical power).
Level two credentialing: pilot measurement of the performance characteristics (i.e., sensitivity and specificity) of the candidate marker in the desired clinical setting to estimate its likelihood of success in a subsequent larger clinical validation study.
Table 1.
Power calculations for case-control studies using PSA as an examplea)
(A) Level 1 credentialing (verification) in a homogeneous disease populationb)
| CV | Replicates | N = 5 | N = 10 | N = 20 |
|---|---|---|---|---|
| A.1 For the pooling of N case samples and N controls samples in a homogeneous disease population | ||||
| 0.8 | 3 | 30.0 | 30.5 | 30.6 |
| 5 | 50.4 | 51.1 | 51.6 | |
| 10 | 78.0 | 79.7 | 81.2 | |
| 15 | 88.2 | 91.7 | 93.7 | |
| 20 | 91.8 | 95.6 | 96.9 | |
| 0.5 | 3 | 51.0 | 52.3 | 52.4 |
| 5 | 79.0 | 81.7 | 83.5 | |
| 10 | 93.1 | 96.8 | 98.3 | |
| 15 | 96.0 | 98.8 | 99.8 | |
| 20 | 97.2 | 99.3 | 99.9 | |
| 0.2 | 3 | 93.0 | 96.0 | 97.0 |
| 5 | 97.4 | 99.6 | 100 | |
| 10 | 98.5 | 99.8 | 100 | |
| 15 | 98.7 | 99.8 | 100 | |
| 20 | 98.7 | 99.9 | 100 | |
| A.2 For comparing N individual case samples and N individual controls samples in a homogeneous disease population | ||||
| 0.8 | 1 | 30.0 | 60.7 | 88.3 |
| 3 | 63.7 | 92.2 | 99.5 | |
| 0.5 | 1 | 47.4 | 83.1 | 97.8 |
| 3 | 67.6 | 94.2 | 99.7 | |
| 0.2 | 1 | 66.0 | 93.0 | 99.7 |
| 3 | 68.6 | 95.0 | 99.8 | |
| 0.1 | 1 | 67.0 | 99.8 | 100 |
| 3 | 70.8 | 100 | 100 | |
As we will demonstrate, it is useful to divide verification into these two levels of credentialing because the technology requirements (sample throughput, assay precision, assay multiplexing) differ between them. For example, we will argue that although biomarker candidates must be measured in individual patient samples for level two credentialing, a pooling strategy is possible for level one credentialing. Pooling is potentially advantageous since pooling plasma samples from multiple individuals provides an opportunity to reduce sample numbers (and hence throughput requirements), reduce the sample volumes required from individual clinical samples, and reduce the cost of verification. Reduced throughput requirements are a major advantage early on in verification, since this allows us to accommodate workflows that are too cumbersome and imprecise for validation studies, but that may provide a fast and relatively cheap way to screen a large number of candidates (see below).
Table 1A–C show the results of simulating different experimental scenarios for level one and level two credentialing, respectively. The statistical power for detecting PSA as a potential biomarker in plasma is calculated for various assay precisions (coefficient of variation, CV), numbers of samples (N), and numbers of replicate assays performed. Here statistical power is defined as our probability of detecting a biomarker with our assay given that the marker is differentially expressed between case and control; ideally, our experimental design should be associated with as high of a statistical power as possible (to avoid false negatives), minimally >90%. For level one credentialing, two study designs are considered: one using pooled plasma from multiple individuals (Table 1A.1, 1B.1) and another using individual plasma samples (i.e., not pooled; Table 1A.2, 1B.2). Several important conclusions can be drawn regarding the performance requirements of our ultimate verification workflow.
(C) Level 2 credentialing (verification) in a homogeneous disease populationd)
| Assay CV | N = 100 | N = 200 | N = 500 | N = 1000 | N = 5000 |
|---|---|---|---|---|---|
| C.1 Estimated sensitivity (%) using different assay CVs and sample sizes | |||||
| 0.5 | 65(4.8) | 65(3.4) | 65(2.1) | 65(1.5) | 65(0.7) |
| 0.3 | 70.5(4.6) | 70.5(3.3) | 70.5(2.1) | 70.5(1.4) | 70.5(0.6) |
| 0.25 | 71.8(4.5) | 71.8(3.2) | 71.8(2) | 71.8(1.5) | 71.8(0.6) |
| 0.15 | 73.7(4.4) | 73.6(3.1) | 73.7(2) | 73.6(1.4) | 73.6(0.6) |
| 0.1 | 74.2(4.4) | 74.1(3.1) | 74.2(2) | 74.2(1.4) | 74.1(0.6) |
| 0.05 | 74.4(4.4) | 74.2(3.1) | 74.3(2) | 74.3(1.4) | 74.3(0.6) |
| C.2 Estimated specificity (%) under different assay CVs and sample sizes | |||||
| 0.5 | 84.4(3.7) | 84.4(2.6) | 84.4(1.7) | 84.4(1.2) | 84.4(0.5) |
| 0.3 | 86.2(3.4) | 86.2(2.4) | 86.1(1.5) | 86.1(1.1) | 86.1(0.5) |
| 0.25 | 86.6(3.4) | 86.6(2.4) | 86.5(1.5) | 86.5(1.1) | 86.5(0.5) |
| 0.15 | 87.3(3.3) | 87.3(2.3) | 87.3(1.5) | 87.2(1) | 87.2(0.5) |
| 0.1 | 87.7(3.2) | 87.5(2.3) | 87.6(1.4) | 87.6(1) | 87.5(0.5) |
| 0.05 | 88(3.2) | 87.9(2.2) | 87.9(1.4) | 87.9(1) | 87.9(0.5) |
| C.3 Power to detect all markers/tests comparable to or better than PSA.4 ng/ml (370% sample sensitivity and 385% sample specificity) using different assay CVs and sample sizes | |||||
| 0.5 | 7.7 | 3.7 | 0.4 | 0 | 0 |
| 0.3 | 36.1 | 40.5 | 45.4 | 56.2 | 76.8 |
| 0.25 | 45.3 | 53.7 | 69.2 | 82.4 | 99.7 |
| 0.15 | 60.6 | 73.2 | 90.3 | 97.8 | 100 |
| 0.1 | 66 | 77.8 | 94.2 | 99.2 | 100 |
| 0.05 | 69 | 82.3 | 96.1 | 99.7 | 100 |
The above data are simulations performed using actual population data for the PSA marker. Specifically, the distributions of PSA levels in the cancer and the normal populations were derived from Table 3 in Schroder et al. [16]. For parts A and C, power calculations were conducted as follows: (i) N individual PSA levels were sampled from the empirical PSA distributions of the cancer and the normal populations, respectively; (ii) experimental noise was simulated with the prespecified assay CV, and this noise was then added to the PSA levels; (iii) based on the simulated measurements, One-sided two-sample t-tests were performed (part A) or the sample sensitivity/specificity were estimated (part C); (iv) steps 1–3 were repeated for 5000 iterations, and the average power (at significance level 0.05) was calculated. (Note that the variability of the empirical distribution in Schroder et al. is larger than the simple biological variability of the PSA assay since the Schroder et al. data include all biological, preanalytical, and analytical variability across the population. Hence, we are actually underestimating our power somewhat in this conservative simulation). For part B, we evaluate the power for the pooling and individual strategies under the scenario where the target biomarker elevated in only a subgroup of the case (disease) population. Specifically, we consider three experimental scenarios: (B.1) pooling strategy; (B.2) individual strategy without prior information regarding relevant disease subtypes; (B.3) individual strategy with prior knowledge of the disease subtypes. We denote the percentage of the subtype with elevated marker levels in the whole disease population as S (10–50%) and perform the following simulation: (i) for the N individuals in the case group, select M individuals to represent the elevated subgroup, where M is from Bernoulli(S,N). Then, sample M values from the empirical PSA distribution of the cancer population and N–M values from the empirical PSA distribution of the normal population; (ii) for the N individuals in the control group, sample N values from the empirical PSA distribution of the normal population; (iii) We add different levels of experimental noise (varying CVs) to the simulated marker levels to generate the experimental measurements; (iv) based on the simulated measurements, one-sided two-sample t-tests were performed; (v) steps 1–4 were repeated for 5000 iterations, and the average power (at significant level 0.05) was calculated.
Shown are the powers to identify PSA as a potential marker (i.e., having a different mean levels in the disease and normal populations) in a homogeneous case population using either pooled (A.1) or individual (A.2) samples.
Shown are the powers to identify a target biomarker (i.e., having a different mean levels in the disease and normal populations) in a heterogeneous case population using either pooled (B.1) or individual (B.2 and B.3) samples.
Analysis of individual samples is required to determine whether a given biomarker candidate might achieve minimally acceptable sensitivity and specificity. The purpose of this simulation is to determine the effects of sample size and assay precision on our ability to estimate the sensitivity and specificity of a biomarker candidate typified by PSA. For this simulation, we use the conventional cutoff value of PSA = 4 ng/mL. At this cutoff, in a case-control study with equal sample sizes in the case and control groups, the sample sensitivity (percent of people having PSA>4 ng/mL among all case samples) for PSA is 73.9%, and the sample specificity (percent of case samples among all the samples having PSA>4 ng/mL) is 88%. (Note, the sample specificity is different from the specificity in the general population, which due to the overall low incidence of the disease is only 27.58% for a cutoff of PSA>4 ng/mL). C.1 and C.2 show our theoretical ability to estimate the sensitivity and specificity for PSA given various assay CV’s and sample sizes. C.3 shows the power to detect all markers/tests comparable to or better than PSA>4 ng/mL (≥70% sample sensitivity and ≥85% sample specificity) using different assay CVs and sample sizes.
Throughout Table 1 all powers ≥90% are shown in boldface.
The number in parentheses represent the standard deviation.
B Level 1 credentialing (verification) in a heterogeneous disease population in which the target biomarker is only elevated in S% of casesc)
| S (%) | CV | Replicates | N = 5 | N = 10 | N = 20 | N = 50 | N = 100 | N = 150 | N = 200 | N = 250 |
|---|---|---|---|---|---|---|---|---|---|---|
| B.1 For the pooling of N case samples and N controls samples and a heterogeneous disease population with no prior knowledge of disease subtypes (S) | ||||||||||
| 10 | 0.8 | 5 | 16.8 | 14.7 | 13 | 12.2 | 12.3 | 11.4 | 10.4 | 10.8 |
| 10 | 0.8 | 10 | 27.4 | 23.6 | 20.3 | 18.4 | 16.8 | 15 | 15.6 | 15.2 |
| 10 | 0.8 | 15 | 33.5 | 30.5 | 28.2 | 23.7 | 21.9 | 21.5 | 19.8 | 20.3 |
| 10 | 0.8 | 20 | 38 | 34.8 | 32 | 28.3 | 25.3 | 25.2 | 24.4 | 24.1 |
| 10 | 0.5 | 5 | 29 | 24.4 | 22.5 | 19.9 | 18.7 | 17.2 | 17 | 16.1 |
| 10 | 0.5 | 10 | 38.7 | 36 | 35.3 | 30.6 | 28.4 | 27 | 26.8 | 25.9 |
| 10 | 0.5 | 15 | 43 | 43.4 | 42.8 | 38.3 | 37.7 | 35.4 | 37.1 | 36.4 |
| 10 | 0.5 | 20 | 46.5 | 45.7 | 47 | 44.9 | 44.1 | 43.2 | 43.1 | 43.4 |
| 10 | 0.2 | 5 | 48.3 | 49.7 | 49.4 | 49.3 | 50.3 | 50.1 | 49.7 | 50.5 |
| 10 | 0.2 | 10 | 54.1 | 54.5 | 57.8 | 61.4 | 66.6 | 68.7 | 70.5 | 71.5 |
| 10 | 0.2 | 15 | 54.7 | 58.4 | 61.3 | 66.6 | 72.8 | 76.6 | 79.2 | 81.1 |
| 10 | 0.2 | 20 | 56.5 | 59 | 63.5 | 71.2 | 76.7 | 80.5 | 84.5 | 85.6 |
| 20 | 0.8 | 5 | 21.1 | 19.6 | 19.8 | 18.3 | 18.1 | 19.4 | 18.3 | 18.2 |
| 20 | 0.8 | 10 | 33.6 | 31.7 | 30 | 30.2 | 28.9 | 28.9 | 29.5 | 28.6 |
| 20 | 0.8 | 15 | 43.3 | 41.3 | 41.1 | 40.5 | 41.1 | 38.9 | 40.1 | 41 |
| 20 | 0.8 | 20 | 46.8 | 47.2 | 47.6 | 47.9 | 47.8 | 47.6 | 48.7 | 48.1 |
| 20 | 0.5 | 5 | 35.9 | 35 | 34.2 | 32.8 | 32.1 | 31.4 | 31.9 | 32.6 |
| 20 | 0.5 | 10 | 49.8 | 48.9 | 50.4 | 49.6 | 52.5 | 52 | 51.2 | 51.8 |
| 20 | 0.5 | 15 | 54.8 | 58.6 | 60.9 | 63.2 | 65.2 | 68.6 | 67.5 | 69.3 |
| 20 | 0.5 | 20 | 57.3 | 62.1 | 63.6 | 70 | 74.2 | 76 | 76.4 | 78.7 |
| 20 | 0.2 | 5 | 59.3 | 62.7 | 68.3 | 75.6 | 79.5 | 82.6 | 83.1 | 84 |
| 20 | 0.2 | 10 | 63.4 | 70.3 | 76.3 | 85.6 | 92.3 | 95.3 | 96.4 | 97.4 |
| 20 | 0.2 | 15 | 65.8 | 72.2 | 78.6 | 89.6 | 95.4 | 98.2 | 98.7 | 99.3 |
| 20 | 0.2 | 20 | 67.7 | 72.5 | 79.4 | 91.6 | 97 | 98.6 | 99.3 | 99.5 |
| 30 | 0.8 | 5 | 24.3 | 25.4 | 24.8 | 25.7 | 24.4 | 25.1 | 26 | 25.2 |
| 30 | 0.8 | 10 | 41.1 | 40.6 | 42.2 | 42.5 | 40.3 | 41.5 | 41.3 | 39.9 |
| 30 | 0.8 | 15 | 50.3 | 52.3 | 53.3 | 55.8 | 57.6 | 56.7 | 56.3 | 55.7 |
| 30 | 0.8 | 20 | 56.3 | 58.1 | 59.5 | 64.6 | 65.2 | 66.3 | 67 | 66.4 |
| 30 | 0.5 | 5 | 43.6 | 43.3 | 44 | 45 | 45.4 | 44.5 | 45.3 | 46.6 |
| 30 | 0.5 | 10 | 57.7 | 61 | 65.1 | 68.5 | 70.7 | 71.6 | 72.1 | 72.7 |
| 30 | 0.5 | 15 | 63.5 | 70.5 | 74.3 | 81.2 | 84.1 | 85.9 | 86.1 | 87.6 |
| 30 | 0.5 | 20 | 66.7 | 72.8 | 79.6 | 86.1 | 90.7 | 92.1 | 92.8 | 94.1 |
| 30 | 0.2 | 5 | 68.8 | 75 | 81.6 | 89.2 | 93.7 | 95.7 | 96 | 96.2 |
| 30 | 0.2 | 10 | 72.6 | 78.4 | 87.4 | 95.8 | 98.9 | 99.5 | 99.8 | 99.9 |
| 30 | 0.2 | 15 | 73.1 | 81.6 | 90.4 | 97.1 | 99.6 | 99.9 | 100 | 100 |
| 30 | 0.2 | 20 | 75.4 | 83 | 91.2 | 97.9 | 99.8 | 100 | 100 | 100 |
| 40 | 0.8 | 5 | 28.3 | 28.8 | 28.7 | 31 | 29.9 | 30.8 | 31.3 | 30.7 |
| 40 | 0.8 | 10 | 47.9 | 48.5 | 49.8 | 51.6 | 51.9 | 52.8 | 51.3 | 52.5 |
| 40 | 0.8 | 15 | 59.2 | 60.6 | 63.3 | 65.6 | 68.6 | 69.1 | 68.9 | 69.2 |
| 40 | 0.8 | 20 | 63.8 | 66.5 | 71.3 | 74.2 | 77.2 | 78.4 | 79.2 | 78.6 |
| 40 | 0.5 | 5 | 49.2 | 50.9 | 52.5 | 54.5 | 55.4 | 55.5 | 56.2 | 55.7 |
| 40 | 0.5 | 10 | 64.6 | 70.3 | 75.6 | 80.2 | 83.6 | 83.8 | 84.7 | 83.2 |
| 40 | 0.5 | 15 | 71.7 | 78.1 | 84.1 | 90 | 93.3 | 94.2 | 94.9 | 95.1 |
| 40 | 0.5 | 20 | 74.4 | 80.6 | 87.7 | 94.4 | 96.8 | 97.5 | 97.8 | 98.1 |
| 40 | 0.2 | 5 | 75.1 | 83.2 | 89.8 | 96.3 | 98.3 | 98.8 | 99 | 99.1 |
| 40 | 0.2 | 10 | 79.9 | 87.4 | 94.1 | 99.1 | 99.8 | 100 | 100 | 100 |
| 40 | 0.2 | 15 | 81.2 | 88.8 | 95.3 | 99.5 | 100 | 100 | 100 | 100 |
| 40 | 0.2 | 20 | 81.8 | 89.1 | 95.6 | 99.6 | 99.9 | 100 | 100 | 100 |
| 50 | 0.8 | 5 | 32.7 | 34.8 | 33.3 | 34.4 | 36.3 | 35 | 36.1 | 35.9 |
| 50 | 0.8 | 10 | 53.7 | 56.4 | 57.1 | 58.1 | 59 | 60.2 | 60 | 59.2 |
| 50 | 0.8 | 15 | 64.5 | 69.4 | 71.3 | 73.9 | 77 | 76.2 | 76.2 | 77.3 |
| 50 | 0.8 | 20 | 70.1 | 75.3 | 79.9 | 83.9 | 86.4 | 85.7 | 86.3 | 87 |
| 50 | 0.5 | 5 | 54.6 | 56.9 | 60.3 | 62.7 | 63.7 | 63.3 | 63.9 | 62.9 |
| 50 | 0.5 | 10 | 72.1 | 78.3 | 82.8 | 88.8 | 88.9 | 89.8 | 90.9 | 90.3 |
| 50 | 0.5 | 15 | 78.2 | 84.8 | 91.2 | 95.7 | 97.1 | 97.5 | 98 | 98 |
| 50 | 0.5 | 20 | 79.6 | 87.9 | 93.3 | 97.7 | 99.1 | 99.5 | 99.5 | 99.6 |
| 50 | 0.2 | 5 | 83 | 89.7 | 94.8 | 98.7 | 99.4 | 99.4 | 99.6 | 99.6 |
| 50 | 0.2 | 10 | 85 | 92.2 | 97.6 | 99.9 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 15 | 86.1 | 93.7 | 98.1 | 99.9 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 20 | 87.8 | 94.5 | 98.3 | 100 | 100 | 100 | 100 | 100 |
| B.2 For comparing N individual case samples and N individual controls samples and a heterogeneous disease population with no prior knowledge of disease subtypes (S) | ||||||||||
| 10 | 0.8 | 1 | 6.1 | 6.7 | 8 | 11.4 | 15.3 | 18.3 | 23.5 | 25.9 |
| 10 | 0.8 | 3 | 6.7 | 7.1 | 11 | 16.2 | 24 | 31.5 | 38.6 | 43.8 |
| 10 | 0.8 | 5 | 7 | 7.8 | 11 | 17.1 | 25.5 | 31.8 | 37.4 | 44 |
| 10 | 0.5 | 1 | 7 | 6.9 | 9.4 | 13.8 | 21.8 | 26.8 | 31.6 | 37.9 |
| 10 | 0.5 | 3 | 7.6 | 7.9 | 10.3 | 16.4 | 23.7 | 33.1 | 39.3 | 44.4 |
| 10 | 0.5 | 5 | 6.9 | 7.6 | 10.1 | 16.9 | 25.5 | 31.4 | 38.5 | 44.8 |
| 10 | 0.2 | 1 | 6.5 | 7.7 | 11 | 15.8 | 24.7 | 32.2 | 37.1 | 45.2 |
| 10 | 0.2 | 3 | 7.3 | 8.3 | 10.4 | 16.1 | 25.1 | 33.2 | 40 | 47.2 |
| 10 | 0.2 | 5 | 7.2 | 8 | 11 | 15.6 | 24.5 | 31.3 | 38.8 | 45.2 |
| 20 | 0.8 | 1 | 8.4 | 9.9 | 13.5 | 22.6 | 36.3 | 46.7 | 56.8 | 64.2 |
| 20 | 0.8 | 3 | 10.1 | 13.3 | 20.7 | 38.3 | 58.5 | 75.1 | 86.2 | 91.4 |
| 20 | 0.8 | 5 | 10.5 | 13.6 | 20.9 | 38.8 | 60.1 | 77.1 | 87.1 | 92.4 |
| 20 | 0.5 | 1 | 9.2 | 10.6 | 18.2 | 32.5 | 50.3 | 65 | 77.2 | 83.7 |
| 20 | 0.5 | 3 | 10.5 | 13.3 | 20 | 38.5 | 60.5 | 76.8 | 86.8 | 92.5 |
| 20 | 0.5 | 5 | 10.1 | 13.4 | 21.7 | 38.8 | 61.8 | 76.2 | 86.9 | 92.9 |
| 20 | 0.2 | 1 | 10 | 12.9 | 20.2 | 37.7 | 61.6 | 76.3 | 85.9 | 92.2 |
| 20 | 0.2 | 3 | 10.1 | 12.9 | 21.8 | 38.3 | 60.8 | 77.4 | 87 | 92.5 |
| 20 | 0.2 | 5 | 10.7 | 13.9 | 20.6 | 39.5 | 61.7 | 76.9 | 86 | 92.8 |
| 30 | 0.8 | 1 | 9.9 | 13.5 | 22.4 | 38.6 | 60.4 | 77 | 85.2 | 92.3 |
| 30 | 0.8 | 3 | 13.8 | 19.8 | 32.9 | 63.8 | 89 | 96.9 | 99.2 | 99.8 |
| 30 | 0.8 | 5 | 13.7 | 20.2 | 35.3 | 65.7 | 89.8 | 97.2 | 99.3 | 99.8 |
| 30 | 0.5 | 1 | 12.2 | 17 | 27.9 | 53.6 | 79.9 | 91.8 | 97.3 | 99.2 |
| 30 | 0.5 | 3 | 13.9 | 21.8 | 34.4 | 65.6 | 89.4 | 97.6 | 99.3 | 99.9 |
| 30 | 0.5 | 5 | 14.8 | 20.5 | 35.7 | 66.1 | 89.5 | 97.9 | 99.4 | 99.9 |
| 30 | 0.2 | 1 | 14.7 | 21.2 | 33.6 | 65.1 | 89.3 | 97.5 | 99.3 | 99.9 |
| 30 | 0.2 | 3 | 14.2 | 20.6 | 36.1 | 65.7 | 90.4 | 97.3 | 99.3 | 99.8 |
| 30 | 0.2 | 5 | 14.4 | 21.6 | 35.2 | 65.5 | 90.5 | 97.6 | 99.4 | 99.9 |
| 40 | 0.8 | 1 | 12.8 | 17.8 | 29.2 | 56.4 | 82.5 | 93.2 | 98.2 | 99.2 |
| 40 | 0.8 | 3 | 18.6 | 30.2 | 50.1 | 85.6 | 98.9 | 99.9 | 100 | 100 |
| 40 | 0.8 | 5 | 18.5 | 30.3 | 51.7 | 86.3 | 99.1 | 100 | 100 | 100 |
| 40 | 0.5 | 1 | 16.9 | 25.5 | 42.9 | 76.1 | 95.7 | 99.2 | 100 | 100 |
| 40 | 0.5 | 3 | 19.4 | 30.5 | 53.6 | 85.8 | 98.9 | 100 | 100 | 100 |
| 40 | 0.5 | 5 | 19.7 | 29.8 | 53.7 | 87.7 | 99 | 100 | 100 | 100 |
| 40 | 0.2 | 1 | 18.9 | 30.3 | 52.8 | 86.5 | 99 | 100 | 100 | 100 |
| 40 | 0.2 | 3 | 20.3 | 31.9 | 55.1 | 87.5 | 99.1 | 99.9 | 100 | 100 |
| 40 | 0.2 | 5 | 20 | 31.2 | 52.6 | 88 | 98.9 | 100 | 100 | 100 |
| 50 | 0.8 | 1 | 14.9 | 24.9 | 40.7 | 73.7 | 94.4 | 98.9 | 99.9 | 100 |
| 50 | 0.8 | 3 | 25.3 | 41.5 | 69.2 | 96.5 | 99.9 | 100 | 100 | 100 |
| 50 | 0.8 | 5 | 27.2 | 42.6 | 70.1 | 96.8 | 100 | 100 | 100 | 100 |
| 50 | 0.5 | 1 | 21.8 | 33.5 | 58.2 | 91 | 99.5 | 100 | 100 | 100 |
| 50 | 0.5 | 3 | 26.6 | 42.2 | 70.6 | 96.4 | 100 | 100 | 100 | 100 |
| 50 | 0.5 | 5 | 26.6 | 43.4 | 70.6 | 97.2 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 1 | 25.6 | 43.2 | 70.9 | 96.6 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 3 | 26.5 | 42.4 | 70.6 | 96.6 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 5 | 26.4 | 43.7 | 71.5 | 97.1 | 100 | 100 | 100 | 100 |
| B.3 For comparing N individual case samples and N individual controls samples and a heterogeneous disease population, given prior knowledge of disease subtypes (S) | ||||||||||
| 10 | 0.8 | 1 | 2.9 | 9.1 | 35.9 | 60 | 83 | 92.4 | 97 | 98.7 |
| 10 | 0.8 | 3 | 3.9 | 15 | 55.9 | 90.8 | 99.5 | 100 | 100 | 100 |
| 10 | 0.8 | 5 | 3.7 | 15.2 | 56.8 | 92.8 | 99.7 | 100 | 100 | 100 |
| 10 | 0.5 | 1 | 3.2 | 10.8 | 47.7 | 80.6 | 96.7 | 99.5 | 99.9 | 100 |
| 10 | 0.5 | 3 | 4.5 | 15.6 | 58.6 | 91.2 | 99.7 | 100 | 100 | 100 |
| 10 | 0.5 | 5 | 4.1 | 14.7 | 58.5 | 91.6 | 99.7 | 100 | 100 | 100 |
| 10 | 0.2 | 1 | 3.8 | 14.5 | 57.9 | 91.6 | 99.6 | 100 | 100 | 100 |
| 10 | 0.2 | 3 | 4.3 | 15.3 | 58.3 | 92.1 | 99.7 | 100 | 100 | 100 |
| 10 | 0.2 | 5 | 4.6 | 15.3 | 57.9 | 91.6 | 99.6 | 100 | 100 | 100 |
| 20 | 0.8 | 1 | 8.8 | 23.8 | 51.3 | 81.4 | 96.4 | 99.5 | 99.9 | 100 |
| 20 | 0.8 | 3 | 13 | 39.4 | 83.1 | 99.4 | 100 | 100 | 100 | 100 |
| 20 | 0.8 | 5 | 14.4 | 39.3 | 84.1 | 99.4 | 100 | 100 | 100 | 100 |
| 20 | 0.5 | 1 | 11.7 | 31.3 | 69.5 | 95.7 | 99.8 | 100 | 100 | 100 |
| 20 | 0.5 | 3 | 13.8 | 39 | 82.9 | 99.5 | 100 | 100 | 100 | 100 |
| 20 | 0.5 | 5 | 14.3 | 41.6 | 85.7 | 99.6 | 100 | 100 | 100 | 100 |
| 20 | 0.2 | 1 | 14.3 | 40.4 | 84.1 | 99.6 | 100 | 100 | 100 | 100 |
| 20 | 0.2 | 3 | 14.3 | 39.7 | 84.3 | 99.5 | 100 | 100 | 100 | 100 |
| 20 | 0.2 | 5 | 14.2 | 39.9 | 84.3 | 99.6 | 100 | 100 | 100 | 100 |
| 30 | 0.8 | 1 | 15.1 | 34.5 | 61.7 | 90.7 | 99.1 | 99.9 | 100 | 100 |
| 30 | 0.8 | 3 | 25.3 | 57.6 | 93 | 99.9 | 100 | 100 | 100 | 100 |
| 30 | 0.8 | 5 | 25 | 59.9 | 94 | 100 | 100 | 100 | 100 | 100 |
| 30 | 0.5 | 1 | 19.9 | 48.3 | 81.7 | 99.3 | 100 | 100 | 100 | 100 |
| 30 | 0.5 | 3 | 26.1 | 62 | 94 | 100 | 100 | 100 | 100 | 100 |
| 30 | 0.5 | 5 | 26.2 | 61.7 | 93.8 | 100 | 100 | 100 | 100 | 100 |
| 30 | 0.2 | 1 | 25.3 | 61.7 | 94.5 | 100 | 100 | 100 | 100 | 100 |
| 30 | 0.2 | 3 | 25.9 | 61.9 | 94.5 | 100 | 100 | 100 | 100 | 100 |
| 30 | 0.2 | 5 | 25.6 | 61.5 | 94.2 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.8 | 1 | 22 | 43.5 | 69.2 | 95 | 99.9 | 100 | 100 | 100 |
| 40 | 0.8 | 3 | 35.6 | 72 | 97.1 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.8 | 5 | 35.7 | 75.1 | 97.6 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.5 | 1 | 29.3 | 61.9 | 89.9 | 99.9 | 100 | 100 | 100 | 100 |
| 40 | 0.5 | 3 | 36.3 | 74.8 | 98 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.5 | 5 | 36.5 | 76.6 | 98.2 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.2 | 1 | 36.9 | 73.8 | 97.6 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.2 | 3 | 38.2 | 76.5 | 98.2 | 100 | 100 | 100 | 100 | 100 |
| 40 | 0.2 | 5 | 37.4 | 76.9 | 97.7 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.8 | 1 | 26.5 | 48.4 | 74 | 97.6 | 100 | 100 | 100 | 100 |
| 50 | 0.8 | 3 | 44.5 | 81.7 | 99.3 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.8 | 5 | 48.1 | 84.8 | 99 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.5 | 1 | 36.2 | 68.4 | 94 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.5 | 3 | 46.9 | 84.3 | 99.2 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.5 | 5 | 46.5 | 85.5 | 99.5 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 1 | 45.5 | 84.5 | 99.3 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 3 | 47.3 | 85.8 | 99.3 | 100 | 100 | 100 | 100 | 100 |
| 50 | 0.2 | 5 | 48 | 85.4 | 99.2 | 100 | 100 | 100 | 100 | 100 |
For level one credentialing, if we want ≥90% power to detect PSA as a potential biomarker worthy of further study (i.e., mean plasma levels significantly differ between case and control populations), we find that:
We can achieve our goal using either pooled samples or by analyzing individual samples.
Pooling is advantageous whenever the sample size is limited since the impact of sample size is much less for pooled samples than for individual analyses.
Pooling is also advantageous wherever the cost and/or throughput of each experiment are limiting. For example, if we fix the CV = 0.5 and N = 10, the pooled strategy would require 20 experimental runs (ten replicates each of one case pool and one control pool) to achieve 96.8% power to detect PSA as a potentially useful marker. In contrast, the individual analyses strategy would require 60 experimental runs (three replicates each of ten cases and ten controls) to achieve comparable power (94.2%).
If adequate numbers of clinical samples are available, and if the cost and throughput of experiments is not a concern, then an individual sample design should be chosen for two reasons: (a) Analyses of individual samples will ultimately be required for clinical validation of candidate markers, so that operating characteristics (e.g., sensitivity and specificity) can be determined (Table 1C); (b) a drawback to pooling lies in the reduction of bimodal populations to a single, more homogenous population. If there is heterogeneity in the disease population (e.g., molecular subtypes of cancer such as hormone-responsive vs. hormone-resistant cancer) resulting in the target biomarker’s being elevated in only a subpopulation (S), this marker could be lost by pooling, depending on the size of the subpopulation. This is demonstrated in Tables 1B.1–B.3 where we repeat the simulation assuming two disease subtypes within the cases with or without prior knowledge of these subtypes. If we have no prior knowledge regarding the presence of disease subtypes in our case population and if sample numbers are small (<200 cases), a pooling strategy works best even if there are two subtypes of disease in the case population. For example, if a given biomarker is elevated in only 20% of the case population and we fix the assay CV = 0.2 and N = 50, a pooling strategy would give us a 75.6% chance of detecting the marker in the subpopulation (assuming five replicate measurements per pool = 10 measurements total; Table 1B.1), whereas an individual strategy would only give us a 37.7% chance of detecting the marker in the subpopulation (assuming one measurement per sample = 100 assays total; Table 1B.2), Although this is initially counterintuitive, the enlarged variation in the case population (due to the presence of two subtypes) makes the individual strategy less favorable until sample size becomes larger (N>200). It is also noteworthy that at sample sizes <200, pooling not only provides greater statistical power but also lower-throughput requirements, as in the example just discussed (10 vs. 100 assays required). If we do have prior knowledge about the two subtypes of cases in our study population (i.e., the clinical samples are annotated to allow us to identify the two subpopulations), then the individual strategy is clearly most advantageous (Table 1B.3).
It is apparent from Table 1 that precision (CV) has a major impact on our statistical power. Hence, to maximize our capacity to test candidates, it is imperative that we optimize our verification assay workflows to ensure the highest precision possible and institute Standard Operating Procedures (SOP) to ensure that we consistently achieve high precision. For example, in the individual sample analyses of a homogeneous disease population (Table 1A.2) where N = 10 and CV = 0.2, 20 sample analyses (one replicate for each of ten cases + ten controls) must be performed to achieve power = 93%; in contrast 60 sample analyses (three replicates for each of ten cases + ten controls) would be required to achieve comparable power (92.2%) using a platform with CV = 0.8. Thus, a platform with CV = 0.2 has 3×greater capacity to test candidates than a platform with CV = 0.8.
Based on the above considerations, in order to achieve level one credentialing (for a candidate typified by PSA and a homogeneous disease population), we will need plasma samples from 20 cases and 20 well-matched controls. Additionally, we need to devise an assay technology with the following characteristics
Capacity to test 1000 candidate biomarker proteins over a several month period;
sensitivity ≤nanogram per milliliter in plasma;
assay CV≤0.5;
throughput to run up to ten replicate measurements per candidate.
If the target biomarker is elevated in only a subset (S) of the case population of which we have no prior knowledge, our requirements are more stringent. In this scenario, we will need >100 samples (depending on the prevalence of the disease subtype in which the biomarker is present; Table 1B) and assay CV ≤ 0.2 to detect a marker elevated in at least 20% of the case population.
For level two credentialing, our goal is to identify the subset of candidate markers most likely to meet the minimally acceptable sensitivity and specificity in a given clinical setting. Hence, we must perform a pilot study to characterize the distribution of the marker in the population, allowing us to estimate its sensitivity and specificity. The success of this step relies on how accurately the sensitivity and specificity can be measured; therefore, assay precision (CV) again plays an important role. As we can see from Table 1C, a large CV will result in underestimation of sensitivity and specificity. For example, for PSA the actual sample sensitivity is 73.9% and sample specificity is 88% [16]. In our simulation, when CV = 0.5 the estimated sensitivity is about 65%, which is 8.9% lower than the true sensitivity of PSA based on the population study [16]. In addition to using a precise assay, a larger number (100s–1000s) of individual patient samples (Table 1C) will be needed compared with level one credentialing. For example, for the estimation of sensitivity (Table 1C.1), around 1000 cases and 1000 controls would be needed to get a 90% confidence interval (CI) spanning less than 5% (i.e., CI = (x − 2.5%, x + 2.5%)); or more than 5000 cases and 5000 controls will be needed to get a CI spanning less than 2% (i.e., (x − 1%, x + 1%)). These requirements can also be viewed from another angle. In order to have 90% power to identify PSA as a good candidate marker worthy of follow up (i.e., 70% sample sensitivity and 85% sample specificity), we would need 500 cases and 500 controls with a CV = 0.15. By comparison, this would require a couple of thousand cases and controls with a CV = 0.25.
Based on the above considerations, in order to achieve level two credentialing for a marker similar to PSA, we will need plasma samples from a minimum of 500 cases and 500 well-matched controls. Additionally, we need to devise an assay technology with the following characteristics:
Capacity to test 100s candidate biomarker proteins over a few month period;
sensitivity ≤ nanogram per milliliter in plasma;
assay CV≤0.2;
throughput to run up to 1000 measurements per candidate biomarker.
Note that level two credentialing is still just a pilot study using limited throughput assays to determine if a candidate is trending toward utility and therefore worthy of making a better high-throughput clinical-grade assay. True clinical validation, however, will require an even larger-scale case-control or cohort study in order to carefully examine the impact of other covariates on the proposed marker test, to determine the positive predictive values and false referral probabilities in real practice, and to compare or combine the new test with existing clinical tests. Although candidates showing promise in pilot level two credentialing studies may still not pass the test of ultimate clinical validation, level two credentialing is important because it allows us to advance only the most promising of candidates forward to clinical validation trials, thereby saving time, money, and clinical specimens and helping to maximize our return on investment.
It should be noted that the power calculations described in Table 1 are based on known distributions of PSA levels in the cancer and the normal populations. Hence, these results can be generalized to other biomarkers showing similar population distributions, but markers with vastly different distributions would require that new calculations be performed based on the specific behavior of that marker. In the absence of knowing the population variation for markers yet to be discovered, it is useful to look at a well-studied example such as PSA to provide general guidance in planning verification studies.
4 Proposed staged approach to biomarker verification and validation
The further along in the biomarker pipeline that a candidate moves, the more time and resources become invested in that candidate. Hence, it is prudent for us to advance candidates through sequential, economical stages, each stage requiring additional credentialing for a given candidate to be advanced (Fig. 1). Based on the above statistical considerations (Table 1), one possible staged workflow for biomarker candidate verification and validation is described below.
4.1 Level one credentialing ($)
The goal is to determine if the mean levels of each of ~1000 candidate protein biomarkers differ between case and control populations to allow selection of a subset of most promising markers for further investment in assay development. Table 2 summarizes the reagents cost, lead time, sensitivity, throughput (working assay), and sample consumption for several existing and emerging technologies for measuring protein levels. If resources were unlimited, we would generate antibodies/immunoassays for each candidate so that we could achieve the highest possible sensitivity for detecting the candidates in plasma. Unfortunately, for the vast majority of protein biomarker candidates there will be no commercially available antibody, and generating antibodies to all of our 1000 candidates is cost- and time-prohibitive (estimated cost >$2 million; lead time 9–12 months).
Table 2.
Summary of available assays for verification of protein biomarker candidates
| Assay | Reagents | Reagents Cost | Lead Time | Sensitivity | Throughput (working assay) | Sample consumption | Notes |
|---|---|---|---|---|---|---|---|
| Western blot | mAb | Mouse $6000; rabbit $8000 Production and purification mAb $2500 |
9–12 months | 5–500 ng/mL [72–74] | 20 antibodies per day | 10 μL; 1 candidate | Not quantitative; low specificity esp for pAb |
| pAb | $1000 (affinity purified) | 6–9 months | |||||
| ELISA | Two antibodies | Make MAbs $6000–16 000 Production and purification 2 mAb’s $5000 |
18–24 months | 10 pg/mL [75] | 25 candidates per day | 10–100 μL; 1 candidate | Autoantibody interference (false negatives) Heterophilic antibodies (false positives) |
| Recombinant protein | Make recombinant protein $5000 | ||||||
| MRM (depleted plasma) | Labeled and unlabeled peptide | Peptides $1000 (5 nmol) | 1–2 months | 100–1000 ng/mL [46, 49] | 150 peptides×5 samples per instrument per day | 100 μL; 150 peptides | Must choose proteotypic peptides |
| MRM (depleted + SCX plasma) | Labeled and unlabeled peptide | Peptides $1000 (5 nmol) | 1–2 months | 1–100 ng/mL [46] | 150 peptides×1 sample per instrument per day | 100 μL; 150 peptides | Must choose proteotypic peptides |
| MRM (SISCAPA eluate) | Labeled and unlabeled peptide | Peptides $1000 (5 nmol) | 6–9 months | 10–100 ng/mL [32] | 10 peptides×20 samples per day | 10–100 μL; 10 candidates | Must choose proteotypic, immunogenic peptides |
| Antibody | PcAb $1500 (affinity purified) Mouse mAb with production $8500 rabbit MAb with production $10 500 |
18 months | TBD TBD |
||||
| MRM (SCX + SISCAPA) | Labeled and unlabeled peptide | Peptides $1000 (5 nmol) | 6–9 months | TBD | 10 peptides×20 samples per day | 10–100 μL; 10 candidates | Must choose proteotypic, immunogenic peptides |
| Antibody | PcAb $1500 (affinity purified) Mouse mAb with production $8500 Rabbit mAb with production $10 500 |
18 months | TBD TBD |
||||
Building assays to follow up candidates emerging from biomarker discovery experiments is a major bottleneck in the pipeline. Shown are some of the working parameters for several conventional and emerging assay formats.
TBD, to be determined; pAb, polyclonal Ab.
One emerging alternative to the immunoassay is to use MRM-MS/MS to verify candidate biomarkers. This targeted mode of MS is well entrenched in clinical chemistry laboratories where it is used to measure “small” molecules such as drug metabolites [39, 40]. MRM differs from the typical shotgun MS/MS-based approaches used in discovering biomarker candidates in that MRM is a targeted technique directed to measure proteotypic peptides with known fragmentation properties. In MRM, a specific precursor ion (corresponding to a proteotypic peptide) and a specific fragment ion are selected by the MS1 and MS2 modes of the mass spectrometer, respectively. The instrument cycles through a number of precursor/fragment ion pairs (dubbed “transitions”) sequentially, and records the signal over time (the chromatographic elution of the analyte) [41]. The combination of precursor/fragment ion masses and retention times of multiple transitions from the same peptide result in high specificity for the targeted peptide. In addition, the instrument is only analyzing a subset of ions present in a complex mixture (reducing the overall chemical background) resulting in a substantial increase in sensitivity. The MRM experiment is ideally suited to triple quadrupole instruments [41, 42]. Recently, LC-MRM-MS/MS has been coupled to stable isotope dilution methods to measure concentrations of proteotypic peptides as surrogates for quantification of biomarker candidates in complex biological matrices such as tissue lysates and plasma [32, 43–50]. Assay linear ranges in plasma typically span four to five orders of magnitude with CVs <20%.
The capability of multiplexing many peptides into a single run is an important advantage in using MRM-MS/MS. A triple quadrupole operated in MRM mode is capable of monitoring 100 peptides or more in a single run, depending on the number of time segments [45]. Recent improvements in acquisition software allows for scheduling MRM transitions at specific time points in a chromatographic separation [51], increasing the number of transitions that can be monitored in a run to around 1000, drastically improving the multiplexability. Recent work demonstrating quantitative MRM using a MALDI source also has the potential to dramatically improve sample throughput [52].
The primary limitation for applying LC-MRM-MS/MS directly to plasma samples for biomarker verification studies is sensitivity. Typical LOQ are in the range of 100–1000 ng/mL of target protein in plasma [32, 45, 48]. Most novel and specific biomarkers are expected to occur at ≤ nanogram per milliliter levels.
Recently, it has been shown that coupling MRM-MS/MS with minimal fractionation of plasma dramatically improves the sensitivity, raising hope for measuring candidates directly in plasma [46, 51]. In these approaches, plasma is subjected to minimal fractionation using N-glycopeptide enrichment [51] or abundant protein depletion and strong cation exchange chromatography at the peptide level [46] prior to LC-MRM-MS/MS. For example, coupled with stable isotope dilution, using abundant protein depletion and SCX is multiplexable and able to achieve LOQ in the 1–10 ng/mL range without immunoaffinity enrichment of either proteins or peptides. However, this workflow is somewhat laborious, and its many steps will likely introduce experimental variation from run-to-run. In addition, the analysis timeframe is lengthened by the number of fractions analyzed by LC-MRM-MS/MS, limiting the sample throughput. Also, although the coefficients of variation for the MRM step range from 3 to 15%, reproducibility of fractionation and/or enrichment steps, such as abundant protein depletion and strong cation exchange chromatography, has not been assessed and will almost certainly introduce additional noise. Nonetheless, as discussed above (Table 1A.1), although a workflow that is limited in sample throughput and is associated with a CV≤0.5 would not be sufficient for level two credentialing or clinical validation, it would be perfectly acceptable for level one credentialing, where pooled samples and higher CVs can be tolerated.
In applying this approach to our hypothetical biomarker candidates, purchasing stable isotope standard (SIS) peptides for each of the 1000 candidate proteins would be cost-prohibitive. Hence, we propose that before investing in costly SIS peptides, LC-MRM-MS/MS can be performed semi-quantitatively by normalizing the amount of total peptides loaded on column across samples, or by using MRM transitions from nonchanging proteins in the sample to normalize the candidate response. As has been demonstrated [32], this semiquantitative look at candidates successfully allows triage of only those showing initial promise for further resource investment, and allows us to test literally hundreds-to-thousands of candidates with a minimum of upfront investment. Ideally (depending on resources), all markers meeting significance (using a statistical test such as the t-test) in level one credentialing will enter the level two credentialing. (For our specific PSA example, markers with ≥4×change would have approximately 90% chance of being truly different between the groups (Table 1)).
Even with limited fractionation, sensitivity remains a major limitation in this stage of the pipeline. For example, some candidates may be present in plasma at too low of abundance for detection in this workflow, yet be useful biomarkers that could be detected with higher sensitivity (affinity based) assays. We have no way of identifying these candidates without investing in high sensitivity assays, so we will still potentially have a high false negative rate for this class of candidates.
4.2 Level two credentialing ($$)
The goal is to estimate each marker’s sensitivity and specificity to determine if the marker shows sufficient promise to warrant a full clinical validation trial. This will require quantitative measurement of the candidate markers in many hundreds of individual patient samples with a CV≤0.2 (Table 1C.3), and hence will require a different workflow than that proposed above for level one credentialing.
Specifically, for level two credentialing an affinity reagent will need to be generated to enrich each candidate in a onestep, highly precise, preferably automatable process. A technique has recently been described that achieves these goals [37, 38, 53]. In this technique, Stable Isotope Standards and Capture by Antipeptide Antibodies (SISCAPA), immobilized affinity-purified antipeptide polyclonal antibodies are used to capture specific peptides of interest [37]. Captured peptides are subsequently eluted and detected by MRM-MS/MS. Quantitative results can be obtained by spiking in SIS peptides at known concentrations prior to immunoaffinity capture. The concentrations of the measured peptides are then used as surrogates for the concentrations of the biomarker protein candidates. This technique, using affinity-purified polyclonal antibodies, has been shown to achieve LOQ in the nanogram per milliliter range in plasma [32, 38]. Furthermore, selection of very high-affinity mAb is expected to further improve the sensitivity of the SISCAPA method.
The need for an antibody as well as a SIS peptide significantly raises the cost of level two credentialing over that for level one credentialing; reagents alone will cost ~$3000 per candidate tested (Table 2). Hence, we will likely be limited to testing 100s of candidates.
4.3 Validation ($$$)
Promising candidate biomarkers identified during level two credentialing will then be validated in real clinical practice, where the impact of other clinical covariates on the proposed test will be investigated. Positive predictive values and false referral probabilities at the population level will be determined. Additionally, panels of markers or perhaps the predictive value of changes in marker levels over time within an individual will need to be assessed. These complexities require a clinical-grade assay capable of high throughput and accurate measurements; hence the assay reagents must be well characterized and renewable, requiring that a mAb be generated. This further increases the cost and time investment in each candidate compared to level two credentialing (Table 2), and it is likely that resources will limit the numbers of candidates that can be tested to 10s.
As discussed above, the immunoassay is the conventional protein concentration assay format in the clinical setting; the ELISA is a well-known example. Despite its widespread use and favorable characteristics (quantitative, sensitive, high throughput), the ELISA does have some disadvantages [54, 55]. First, the creation of a sandwich immunoassay requires generating two different antibodies that both recognize the native protein and are free from steric interference with one another. Second, interfering autoantibodies can mask the surface features recognized by reagent antibodies [56, 57], a rarely appreciated problem in the clinical laboratory. Third, endogenous, nonspecific heterophilic antireagent antibodies can cause falsely elevated protein concentrations in as many as 3% of human samples [56, 58–60]. While it has less significance in the verification of potential biomarkers, a fourth disadvantage that plagues immunoassays in clinical settings is a lack of standardization. It is extremely uncommon for the clinical community to have access to truly useful standard materials that permit comparisons between the assays that were used to validate biomarkers and the many assays that might be used clinically at different centers [61].
The SISCAPA technology described above is one potential alternative to the ELISA. Advantages of SISCAPA are that (i) it only requires one antibody and so is cheaper; (ii) the antibody need not recognize the native protein, only a proteotypic peptide, so is easier to generate; (iii) the mass spectrometer essentially acts as the secondary antibody, so specificity is absolute; (iv) it is highly multiplexable and consumes small volumes (microliters) of clinical plasma specimens; (v) it directly detects antigen-derived peptide normalized to a stable isotope-labeled internal standard peptide, which could be easily standardized across laboratories. A disadvantage of SISCAPA is that the use of quantitative targeted MS methods requires that a proteotypic peptide be an accurate surrogate for measuring protein biomarker abundance. The validity of this assumption is threatened by the imperfect nature of trypsin digestion, for which no current standards exist. To avoid this source of error, some recent work has demonstrated the use of stable isotope-labeled proteins as standards in immunoaffinity-enrichment coupled to quantitative MS [62]. The verdict is still out as to whether, after further development, the SIS-CAPA-MRM technology will ultimately replace (or complement) the ELISA as a gold standard for clinical diagnostics, or whether it will “simply” provide a desperately needed bridging technology between verification and validation studies [36, 37].
5 Unmet needs
Given conventional capabilities, it is imperative that we develop a practical biomarker pipeline allowing pilot testing (verification) of thousands of protein candidates in hundreds of patient samples in a reasonable timeframe (<1 year) so that only the most promising candidates are triaged for lengthy and costly clinical validation studies. There are many unmet needs that could dramatically impact our success in assembling such a pipeline.
For example, the poor availability and often unacceptable quality of commercially available antibodies necessitates expensive and time-consuming de novo reagent generation for most candidates. As is being addressed (http://proteomics.cancer.gov/programs/reagents_resource/), there is a tremendous opportunity to partner with industry as well as academic efforts [63] (http://www.proteinatlas.org/) to generate well-characterized affinity reagents to the human proteome. If the immunogens were properly designed to support MS applications, these reagents would be invaluable for the biomarker pipeline described herein.
Additionally, ongoing efforts to clone, tag, and purify human proteins [64, 65] have the potential to greatly facilitate MRM-based biomarker candidate verification. The choice of the proteotypic peptide for monitoring is critical in constructing a successful assay. Small-scale purification of biomarker candidate proteins would allow LC-MS/MS analysis of the candidate proteins and thereby facilitate empirical selection of high-performing proteotypic peptides and transitions for targeted MS/MS analyses in complex human specimens. Without the ability to generate these empirical data, one must rely on mining of large proteomic databases (e.g., PeptideAtlas [66], Global Proteome Machine Database [67], PRIDE [68]) for peptides seen frequently or at relatively high intensity. Unfortunately, not all proteins of interest are represented in the databases and the extent of variability in manufacturer/instrument platforms for choosing proteotypic peptides remains to be determined. Recent attempts have been described to computationally predict proteotypic peptides, but the generality of this approach remains untested [69].
Technological improvements that increase the sensitivity of targeted LC-MS-based proteomic measurements of candidate proteins [70, 71] will also greatly improve our success rate by decreasing our false negative rate during level one credentialing. It is also conceivable that if the sensitivity of the instrument platforms can be improved ≥104, we may no longer rely on the generation of antibodies. This would tremendously decrease the cost and lead time for testing candidates and allow the number of candidates that can be tested in validation studies to be increased ≥100-fold. In the meantime, the generation of high-throughput, affordable, highly reproducible depletion, or fractionation technologies that further improve our sensitivity for measuring low abundance analytes will further improve the pipeline’s success.
On a biological front, the acute phase (aka host or inflammatory) response has been extensively slandered in the biomarker world, since the predominantly abundant proteins whose levels change as part of this response are not altered in a disease-specific pattern. Hence, due to their low specificity, they are considered to be diagnostically of little or no value. However, little is known about this host response, except for a handful of proteins. An organized effort to systematically characterize this response on a more global level would aid the biomarker field either by allowing us to eliminate these proteins from further consideration or alternatively by revealing that when they are considered more comprehensively they may actually have diagnostic value in some clinical settings.
As targeted proteomic platforms become more sensitive and as high-quality reagents become available for all human proteins, it will someday become possible to build sensitive, targeted assays for the entire proteome, merging the discovery and verification stages of the biomarker pipeline and allowing us to truly comprehensively test for protein biomarkers. Until this time, we will be dependent on a hypothesis-driven approach to selecting biomarker candidates for testing. Genomic technologies provide excellent sources of biomarker candidates via gene expression profiling and DNA copy number measurements. Sequencing and tiling arrays provide the opportunity to discover disease-associated mutations, novel fusion proteins, or splice variants that may show high specificity as biomarkers. Additionally, for many diseases, well-characterized disruptions in normal physiology or cell biology may provide a source of hypothesis-driven candidate selection such as angiogenesis in cancer.
Finally, the daunting, costly, complex, interdisciplinary effort required to move a candidate through from discovery to validation creates a situation where there is no feedback loop because those doing the actual discovery are often unaware of the outcome of the downstream follow up. Oftentimes, proteomic core facilities are paid (or collaborate) to generate biomarker discovery datasets for investigators studying a particular disease of interest. Lists of identified protein candidates, with some estimate of their relative abundances in cases versus controls, are then passed back to the primary investigators for follow-up studies that largely involve a painfully uninformed prioritization of candidates followed by costly and lengthy assay generation. Not surprisingly, the success rate of this approach is abysmal. These failures should not be misinterpreted as evidence that proteomics is not a worthwhile endeavor; rather, they are evidence that we need better integration. Currently, there is a disconnect between the discovery, verification, and clinical validation stages, making it impossible for paradigms to emerge that will iteratively improve performance. The recent application of LC-MRM-MS/MS methods to candidate verification provides us a new and exciting opportunity to keep proteomic centers engaged in the biomarker pipeline beyond basic discovery, and will thereby provide them with a valuable feedback loop about the unique issues of biomarker discovery proteomics compared to the more familiar protein-cataloging proteomics, and thereby facilitate iterative changes to improve overall success rates.
Acknowledgments
The authors are thankful for the generous support of the National Cancer Institute’s Clinical Proteomic Technologies for Cancer, the Entertainment Industry Foundation, and the Paul G. Allen Family Foundation.
Abbreviations
- CI
confidence interval
- MRM
multiple reaction monitoring
- PSA
prostate-specific antigen
- SIS
stable isotope standards
- SISCAPA
stable isotope standard with capture by antipeptide antibody
Footnotes
The authors have declared no conflict of interest.
References
- 1.Schrag D, Kuntz KM, Garber JE, Weeks JC. Life expectancy gains from cancer prevention strategies for women with breast cancer and BRCA1 or BRCA2 mutations. JAMA. 2000;283:617–624. doi: 10.1001/jama.283.5.617. [DOI] [PubMed] [Google Scholar]
- 2.Etzioni R, Urban N, Ramsey S, McIntosh M, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 3.Yu D, Hung MC. Overexpression of ErbB2 in cancer and ErbB2-targeting strategies. Oncogene. 2000;19:6115–6121. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- 4.Hughes T, Deininger M, Hochhaus A, Branford S, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Baggerly KA, Morris JS, Edmonson SR, Coombes KR. Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005;97:307–309. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- 7.Liotta LA, Lowenthal M, Mehta A, Conrads TP, et al. Importance of communication between producers and consumers of publicly available experimental data. J Natl Cancer Inst. 2005;97:310–314. doi: 10.1093/jnci/dji053. [DOI] [PubMed] [Google Scholar]
- 8.Ransohoff DF. Lessons from controversy: ovarian cancer screening and serum proteomics. J Natl Cancer Inst. 2005;97:315–319. doi: 10.1093/jnci/dji054. [DOI] [PubMed] [Google Scholar]
- 9.Check E. Proteomics and cancer: running before we can walk? Nature. 2004;429:496–497. doi: 10.1038/429496a. [DOI] [PubMed] [Google Scholar]
- 10.Master SR. Diagnostic proteomics: back to basics? Clin Chem. 2005;51:1333–1334. doi: 10.1373/clinchem.2005.053686. [DOI] [PubMed] [Google Scholar]
- 11.Ulrich R, Friend SH. Toxicogenomics and drug discovery: will new technologies help us produce better drugs? Nat Rev Drug Discov. 2002;1:84–88. doi: 10.1038/nrd710. [DOI] [PubMed] [Google Scholar]
- 12.Dimasi JA. Risks in new drug development: approval success rates for investigational drugs. Clin Pharmacol Ther. 2001;69:297–307. doi: 10.1067/mcp.2001.115446. [DOI] [PubMed] [Google Scholar]
- 13.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 14.Adams CP, Brantner VV. Estimating the cost of new drug development: is it really 802 million dollars? Health Aff (Millwood) 2006;25:420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- 15.Phillips KA, Van Bebber S, Issa AM. Diagnostics and biomarker development: priming the pipeline. Nat Rev Drug Discov. 2006;5:463–469. doi: 10.1038/nrd2033. [DOI] [PubMed] [Google Scholar]
- 16.Schroder FH, Carter HB, Wolters T, van den Bergh RC, et al. Early detection of prostate cancer in 2007. Part 1: PSA and PSA kinetics. Eur Urol. 2008;53:468–477. doi: 10.1016/j.eururo.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 17.Ross JS. Financial determinants of outcomes in molecular testing. Arch Pathol Lab Med. 1999;123:1071–1075. doi: 10.5858/1999-123-1071-FDOOIM. [DOI] [PubMed] [Google Scholar]
- 18.Jemal A, Tiwari RC, Murray T, Ghafoor A, et al. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 19.Etzioni R, Penson DF, Legler JM, di Tommaso D, et al. Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst. 2002;94:981–990. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 20.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 21.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizuka S, Charboneau L, Young L, Major S, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Washburn MP, Koller A, Oshiro G, Ulaszek RR, et al. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 27.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Liu AY, Loriaux P, Wollscheid B, et al. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.States DJ, Omenn GS, Blackwell TW, Fermin D, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 30.Calvo S, Jain M, Xie X, Sheth SA, et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 31.Rinner O, Mueller LN, Hubalek M, Muller M, et al. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
- 32.Whiteaker JR, Zhang H, Zhao L, Wang P, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 33.Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schadt EE, Lamb J, Yang X, Zhu J, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, Wiener MC, Zhang C, Fridman A, et al. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson NL. The roles of multiple proteomic platforms in a pipeline for new diagnostics. Mol Cell Proteomics. 2005;4:1441–1444. doi: 10.1074/mcp.I500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Anderson NL, Anderson NG, Haines LR, Hardie DB, et al. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 38.Whiteaker JR, Zhao L, Zhang HY, Feng LC, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroder FH, Carter HB, Wolters T, van den Bergh RC, et al. Early detection of prostate cancer in 2007 Part 1: PSA and PSA kinetics. Eur Urol. 2007 doi: 10.1016/j.eururo.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 40.Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 41.Chace DH, Kalas TA. A biochemical perspective on the use of tandem mass spectrometry for newborn screening and clinical testing. Clin Biochem. 2005;38:296–309. doi: 10.1016/j.clinbiochem.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 43.MacCoss MJ, Matthews DE. Quantitative MS for proteomics: teaching a new dog old tricks. Anal Chem. 2005;77:294A–302A. doi: 10.1021/ac053431e. [DOI] [PubMed] [Google Scholar]
- 44.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, et al. Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 45.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguiar M, Masse R, Gibbs BF. Mass spectrometric quantitation of C-reactive protein using labeled tryptic peptides. Anal Biochem. 2006;354:175–181. doi: 10.1016/j.ab.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 49.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3:644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 50.Kuhn E, Wu J, Karl J, Liao H, et al. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4:1175–1186. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 51.Wu SL, Amato H, Biringer R, Choudhary G, et al. Targeted proteomics of low-level proteins in human plasma by LC/MSn: using human growth hormone as a model system. J Proteome Res. 2002;1:459–465. doi: 10.1021/pr025537l. [DOI] [PubMed] [Google Scholar]
- 52.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Melanson JE, Chisholm KA, Pinto DM. Targeted comparative proteomics by liquid chromatography/matrix-assisted laser desorption/ionization triple-quadrupole mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:904–910. doi: 10.1002/rcm.2391. [DOI] [PubMed] [Google Scholar]
- 54.Berna M, Schmalz C, Duffin K, Mitchell P, et al. Online immunoaffinity liquid chromatography/tandem mass spectrometry determination of a type II collagen peptide biomarker in rat urine: Investigation of the impact of collision-induced dissociation fluctuation on peptide quantitation. Anal Biochem. 2006;356:235–243. doi: 10.1016/j.ab.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 55.Hoofnagle AN, Wener MH. Serum thyroglobulin: A model of immunoassay imperfection. Clin Lab Int. 2006;12:12–14. [Google Scholar]
- 56.Spencer CA, Lopresti JS. Measuring thyroglobulin and thyroglobulin autoantibody in patients with differentiated thyroid cancer. Nat Clin Pract Endocrinol Metab. 2008;4:223–233. doi: 10.1038/ncpendmet0757. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe M, Uchida K, Nakagaki K, Kanazawa H, et al. Anti-cytokine autoantibodies are ubiquitous in healthy individuals. FEBS Lett. 2007;581:2017–2021. doi: 10.1016/j.febslet.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 58.Hennig C, Rink L, Fagin U, Jabs WJ, Kirchner H. The influence of naturally occurring heterophilic anti-immunoglobulin antibodies on direct measurement of serum proteins using sandwich ELISAs. J Immunol Methods. 2000;235:71–80. doi: 10.1016/s0022-1759(99)00206-9. [DOI] [PubMed] [Google Scholar]
- 59.Kricka LJ, Schmerfeld-Pruss D, Senior M, Goodman DB, Kaladas P. Interference by human anti-mouse antibody in two-site immunoassays. Clin Chem. 1990;36:892–894. [PubMed] [Google Scholar]
- 60.Nahm MH, Hoffmann JW. Heteroantibody: phantom of the immunoassay. Clin Chem. 1990;36:829. [PubMed] [Google Scholar]
- 61.Sapin R. Insulin immunoassays: fast approaching 50 years of existence and still calling for standardization. Clin Chem. 2007;53:810–812. doi: 10.1373/clinchem.2006.084012. [DOI] [PubMed] [Google Scholar]
- 62.Brun V, Dupuis A, Adrait A, Marcellin M, et al. Isotope-labeled Protein Standards: Toward Absolute Quantitative Proteomics. Mol Cell Proteomics. 2007;6:2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Uhlen M. Mapping the human proteome using antibodies. Mol Cell Proteomics. 2007;6:1455–1456. [PubMed] [Google Scholar]
- 64.Lamesch P, Li N, Milstein S, Fan C, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brizuela L, Braun P, LaBaer J. FLEXGene repository: from sequenced genomes to gene repositories for high-throughput functional biology and proteomics. Mol Biochem Parasitol. 2001;118:155–165. doi: 10.1016/s0166-6851(01)00366-8. [DOI] [PubMed] [Google Scholar]
- 66.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J Proteome Res. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- 68.Jones P, Cote RG, Martens L, Quinn AF, et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 2006;34:D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mallick P, Schirle M, Chen SS, Flory MR, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 70.Ibrahim Y, Tang K, Tolmachev AV, Shvartsburg AA, Smith RD. Improving mass spectrometer sensitivity using a high-pressure electrodynamic ion funnel interface. J Am Soc Mass Spectrom. 2006;17:1299–1305. doi: 10.1016/j.jasms.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baker ES, Clowers BH, Li F, Tang K, et al. Ion mobility spectrometry-mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J Am Soc Mass Spectrom. 2007;18:1176–1187. doi: 10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Combs TP, Wagner JA, Berger J, Doebber T, et al. Induction of adipocyte complement-related protein of 30 kilodaltons by PPARgamma agonists: a potential mechanism of insulin sensitization. Endocrinology. 2002;143:998–1007. doi: 10.1210/endo.143.3.8662. [DOI] [PubMed] [Google Scholar]
- 73.Tomlinson S, Muranjan M, Nussenzweig V, Raper J. Haptoglobin-related protein and apolipoprotein AI are components of the two trypanolytic factors in human serum. Mol Biochem Parasitol. 1997;86:117–120. [PubMed] [Google Scholar]
- 74.Ghaemmaghami S, Huh WK, Bower K, Howson RW, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 75.Imagawa K, Matsumoto Y, Numata Y, Morita A, et al. Development of a sensitive ELISA for human leptin, using monoclonal antibodies. Clin Chem. 1998;44:2165–2171. [PubMed] [Google Scholar]