Abstract
Background
Heart failure results from abnormalities in multiple biological processes that contribute to cardiac dysfunction. We tested the hypothesis that inherited variation in genes of known importance to cardiovascular biology would thus contribute to heart failure risk.
Methods and Results
We utilized the ITMAT/Broad/CARe (IBC) cardiovascular SNP-array to screen referral populations of advanced heart failure patients for variants in ~2,000 genes of predicted importance to cardiovascular biology. Our design was a two-stage case-control study. In Stage 1, genotypes in Caucasian heart failure patients (n=1,590; ejection fraction 32±16%) were compared to those in unaffected controls (n=577; ejection fraction 67±8%) recruited from the same referral centers. Associations were tested for independent replication in Stage 2 (n=308 cases, 2,314 controls). Two intronic SNPs showed replicated associations with all-cause heart failure: rs1739843 in HSPB7 (combined P=3.09×10−6) and rs6787362 in FRMD4B (P=6.09×10−6). For both SNPs the minor allele was protective. In subgroup analyses, rs1739843 associated with both ischemic and nonischemic heart failure, whereas rs6787362 associated principally with ischemic heart failure. Linkage disequilibrium surrounding rs1739843 suggested that the causal variant resides in a region containing HSPB7 and a neighboring gene, CLCNKA, whereas the causal variant near rs6787362 is probably within FRMD4B. Allele frequencies for these SNPs were substantially different in African Americans (n=635 cases, 714 controls) and showed no association with heart failure in this population.
Conclusions
Our findings identify regions containing HSPB7 and FRMD4B as novel susceptibility loci for advanced heart failure. More broadly, in an era of genome-wide association studies, we demonstrate how knowledge of candidate genes can be leveraged as a complementary strategy to discern the genetics of complex disorders.
Keywords: cardiomyopathy, genetics, heart failure
Heart failure in the United States will strike one of every five Americans and accounts annually for approximately one million hospitalizations, over 50,000 deaths, and almost $35 billion in health care costs1. Although heart failure risk is heritable, it is estimated that at most 40% of idiopathic cardiomyopathy is monogenetic2. The genetic architecture of sporadic heart failure is poorly characterized, but is believed to be driven by multiple common genetic variants that combine with environmental factors to determine individual susceptibility. Consistent with this idea, well-validated susceptibility genes or loci have been identified for clinical precursors of heart failure, but not for heart failure itself3. Identification of common variants associated with heart failure would inform our understanding of the disease and provide genetic risk markers to better direct disease surveillance and prevention in asymptomatic individuals.
Most studies reporting genetic associations in heart failure have focused on small numbers of candidate genes3. Because the resulting associations have frequently failed to be replicated4, genome-wide association studies (GWAS) have supplanted candidate gene studies in large-scale comparative genomics research5. The unbiased GWAS approach has identified genetic risks for clinical antecedents of heart failure, such as hypertension, atherosclerosis, and diabetes mellitus6, but genome-wide studies in heart failure are few7, and have yet to be replicated. Furthermore, these studies typically implicate relatively large genetic loci, and so assignment of causality has not often been achieved. Consequently, the full import of GWAS findings on disease pathology can be difficult to determine.
Here, we combined the analytical advantages of a large-scale multi-gene approach with the mechanistic advantages of examining biologically relevant genes in a large-scale candidate gene case-control study of advanced heart failure. We utilized the ITMAT/Broad CARe (IBC) cardiovascular SNP-array8 to profile common single nucleotide polymorphisms in ~2,000 genes of predicted importance to cardiovascular disease in referral populations of patients with 4 advanced heart failure. Our analysis identifies common variants in HSPB7 and FMRD4B that show replicated associations in Caucasians with advanced heart failure.
Methods
Overview
Our design was a two-stage case-control study in referral populations of patients with advanced heart failure. In Stage 1, genotypes in Caucasian heart failure patients were compared with those in unaffected Caucasian controls derived from the same referral centers. We performed analyses comparing all-cause heart failure with controls, as well as stratified analyses examining ischemic and non ischemic subgroups separately. SNPs surpassing a pre-specified threshold for statistical significance were then tested for replication in Stage 2 using an independent population of Caucasian cases and controls. We also tested whether SNPs associated with heart failure in Caucasians were associated with heart failure in African Americans.
Study Populations
Stage 1: Caucasian Discovery Population
Caucasian heart failure cases (n=1590) were recruited from the Penn Heart Failure Study and the Cincinnati Heart Study, two ongoing NIH-sponsored (HL077101 and HL088577) prospective observational studies of patients with advanced heart failure referred for subspecialty care at the University of Pennsylvania Health System (Philadelphia, Pennsylvania) and at the University of Cincinnati Medical Center (Cincinnati, Ohio). For both studies, the primary inclusion criterion is a clinical diagnosis of heart failure with abnormal left ventricular function. Extensive clinical data were collected at enrollment as previously described9, 10. Caucasian controls (n=577) were recruited from among the population of patients referred for cardiovascular evaluation at Cincinnati or at Penn. Criteria for being a control were no history of heart failure, normal ventricular function on cardiac imaging, and no evidence of coronary artery disease as determine either by a negative treadmill exercise test or by maximal coronary stenoses of 20% or less on coronary angiography. Our recruitment strategy aimed to frequency-match cases and controls based on gender and age.
Stage 2: Caucasian Replication Population
To test for independent replication of associations identified in Stage 1, we recruited additional Caucasian heart failure cases (n=308) from the University of Cincinnati using the same inclusion criteria. These were compared to additional Caucasian controls who were referred to Penn or to Cincinnati for medical evaluation and who were free of clinical heart disease by history and physical examination (n=2,314). Unlike controls used in Stage 1, phenotyping of Stage 2 controls did not include assessment of ventricular function, stress testing, or coronary angiography.
African Americans
To explore whether SNPs associated in Caucasians were also associated in African Americans, we recruited African American heart failure cases from the University of Cincinnati using the same inclusion criteria. These subjects were compared to African American controls referred to Cincinnati or to Penn and who were free of heart disease by history and physical examination. Again, phenotyping of controls did not include assessment of ventricular function, stress testing, or coronary angiography.
Study protocols from all centers were approved by their respective institutional review boards. All patients provided written informed consent for genetic testing.
Genotyping
We used the IBC cardiovascular SNP-array to compare genotype frequencies in cases and controls in the Caucasian discovery population. The IBC array was designed by a consortium of experts who developed a prioritized list of candidate genes likely to be involved in cardiovascular disorders8. Ancestry-informative markers were included. Tagging SNPs for loci of interest were determined using a “cosmopolitan” approach appropriate for populations of different ancestry. For the Caucasian replication population and for African Americans, a minority of subjects was genotyped using the IBC array and the majority was genotyped at selected SNPs using Pyrosequencing. To ensure that both technologies provided similar results, we genotyped our top two associated SNPs using both approaches in 1000 individuals and found a >98% concordance of genotypes (see Supplemental Methods).
Data Analysis
Unlike GWAS, there are no broadly accepted standards regarding the level of statistical significance for large-scale candidate gene association studies where SNPs are tightly clustered in genes of high expected relevance to disease, and where association tests are not independent due to a high degree of linkage disequilibrium (LD). Hence, we adapted the two-stage approach utilized in most GWAS studies, and we regarded independent replication as the most reliable measure of true association. For Stage 1, SNPs were selected based on a pre-specified P-value threshold and then tested for independent replication in Stage 2. GWAS studies using current platforms perform approximately 1 million association tests, and a P-value threshold of 5×10−7 utilized by the Wellcome Trust Case-Control Consortium11 and by a recent GWAS of echocardiographic phenotypes12 has become an accepted threshold for selection of Stage 1 SNPs. After quality control and after accounting for the high degree of LD among SNPs on the IBC array, we estimate that our Stage 1 analysis conducts ~10,000 association tests, which is 100-fold fewer than a typical GWAS. Hence we a priori selected P<5×10−5 as the threshold for significance in Stage 1. All subjects in the Stage 1 population had genetically-inferred Caucasian Ancestry based on multi-dimensional scaling (MDS) of all IBC genotypes (Supplemental Figure 1). Our primary analyses compared genotype frequencies between cases and controls using PLINK using an additive genetic model adjusting for age, gender, and study site. Though all subjects were genetically inferred Caucasians, we also adjusted for 2 principal components of race derived from MDS to account for any residual differences in ancestry. Secondary analyses included additional adjustments for hypertension and diabetes. We first used these analyses to compare all cases with controls, and repeated them after stratifying the cases into ischemic and nonischemic subgroups. Ischemic heart failure was defined by at least one 50% narrowing on coronary angiography, a positive stress test, history of an acute coronary syndrome, or prior coronary revascularization. Patients free of these criteria were classified as nonischemic. Patients that could not be clearly classified clearly were labeled as “other” and were not included in subgroup analyses.
SNPs that met criteria for association in Stage 1 were tested for replication in Stage 2, adjusting for age, sex, and study site. To be considered replication, we required that the direction of association for allele frequencies be the same as in Stage 1. Using a 1-sided P-value distribution is therefore necessary in order for the P-value distribution to be correct under the null hypothesis, and we required 1-sided P-value <0.05 as the criterion for replication12. To obtain better risk estimates, we conducted combined analyses of all available Caucasian cases and controls for the replicated SNPs, adjusting for age, sex, site, and analysis stage
To explore loci tagged by our most strongly associated SNPs, we returned to the IBC data used in Stage 1 and imputed IBC genotypes with HapMap to increase genotype coverage. We 9 then used these data to explore LD at the loci of interest using SNAP13. For a variant with minor allele frequency or 0.4, we had 80% power to detect an odds ratio of 1.20 in the combined Stage 1 and Stage 2 populations. Further details regarding our analysis, including additional power calculations and tests for Hardy-Weinberg equilibrium, are provided in the Supplemental Material.
Results
Study Populations
In the Caucasian discovery population (Table 1), cases and controls were of similar age and most were male. Based on selection criteria, controls had no heart failure, no significant coronary artery disease on coronary angiography or stress testing, and a normal ejection fraction of 67±8%. By contrast, heart failure cases had an advanced systolic heart failure phenotype with an average ejection fraction of 32±16%, and were treated accordingly with neurohormonal antagonists, cardiac resynchronization, and implanted cardioverter-defibrillators. The prevalence of ischemic and nonischemic etiologies was roughly equivalent. Hypertension and diabetes were more common among cases than controls, as expected14, 15.
Table 1.
Study populations (total n=6138 subjects)*
| Stage1: Caucasian Discovery |
Stage 2: Caucasian Replication |
African Americans |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases† |
P | Controls | Cases | P | Controls | Cases | P | |||
| Penn | Cincinnati | Combined | |||||||||
| N | 577 | 755 | 835 | 1590 | - | 2314 | 308 | - | 714 | 635 | - |
| Age, years | 55±12 | 58±13 | 56±13 | 57±13 | <0.001 | 55±10 | 57±13 | 0.002 | 53±11 | 54±13 | 0.12 |
| Male Gender | 394 (68) | 528 (70) | 564 (68) | 1092 (69) | 0.86 | 1287 (56) | 204 (66) | <0.001 | 292 (41) | 345 (54) | |
| Ejection Fraction, percent | 67±8 | 33±16 | 32±15 | 32±16 | <0.001 | - | 32±12 | - | - | 32±14 | - |
| Subgroup | |||||||||||
| Ischemic | - | 334 (44) | 492 (59) | 826 (52) | - | - | 180 (58) | - | - | 300 (47) | - |
| Nonischemic | - | 361 (48) | 312 (37) | 673 (42) | - | - | 118 (38) | - | - | 324 (51) | - |
| Other | - | 60 (8) | 31 (4) | 91 (6) | - | - | 10 (4) | - | - | 11 (2) | - |
| Hypertension | 147 (25) | 255 (34) | 565 (68) | 820 (52) | <0.001 | 835 (36) | 213 (69) | <0.001 | 312 (44) | 556 (87) | <0.001 |
| Diabetes | 39 (7) | 207 (27) | 328 (39) | 535 (34) | <0.001 | 754 (33) | 123 (40) | 0.01 | 355 (49) | 258 (41) | 0.001 |
| Treatments | |||||||||||
| Beta-blocker | 148 (25) | 627 (83) | 647 (77) | 1274 (80) | <0.001 | 172 (7) | 245 (80) | <0.001 | 61 (9) | 552 (87) | <0.001 |
| ACEI or ARB | 47 (8) | 638 (84) | 651 (78) | 1292 (81) | <0.001 | 533 (23) | 258 (87) | <0.001 | 240 (34) | 582 (92) | <0.001 |
| Aldosterone Antagonist | - | 233 (31) | 222 (27) | 455 (29) | <0.001 | - | 62 (20) | <0.001 | - | 180 (28) | <0.001 |
| Cardiac Resynchronization | - | 221 (30) | 116 (14) | 337 (21) | <0.001 | - | 45 (15) | <0.001 | - | 41 (6) | <0.001 |
| Defibrillator | - | 337 (45) | 257 (31) | 594 (37) | <0.001 | - | 76 (25) | <0.001 | - | 143 (23) | <0.001 |
Data are displayed as mean±SD or frequency(percent). ACEI, angiotensin converting enzyme inhibitor; ARB, angiotensin II receptor blocker. P-values are for comparisons of continuous (t-test) and categorical (χ2-test) variables between cases and controls.
Characteristics of cases are presented for each center individually and for all Stage 1 cases combined.
In the Caucasian replication population, cases again had severe systolic heart failure with an average ejection of 32±12%. Controls had no heart failure based on selection criteria. There was a higher prevalence of diabetes and hypertension among controls in the replication population than in the discovery population. African American cases had severe systolic heart failure with average ejection fraction of 32±14%. The prevalence of hypertension and diabetes was higher among both cases and controls compared to the Caucasian populations, as expected based on general population studies16.
Stage 1 Analysis Using IBC Data
After eliminating SNPs with minor allele frequency <1%, call rates <95%, or not in Hardy-Weinberg equilibrium, 31,682 autosomal SNPs (of 44,720 on the IBC array) were called in the Caucasian discovery population. The Q-Q plot (Supplemental Figure 2) showed a relatively small inflation factor (λ=1.065) that is similar to or lower than those found in several successful GWAS studies11 and revealed a number of SNPs with lower P-values than expected by chance. Twenty-two genes were associated with all-cause heart failure at P<0.001 (Supplemental Table 1). Two of these SNPs surpassed our threshold for significance in primary analyses (rs1738943[HSPB7], P =2.8×10−5 and rs6787362[FRMD4B], P=4.5×10−5), and two SNPs approached but did not surpass this threshold (rs7174882[PCSK6], P=5.7×10−5 and rs16877169[PDE4D], P=5.9×10−5). Subgroup analyses yielded only one association that met criteria for statistical significance (rs4581654[LIPC], P=4.9 × 10−5) associated with nonischemic heart failure (Supplemental Table 2). LIPC is a well characterized gene involved in hepatic lipid metabolism17. As such, we reasoned this association was likely attributable to differences in unmeasured lipid phenotypes between cases and controls, and we chose not pursue this association any further. Thus, four SNPs were selected for Stage 2: rs1738943, rs6787362, rs7174882, and rs16877169.
Stage 2 Replication
Table 2 summarizes association results for the four SNPs selected for Stage 2. Minor allele frequencies in controls were similar to those in the HapMap CEU population. Two of the four SNPs showed significant associations in the Caucasian replication population: rs1738943(HSPB7) and rs6787362(FRMD4B). As expected18, the associations in Stage 2 were less marked than in Stage 1 with minor allele frequencies that were less divergent between cases and controls. For both SNPs, the minor allele frequency was smaller in cases, consistent with a protective effect of the minor allele. In a combined analysis of all available genotypes, P-values were 3.09×10−6 for rs1739843 and 6.09×10−6 for rs6787362.
Table 2.
Replication of top associations and combined analysis of all Caucasian genotypes
| Gene | Chr | SNP | SNP type | Minor/Major allele | HapMap CEU MAF | Stage 1 Discovery (IBC Array)* |
Stage 2 Replication† |
Combined‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control MAF | Case MAF | OR | P-value | Control MAF | Case MAF | OR | P-value | OR (95%CI) | P-value | ||||||
| HSPB7 | 1 | rs1739843 | Intronic | A/G | 0.450 | 0.436 | 0.367 | 0.75 | 2.8E-05 | 0.421 | 0.375 | 0.82 | 0.018 | 0.77 (0.69, 0.86) | 3.09E-06 |
| FRMD4B | 3 | rs6787362 | Intronic | G/A | 0.104 | 0.140 | 0.098 | 0.65 | 4.51E-05 | 0.116 | 0.084 | 0.74 | 0.025 | 0.67 (0.57, 0.80) | 6.90E-06 |
| PCSK6 | 15 | rs7174882 | Intronic | C/A | 0.500 | 0.511 | 0.449 | 0.76 | 5.70E-05 | 0.484 | 0.497 | 0.94 | 0.28 | - | - |
| PDE4D | 5 | rs16877169 | Intronic | A/C | 0.076 | 0.119 | 0.075 | 0.63 | 5.90E-05 | 0.089 | 0.080 | 0.88 | 0.23 | - | - |
N= 1590 cases, 577 controls
N= 308 cases, 2314 controls
from a pooled analysis of both datasets (N = 1898 cases, 2891 controls)
OR, odds ratio for the minor allele; MAF, minor allele frequency; CI, confidence interval
Association in African Americans
For each of the 4 SNPs identified in our Caucasian discovery population, we also explored associations in African Americans with heart failure (Supplemental Tables 3 and 4). For three of the SNPs, minor allele frequencies in African Americans were lower than those observed in Caucasians and these SNPs were not associated with African American heart failure. One SNP, rs16877169, showed a higher minor allele frequency in African Americans and showed modest association with African American heart failure (P=0.039).
Subgroups and Intermediate Phenotypes
We utilized all available data to explore whether the strength of association for our top findings varied with the underlying cause of heart failure (Table 3). The association with HSPB7 was noteworthy in both subtypes, but appeared slightly stronger in nonischemic heart failure, as indicated by a slightly lower P-value and odds ratio. The association with FRMD4B appeared stronger in ischemic heart failure. We also considered whether the risk alleles might lead to heart failure by increasing risk for intermediate phenotypes that contribute to heart failure risk over time. Diabetes and hypertension are well-established risk factors for heart failure and also have a genetic basis. We therefore included history of diabetes and history of hypertension as additional covariates in our regression models to test whether this adjustment attenuated the SNP associations. As shown in Table 3, the associations remained strong, indicating that these intermediate phenotypes are unlikely to play a major role in the causal pathway from risk allele to development of heart failure.
Table 3.
Analysis of heart failure subgroups and intermediate phenotypes in Caucasians
| Gene | SNP | Subgroup | OR | P | ORDM, HTN | PDM, HTN |
|---|---|---|---|---|---|---|
| HSPB7 | rs1739843 | All | 0.77 (0.69, 0.86) | 3.09E-06 | 0.77 (0.68, 0.86) | 6.34E-06 |
| Ischemic | 0.78 (0.68, 0.89) | 2.56E-04 | 0.75 (0.65, 0.87) | 9.14E-05 | ||
| Nonischemic | 0.75 (0.65, 0.87) | 9.2E-05 | 0.76 (0.65, 0.88) | 2.4E-04 | ||
| FRMD4B | rs6787362 | All | 0.67 (0.57, 0.80) | 6.90E-06 | 0.66 (0.56, 0.80) | 8.06E-06 |
| Ischemic | 0.66 (0.54, 0.82) | 1.41E-04 | 0.68 (0.55, 0.85) | 5.9E-04 | ||
| Nonischemic | 0.69 (0.55, 0.86) | .001 | 0.67 (0.53, 0.85) | .001 |
n = 1898 cases (1006 ischemic, 791 nonischemic, 101 other), 2891 controls
OR is for the minor allele, adjusted for age, sex, and site. ORDM, HTN is additionally adjusted for history of diabetes and history of hypertension
HSPB7 and FRMD4B Loci
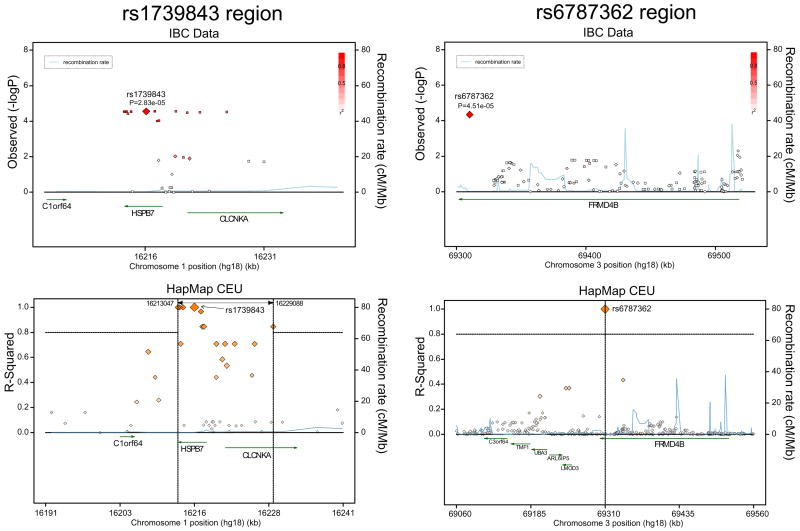
Next we explored the loci tagged by the two replicated SNP-associations in Caucasians. We imputed our IBC genotypes with HapMap CEU to increase coverage in these regions and constructed association plots (Figure 1, top panels). For comparison, we also explored the LD structure at these two loci using HapMap data alone (Figure 1, bottom panels). As shown, rs1739843 is located within a region of strong LD that spans two genes: HSPB7 and CLCNKA. As such, the causal genetic variant responsible for our association could be anywhere in this LD block and could thus reside within either of these two genes or within intervening regulatory regions. By contrast, rs6787362 is located in the 3’ end of FRMD4B in an area of low LD. This was evident both in our own data and in HapMap, which showed no nearby SNPs in strong correlation with rs6787362. Thus the causal variant is likely to be close to rs6787362 and may lie within FRMD4B itself.
Figure 1. Genomic regions containing our top associated SNPs.
The upper panels show strength of association in the Stage 1 Caucasian discovery population using IBC data; genotyped IBC SNPs are indicated with diamonds and imputed SNPs with squares. The lower panels demonstrate LD(R 2) and recombination rates using HapMap CEU data alone; vertical dashed lines demarcate an LD block in which R2> 0.8.
Discussion
We report the first large-scale candidate gene case-control study of advanced heart failure using the high density IBC cardiovascular SNP-array. Approximately 30,000 SNPs were studied in 2,000 candidate genes with a high a priori probability of cardiovascular involvement, thereby leveraging knowledge of cardiovascular biology and the results of prior genome-association and expression quantitative trait studies8. By so doing, we identified and replicated two common genetic variants that are significantly associated with advanced heart failure in Caucasians.
These findings provide specific genetic data supporting the observation that clinical heart failure, though a complex and heterogeneous phenotype, results in part from an inherited predisposition19. For both SNPs identified (rs1738943 and rs6787362) the minor allele is less frequent in cases than in controls, indicating a protective effect of the minor allele. However, the magnitude of these protective effects are too small to be of any real clinical value by themselves, which is similar to risk or protective effects associated with many common genetic variants11. Friedrichs et al recently published a candidate-gene association study of inflammatory genes that identified variants associated with dilated cardiomyopathy20. As more such variants are identified, it may be possible to develop a composite measure or score of multiple risk and protective alleles with more predictive ability and thus more clinical use, as has been attempted for lipid disorders21.
The most important implication of our findings is the identification of specific genes that may contribute to heart failure pathogenesis in humans. Our top association was rs1738943 (combined P=3.09×10−6), which is located in a 5′ intronic region of the gene encoding the heat shock protein B7 (HSPB7). HSPB7, also known as the cardiovascular heat shock protein (cvHSP), is a member of the small heat shock protein family and is expressed almost exclusively in cardiac and skeletal muscle. One of the most abundant myocardial transcripts22, HSPB7 preserves contractile integrity by binding to and stabilizing sarcomeric proteins23, 24. Mutations in another small heat shock protein, CRYAB (also known as HSPB5), cause a rare form of familial cardiomyopathy23, and common variation in HSPB7 may similarly alter susceptibility to adverse cardiac remodeling. Further scrutiny of the locus containing rs1738943 shows that although this SNP is located within HSPB7, it resides in a block of high LD that spans HSPB7 and a nearby gene, CLCNKA, which encodes a voltage-sensitive chloride channel expressed mainly in the kidney25. Variation in CLCNKA has been associated with alterations in renal sodium re-absorption and salt-sensitive hypertension25, 26. Thus, an alternative explanation for our finding is that rs1738943 may serve as a marker for pathogenic variants in CLCNKA that lead to excess sodium retention that, in turn, contribute to heart failure risk. Discerning the underlying causal variants will require resequencing of the entire LD block spanning both HSPB7 and CLCNKA.
We also found a replicated association between rs6787362 and all-cause heart failure (combined P=6.09 × 10−6). This SNP is located in a 3′ intronic region of the FRMD4B gene. FRMD4B27, FERM- domain containing protein 4B, was identified in a screen for proteins that physically interact with CYTH3, a downstream effector of PI-3 kinase signaling. PI3-kinase is a mediator of many different signaling pathways and it is difficult to speculate a specific mechanism based on our data alone. Unlike the HSPB7 locus, the locus containing rs6787362 is in an area of weak LD. The causal variant may thus be within the FRMD4B gene, and re-sequencing of a narrower region may be sufficient for it to be identified. We note that this association was strongest in patients with ischemic heart failure (Table 3). Thus it is possible that FRMD4B contributes to heart failure risk by conferring risk for coronary artery disease. This is in contrast to the HSPB7 association, which was noteworthy in both ischemic and nonischemic subtypes.
SNP frequencies were substantially different in African Americans, and the associations identified in Caucasians did not replicate in African Americans with heart failure, with the possible exception of the rs16877169 variant in PDE4D (Supplemental Table 3). These observations are consistent with previous studies that demonstrate differences in heart failure risk factors16, therapeutic response28, 29, and clinical outcomes30 in African Americans and Caucasians. Further work in larger cohorts is required to identify the extent to which ancestry-specific genetic variants explain these observations.
The use of a large-scale candidate gene approach stands in contrast to numerous GWAS studies that have rapidly emerged over the past few years and that have identified common variants associated with diabetes mellitus, hypertension, dyslipidemia, coronary artery disease, and echocardiographic traits31. The strength of the “hypothesis-free” GWAS approach is an unbiased look at the entire genome, with the consequence that all genetic variation is regarded equally. However, for any given disease process, each gene has a different likelihood of involvement in pathogenesis, which is not considered in most whole-genome approaches. Thus, the chance of false positive associations is enhanced, and the true associations identified are dominated by variants with modest effect sizes, such as the 9p21 locus associated with myocardial infarction11. By contrast, our approach sacrifices an unbiased view for one that leverages years of accumulated knowledge regarding the biology of cardiovascular disorders. This may reduce the likelihood of false-positive associations and identifies smaller genic regions that suggest specific pathogenetic mechanisms, but at a cost of ignoring regions of the genome that may contain important but unanticipated variants that predispose to heart failure. We thus regard GWAS and large-scale candidate gene studies as complementary approaches.
We studied patients with advanced heart failure and substantial ventricular dysfunction recruited from referral populations. By focusing on an extreme phenotype we are more likely to identify genetic variants that relate to myocardial dysfunction per se, rather than genetic variants that influence its clinical antecedents. Indeed, adjusting for hypertension or diabetes had little influence on our findings. We note that our findings may not be generalizable to early-stage heart failure in population-based studies, in which nearly half have preserved systolic function32. However, a comparison of the Framingham 100K heart failure GWAS findings with our own data suggests that there will be some overlap. Two heart failure genes (C9orf39 and RYR2) identified in Framingham had adequate proxy SNPs on the IBC array (Supplemental Table 6). Although C9orf39 showed no association with heart failure in our data, several variants in RYR2 showed modest associations with all-cause heart failure (P=0.016) and nonischemic heart failure (P =8×10−4). We thus provide further evidence to support RYR2 as a heart failure susceptibility gene, which is of particular interest given its well established role in excitation-contraction coupling. As large-scale heart failure genetic studies proceed, it will be instructive to compare findings from referral-based cohorts with those from population-based studies such as the CHARGE33 and CARe consortia8.
We acknowledge several limitations. As in all case-control studies, misclassification of cases and controls and limitations in sample size may reduce power to detect associations. Survival bias may also influence the composition of the cases, since some heart failure patients will die at an earlier stage and not have the opportunity to be recruited. Here we are reassured by independent replication of our principal findings. Like many common phenotypes, heart failure arises via derangements in a myriad of biological pathways and is rightly termed “complex.” Beyond this biological complexity, there is also substantial heterogeneity in the definition and diagnosis of heart failure, and this remains a particular challenge for studies of heart failure genetics. In this regard, detailed phenotyping in our discovery population, including clinical evaluation, cardiac imaging, and assessment for coronary artery disease in both cases and in controls, is a major strength. Finally, although we replicated our top SNPs identified in primary analyses, other promising variants identified in secondary analyses, such as ARHGAP26 and LIF (Supplemental Table 1), could benefit from further study in larger populations.
In conclusion, we have utilized a large-scale candidate gene approach to identify and replicate two common genetic variants associated with risk of advanced heart failure in Caucasians. These findings implicate regions containing HSPB7 and FRMD4B as novel susceptibility loci for advanced heart failure. In an era of genome wide association studies, our study thus demonstrates how knowledge of candidate genes can be leveraged as a complementary strategy to discern the genetics of complex disorders.
Supplementary Material
Acknowledgments
Acknowledgements: none.
Funding sources: This work was funded by NIH R01HL088577, R21HL092379, R01HL087871, P50 HL077101, P50HL077113, RC2 HL102222, and the Penn Cardiovascular Institute.
Footnotes
Disclosures: none.
References
- 1.Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O’Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y. Heart disease and stroke statistics--2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 2.Mestroni L, Rocco C, Gregori D, Sinagra G, Di LA, Miocic S, Vatta M, Pinamonti B, Muntoni F, Caforio AL, McKenna WJ, Falaschi A, Giacca M, Camerini Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity. Heart Muscle Disease Study Group. J Am Coll Cardiol. 1999;34:181–90. doi: 10.1016/s0735-1097(99)00172-2. [DOI] [PubMed] [Google Scholar]
- 3.Dorn GW, Cresci S. Genome-wide association studies of coronary artery disease and heart failure: where are we going? Pharmacogenomics. 2009;10:213–23. doi: 10.2217/14622416.10.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freely associating. Nat Genet. 1999;22:1–2. doi: 10.1038/8702. [DOI] [PubMed] [Google Scholar]
- 5.Topol EJ, Murray SS, Frazer KA. The genomics gold rush. JAMA. 2007;298:218–21. doi: 10.1001/jama.298.2.218. [DOI] [PubMed] [Google Scholar]
- 6.O’Donnell CJ, Nabel EG. Cardiovascular Genomics, Personalized Medicine, and the National Heart, Lung, and Blood Institute: Part I: The Beginning of an Era. Circ Cardiovasc Genet. 2008;1:51–7. doi: 10.1161/CIRCGENETICS.108.813337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D’Agostino RB, Sr, Fox CS, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Murabito JM, Newton-Cheh C, O’Donnell CJ, Seshadri S, Vasan RS, Wang TJ, Wolf PA, Levy D. Framingham Heart 21 Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8 (Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, Spertus JA, Koch WJ, Kardia SL, Dorn GW. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–7. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ky B, Kimmel SE, Safa RN, Putt ME, Sweitzer NK, Fang JC, Sawyer DB, Cappola TP. Neuregulin-1{beta} Is Associated With Disease Severity and Adverse Outcomes in Chronic Heart Failure. Circulation. 2009 doi: 10.1161/CIRCULATIONAHA.109.856310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. 22. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, Watzinger N, Larson MG, Smith NL, Dehghan A, Grosshennig A, Schillert A, Teumer A, Schmidt R, Kathiresan S, Lumley T, Aulchenko YS, Konig IR, Zeller T, Homuth G, Struchalin M, Aragam J, Bis JC, Rivadeneira F, Erdmann J, Schnabel RB, Dorr M, Zweiker R, Lind L, Rodeheffer RJ, Greiser KH, Levy D, Haritunians T, Deckers JW, Stritzke J, Lackner KJ, Volker U, Ingelsson E, Kullo I, Haerting J, O’Donnell CJ, Heckbert SR, Stricker BH, Ziegler A, Reffelmann T, Redfield MM, Werdan K, Mitchell GF, Rice K, Arnett DK, Hofman A, Gottdiener JS, Uitterlinden AG, Meitinger T, Blettner M, Friedrich N, Wang TJ, Psaty BM, van Duijn CM, Wichmann HE, Munzel TF, Kroemer HK, Benjamin EJ, Rotter JI, Witteman JC, Schunkert H, Schmidt H, Volzke H, Blankenberg S. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–78. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutter MK, Parise H, Benjamin EJ, Levy D, Larson MG, Meigs JB, Nesto RW, Wilson PW, Vasan RS. Impact of glucose intolerance and insulin resistance on cardiac structure and function: sex-related differences in the Framingham Heart Study. Circulation. 2003;107:448–54. doi: 10.1161/01.cir.0000045671.62860.98. [DOI] [PubMed] [Google Scholar]
- 15.Levy D, Larson MG, Vasan RS, Kannel WB, Ho KK. The progression from hypertension to congestive heart failure. JAMA. 1996;275:1557–62. 23. [PubMed] [Google Scholar]
- 16.Bibbins-Domingo K, Pletcher MJ, Lin F, Vittinghoff E, Gardin JM, Arynchyn A, Lewis CE, Williams OD, Hulley SB. Racial differences in incident heart failure among young adults. N Engl J Med. 2009;360:1179–90. doi: 10.1056/NEJMoa0807265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gieger C, Geistlinger L, Altmaier E, Hrabe de AM, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, Illig T, Suhre K. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraft P. Curses--winner’s and otherwise--in genetic epidemiology. Epidemiology. 2008;19:649–51. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 19.Lee DS, Pencina MJ, Benjamin EJ, Wang TJ, Levy D, O’Donnell CJ, Nam BH, Larson MG, D’Agostino RB, Vasan RS. Association of parental heart failure with risk of heart failure in offspring. N Engl J Med. 2006;355:138–47. doi: 10.1056/NEJMoa052948. [DOI] [PubMed] [Google Scholar]
- 20.Friedrichs F, Zugck C, Rauch GJ, Ivandic B, Weichenhan D, Muller-Bardorff M, Meder B, El Mokhtari NE, Regitz-Zagrosek V, Hetzer R, Schafer A, Schreiber S, Chen J, Neuhaus I, Ji R, Siemers NO, Frey N, Rottbauer W, Katus HA, Stoll M. HBEGF, SRA1, and IK: Three cosegregating genes as determinants of cardiomyopathy. Genome Res. 2009;19:395–403. doi: 10.1101/gr.076653.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 22.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–11. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krief S, Faivre JF, Robert P, Le DB, Brument-Larignon N, Lefrere I, Bouzyk MM, Anderson KM, Greller LD, Tobin FL, Souchet M, Bril A. Identification and characterization of cvHsp. A novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J Biol Chem. 1999;274:36592–600. doi: 10.1074/jbc.274.51.36592. [DOI] [PubMed] [Google Scholar]
- 24.Golenhofen N, Perng MD, Quinlan RA, Drenckhahn D. Comparison of the small heat shock proteins alphaB-crystallin, MKBP, HSP25, HSP20, and cvHSP in heart and skeletal muscle. Histochem Cell Biol. 2004;122:415–25. doi: 10.1007/s00418-004-0711-z. [DOI] [PubMed] [Google Scholar]
- 25.Barlassina C, Dal FC, Lanzani C, Manunta P, Guffanti G, Ruello A, Bianchi G, Del VL, Macciardi F, Cusi D. Common genetic variants and haplotypes in renal CLCNKA gene are associated to salt-sensitive hypertension. Hum Mol Genet. 2007;16:1630–8. doi: 10.1093/hmg/ddm112. [DOI] [PubMed] [Google Scholar]
- 26.Sile S, Velez DR, Gillani NB, Alexander CA, George AL, Jr, Williams SM. Haplotype diversity in four genes (CLCNKA, CLCNKB, BSND, NEDD4L) involved in renal salt reabsorption. Hum Hered. 2008;65:33–46. doi: 10.1159/000106060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klarlund JK, Holik J, Chawla A, Park JG, Buxton J, Czech MP. Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J Biol Chem. 2001;276:40065–70. doi: 10.1074/jbc.M105260200. [DOI] [PubMed] [Google Scholar]
- 28.Dries DL, Exner DV, Gersh BJ, Cooper HA, Carson PE, Domanski MJ. Racial differences in the outcome of left ventricular dysfunction. N Engl J Med. 1999;340:609–16. doi: 10.1056/NEJM199902253400804. [DOI] [PubMed] [Google Scholar]
- 29.Taylor AL, Ziesche S, Yancy C, Carson P, D’Agostino R, Jr, Ferdinand K, Taylor M, Adams K, Sabolinski M, Worcel M, Cohn JN. Combination of isosorbide dinitrate and hydralazine in blacks with heart failure. N Engl J Med. 2004;351:2049–57. doi: 10.1056/NEJMoa042934. [DOI] [PubMed] [Google Scholar]
- 30.Cresci S, Kelly RJ, Cappola TP, Diwan A, Dries D, Kardia SL, Dorn GW. Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–44. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter DJ, Altshuler D, Rader DJ. From Darwin’s finches to canaries in the coal mine--mining the genome for new biology. N Engl J Med. 2008;358:2760–3. doi: 10.1056/NEJMp0804318. [DOI] [PubMed] [Google Scholar]
- 32.Redfield MM, Jacobsen SJ, Burnett JC, Jr, Mahoney DW, Bailey KR, Rodeheffer RJ. Burden of systolic and diastolic ventricular dysfunction in the community: appreciating the scope of the heart failure epidemic. JAMA. 2003;289:194–202. doi: 10.1001/jama.289.2.194. [DOI] [PubMed] [Google Scholar]
- 33.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JCM, Boerwinkle E on Behalf of the CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of Prospective Meta-Analyses of Genome-Wide Association Studies From 5 Cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.