Abstract
The etiology of non-syndromic hydrocephalus is poorly understood. Via positional cloning in a consanguineous family with autosomal recessive hydrocephalus we have now identified a homozygous splice site mutation in the CCDC88C gene as a novel cause of a complex hydrocephalic brain malformation. The only living patient showed normal psychomotor development at the age of 3 years and 3 months and her deceased aunt, who was assumed to suffer from the same condition, had mild mental retardation. The mutation in the affected patients, a homozygous substitution in the donor splice site of intron 29, resulted in a shorter transcript due to exclusion of exon 29 and loss of functional protein, as shown by Western blotting (p.S1591HfsX7). In normal human tissue panels, we found CCDC88C ubiquitously expressed, but most prominently in the fetal brain, especially in pons and cerebellum, while expression in the adult brain appeared to be restricted to cortex and medulla oblongata. CCDC88C encodes DAPLE (HkRP2), a Hook-related protein with a binding domain for the central Wnt signalling pathway protein Dishevelled. Targeted quantitative RT-PCR and expression profiling of 84 genes from the Wnt signalling pathway in peripheral blood from the index patient and her healthy mother revealed increased mRNA levels of CCDC88C indicating transcriptional upregulation. Due to loss of CCDC88C function β-catenin (CTNNB1) and the downstream target LEF1 showed increased mRNA levels in the patient, but many genes from the Wnt pathway and transcriptional target genes showed reduced expression, which might be explained by a complex negative feedback loop. We have thus identified a further essential component of the Wnt signalling pathway in human brain development.
Key Words: Brain malformation, CCDC88C, Daple, Hydrocephalus, Wnt signalling
Introduction
A prenatal hydrocephalus is observed in about 1:2,000 pregnancies. The etiology is heterogeneous including intrauterine infections, neural tube defects, chromosomal aberrations and a wide variety of monogenic syndromes, twinning as well as CNS tumors, but remains unsolved in the majority of non-syndromic cases [Garne et al., 2010]. Prognosis critically depends on the presence of additional cerebral and extracranial malformations, which are observed in about 70−80% of the cases [Schrander-Stumpel and Fryns, 1998]. Important syndromal forms of congenital hydrocephalus include the autosomal recessive syndromal lissencephalies type 2 Walker-Warburg syndrome and Muscle-eye-brain disease with additional observation of a hypoplastic cerebellum and brainstem together with cobblestone lissencephaly and agenesis or hypoplasia of the corpus callosum as well as postnatal congenital muscular hypotonia, various eye abnormalities and global developmental delay, which in about half of the cases result from mutations in one of currently 6 genes associated with the O-glycosylation of α-dystroglycan [Bouchet et al., 2007]. The most frequent monogenic form of congenital hydrocephalus is due to hemizygous mutations in the L1CAM gene (about 7−15%) and clinically characterised by the distinct combination of a hydrocephalus due to stenosis of the aqueduct of Sylvius commonly in association with adducted thumbs (HSAS) [Tapanes-Castillo et al., 2010; Vos et al., 2010].
For siblings of sporadic patients with non-syndromic hydrocephalus a recurrence risk of about 1−4% has been determined and together with several monogenic animal models reflects the contribution of genetic factors to the pathogenesis of hydrocephalus [Schrander-Stumpel and Fryns, 1998; Zhang et al., 2006]. However, besides L1CAM, no further genes underlying human non-syndromic congenital hydrocephalus have yet been identified. By autozygosity mapping, we now revealed a homozygous mutation in the CCDC88C gene as underlying an autosomal recessive inherited form of non-syndromic complex hydrocephalus with apparently normal or at most mildly retarded psychomotor development.
Patients and Methods
Patients
The study was approved by the ethical review board of the medical faculty of the Friedrich-Alexander University of Erlangen-Nuremberg and informed consent was obtained from the parents. We studied a consanguineous family; 58 patients with schizencephaly, 20 of which had additional brain malformations including 3 patients with solitary cerebral cysts, and 5 patients with variable brain malformations with enlarged cerebrospinal fluid spaces.
Family History
The simplified pedigree of this consanguineous family is depicted in figure 1A. IV:1, a healthy woman of Algerian origin presented during her first pregnancy at 26 weeks of gestation with abnormal fetal ultrasound showing enlarged cerebral ventricles (fig. 1B). Fetal karyotyping from amniotic fluids revealed a normal female karyotype at a 500 GTG-banding resolution. At 29 weeks of gestation schizencephaly was suspected and after exclusion of maternal contamination using 13 microsatellite markers (PowerPlex16; Promega), EMX2 sequencing was performed on fetal DNA, which showed normal results. The pregnancy was eventually terminated and no post-mortem examination was performed. Similar ultrasound findings were detected in the second pregnancy of IV:1 at 25 weeks of gestation, but this time the parents decided to give birth to the girl. V:2 was born at 38 weeks of gestation with a weight of 2,490 g, a length of 45 cm and a head circumference of 34 cm. Apgar scores were 9-10-10, umbilical cord pH was 7.18 and base excess was −8. Cerebral MRI scans at the age of 3 days revealed marked dilatation of the lateral ventricles, while the third and fourth ventricles were not enlarged (fig. 1C, D, E). No cerebrospinal fluid flow void was evident in the aqueduct which may indicate either primary or secondary stenosis. Since no follow-up MRI was available, this cannot be further evaluated. A diverticulum-like pouch extended from the medial-posterior aspect of the left lateral ventricle into the interhemispheric space, extending through the tentorium into the infratentorial space, leading to mild compression of the upper cerebellar vermis. The posterior fossa was markedly enlarged with supra- and retrocerebellar fluid accumulation. The vermis was of normal shape, not elevated, and the communication between the fourth ventricle and the cisterna magna was of normal dimension. Brainstem morphology was not remarkable. Electroencephalography at the age of 5 days showed bitemporal and mild central hypersynchronous activity with sharp waves and spikes. The child showed no other malformations, dysmorphism or neurological anomalies. Because of increasing head circumference (40.3 cm) a ventriculo-peritoneal shunt was placed at the age of 4 weeks in the absence of any signs of increased intracranial pressure. At the age of one year the girl was hospitalized because of a tonic-clonic seizure with fever of 40.2°C after a few days of gastroenteritis with diarrhea and vomiting. At the age of 12.5 months she was found normally developed. She was able to walk assisted and produced sounds. Body weight was 9.4 kg (50th centile), height was 72.5 cm (10th centile) and head circumference was 45.5 cm (25−50th centile). At the age of 3 years and 3 months she spoke Arabic and German in 3-word sentences with grammatical errors and dyslalia. Communication, playing behaviour, coordination and fine motor skills were normal. Bayley Scales of Infant development testing revealed 156 points corresponding to a developmental quotient of 95. She had convergent strabismus which was treated with plasters, which she did not tolerate well. Her height was 94 cm (10th centile), her weight was 13 kg (10th centile) and her head circumference was 48.7 cm (25th−50th centile).
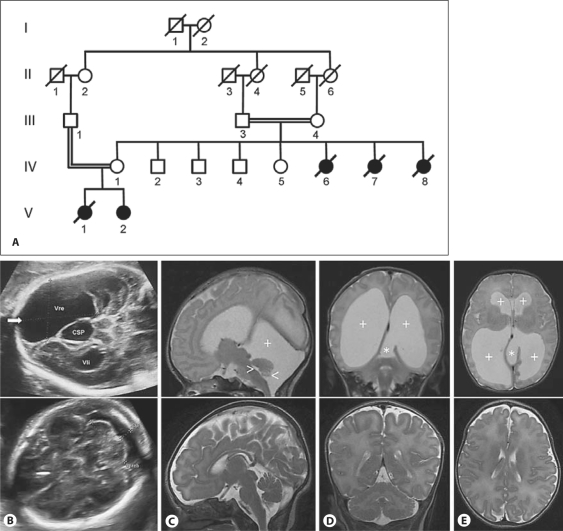
Fig. 1.
Pedigree and clinical findings in the investigated family. A Pedigree of the consanguineous family. IV:6 and IV:7 died neonatally of unknown cause. IV:8 died at the age of 22 years and was reported to have suffered from mild mental retardation. V:1 and V:2 showed a similar hydrocephalic brain malformation. III:1, IV:1, V:1 and V:2 were clinically investigated by the authors. B−E Ultrasound and MRI in affected individuals and normal scans for comparison in the lower row. B Ultrasound examination of V:1 at 29 weeks of gestation, demonstrating marked ventricular dilatation and an interhemispheric cystic structure in the posterior aspect (CSP). C, D, E T2w MRI of V:2 at 5 days. C Midsagittal MRI demonstrating a small vermis in normal position (<), a small 4th ventricle (>), marked retrocerebellar fluid accumulation in an enlarged posterior fossa (+), and no flow void in the aqueduct (data not shown). D Coronal section demonstrating pronounced ventricular dilatation (+). There is a diverticular-like extension from the medial aspect of the left lateral ventricle (trigone) extending through the tentorium into the posterior fossa (*). E Axial view demonstrating marked dilatation of the lateral ventricles (+). The diverticular-like extension is visible at the medial aspect of the left trigone (*). The lower row of ultrasound and MRI figures represents comparable normal sections.
The family history was remarkable for parental consanguinity (first cousins once removed). The mother of V:I and V:II, who's parents were first cousins, in addition to 4 healthy siblings had two sisters who died neonatally of unknown reason. A further sister IV:8 died at the age of 22 years. She was reported mentally retarded with a developmental age of 8−10 years.
Patient Samples
Genomic DNA from whole EDTA blood, saliva or amniotic fluid samples was extracted using a Flexigene AGF3000 technology (Qiagen, Hilden, Germany) on an automated Autogenflex 3000 isolator (Autogen, Holliston, Mass., USA) according to the protocols supplied by the provider. cDNA was synthesized from RNA extracted from fresh blood samples collected in PAXgene tubes (Becton Dickinson, Heidelberg, Germany) using the Superscript II Reverse Transcriptase Kit with random hexamer primers (Invitrogen, Carlsbad, Calif., USA).
Linkage Analysis
We performed genome-wide linkage analysis and homozygosity mapping in the consanguineous family (fig. 1A). All living individuals shown in the pedigree were available for linkage analysis. In addition DNA from amniotic fluid of V:1 was included in the genetic studies. Thus, DNA samples of the 2 affected children, both unaffected parents and seven further unaffected relatives were genotyped with the GeneChip Human Mapping 250K Nsp Array (Affymetrix, Santa Clara, Calif., USA). Genotypes were called by the GeneChip Genotyping software (GTYPE v4.0; Affymetrix). Simulation analysis for expected LOD scores (ELOD) for the given family pedigree was performed with a parametric 2-point analysis with the software FastSLink [Weeks et al., 1990]. Relationships of family members were verified by checking the genotype data with the software Graphical Representation of Relationships (GRR) [Abecasis et al., 2001]. Mendelian errors due to genotype errors were checked with the software Pedcheck [O'Connell and Weeks, 1998] and erroneous genotypes were removed from the data file. Parametric multipoint linkage analysis and haplotype construction were done with the programs Allegro [Gudbjartsson et al., 2000, 2005] and Merlin [Rampersaud et al., 2005]. Data handling was done with the software easyLinkage-Plus [Hoffmann and Lindner, 2005] under the assumption of an autosomal recessive mode of inheritance with 100% penetrance, disease allele frequency of 0.01% and at least 0.01 cM intermarker distance. Visualisation of haplotype data was performed using the software haplopainter [Thiele and Nürnberg, 2005].
Mutation Analysis of CCDC88C
Primers for all 30 exons, the complete 3′ and 5′ UTR, including 1,500 bp flanking promotor region were designed with the software ExonLocator [Schageman et al., 2004] and Primer3 [Rozen and Skaletsky, 2000]. Primer sequences are summarized in table 1. Genomic DNA of participants was whole genome amplified with Phi29 polymerase (GenomiPhi v2; GE Healthcare, UK) for mutation analysis. Subsequently all sequence aberrations were verified with original genomic DNA. Genomic DNA (20−50 ng) was amplified by PCR in 20-μl reactions with intronic primers according to standard procedures. The PCR products were purified with the AMPure Kit (Agencourt, Beverly, Mass., USA) on a Biomek NX instrument (Beckman Coulter, Fullerton, Calif., USA). Sequencing reactions were performed on both strands using the BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems, Foster City, Calif., USA) according to the manufacturer's instructions. After purification using the CleanSEQ Kit (Agencourt), the products were analyzed on an ABI Genetic Analyzer 3730 (Applied Biosystems) and the traces were evaluated using the SeqPilot 3.2.1.5 software (JSI medical systems, Costa Mesa, Calif., USA).
Table 1.
Primers used for sequencing CCDC88C
| Primer forward | Sequence | Primer reverse | Sequence |
|---|---|---|---|
| 5′UTR_p1_f | agcgatggcacacactcc | 5′UTR_p1_r | gtacagggttgacccgaagg |
| 5′UTR_p2_f | agcgctgggctgtaatacc | 5′UTR_p2_r | caggaagcaggtgatgtgg |
| 5′UTR_p3_f | ttgccctttttcctgtcg | 5′UTR_p3_r | ctccgaggttgctctgc |
| 5′UTR_p4_f | ggggcctcttgactgtacc | 5′UTR_p4_r | ctcggggaggaacaatacg |
| Ex1n_f | cctcgggccaccagtcc | Ex1n_r | ggctccagagttacaaagc |
| Ex2_f | tcctcctgtctcgtgtctgtt | Ex2_r | cgtctccaccttcagagaactac |
| Ex3_f | ctgcactgatatgtggttggtt | Ex3d_r | cctaaacataagcaccgtttcc |
| Ex3b_p1_f | cacccctgtaaggagaaagg | Ex3b_p1_r | cctccaggggatcacacg |
| Ex3b_p2n_f | atcactgtgggctcttctgg | Ex3b_p2n_r | tgttactggtcccttgacagg |
| Ex3b_p3_f | actgcaacctcatcctctgc | Ex3b_p3_r | atcccttctcccactgatgg |
| Ex3b_p4n_f | gcaagtttttgctgttctgc | Ex3b_p4n_r | gctagggccttgacaataacc |
| Ex4_f | gtttgcatcttacgtgctcctt | Ex4_r | ctcagggtaatgaacccactctt |
| Ex5_f | tggaagatcgatggagtgtgta | Ex5_r | gtactagaaacagctgccacgtaa |
| Ex6 f | agacacatcagtgtaaggctggt | Ex6_r | caatgtgtctgtggtgtctgaa |
| Ex7_f | ctgaaagcagaagtggttcgta | Ex7_r | ctgaagatgaagggagaggagat |
| Ex8_f | accttcctcctcgtttcttagtc | Ex8_r | agctagtccgaggtcaaagagta |
| Ex9_f | agtggcggatggacatagttta | Ex9_r | atctccacctgctgtcactaaga |
| Ex10_i_r | aatttccagcagccccatc | ||
| Ex10_a_r | ggagaaaccctgaatcactgc | ||
| Ex10_f | cctgtttcttagtgacagcaggt | Ex10_r | acaggtcagtctgaatgtgctt |
| Ex11_f | ctcggacttccctctagattgtt | Ex11_r | caagtagatggaactacaggctca |
| Ex12_f | agtgagccagtacatcttcatcc | Ex12_r | caagtggagcttgaaggaaatc |
| Ex13_f | tgatgaccatctcggagctt | Ex13d_r | tgacacctgcatgctctacc |
| Ex13bf | gtgggttctggcataagacc | Ex13br | tgtagcaactccccacacc |
| Ex14_f | gctacagaatgatgtcaggaggt | Ex14_r | cagtaccaggaatggctgtaatc |
| Ex15_p1_f | tctaagtggtggtgtctgtgc | Ex15_p1_r | ttctcggtggctgtctcc |
| Ex15_p2_f | agctgcacagggacttgg | Ex15_p2_f | ctgctctccaggctctgc |
| Ex15_p3_f | gaggaagaacgtggatcagcgctcaagg | Ex15_p3_r | ggtgacttgcttggtgagg |
| Ex15_p4_f | aggtggagctcaaggatgc | Ex15_p4d_r | aaagcatttaaggaggacacaga |
| Ex16_f | atgtagccacctggtctggtt | Ex16_r | actgaacagtggaggaagtgct |
| Ex17_f | gaacttgcggagaggaagtagat | Ex17_r | gtatcttgcctggtggacttg |
| Ex18_f | cgtctggcttacacagacca | Ex18d_r | aggacctgcaactcaagacg |
| Ex19_f | agtacctgtggccagttagcat | Ex19_r | gatcactgtttacagcgggtct |
| Ex20_f | gtaccctctgggtcttctgc | Ex20_r | caggccccacctttcc |
| Ex21_f | ctctgggatgtgagttacctgtc | Ex21_r | ggacttgaacttctcagtggttg |
| Ex22_f | gttacctgtttgtcaggctcttc | Ex22_r | cctccacacatttctgtctttg |
| Ex23_f | ttcttcctttgggagctaggtt | Ex23_r | agaagtttctggtggtggaaag |
| Ex24_f | ttccaaactcagtgaccatgac | Ex24d_r | gattctgaactctctctgatgtgg |
| Ex25_f | aacatcccagcccatgttag | Ex25_r | ctcgtaacacatctaagcgaagg |
| Ex25b_f | cacccagctctctctaaacg | Ex25b_r | ttttaggaagtgcagcaacc |
| Ex26b_f | cctctgacaccacctgaagc | Ex26b_r | agactgagagggagagcaacc |
| Ex27_f | agcagaggtggaagtgtgattt | Ex27_r | cttaaaggaaaggtaccgtgga |
| Ex27b_f | tccgtaaccatgactcactagc | Ex27b_r | tcgaatccaatgacaagtgc |
| Ex28_f | ctgggatatacctggtcattgg | Ex28_r | gtagcatgatcacagcccactat |
| Ex29_f | gaagcctctagaagccacca | Ex29n_r | ttgcggtgacttgtcagcta |
| Ex30_p1n_f | acatgtgcctcggagaagg | Ex30_p1n_r | ctgaggggactcctgtttgc |
| Ex30_p2_f | aagaaggggccaagatgc | Ex30_p2d_r | ggagctgccagcctttcc |
| Ex30_p3_f | cctgtctctgggcagacc | Ex30_p3_r | ctgcagcaggtgagaagtgc |
| Ex30_p4_f | gacacgaggcgcttctcc | Ex30_p4d_r | cggactgctctttgagacg |
| Ex30_p5_f | gtcggtctcccgatttgg | Ex30_p5_r | ttgcaaatgagtgttctactgg |
| 3′UTR_p1_f | tgtaacgtgttttctgtgtctgc | 3′UTR_p1_r | ctgtgtggtccccaaacc |
| 3′UTR_p2_f | aagtgagagtgctgctgttcc | 3′UTR_p2_r | gaatcttgaccaaagacacaaagc |
| 3′UTR_p3_f | gggtcatccgcacagagc | 3′UTR_p3_r | acccgatgctaacaatgtgc |
| 3′UTR_p4_f | ctgctgctttgtgtctttgg | 3′UTR_p4_r | gcagggagccccaagtcc |
Characterization of Splicing Pattern of Intron 29
To ascertain the effect of the IVS29 + 1 splice site mutation we amplified exons 27−30 and 28−30 from cDNA to generate wt amplicons of 435 bp and 541 bp, respectively. Amplified PCR products were evaluated by sizing in agarose gels and direct sequencing.
Western Blotting
Western blot analysis using protein lysates extracted from Epstein-Barr virus-transformed lymphoblastoid cell lines (LCLs) of patient V:II, her mother and controls were performed using a rabbit monoclonal KIAA1509 (CCDC88C) antibody which detects aa1566 to 1693 (encoded by exons 28 and 29) (Sigma Aldrich, St. Louis, Mo., USA). A β-actin antibody (ab8227) was used as control and a HRP-conjugated goat anti-rabbit IgG antibody (170−6515; Bio-Rad, Munich, Germany) as secondary antibody. Electrophoresis was performed on a 3−8% Tris-Acetate gel using the NuPage Novex system from Invitrogen.
Expression Studies of CCDC88C in Human Tissue Panels
The expression profile of CCDC88C was investigated by isoform specific RT-PCR (tables 2 and 3) and quantitative RT-PCR with the CCDC88C pre-designed TaqMan Gene Expression Assay Hs00929473_m1 (Applied Biosystems) using following cDNA panels: Human Multiple Tissue cDNA (MTC) Panel I (Clontech), Human Fetal Multiple Tissue cDNA (MTC) Panel (Clontech), Human Adult Neural Tissue II (BioChain) and Human Fetal Neural Tissue II (BioChain).
Table 2.
Primers used for isoform-specific RT-PCR of CCDC88C
| Primer forward | Sequence | Primer reverse | Sequence |
|---|---|---|---|
| RT_IF1f | aaagccaggcagatcaagg | RT_IF1r | aggcgctctgacttcttgc |
| RT_IF2f | gccaggatgtccgtactaagc | RT_IF2r | cgaggcaaggggtactcg |
| RT_IF3af | cagcctcatttctcaatgtcc | RT IF3ar | tgcttgaaggcatgaagagg |
| RT_IF3bf | ctgctgccctgcttctagg | RT_IF3br | tgctggatgtgtttctactgc |
| RT_IF3f | ctacagccccttccaactcc | RT_IF3r | ctggggatcaggtcttcg |
| RT_IF4nf | ggacggcatctttttgaacc | RT_IF4nr | aatgcagacactcccagacc |
Table 3.
Exon localisation and expected fragment length of isoform-specific primers of CCDC88C
| Product | Starting exon | Ending exon | Length, bp |
|---|---|---|---|
| IF1 | 14/15 | 15 | 617 |
| IF2 | 25b | 29 | 488 |
| IF3a | 26b | 27b | 403 |
| IF3f3ar | 26 | 27b | 234 |
| IF3b | 26b | 28 | 540 |
| IF4 | 2 | 3b | 540 |
Expression Studies in Patient Samples
To measure the expression of CCDC88C and β-catenin pre-designed TaqMan Gene Expression Assays were used (Hs00929473_m1, Hs00170025_m1; Applied Biosystems). Expression levels in patients’ RNA were quantified by real-time PCR in 4 replicates each as previously described [Rauch et al., 2008] and results were normalized to the mean of 3 endogenous controls (β-2-microglobulin (huB2M), β-actin (huACTB), acidic ribosomal protein (huPO)). Normalized expression levels were set in relation to 8 controls.
Expression levels of 84 genes from the Wnt pathway were analysed using the Wnt Signaling Pathway PCR Arrays (SABioscience, Frederick, Md., USA) which is based on quantitative RT-PCR using SYBR green detection on an ABI PRISM 7900HT device (Applied Biosystems) according to the supplier's instructions. Six different healthy, unrelated controls were analysed once, while the patient and her mother were analysed four times each.
Results
Linkage and Mutation Analysis
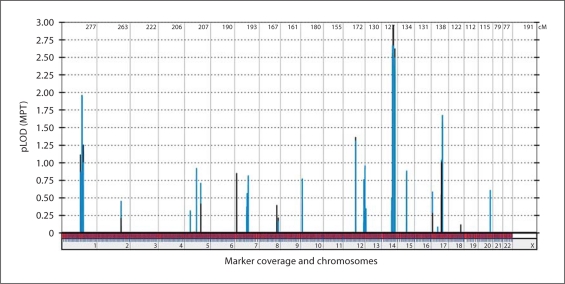
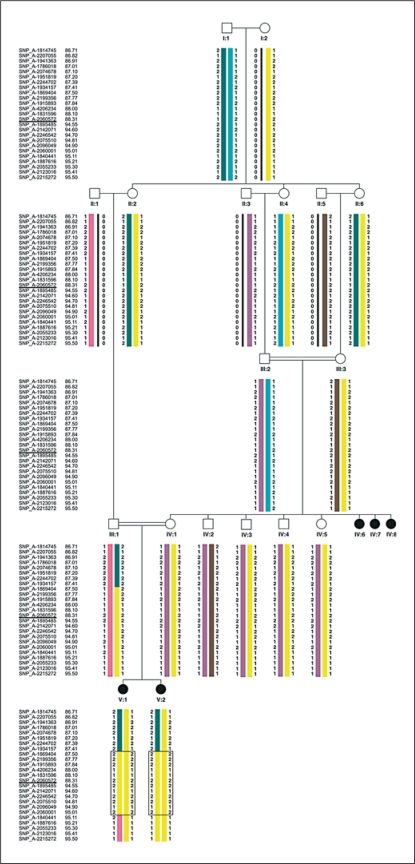
Whole genome LOD score analysis under the assumption of full penetrance and a disease allele frequency of 0.01% revealed a maximum LOD score of 2.96 on chromosome 14q (fig. 2). This finding was in line with the simulation analysis and the obtained ELOD score of 2.56. Due to the complexity of the pedigree, though, it was impossible to calculate LOD-scores using the computer program Allegro on any of our computers. Only when we portioned the pedigree we were able to perform the analysis. For the first calculation we excluded the healthy individuals IV:2 and IV:3, for the second IV:4 and IV:5. Both calculations indicated linkage to a single locus on chromosome 14 with a maximum LOD-score close to 3. As both subsets coincided but could not be calculated together we assume that the overall LOD-score clearly reaches genome-wide significance. Haplotype analysis revealed a linked region of 3.37 Mb on chromosome 14 flanked by the markers SNP_A-1934157 (rs41463644) and SNP_A-1840441 (rs7148382) (fig. 3) containing 25 positional candidate genes, including all RefSeq, orthologuous and predicted genes. In the analysis for potential candidate genes, we focused on genes which were expressed in the brain and/or found in signal transduction pathways in which genes are involved in cerebral cortex malformations [Brunelli et al., 1996; Granata et al., 1997; Cecchi, 2002; Theil et al., 2002; Muzio et al., 2005]. The most promising gene was CCDC88C (NM_001080414). The gene encompasses 30 exons and codes for the DVL-binding protein DAPLE. It interacts as a homooligomer with the PDZ domain of DVL1 and functions as a negative regulator of the canonical Wnt signalling pathway to inhibit CTNNB1/β-catenin stabilization [Oshita et al., 2003]. By direct sequencing of CCDC88C exons of the affected child V:2 we identified a homozygous splice site mutation at position IVS29 + 1 which changes the splice site donor G-U to A-U (c.5058 + 1G>A). The affected fetus V:1 was also homozygous for this mutation, while, as expected, both parents were heterozygous. All other unaffected family members were homozygous for the wild-type allele or heterozygous (fig. 4A). The mutation was not detected in 224 German healthy controls.
Fig. 2.
Results of whole genome linkage analysis. Parametric multipoint LOD-scores of pedrigree subset A are in blue and those of subset B are in black. Both pedigree parts indicate linkage to a single locus on chromosome 14 with a maximum LOD-score close to 3. As both subsets coincide but could not be calculated together we assume that the overall LOD-score clearly reaches genome-wide significance.
Fig. 3.
Haplotypes of the linked interval in the pedigree. The linked region of 3.37 Mb on chromosome 14 is flanked by the markers SNP_A-1934157 (rs41463644) and SNP_A-1840441 (rs7148382).
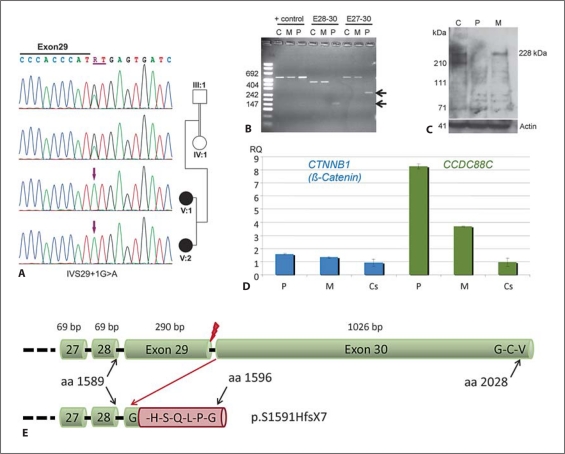
Fig. 4.
Characterization of the detected splice site mutation IVS29 + 1G>A. C = Control, M = Mother, P = Patient. A Electropherograms of the homozygous mutation detected in both affected siblings and heterozygous carrier status of both parents. B Confirmation of an aberrant transcript by reverse transcription PCR. The aberrant splicing in the patient caused by the mutation in intron 29 results in deletion of exon 29, which was confirmed by sequencing of the aberrant transcript. Deletion of exon 29 (290 bp) results in a frameshift in the last exon and subsequent loss of the terminal Dishevelled binding site. C Western blotting using an antibody against the epitope encoded by exons 28 and 29 confirmed absence of wild-type protein in the patient. D Results of quantitative RT-PCR using minor grove binding probes for CTNNB1 (β-catenin) and CCDC88C on cDNA derived from PAXgene RNA tube samples from the patient and her mother in comparison to 8 unrelated healthy controls. In line with the assumed loss of function without nonsense-mediated mRNA decay, we found significantly increased expression of both CCDC88C and β-catenin in the patient. The patient's heterozygous mother showed also increased CCDC88C expression levels but no statistical significant alteration of β-catenin expression (p < 0.028 in the patient, p < 0.148 in the mother; Wilcoxon two-sample test). E Scheme of the effect of the splice site mutation on the protein level. Upper cartoon showing the terminal part of the wild-type protein ending with the G-C-V Dishevelled binding site. Lower cartoon indicates the frameshift resulting from loss of exon 29 after the first amino acid from exon 30, the addition of 6 amino acids and the loss of the Dishevelled binding site (p.S1591HfsX7). Red flash indicates site of splice site mutation.
To further evaluate the splice site mutation we amplified two different exon 29 spanning fragments (exons 27−30 and 28−30) from the cDNA of the affected child V:2 and the mother IV:1. The mother and a control sample showed solely the wt amplicons of 435 bp and 541 bp, respectively, by sizing in agarose gel. Shortened amplicons were detectable only in the affected child V:2 (fig. 4B). Lack of amplification of the abnormal transcript in the mother may be explained by privileged amplification of the wild-type allele which is absent in the patient. By direct sequencing of the shortened transcript we could confirm the deletion of 290 bp corresponding to exon 29. Since exon 29 does not end with a codon this results in a frame shift in exon 30 and destroys the Dishevelled binding site which is encoded by the 3 terminal codons (p.S1591HfsX7). Western blotting confirmed the absence of wild-type protein in the patient, but could not exclude the presence of an abnormal protein, since the antibody binds to the amino acids encoded by the deleted exon 29 (fig. 4C).
While none of the 58 patients with schizencephaly showed a potential mutation, one of the 5 patients with enlarged cerebrospinal fluid spaces, abnormal gyral pattern and profound developmental delay was found to harbour two compound heterozygous variants. The transition c.1715C>T in exon 15 leads to the amino acid exchange p.S572L in a highly conserved position. The transversion c.5954C>G (rs3814839) in exon 30 also results in an amino acid exchange (p.S1985C). While the exon 30 variant was not detected in 504 controls, the variant in exon 15 was detected twice in heterozygous status. The parents were heterozygous for one of both variants, respectively, but the healthy sister was heterozygous for both mutations. Therefore the mutations were not considered to be the cause of the disease in the patient.
Expression Studies in the Patient and Her Heterozygous Mother
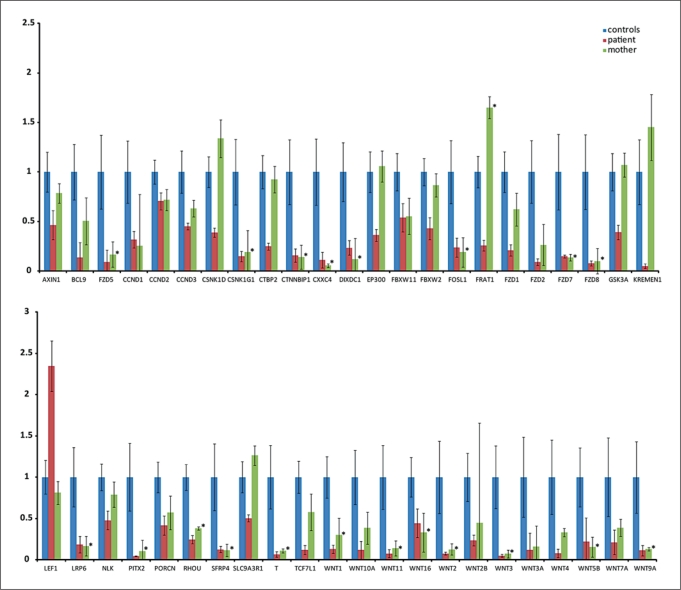
Quantitative RT-PCR studies in RNA derived from PAXgene samples of patient V:2 and her heterozygous carrier mother revealed an 8.3-fold increased CCDC88C expression in the patient and an 3.7-fold increased expression in the mother. These findings confirm the absence of nonsense mediated mRNA decay and indicate increased transcription due to loss of CCDC88C function. β-catenin expression showed a significant increased expression in the patient only, while the expression level in the mother was not significantly altered (fig. 4D). Expression profiling for the Wnt pathway genes revealed significant down-regulation of many genes in the patient, some of which, though, were also diminished in the healthy heterozygous mother (fig. 5). Genes that were significantly down-regulated in the patient, but not in the mother, were: AXIN1, CSNK1D, CTBP2, EP300, FBXW2, FRAT1, FZD1, GSK3A, KREMEN1, SLC9A3R1 and TCF7L1. LEF1 was the only gene that apart from β-catenin was significantly up-regulated in the patient and did not show a significant change in the mother (fig. 5, 6).
Fig. 5.
Diagram of relative changes in statistically significant differentially expressed genes in blood of the patient (red) using the Human Wnt Signalling Pathway PCR Arrays (SABioscience). Values of the healthy carrier mother (green) that also show statistically significant alterations are marked with an asterisk. Note significant over-expression of LEF1 and down-regulation of many WNT signalling proteins including the Frizzled receptor and transcriptional target genes in the patient. Standard deviations in the patient and the mother are derived from 4 different experiments each, while the standard deviations in controls are derived from single experiments in six different individuals. The commonly narrow standard deviations in the patient and her mother reflect the high reliability of the measurements, while the quite big standard deviations in the controls indicate a broad biological variability in expression levels for many of the genes in blood.
Fig. 6.
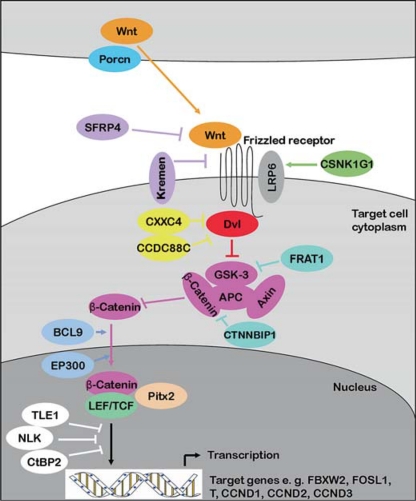
Scheme of the Wnt signalling pathway including proteins from the RT-PCR array. Porcn, a transmembrane protein is involved in the secretion of Wnt proteins, which represent intercellular signalling molecules [Rocheleau et al., 1997]. Reaching their target cells, Wnt proteins bind to Frizzled transmembrane receptors. The secreted protein SFRP4 shows homology to the cystein-rich ligand binding domain of the Frizzled receptors and thus functions as an antagonist of Wnt binding [Rattner et al., 1997]. Kremen also has an antagonistic function, since it represents the receptor of the negative regulator Dkk [Mao et al., 2002]. LRP6 represents a co-receptor of Wnt proteins, which is regulated by CSNK1G1 [Davidson et al., 2005]. Dvl, which is inhibited by CXXC4 and CCDC88C, mediates Wnt signalling via inhibition of GSK-3. GSK-3 is also inhibited by FRAT1 [Yost et al., 1998] and β-catenin is inhibited by CTNNBIP1 [Tago et al., 2000]. By inhibition of the β-catenin degrading GSK-3/APC/Axin complex β-catenin accumulates in the cytoplasm and is transferred to the nucleus, where it associates with LEF, TCF or Pitx2 proteins to activate transcription of target genes. BCL9 and EP300 represent transcriptional co-activators [Sun et al., 2000; Hoffmans et al., 2005] and TLE1, NLK and CtBP2 represent negative regulators [Chen et al., 1999; Furusawa et al., 1999; Ishitani et al., 2003].
Expression Profiling of CCDC88C
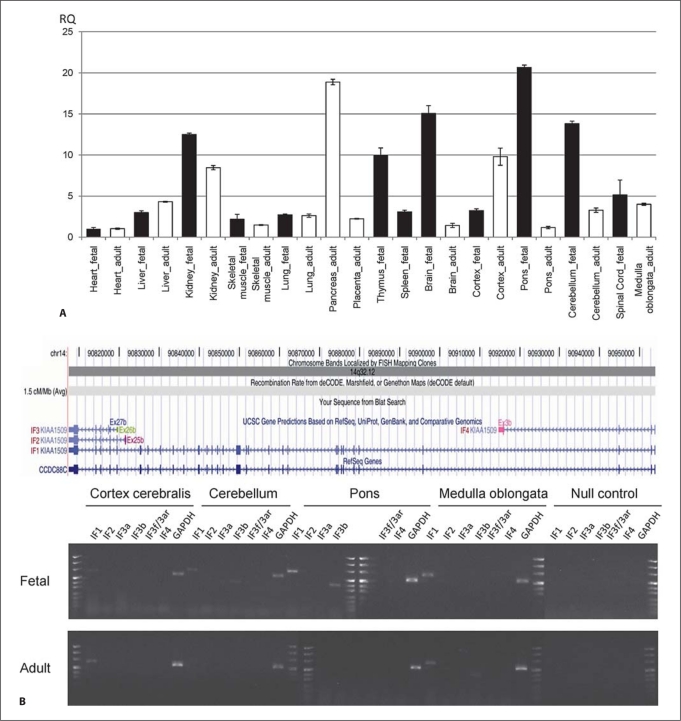
In order to gain further insight into the developmental role of CCDC88C and its different isoforms in humans, we performed isoform-specific expression profiling by quantitative RT-PCR from cDNA panels of various human fetal and adult tissues. These analyses revealed highest fetal expression in the brain, especially in the pons and cerebellum, while in adult brain tissues expression was highest in the cortex. The highest expression in adult tissues was found in the pancreas. Expression was also relatively high in fetal and adult kidney and in fetal thymus (fig. 7A). Isoform specific RT-PCR revealed the expected products for isoforms IF1 (exons 14−15), IF2 (exons 25b−29), IF3b (exons 26b−28), IF3f3ar (exons 26−27b) and IF4 (exons 2−3b) in blood (fig. 7B and data not shown). IF3a (exons 26b−27b) could not be amplified. Isoform-specific RT-PCR from human brain cDNA panels revealed that in adult brain only the longest isoform 1 was expressed and only in the cortex and medulla oblongata. In fetal brain isoform 1 was detected in the cortex cerebralis, cerebellum, pons and spinal cord. In addition isoform 3b was expressed in fetal cerebellum and pons and in adult medulla oblongata (fig. 7B).
Fig. 7.
CCDC88C expression studies by quantitative RT-PCR in cDNA from various human fetal and adult tissues showed highest relative expression in fetal brain and highest relative expression in adult pancreas (A). B Upper part represents a scheme of the various isoforms and the lower part shows the results of isoform-specific CCDC88C RT-PCR on fetal and adult brain cDNA panels. Size standard is puC Mix 8 (Fermentas) and GAPDH primers were used as positive control. IF1 is expressed in all fetal brain tissues investigated, but only in adult cortex and medulla. From the other isoforms only IF3b was found expressed in brain tissues: in fetal cerebellum and pons and in adult medulla. Primers for isoform specific RT-PCR were located in exon 14/15 and exon 15 for IF1 (617 bp), exon 25b and exon 29 for IF2 (488 bp), in exon 26b and 27b for IF3a (403 bp), in exon 26 and 27b for IF3f3ar (234 bp), in exon 26b and exon 28 for IF3b (540 bp) and in exon 2 and 3b for IF4 (540 bp).
Discussion
Via genome-wide linkage analysis we revealed a homozygous CCDC88C splice site mutation as underlying a distinct autosomal recessive complex hydrocephalic brain malformation with impressive prenatal enlargement of the lateral ventricles and the posterior fossa, and a diverticulum-like pouch. Despite the severe prenatal cerebral phenotype the affected patient V:2 showed normal psychomotor development at the age of 3 years and 3 months after ventriculo-peritoneal shunting and besides one febrile seizure no further organic or neurologic abnormality. One aunt of this patient, who was also an offspring of a consanguineous mating, was reported to have been mildly mentally retarded. Assuming the same condition in her, this may indicate that V:2 might show mild intellectual disabilities only later in life, or that this aunt might have developed normally if her hydrocephalus had been detected and treated accordingly. The diverticulum-like pouch in V:2 was initially considered to resemble schizencephaly, which is defined as true clefts formed in the brain as the result of failure of development of the cerebral mantle in the zones of cleavage of the primary cerebral fissures, which must be lined by abnormal gray matter (microgyria) [Leventer et al., 2008]. Mutations in EMX2, which is also involved in Wnt signalling, was reported to cause schizencephaly in 1996, however, this association has not since been confirmed [Merello et al., 2008]. Nevertheless, we sequenced CCDC88C in 58 patients with a radiological diagnosis of schizencephaly with or without reported associated brain malformations, but could not detect any mutation, underscoring that the brain malformation observed in the presented consanguineous family is distinct from schizencephaly. We then sequenced 5 additional patients with variable unclassified brain malformations with enlarged lateral ventricles, but did not find disease causing mutations. Therefore, CCDC88C mutations seem to cause a distinct non-syndromic complex hydrocephalus with apparently normal psychomotor development despite the impressive prenatal cerebral phenotype.
CCDC88C encodes a member of the hook-related proteins also known as Hook-related protein 2 [Enomoto et al., 2006]. This protein family is characterized by a microtubuli binding aminoterminal domain and a central coiled-coil domain. Hook-related protein 2 has in addition a carboxy-terminal domain with residues Glycin-Cystein-Valin which can bind to the PDZ-domain of Dishevelled (Dvl), an important scaffold protein involved in the regulation of the Wnt signalling pathway. Therefore it is also known as DAPLE which stands for ‘Dishevelled associating protein with a high frequency of leucin residues’. The Wnt protein family transmits a plethora of intercellular signals crucial for the development and homeostasis of metazoan animals [Logan and Nusse, 2004]. Dependent on the context, a given Wnt signal may denote cell proliferation, apoptosis, differentiation, migration, polarization, cell-fate specification, or other cellular processes. During early embryonal development WNTs are important caudalizing factors for the establishment of the initial anterior-posterior polarity of the brain. Disruption of Wnt-1 has been shown to disturb development of midbrain and anterior hindbrain structures including the cerebellum. Furthermore, an implication of Wnt signalling in the pathogenesis of hydrocephalus and cystic malformations of the posterior fossa is suggested by the observation of a complex phenotype including posterior cyst and postnatal hydrocephalus in transgenic mice with ectopically expressed Engrailed-1, a homeodomain transcription factor downstream of Wnt-1 [Rowitch et al., 1999]. Likewise, the distinct phenotype of the index patient in this study includes a hydrocephalus, a medial diverticulum and marked retrocerebellar fluid accumulation in an enlarged posterior fossa (fig. 1C).
Interestingly, cerebral midline cysts with or without enlarged ventricles are commonly observed as ‘posterior cysts’ within the phenotypic spectrum of holoprosencephaly and/or as ‘interhemispheric cysts’ in patients with agenesis of the corpus callosum [Simon et al., 2001], which assumingly may result from heterozygous mutations in the 2 genes SIX3 and SHH. Both genes as well as L1CAM have been shown to interact with Wnt signalling during early embryonal development [Lacbawan et al., 2009; Grana et al., 2010; Sylvester et al., 2010].
Wnt signals are not only crucial for development, but are also required for adult tissue maintenance including synaptic function, and perturbations promote human degenerative diseases and cancer. Wnt signalling is mediated through the seven-transmembrane Wnt receptor, Frizzled, which transmits the signal downstream via the cytoplasmic protein Dvl. There are two distinct pathways downstream of Dvl, the canonical β-catenin pathway and the non-canonical JNK pathway. Binding of Wnt to Frizzled leads to the accumulation, nuclear translocation, and interaction of β-catenin with the Tcf/Lef family of transcription factors. Therefore, β-catenin functions as a co-activator of transcription of Wnt-responsive genes. In the mouse, Daple inhibits Wnt3a induced accumulation of β-catenin and activation of T-cell factor transcriptional activity [Oshita et al., 2003]. In contrast to murine Daple, the Xenopus homologue of Daple activates Wnt signalling during development [Kobayashi et al., 2005].
The splice site mutation in the presented family leads to a premature stop codon within the last exon of CCDC88C, therefore assumingly not resulting in nonsense mediated mRNA decay. This was confirmed by quantitative RT-PCR in the patient's blood sample which indeed showed significantly increased CCDC88C mRNA levels, probably as a feedback mechanism from loss of function. As expected from the loss of function of the negative regulator CCDC88C through failure to inhibit Dvl which itself inhibits the GSK3-APC complex which normally inactivates β-catenin, we also found increased levels of β-catenin and the downstream protein LEF1 in the patient. Nevertheless, many upstream and downstream inhibiting as well as promoting Wnt proteins showed reduced expression, which could be explained by a complex negative feedback loop (fig. 5, 6). Therefore, loss of human DAPLE might reduce Wnt signalling mediated gene transcription due to a negative feedback loop. However, since the healthy mother also showed diminished expression of some of the genes, expression studies in patient's blood may not reflect the mechanisms during development and neuronal maintenance. Therefore, further studies in model systems are necessary to elucidate these findings. Nevertheless, these data indicate that disturbed Wnt signalling would have been detectable as an underlying disease cause in the patient by initial expression profiling. Since expression profiling is increasingly considered as a screening method to narrow down the underlying cause in patients with clinically and genetically hetergeneous conditions, this expression profile may help to identify further patients with CCDC88C mutations.
The expression pattern of DAPLE in embryonic mammals has not been examined, but expression of Xenopus Daple was found in various developing tissues including the mesencephalic region of the neural plate, the tail, eye and otic vesicles, olfactory placode, and the pharyngeal cavity [Kobayashi et al., 2005]. We investigated expression of human CCDC88C in a panel of embryonic and adult tissues by quantitative RT-PCR and found highest expression in fetal pons, cerebellum, thymus and kidney, as well as in adult pancreas, kidney and cortex. Nevertheless, loss of functional DAPLE in humans seems to affect only brain development. This finding may be related to isoform specific functions supported by our finding of nearly exclusive expression of isoform 1 in various fetal and adult brain tissues. The high CCDC88C expression in fetal pons and cerebellum corresponds well to the apparently abnormal development of the posterior fossa observed in our patient.
In conclusion, we report the first human phenotype associated with a mutation of the CCDC88C gene which encodes the Wnt signalling inhibitor DAPLE. Despite a dramatic prenatal hydrocephalic brain malformation, the affected living patient develops within the normal range and appears to have no further health issues.
Acknowledgement
This study was supported by the ‘German Mental Retardation Network’ (MRNET) funded by the German Federal Ministry of Education and Research (BMBF) as a part of the National Genome Research Network (NGFNplus) to A. Reis and A. Rauch.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Bouchet C, Gonzales M, Vuillaumier-Barrot S, Devisme L, Lebizec C, et al. Molecular heterogeneity in fetal forms of type II lissencephaly. Hum Mutat. 2007;28:1020–1027. doi: 10.1002/humu.20561. [DOI] [PubMed] [Google Scholar]
- Brunelli S, Faiella A, Capra V, Nigro V, Simeone A, Cama A, Boncinelli E. Germline mutations in the homeobox gene EMX2 in patients with severe schizencephaly. Nat Genet. 1996;12:94–96. doi: 10.1038/ng0196-94. [DOI] [PubMed] [Google Scholar]
- Cecchi C. Emx2: a gene responsible for cortical development, regionalization and area specification. Gene. 2002;291:1–9. doi: 10.1016/s0378-1119(02)00623-6. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Ping J, Takahashi M. Girdin, a novel actin-binding protein, and its family of proteins possess versatile functions in the Akt and Wnt signaling pathways. Ann NY Acad Sci. 2006;1086:169–184. doi: 10.1196/annals.1377.016. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Moribe H, Kondoh H, Higashi Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor deltaEF1. Mol Cell Biol. 1999;19:8581–8590. doi: 10.1128/mcb.19.12.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garne E, Loane M, Addor MC, Boyd PA, Barisic I, Dolk H. Congenital hydrocephalus–prevalence, prenatal diagnosis and outcome of pregnancy in four European regions. Eur J Paediatr Neurol. 2010;14:150–155. doi: 10.1016/j.ejpn.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Grana TM, Cox EA, Lynch AM, Hardin J. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev Biol. 2010 doi: 10.1016/j.ydbio.2010.05.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata T, Farina L, Faiella A, Cardini R, D'Incerti L, Boncinelli E, Battaglia G. Familial schizencephaly associated with EMX2 mutation. Neurology. 1997;48:1403–1406. doi: 10.1212/wnl.48.5.1403. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Thorvaldsson T, Kong A, Gunnarsson G, Ingolfsdottir A. Allegro version 2. Nat Genet. 2005;37:1015–1016. doi: 10.1038/ng1005-1015. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH. easyLINKAGE-Plus–automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Stadeli R, Basler K. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol. 2005;15:1207–1211. doi: 10.1016/j.cub.2005.05.054. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Michiue T, Yukita A, Danno H, Sakurai K, et al. Novel Daple-like protein positively regulates both the Wnt/beta-catenin pathway and the Wnt/JNK pathway in Xenopus. Mech Dev. 2005;122:1138–1153. doi: 10.1016/j.mod.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Lacbawan F, Solomon BD, Roessler E, El-Jaick K, Domene S, et al. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function. J Med Genet. 2009;46:389–398. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventer RJ, Guerrini R, Dobyns WB. Malformations of cortical development and epilepsy. Dialogues Clin Neurosci. 2008;10:47–62. doi: 10.31887/DCNS.2008.10.1/rjleventer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Merello E, Swanson E, De Marco P, Akhter M, Striano P, et al. No major role for the EMX2 gene in schizencephaly. Am J Med Genet A. 2008;146A:1142–1150. doi: 10.1002/ajmg.a.32264. [DOI] [PubMed] [Google Scholar]
- Muzio L, Soria JM, Pannese M, Piccolo S, Mallamaci A. A mutually stimulating loop involving emx2 and canonical wnt signalling specifically promotes expansion of occipital cortex and hippocampus. Cereb Cortex. 2005;15:2021–2028. doi: 10.1093/cercor/bhi077. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshita A, Kishida S, Kobayashi H, Michiue T, Asahara T, Asashima M, Kikuchi A. Identification and characterization of a novel Dvl-binding protein that suppresses Wnt signalling pathway. Genes Cells. 2003;8:1005–1017. doi: 10.1111/j.1365-2443.2003.00692.x. [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Scott WK, Hauser ER, Speer MC. Potential for expanded power in linkage studies using the ALLEGRO and MERLIN software programs. J Med Genet. 2005;42:e68. doi: 10.1136/jmg.2005.032029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA. 1997;94:2859–2863. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, et al. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Danielian PS, McMahon AP, Zec N. Cystic malformation of the posterior cerebellar vermis in transgenic mice that ectopically express Engrailed-1, a homeodomain transcription factor. Teratology. 1999;60:22–28. doi: 10.1002/(SICI)1096-9926(199907)60:1<22::AID-TERA7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Schageman JJ, Horton CJ, Niu S, Garner HR, Pertsemlidis A. ELXR: a resource for rapid exon-directed sequence analysis. Genome Biol. 2004;5:R36. doi: 10.1186/gb-2004-5-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrander-Stumpel C, Fryns JP. Congenital hydrocephalus: nosology and guidelines for clinical approach and genetic counselling. Eur J Pediatr. 1998;157:355–362. doi: 10.1007/s004310050830. [DOI] [PubMed] [Google Scholar]
- Simon EM, Hevner RF, Pinter J, Clegg NJ, Delgado M, et al. The dorsal cyst in holoprosencephaly and the role of the thalamus in its formation. Neuroradiology. 2001;43:787–791. doi: 10.1007/s002340100567. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kolligs FT, Hottiger MO, Mosavin R, Fearon ER, Nabel GJ. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc Natl Acad Sci USA. 2000;97:12613–12618. doi: 10.1073/pnas.220158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester JB, Rich CA, Loh YH, van Staaden MJ, Fraser GJ, Streelman JT. Brain diversity evolves via differences in patterning. Proc Natl Acad Sci USA. 2010;107:9718–9723. doi: 10.1073/pnas.1000395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K, Nakamura T, Nishita M, Hyodo J, Nagai S, et al. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- Tapanes-Castillo A, Weaver EJ, Smith RP, Kamei Y, Caspary T, et al. A modifier locus on chromosome 5 contributes to L1 cell adhesion molecule X-linked hydrocephalus in mice. Neurogenetics. 2010;11:53–71. doi: 10.1007/s10048-009-0203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Aydin S, Koch S, Grotewold L, Ruther U. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129:3045–3054. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- Thiele H, Nürnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- Vos YJ, de Walle HE, Bos KK, Stegeman JA, Ten Berge AM, et al. Genotype-phenotype correlations in L1 syndrome: a guide for genetic counselling and mutation analysis. J Med Genet. 2010;47:169–175. doi: 10.1136/jmg.2009.071688. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM. SLINK: a general simulation program for linkage analysis. Am J Hum Genet. 1990;47:A204. [Google Scholar]
- Yost C, Farr GH, 3rd, Pierce SB, Ferkey DM, Chen MM, Kimelman D. GBP, an inhibitor of GSK-3, is implicated in Xenopus development and oncogenesis. Cell. 1998;93:1031–1041. doi: 10.1016/s0092-8674(00)81208-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Williams MA, Rigamonti D. Genetics of human hydrocephalus. J Neurol. 2006;253:1255–1266. doi: 10.1007/s00415-006-0245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]