Abstract
Over the past three decades, the outcome of Ewing sarcoma family tumor (ESFT) patients who are nonmetastatic at presentation has improved considerably. The prognosis of patients with metastatic disease at the time of diagnosis and recurrence after therapy remains dismal. Drug-resistant disease at diagnosis or at relapse remains a major cause of mortality among patients diagnosed with ESFT. In order to improve the outcome for patients with potential relapse, there is an urgent need to find reliable markers that either predict tumor behaviour at diagnosis or identify therapeutic molecular targets at the time of recurrence. An improved understanding of the cell of origin and the molecular pathways that regulate tumorigenicity in ESFT should aid us in the search for novel therapies for ESFT. The purpose of this paper is thus to outline current concepts of sarcomagenesis in ESFT and to discuss ESFT patterns of differentiation and molecular markers that might affect prognosis or direct future therapeutic development.
1. Introduction
Since James Ewing in 1921 first described a diffuse hemangioendothelioma of bone [1], several studies have attempted to elucidate the histogenesis of Ewing's sarcoma (ES) [2, 3]. Based on histology, electron microscopy, and immunostains, it was postulated that ES represents a primitive mesenchymal neoplasm with the potential for multidirectional differentiation [2]. Neural and neuroectodermal differentiation was demonstrated in cell lines [4, 5] and tissue sections [6, 7], and tumors with the histology of Ewing sarcomas were described in soft tissue [8–10]. Lesions with atypical features also appeared [11]. Askin et al. [12] described a distinctive neoplasm in the chest wall that we now recognize as belonging to this group of cancers. Originally, neuroectodermal tumors of bone [13] and soft tissue [8] were described as a separate entities, but some overlap became apparent [14]. The term “primitive neuroectodermal tumor” (PNET) was given to these latter lesions, and the sobriquet “peripheral PNET” (pPNET) was later applied to separate them from unrelated lesions of the central nervous system [15]. With the finding of specific translocations t(11;22) and t(21;22) and accompanying genetic fusions, it became generally accepted that ES of bone and soft tissue, pPNET of bone and soft tissue, Askin's tumor, and ES with neural differentiation represent a single tumor entity with common antigenic profiles, cytogenetic aberrations, and protooncogene expression. This common entity, which has been designated the Ewing sarcoma family of tumors (ESFT), possesses a limited capacity for several different degrees of differentiation [16–18].
The cell of origin for ESFTs has been hotly debated ever since their discovery [16, 19, 20]. Candidates have included neuroectodermal and mesenchymal cells of hematopoietic and nonhematopoietic origin. Human marrow mesenchymal stem cells (MSCs) currently appear to be a strong candidate as the cell of origin of ESFT. MSCs are permissive for the characteristic EWS-FLI1 fusion and can initiate reprogramming toward ES cancer stem cells [20, 21].
The overall prognosis of ESFT has improved with the improved use of chemotherapy for localized tumors. However, metastatic and recurrent disease still has a dismal prognosis. A number of pathological and molecular characteristics have been linked to the clinical outcome of ESFT. In this paper, we will outline the pathological features of ESFT, review the current concepts about their sarcomagenesis, discuss their capacity for multipotential differentiation, and focus on the controversies and impact of neural differentiation and other pathobiological parameters in patient outcome.
2. Morphology
Before cytogenetics established that ES and pPNET share the balanced translocation t(11;22) (q24;q12) in over 90% of cases [22], ES and pPNET were regarded as totally separate entities distinguished by their morphologic and ultrastructural characteristics. ES occurred primarily in bone and pPNET in soft tissue. However, these boundaries crumbled with reports of discrepant features, such as an extraskeletal neoplasm resembling ES [9], a neuroectodermal tumor of bone [13], and ES with neural features [14]. Thanks to cytogenetic and molecular studies, it is now accepted that ES and pPNET comprise opposite poles of a histological continuum of a single group of neoplasms. In between lie lesions with varying degrees of atypical features (atypical ES) and differentiation. With the use of genetic tools, evidence emerged that morphological and immunohistochemical spectrum of ESFT may be broader than has traditionally been accepted [23], and additional histological patterns of ESFT have now been described [23, 24]. The histology of ESFT can be described as “classical”, “atypical”, or “variant”. The following section will detail these histological features.
2.1. Classical Ewing Sarcoma Histology
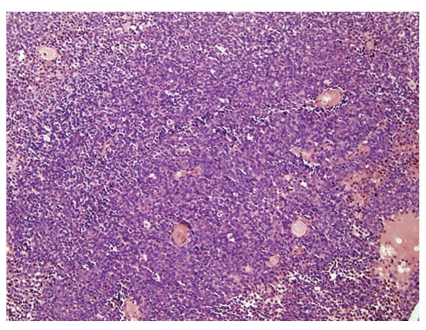
Microscopically, classical (typical) ES is the prototypic undifferentiated small round cell tumor, composed of sheets of monotonous round or oval cells with primitive-appearing nuclei and moderate amount of clear to amphophilic cytoplasm (Figure 1). The nuclear characteristics of the prototypic ES are those of a primitive uncommitted cell, that is an oval configuration with smooth contours, single inconspicuous nucleoli, and finely dispersed chromatin. The nuclei are generally centrally located and surrounded by a thin rim of clear to vacuolated cytoplasm. The lesions are highly cellular with little intervening intercellular space. Paradoxically, typical cases contain few mitoses. Cytologically, these tumors comprise a mixture of lightly staining “primary” cells and darkly staining “secondary” cells. Some feel that the “secondary” cells are effete versions of the “primary cells” that result from degenerative changes that lead to nuclear hyperchromasia or basophilia, but some ultrastructural studies show no signs of degenerative changes [25]. Sixty to seventy-five per cent of ESFTs [23, 24] are reported to have this classical histology (Table 1).
Figure 1.
Classical Ewing's sarcoma, microscopic image of typical histomorphology. The lesion comprises patternless sheets of small blue cells with round, regular nuclei, even nuclear margins, minimal cytoplasm, and indistinct boundaries.
Table 1.
| Number of cases | Classical | pPNET | Atypical | Spindle | Sclerosing | Adamantinoid | Reference |
|---|---|---|---|---|---|---|---|
| 66 | 46 | 9 | 3 | 3 | 2 | 3 | [23] |
| 415 | 280 | 53 | 82 | nr | nr | nr | [24] |
Nr: not reported.
2.2. Variant Histologies
2.2.1. Atypical ES (or Large Cell ES)
The cells of atypical ES are larger and more pleomorphic. They contain pleomorphic nuclei with indented, irregular nuclear membranes, conspicuous nucleoli, and increased amounts of cytoplasm that is often more eosinophilic than clear. The cells may contain abundant intracytoplasmic glycogen that gives them a clear appearance. A spindle-like configuration with fusiform nuclei can affect more than 50% of the tumor field. Atypical variants with epithelioid or clear cells [24] and a synovial-like pattern have been described [23]. Coincident with the appearance of atypical features, increased mitotic activity appears. Although frank neural differentiation by definition should be lacking, subtle signs may appear by electron microscopy or single immunostains.
2.2.2. pPNET
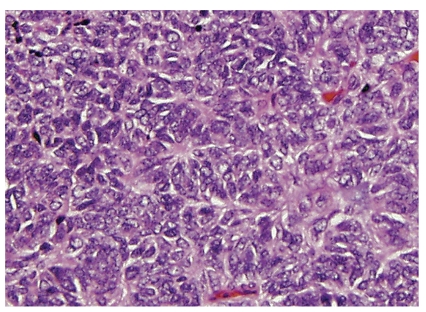
One of the best descriptions of pPNET was written by Jaffe et al. [13], who described neuroectodermal tumor of bone with prominent Homer Wright rosettes (previously called “malignant neuroepithelioma” or “peripheral neuroepithelioma”). They proposed this entity to be a distinct neoplasm, although the Intergroup Ewing Sarcoma Study had included rosette-forming lesions as a form of ES [26]. Jaffe et al. noted that no attempt had been made in prior publications to distinguish Homer Wright rosettes from perivascular or apoptotic pseudorosettes, and they postulated that Homer Wright rosettes are a marker for pPNET. This observation was later confirmed by Llombart-Bosch and Schmidt [7, 27]. As per the original description of neuroblastoma by James Homer Wright, these rosettes should consist of a closely apposed circle of cells with a peripheral wreath of nuclei and a central core of neuropil (Figure 2). The number of rosettes found in pPNETs varies from isolated to numerous. If isolated, they generally contrast with the background of monotonous fields of small round blue cells, but at times the histology can be subtle. Lesions are termed pPNET independent of the degree of differentiation and the number of rosettes. Jaffe presciently proposed that ES may be the most undifferentiated form of pPNET. Subsequently, several retrospective studies indicated that by immunohistochemistry features of neural differentiation can also be revealed in ES, using antibodies against proteins such as neuron-specific enolase, Leu 7 (later termed CD57), and S100 [7, 28, 29]. In 1991 Schmidt et al. [27] proposed that pPNET could be defined as tumors having Homer-Wright rosettes or expressing two different neural immunomarkers. Their retrospective study, which covered several decades of treatment, indicated that pPNETs had a worse clinical outcome than other forms of ESFT. Using Schmidt's criteria, the reported incidence of pPNET varies from 12% to 23%, compared to other ESFTs (Table 2).
Figure 2.
Peripheral primitive neuroectodermal tumor with Homer Wright rosettes. The rosettes contain a circular wreath of oval nuclei that surround a pale eosinophilic, fibrillary core.
Table 2.
Immunohistochemical expression of ESFT markers.
| Reference | CD99 | FLI1 | CK (AE1/3) | HMWCK | desmin | CD117 | CAV1 | CD57 (HNK1) |
|---|---|---|---|---|---|---|---|---|
| Folpe et al. [23] | 100% | 94% | 32% | 5% | 2% | 24% | ND | ND |
| Llombart-Bosch [24] | 99% | 89% | ND | ND | ND | ND | 96% | 53% |
ND: not done; CK: cytokeratin; HMWCK: high molecular weight cytokeratin; CAV: caveolin 1; HNK1: human natural killer antigen 1.
2.2.3. Adamantinoma-Like Pattern
This is a recently described variant of ESFT described by Bridge et al. as an example of “phenotypic drift” [30]. This pattern comprises well-formed nests of moderately hyperchromatic cells with striking peripheral palisading and a prominent host desmoplastic response. Adamantinoma-like ESFTs are positive for CD99, FLI1, pan-cytokeratin, and in distinction from other forms of ESFT, high molecular weight cytokeratin [23]. Note that cytogenetic features characteristic of ESFT distinguish these lesions from true adamantinoma of bone [30, 31].
2.2.4. Spindle Cell Sarcoma-Like Pattern
This is another recently described pattern of ESFT, composed of intersecting fascicles of spindle cells. It should however be noted that brief mention of a similar lesion appeared in 1983 in the description of “peripheral neuroepithelioma” in the first edition of Enzinger and Weiss's classic textbook [32]. Rare cases were also described in 1992 by Cavazzana et al. [33], although genetic confirmation was lacking at that time. There are well-developed, branching intratumoral blood vessels, reminiscent of hemangiopericytoma-like spindle cell sarcomas such as synovial sarcoma. CD99 and FLI1 are positive in all cases, and pan-cytokeratin is positive in two-thirds. Confirmatory genetic testing is required for this diagnosis (see below).
2.2.5. Sclerosing
This new pattern is described as composed of abundant hyalinised matrix. Two cases described by [23] were felt to be reminiscent of sclerosing epithelioid fibrosarcoma or sclerosing rhabdomyosarcoma. The neoplastic cells generally resemble the cells of typical ESFT. CD99 and FLI1 were positive in all cases, pan-cytokeratin was positive in 2 cases, and all were negative for high molecular weight cytokeratin. This lesion must be distinguished from desmoplastic small round cell tumor (DSRCT), which has a different cytogenetic aberration. However, rare tumors may resemble DSRCT histologically and immunophenotypically but contain cytogenetic features typical of ESFT [34].
2.2.6. Vascular-Like
Similar to Ewing's original report of “hemangioendothelioma of bone”, the observation that some ESFTs resemble vascular lesions persists. In Llombart-Bosch's series [24], there were 9 such cases (2%). ESFTs with this pattern contain pseudoendothelial elements that produce angiomatoid spaces lined by elongate tumor cells surrounding lacunae filled with erythrocytes and plasma. Unlike true vessels, the cells lining the surface of the spaces lack basal lamina and are in continuity with the surrounding typical ES. Of note, structures resembling Weibel-Palade bodies have been described in some ultrastructural descriptions [35], although genetic confirmation was lacking in these early reports.
2.3. Current Morphological Controversies
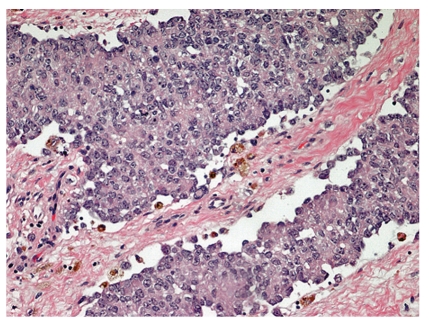
Two large recent retrospective series of genetically confirmed ESFT cases [23, 24] outline the heterogeneity of histological patterns in these lesions (Table 1). It is unclear in these reports whether the patterns were homogeneous or mixed in individual tumors, and current limited pretreatment biopsies preclude extensive histological assessment. In one of the series [24], all the unusual histological subtypes were grouped under the rubric of “atypical variant”. Patterns described as synovial-sarcoma-like, sclerosing, and adamantinomatous were not considered independent entities. Noteworthy is that unlike Llombart-Bosch et al., the Folpe series did not report any hemangioendotheliomatous cases, further emphasizing their rarity as a subtype. Questions remain as to whether or not unusual patterns may be seen in combination with the classical pattern and whether the morphological phenotype of recurrent tumors can be modified by chemotherapy. In our experience, we have seen one case of classical ESFT histology that recurred with an atypical epithelioid histological pattern (Figure 3). Others have described lesions with ganglioneuroma-like features, both before [36] and after [37] chemotherapy.
Figure 3.
Atypical Ewing sarcoma, postchemotherapy and recurrence. This recurrent ESFT displays epithelioid features, such as increased eosinophilic cytoplasm and cohesive nests.
3. Immunohistochemistry
Traditionally, the diagnosis of ESFT has been made by exclusion, but this situation considerably improved over the last 20 years with the introduction of new immunohistochemical markers. These markers include CD99 [38], FLI1 [39], and caveolin1 (CAV1) [24], which are commonly expressed in ESFTs and differentiate them from other small round cell tumors (Table 2). In addition, markers of neural, epithelial, and mesenchymal differentiation have been described in various subsets of ESFT. In the following section, we will briefly describe the immunohistochemical markers associated with ESFT.
3.1. CD99
CD99 is a membrane associated protein that is closely related to the murine THY-1 antigens, major cell surface glycoproteins of murine brain, and thymus cells [40]. It is a 32 KDa membrane glycoprotein that is highly expressed in most cases of ESFT [41]. This protein is encoded by a pseudoautosomal gene found in both X and Y chromosomes. CD99 is also known as MIC2 and is recognized by monoclonal antibodies 12E7, HBA71, O13, and HO36-1.1. It has a key role in several biological processes, including: cell adhesion, migration, and apoptosis; differentiation of T thymocytes; diapedesis of lymphocytes to inflamed vascular endothelium; maintenance of cellular morphology; and regulation of intracellular membrane protein trafficking [42, 43]. Modulation of CD99 expression significantly modifies cell growth in anchorage-independent conditions, and it affects cell migration, tumorigenesis, and metastatic activity in other models.
In ESFT, CD99 appears to prevent terminal neural differentiation [44]. This process is dependent on changes in mitogen-activated protein kinase pathway signalling. [44]. CD99 contributes to cell proliferation, migration, and metastasis of ESFT cells [44]. Questions remain as to whether CD99 is a downstream target of EWS/FLI1.
3.2. FLI1
As will be described below in more detail, the reciprocal translocation t(11;22) results in juxtaposition of the amino terminal domain of EWS to the carboxyl terminus of FLI1 [45]. FLI1 is normally expressed in endothelial cells and hematopoietic cells, including T lymphocytes. It is a member of the ETS (erythroblastosis virus-associated transforming sequences) family of DNA-binding transcription factors, and it is involved in cellular proliferation and tumorigenesis [46]. Folpe et al. [39] showed that a polyclonal antibody to the carboxyl terminus of FLI1 protein was a relatively sensitive (71%) and highly specific (92%) marker of ESFT.
These results and those of Nilsson et al. [47] indicate that FLI1 antibodies may play a valuable role in the immunohistochemical diagnosis of small blue round cell tumors. The combination of CD99 and FLI1 immunostaining appears to improve the specificity of these markers for diagnosis of EWS/FLI1 fusion-positive ESFT [48].
3.3. Caveolin
Caveolae are plasma membrane invaginations that regulate several intracellular signaling pathways. The defining components of caveolae are from 21 to 24 kDa molecules termed caveolins (CAV). There are several types of caveolins. CAV-1 and CAV-2 are ubiquitously expressed, whereas CAV-3 is only expressed in muscle tissue [49]. CAV-1 is the only member of the family required for caveolar formation. CAV-1 acts as a tumor suppressor in breast cancer and other tumors [50], and it is a direct EWS-FLI1 transcriptional target [51]. In ESFT, CAV-1 is a key determinant of tumorigenicity, which implies that it may be a molecular target for new therapeutic strategies [51]. It has also been proposed as a potential diagnostic marker for ESFT [24], but this observation needs additional study for confirmation.
3.4. Markers of Neural Differentiation
As noted above, a neural phenotype has been detected in many ESFT by immunohistochemistry and ultrastructure. In 1982, Schmidt et al. [14] noted that neural features such as processes, dense core granules, and microtubules could be noted in cases diagnosed as Ewing sarcoma. In 1986, Dickman and Triche [52] established diagnostic criteria for differentiation of extraosseous Ewing sarcoma (EOE) versus primitive rhabdomyosarcoma by electron microscopy. In a study of EOE treated with Intergroup Rhabdomyosarcoma Study protocols, Shimada et al. [53] identified 14 cases as pPNET based on neural and schwannian differentiation detected by immunohistochemistry and ultrastructure.
Neural differentiation can be demonstrated in ESFT by antibodies against neuron-specific enolase (NSE), CD57 (Leu-7, HNK1), S-100 [28, 29], neurofilaments (NF), and synaptophysin [54]. Schmidt et al. [27] proposed that pPNET diagnosis could be based on two different neural markers; before their study, a diagnosis of pPNET was made on only one neural marker (NSE, Leu-7, S-100, NF). Partial neural differentiation appears to be a frequent event in ESFT, judging from immunohistochemical and electron microscopic studies [54].
Both ES and pPNET are forms of ESFT, differing only in extent of neuroectodermal phenotype and morphological differentiation [18]. Literature review of older publications indicated an overall survival of only 30% for PNET and 65%–70% for EOE [16].
3.5. Miscellaneous Markers of Differentiation in ESFT
Positivity for various other phenotypic markers has been reported in ESFT. Markers such as desmin [55] and cytokeratins [23, 24] have been described. CD99-negative small round cell tumors with polyphenotypic expression have been reported [56]. Katz et al. [34] reported a polyphenotypic intraabdominal neoplasm with features of both ESFT and DSRCT and EWS/FLI1 transcript. Thorner et al. [56] described a DSRCT-like polyphenotypic small round cell tumor with an EWS/ERG fusion transcript.
Sorensen et al. [57] reported t(11;22) in 1 case of embryonal rhabdomyosarcoma and 4 cases of alveolar rhabdomyosarcoma. This raises questions about the genetic nature of so-called “primitive ectomesenchymomas”, lesions with a mixed myogenic-neural phenotype. These lesions are currently treated as rhabdomyosarcomas [58].
3.6. CD133 in Recurrent ESFT
In recent years, putative tumor-initiating cancer stem cells (tiCSC) have been isolated from human tumors [59]. TiCSC may be more resistant to standard chemo- and radiation-based therapies than bulk tumor cells [60]. CD133 has been described as putative marker of tiCSCs in ESFT [61]. It is thus proposed that CD133 may be a marker of chemoresistance in at least some cases of primary ESFT [61].
4. Cytogenetics and Molecular Features of ESFT
Specific balanced chromosomal translocations cytogenetically characterize ESFT [45] and create aberrant chimeric fusion oncogenes with different transactivation activities [62]. Currently, the gold standard for the diagnosis of ESFT is confirmation of histological diagnosis by cytogenetics/molecular studies. In general, tumors lacking genetic confirmation and signs of specific differentiation are termed “undifferentiated sarcomas” and are currently treated as nonrhabdomyosarcomatous soft tissue sarcomas by the Children's Oncology Group (COG) [63].
4.1. Translocations Involving EWS (See Table 3)
Table 3.
| Translocation | Fusion gene | Per cent positive |
|---|---|---|
| t(11;22)(q24;q12) | EWS-FLI1 | >85 |
| t(21;22)(q22;q12) | EWS-ERG | 5–10 |
| t(19;der)inv(21;22) | EWS-ERG | <1 |
| t(7:22)(p22;q12) | EWS-ETV1 | <1 |
| t(17;22)(q12;q12) | EWS-ETV4 | <1 |
| t(2;22)(q33;q12) | EWS-FEV | <1 |
| t(6;22)(p21;q12) | EWS-POU5F1 | <1 |
| t(1;22)(q36.1;q12) | EWS-PATZI | <1 |
| t(2;22)(q31;q12) | EWS-SP3 | <1 |
| t(20;22)(q13;q12) | EWS-NFATc2 | <1 |
| t(16;21)(p11;q22) | FUS-ERG | <1 |
| t(2;16)(q35;p11) | FUS-FEV | <1 |
| t(4;19)(q35;q13) | CIC-DUX4 | <1 |
In 99% of cases, molecular fusions of ESFT involve the EWS gene (also known as EWSR1; located on chromosome 22) and a member of the ETS family of transcription factors, which includes FLI1 (on chromosome 11) and ERG (in chromosome 21). In 5%–10% of ESFT, EWS is fused to other ETS members such as ETV1, ETV4, or E1AF [64]. Although it is a promiscuous gene that creates chimeras in a variety of neoplasms, fusions of EWS to ETS family genes are unique to ESFT. The list of variants of ETS family that genes may substitute for FLI1 continues to grow. Regardless of the ETS partner involved, variant translocations do not alter the tumor phenotype [65]. EWS fuses to non-ETS genes in other types of tumors, such as desmoplastic small round cell tumor, angiomatoid fibrous histiocytoma, clear cell sarcoma of soft tissue, and extraskeletal myxoid chondrosarcoma [66, 67]. Therefore, the partner gene, rather than EWS per se, appears to specify the tumor type.
Within the t(11;22), the chromosome breakpoint sites vary among 4 introns in EWS gene and six introns in the FLI1 gene and yield a large number of possible EWS-FLI1 fusion combinations [69]. EWS-FLI1 is the most heterogeneous gene fusion in cancer [70]. Different combinations of exons from EWS and FLI1 create up to 18 possible types of in-frame EWS-FLI1 chimeric transcripts [69]; these have been termed “type 1”, “type 2”, and so forth, fusions. Some reports indicate that fusion heterogeneity has functional and clinical significance [71]. Zoubek et al. [72] and De Alava et al. [73] reported that respective subgroups of 55 and 99 patients with localized ESFT and type 1 EWS-FLI1 fusions had longer relapse-free survival than those with nontype 1 fusions. Two more recent prospective studies [15, 72] however indicated that with more effective therapy there is no survival advantage of type 1 fusions in ESFT.
The alternate ESFT translocation t(21;22) causes a fusion of EWS to ERG, which encodes a transcription factor highly related to FLI1. EWS/ERG induces hematopoietic tumors in knock-in mice, suggesting that it can operate in targets other than mesenchymal cells [74].
With the t(2;22), t(7;22), and t(17;22) [70], EWS respectively fuses to FEV, ETV1, and EIAF (additional members of the ETS transcription factor family). These translocations occur only rarely and have not been the subject of detailed study. However, note that their presence would not be detected in ESFT if only EWS/FLI1 and EWS/ERG RTPCR were performed.
In three reported cases of CD99-negative small round cell tumors resembling ESFT, EWS juxtaposes to SP3, 2NF278, and POU5FI genes, which encode transcription factors not been previously implicated in ESFT [75]. Although extremely rare, lesions with the morphology and phenotypic of polyphenotypic small round cell tumor, rhabdomyosarcoma, or desmoplastic small round cell tumor have been reported to contain either EWS-FLI1 [56] or EWS-ERG fusion genes [76].
4.2. ESFT Translocations Not Involving EWS (See Table 3)
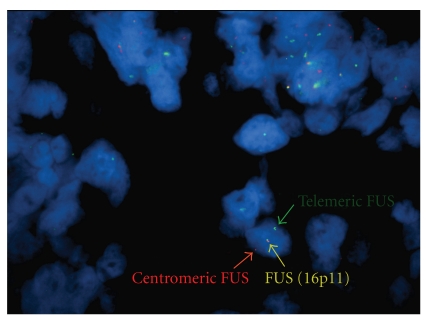
Variant translocations involving the TET family to which EWS belongs have also recently been described. These “promiscuous” molecular partnerships may cause false-negative results during diagnostic evaluation if the appropriate probes are not used (Figure 4) [70]. FUS (also a member of the TET family of RNA-binding proteins) shows considerably homology to EWS and rarely substitutes for EWS in ESFT fusion formation (reviewed by Barr [70]). The resultant t(16:21)(p11;q24) produces an FUS-ERG fusion with no EWS rearrangement [77]. In the t(2;16)(q35;p11) balanced translocation, there is an in-frame fusion of FUS to FEV [78].
Figure 4.
Ewing's sarcoma with FUS translocation. This CD99-positive lesion arose from the femur and had typical Ewing's sarcoma morphology. It was negative for an EWS rearrangement by both RT-PCR and FISH studies. In this interphase nucleus, note that one copy of the FUS locus has been divided, as evidenced by separate centromeric and telomeric gene signals, whereas the other copy has an intact, conjoined signal. Since breakapart FISH only tests a single gene locus, the partner gene in this translocation is unknown (fluorescence in situ hybridization of the FUS locus; courtesy of Dr. Ji-Yun Lee, University of Oklahoma).
4.3. Translocation-Negative ESFT
Some ESFT-like cases with typical morphology and immunohistochemistry may be negative for the common translocations [70]. These lesions constitute genuinely challenging cases, as EWS fusion negativity casts doubts about the diagnosis. Many of these are being treated as “undifferentiated sarcomas” until new information can clarify their biologic nature.
ESFT-like tumors showing similar morphology but lacking CD99 expression may have novel gene fusions. A completely novel gene fusion CIL-DUX4 that does not involve genes related to EWS or ETS family members has been described in two cases [75]. The exact relationship of these lesions to ESFT is undetermined.
The growing complexity of ESFT-related genetic rearrangements indicates that individual cases may have ESFT morphology and phenotype but lack evidence of EWS fusions by standard methodology [70]. As noted above, both partner genes may be separately involved in variant translocations [77]. Thus, a negative result generated by genetics testing should not absolutely preclude a diagnosis of ESFT in the context of typical morphological and immunophenotypic features. This situation underscores the value of using classical karyotype analysis in sarcoma diagnosis, because of the ability of standard cytogenetics to interrogate the cancer genome for balanced chromosomal translocations and other genomic aberrations. On the other hand, one should remember that other round cell sarcomas, particularly synovial sarcoma, alveolar rhabdomyosarcoma, and T-cell lymphoma, may closely mimic ESFT and should be vigorously excluded in these situations.
5. Secondary Genetic Changes of ESFT
Recent studies have focussed on secondary changes of ESFT, such as mutations and epigenetic alterations, particularly as a means of predicting clinical outcome. This work detects molecular targets that could be used in high risk patients. However, most currently available studies suffer from limited sample size, overly complex amounts of data, and lack of multivariate analyses that consider clinical prognostic indicators such as site, age, and stage. Nevertheless, these recent reports offer exciting glimpses into future avenues for exploration and include studies at the DNA, RNA, and protein level.
5.1. Comparative Genomic Hybridization
Comparative genomic hybridization (CGH) has been used to detect numerical gene abnormalities in ESFT [79, 80]. Numerical abnormalities with gains in chromosomes 1q21-22, 8, and 12 have been reported [79, 80]. In a study of 62 ESFT by CGH, univariate analysis showed that patients with gains of chromosomes 1q, 2q, 12, and 20 or losses of 16q and 17p have significantly lower overall survival than those without such aberrations [81]. A separate CGH study by Savola et al. of 31 ESFT [82] reported that the most frequent copy changes were gains at 1q, 2, 8, and 12 and losses at 9p and 16q. In this study, patients whose tumors had three or fewer copy number changes had better survival than patients with tumors having a higher number of copy number aberrations. Expression array studies of 16 of Savola et al.'s cases identified 20 novel ESFT-associated oncogenes and tumor suppressor genes. This study adds new information regarding alterations in ESFT gene copy number and provides valuable data for future analyses.
5.2. Gene Expression Profiles
Other expression array studies have also yielded data of interest. Based on microarray study of a small population of 20 metastatic and primary ESFTs, Ohali et al. [83,] claimed that gene expression at diagnosis might be used to predict recurrence and metastases. In a similar study of 27 ESFT, Schaeffer [84] identified genes that were differentially regulated between metastatic and localized tumors and described characteristic gene expression signatures associated with metastases, including signalling pathways for activation of PDGF and WNT1, apoptosis, angiogenesis, alteration of p53, and resistance to chemotherapy.
The expression profile of EWS/FLI in cell lines has been modulated by use of inhibitory RNA segments (RNAi) [85–87]. This work identifies target genes of the fusion protein and suggests that the phenotype is mediated by the t(11;22)(q24;q12). With this model, there were 3-4 times as many genes that were down-regulated by EWS-FLI1 as were up-regulated. One target gene of EWS/FLI appears to be NKX2.2, whose expression is critical for the transformed phenotype of Ewing's sarcoma cells. NKX2.2 plays a normal role in central nervous system and neuronal development, but its role in oncogenesis is unknown.
Signalling [22] and metabolic pathways [88] are being actively investigated to identify potential candidates for therapeutic intervention and predictive prognostic factors.
5.3. Cell Cycle Genes
Studies involving translocations have generally overshadowed the relevance of secondary mutations in ESFT biology. However, genetic/epigenetic alteration of factors such as p53 or p16 may determine prognosis. p53 alterations appear to define a small clinical sunset of ESFT with a markedly poor outcome [89, 90]. Significant correlation exists between a good chemotherapeutic effect (as determined by tumor necrosis) and the absence of genetic alterations in p53 or p16/p14ARF [91]. p16 data appear contradictory. Loss of heterozygosity of the cell cycle regulatory gene CDKN2A (which codes for p16) has no prognostic significance in primary ESFT, suggesting no role for epigenetic modification of this gene [92]. High levels of p16/p14ARF mRNA predict poor event-free survival, but the results need further confirmation [92]. The inconsistency of data obtained by profiling p16 and p14ARF gene status, mRNA, and protein expression emphasizes the importance of investigating cell cycle regulatory genes in identical clinical samples, using standardized methods, and employing a prospective clinical outcome study. Data regarding p53 pathway impairment and poor ESFT prognosis suggests that this parameter should be investigated in future prospective studies to determine whether it should be added to routine staging analyses. Increased Ki-67 expression has also been reported as a valuable indicator of poor prognosis in localized ESFT [93].
5.4. Microsatellite Instability
Microsatellite instability (MI) has been of great interest in human cancer, particularly colon carcinoma. The prevalence of MI in ESFT and how this factor relates to prognosis are not clear. Aldinger et al. [94] retrospectively analyzed MI in ESFT and concluded that loss of mismatch repair protein expression is not prevalent in ESFT, but the nature of that instability differs from the form observed in colorectal carcinoma. Microsatellites have been found to be responsive elements to EWS-FLI1 [22, 95, 96]. EWS-FLI1 interacts with GGAA-microsatellites to regulate some target genes, including NROB1, an EWS-FLI1-regulated gene that is required for the oncogenic phenotype of ESFT [97].
6. Ontogenesis of ESFT
Since James Ewing in 1921 first described a diffuse hemangioendothelioma of bone [1], several studies have attempted to elucidate the histogenesis of ESFT (reviewed by Yunis [17]). Initial morphological and immunopathological data suggested that ESFTs arise from primary mesenchymal stem cells with potential for multilineage differentiation [40]. The expression of neural markers in ESFT suggests either a neuroectodermal origin or a potential mesenchymal cell origin with secondary expression of a partial neural phenotype.
Several recent articles focusing on expression profiles of ESFT cell lines have given new clues on molecular mechanisms related to the origin, development, and progression of ESFT [79–87]. A large number of EWS/FLI1 target genes have been identified, some of which are upregulated and others are downregulated.
Mesenchymal stem cells (MSCs) are currently considered strong candidates for the cell of origin of ESFT [20, 98, 99]. Among primary human cells, only MSCs display permissiveness for stable EWS-FLI1 expression without undergoing growth arrest [20, 98]. In cell cultures, EWS-FLI1 behaves as an aberrant transcription factor that transforms MSCs by deregulating their gene expression [98]. In human pediatric MSCs (hpMSC), EWS-FLI1 induces the expression of embryonic stem cell genes OCT4, SOX2, and NANOG [100] and numerous genes involved in neural and neuroectodermal differentiation. In special media, transfection of EWS/FLI1 and repression of the inhibitory effect of miRNA cause hpMSC to assume a neural crest stem cell phenotype and to generate a subpopulation of cells that display ESFT features [100]. The target gene SOX2 appears to be a key player in determining ESFT cell differentiation and tumorigenicity [100].
The expression of neural markers in ESFT has also been intensely investigated. In 2005, Hu-liekovan [101] showed that EWS-FLI1 has a profound effect on cell differentiation as well as proliferation. They hypothesized that EWS-FLI1 acts as a lineage determinator rather than a pure oncogene, and they speculated that ESFT probably originates from primitive multipotent progenitor cells that are capable of differentiating into neural crest derivatives. EWS-FLI1 subsequently may impose a neural crest parasympathetic direction to the cells, but it inhibits terminal differentiation. Eventual secondary genetic alterations may be the oncogenic events that lead to cancer.
Baliko et al. in 2007 [102] showed that Notch signalling is active in ESFT tumors and cell lines, which express Notch receptors, ligands, and the Notch target gene HES1. Notch signalling inhibition causes ES cells to assume a more differentiated neuroid phenotype, also supporting the notion that ESFTs are derived from neuroectodermal precursors whose differentiation is inhibited by oncogenesis. Notch signalling inhibition induces neural differentiation but effects only minor changes in tumor growth, suggesting that it should not affect clinical outcome [29, 54, 103].
In 2010 [104] Gayscone reported that the neuronal marker BRN3A is expressed abundantly at the protein level in primary ESFT but not in neuroblastoma or rhabdomyosarcoma. Therefore, it appears that EWS/ETS proteins induce expression of neuronal markers and stimulate early stages of neuronal development but prevent terminal neuronal differentiation. EWS/ETS may regulate neuronal differentiation by controlling selective BRN3A transcriptional function.
7. Predictive Prognostic Factors in ESFT
A variety of predictive factors have been linked to survival of ESFT patients. These include clinical factors as well as the biological markers discussed above. The following section outlines our current understanding of prognostic factors and puts them into a clinical context.
7.1. Metastases
Presence of metastases is the most prominent adverse prognostic factor in ESFT; patients presenting with metastatic tumor have an estimated survival of only 20–25%. [19, 105–109]. Recurrence after therapy also portends a dismal outcome [110]. Recently, it has been reported that patients with primary disseminated multifocal ESFT may survive with intensive multimodal therapy [111].
Site of metastasis appears to be an important prognostic factor in MD. Patients with primary pulmonary metastases fare better than those with nonpulmonary tumor spread, especially following bilateral lung irradiation or myeloablative high-dose therapy (HDT) [107, 111–114], as compared to patients with primary bone and/or bone marrow involvement [108, 111, 113, 115–117].
In the Euro-Ewing 99 trial for patients with disseminated multifocal ESFT (excluding those with isolated pulmonary metastases) [111], increased risk was associated with the number of skeletal metastatic lesions. Of patients with more than 5 metastatic lesions at diagnosis, only 16% survived.
7.2. Localized Disease
Because of their higher survival rate, patients with localized disease at diagnosis offer a better opportunity to study prognostic factors in ESFT. In these patients, cooperative group and single institution studies have associated adverse outcome with older age at presentation (≥14 years [114, 118] or ≥18 years [106]), large tumor volume [119], poor response to induction therapy [120], axial location [114, 120], elevated LDH [121], secondary genetic abnormalities [122], deletion of p16 [123], and mutation of P53 [89, 90]. Prediction results vary widely among studies; they used different criteria (e.g., a large tumor may be defined as one >100 mL, >200 mL, or >8 cm) and treatments.
7.3. Age
The impact of age on the prognosis of ESFT remains controversial [124]. In different studies, the cut off for age analysis is variable: 14 [114, 117], 15 [108], or 18 years [106]. Ferrari [125] stratified ages as up to 14 y, from 15 to 18 y, and >18 y. In certain series, older age has been associated with inferior clinical outcome [106–108, 126], yet others have been unable to demonstrate a significant difference based on age alone [114, 124, 127, 128]. The impact of age can be confounded by the greater proportion of large pelvic primary tumours and more advanced disease in adult patients, and specific biological differences may also play a role [108, 124]. Older age remains an adverse prognostic factor despite the addition of ifosfamide and etoposide to the therapeutic regimen [106]. In a study to assess the impact of different therapies in children and adults, Gupta et al. [124] reported that adults with localized ESFT had an inferior outcome compared to pediatric patients, possibly due to dosage of alkylating agents and timing of local therapy.
7.4. Gender
Gender of patients is not significantly related to ESFT survival [106, 129].
7.5. Size (Tumor Volume)
Classically, large tumors have been associated with a worse prognosis. The size of the tumors has been assessed by longitudinal and volumetric measurements. Tumors greater than 8 cm are associated with a worse prognosis by univariate analysis [118, 130]. However, tumor size disappeared as a prognostic factor in the more intensive EW92 protocol [131]. In the CESS81 study, tumors greater than 100 cm3 were originally associated with worse outcome by univariate analysis [115]. However, with subsequent treatment improvements, the tumor size associated with a worse prognosis increased to greater than 200 cm3 [119]. In a multivariate analysis, Lee [129] reported that metastatic disease and large tumor size remained independent predictor factors, while pelvic involvement did not.
EICESS-92. the largest attempt to stratify ESFT therapy on the basis of tumor size [132] found no difference in survival between standard risk (localized small tumors) and high risk (large localized tumors or patients with metastatic disease) patients receiving standard therapy. Patients in the high risk arm with large localized tumors showed a trend toward improvement with the addition of etoposide, but this trend did not reach statistical significance [132, 133]. Because patients with metastases were in the same risk group as those with large tumors, the results cannot be interpreted based on size alone [133].
Primary tumor volume is a strong independent prognostic factor, even in patients with primary disseminated multifocal ESFT, and is more important than site of primary tumor. A trend toward worse outcome for central tumors has been seen, which may reflect problems in local control [111]. Larger tumor size will always represent a therapeutic challenge because of the problems with local control. However, without more standardized criteria for assessment of tumor size (longitudinal versus volumetric measurements), the value of volume as a prognostic factor will be imperfect [118].
7.6. Site
Axial tumors showed more aggressive behaviour in the early IESS (IESS-I) [134] and UKCCSG/MRC (ET-1) [107] studies. However, the same cooperative groups reported minimal or insignificant differences in outcome based on site (IESS-II and ET-2) [118, 135, 136]. The first POG-CCG Ewing trial (POG-8850/CCG-7881) reported that addition of ifosfamide-etoposide to the standard VACD regimen abrogated the negative prognostic implications of large tumor size (>8 cm) and pelvic location [118, 137]. Multivariate analysis has been controversial. Some studies have found pelvic location and time to local therapy to be associated with outcome [124], while other studies [129] reported that pelvic involvement was not independently associated with risk of death.
The increased prevalence of pelvic ESFT in adult patients is often cited as one of the main contributors to poor outcome [124]. It is likely that pelvic tumors are diagnosed later than tumors of the extremities because of subclinical extension into the pelvic cavity. This factor is consistent with the greater tumor volume of pelvic tumors and their significantly higher proportion of metastases at diagnosis [109, 129].
7.7. Neural Differentiation
The effect of neural differentiation on ESFT behaviour has been a source of conflicting results. For this purpose, ES must be consistently separated from pPNET, but tumoral heterogeneity makes this a difficult task. In spite of initial enthusiasm for neural differentiation as a prognostic factor (reviewed by Dehner [16]), subsequent retrospective studies have found it has no significant impact on clinical behaviour [28, 29, 103, 138].
Of interest, however, is the observation that outcome of osseous and extraosseous ESFTs did not differ in a recent Children's Oncology Group study [139]. An unpublished analysis of osseous ESFT patients by the Children's Cancer Study Group and Pediatric Oncology Group found that in nonmetastatic patients, neural differentiation even offered a survival advantage [140]. Similarly, Parham et al. [54] in 1999 found that neuroectodermal differentiation did not predict tumor behaviour in a retrospective study of 60 cases treated by modern therapy in two large institutions. They suggested that with improved multiagent chemotherapy, surgery, and radiation, differences in outcome between pPNET and ES no longer exist. This was also been reported in other studies [103]. Because of these conflicting results, current studies no longer separate ES from pPNET for prognostication or treatment.
However, Bacci et al. in 2000 [141] indicated that patients with PNET of bone fare significantly worse than patients with classical ES. Of note, cytogenetic results were not included in this study, and the same authors made no claims regarding neural differentiation in two subsequent outcome analyses of nonmetastatic ESFT [141, 142].
Atypical ESFT has been recently claimed in retrospective study to be the only histological variety associated with a less favourable clinical outcome [24], although this was not the case in the initial reports [10, 11].
7.8. Chemotherapy Response
During the past 30 years, improvements in systemic and local therapies have increased survival from less than 5% overall to 65%–70% for localized ESFT and to 25%–30% for metastatic tumors [114, 118, 143]. Alkylating agents (ifosfamide and cyclophosphamide) and anthracyclines (doxorubicin) are the two groups of agents most effective against ESFT. High-risk patients seem to have benefited from intensified treatment that incorporated ifosfamide [106, 117, 119].
Current chemotherapy regimens use the same drugs (vincristine, doxorubicin, cyclophosphamide, ifosfamide, and etoposide), although the exact regimen differs between Europe and North America [19, 105, 144]. Chemotherapy dose intensification and the use of megatherapy with autologous hematopoietic stem cell transplant seem valid alternatives for high risk patients.
Many factors can affect the feasibility, modalities, and timing of proposed treatments. Factors affecting the pharmacokinetics of cancer chemotherapy have important consequences in terms of therapeutic efficacy and safety. Cytochrome P450 [145] plays an important role in the biotransformation of anticancer agents [146] and metabolizes cyclophosphamide and ifosfamide. Factors such as age, genetic polymorphism, and intake of drugs and food can modify the activity of cytochrome P450, [147]. An influence of age on pharmacokinetics of ifosfamide has been reported [148].
Scotlandi (2009) [149] analysed the molecular factors that reflect tumor resistance to chemotherapy in ESFT and found that glutathione metabolism is a major pathway regulating ESFT chemoresistance. Glutathione S-transferases (GSTs) are a family of detoxification enzymes that catalyze the conjugation of glutathione to a wide variety of endogenous and exogenous compounds. GSTs have been implicated in the development of resistance toward chemotherapeutic agents [150]. Low expression of MGST1 (microsomal glutathione S-transferase 1) is significantly associated with better prognosis.
7.9. Chemotherapy-Induced Necrosis
In patients treated with primary chemotherapy followed by surgical excision, assessment of chemotherapy-induced necrosis allows reliable evaluation of tumor chemosensitivity. Good histological response is strongly related to good clinical outcome [119, 125, 126, 128]. The degree of necrosis seems to be independent of the drugs used and the length of preoperative chemotherapy [125]. Histological response is determined by the method of Salzer-Kuntschick et al. [151], in which 10% or fewer viable tumor cells in the surgical specimen are classified as “good response” and greater that 10% viability is regarded as “poor.” Ferrari et al. (2007) [125] investigated the influence of traditional factors (age, sex, site and size of the tumor, fever, and lactate dehydrogenase levels) on histological response of nonmetastatic ESFT; age and sex appear to influence the degree of necrosis.
8. Conclusions
Progress in the treatment of ESFT in the last three decades has derived largely from American and European cooperative trials using chemotherapy intensification and improved local control [19, 105, 118, 152]. Chemotherapy intensification and optimal local control have overcome differences in classical clinical, and biological prognostic factors that formerly portended a poor outcome [105]. With most modern treatment regimens, the disease-free survival for patients with localized disease may approach 70%, while overall survival may exceed 80% [19, 105, 117, 118]. However, a 30% relapse rate is still unacceptably high, considering that most relapsed patients do not survive [153].
The improvement of ESFT therapy is linked to the discovery of new strategies to select patients with poor and good prognosis. In the COG studies in North America, there are 3 risk groups: patients with localized tumours, patients with lung metastases only, and patients with other multiple metastases. Euro E.W.I.N.G. 99 study uses metastatic sites (none, lung, or other) and initial tumor size (< or > than 200 mL) for initial stratification and subsequently considers resectability and histological response to initial chemotherapy to assign patients to treatment randomization [144]. Therefore, excepting metastases at diagnosis, tumor burden, and response to chemotherapy, little is known about factors that determine disease progression or relapse [19].
Drug-resistant disease remains a major cause of mortality among patients diagnosed with ESFT. An improved understanding of the mechanisms of drug resistance and discovery of biomarkers associated with chemoresistance are needed for improved treatment of ESFT patients. Molecular signatures [149] offer a novel means of classification of ESFT patients into high- and low-risk groups, The glutathione metabolism pathway has emerged as one of the most significant pathways associated with prognosis. A small molecule inhibitor of GST enzymes 6-(7-nitro-2, 1, 3-benzoxadiazol-4-ylthio) hexanol NBDHEX has been proposed as a new potential therapeutic agent [149, 154].
The elucidation of the cell of origin is a crucial issue for discovering the mechanisms involved in the genesis of ESFT and for identification of reliable molecular markers and possible therapeutic agents. Mesenchymal stem cells or progenitor cells are currently considered strong candidates as the cell of origin of ESFT [20, 98, 99].
Preliminary data suggest that assessment of CD133 or Ki67 expression in diagnostic biopsies may identify tumors with poor outcome [61, 93], but confirmation is needed. The claim that atypical ES is associated with less favourable outcome needs to be tested in future trials, using clear-cut, reproducible criteria. The pathological criteria to differentiate ES versus pPNET also need to be clearly defined, so that we can establish strict criteria for diagnosis that permit reliable assessment of these features. Future research in the histopathological characterization of ESFT is thus warranted.
Efforts are ongoing to define the mechanisms of tumorigenesis molecular interactions of EWS/FLI1. Target genes that may be accessible for therapeutic agents are receptor tyrosine kineses, histone deacetylases, heat shock proteins, and mTOR derivatives [144, 153, 155].
Secondary mutations that could be of relevance in the biology of ES have often been usually overshadowed by studies of translocations. Secondary events could have relevance not only for prognosis but also for understanding the steps involved in cell transformation. Losses of p161INK4A, mutation of TP53, and deletion of CKDN2A appear to correlate with a poor prognosis [90, 91, 123]. Several groups [22, 115, 156] have identified gains and losses of various chromosomes that may be associated with a poor prognosis [82]. However, none of these parameters has been tested prospectively in a meaningful cohort of patients.
In summary, a better understanding of the molecular pathogenesis and biology of ESFT is leading to a new definition of potential targets for antitumour therapy. As the regulatory pathways responsible for transformation, growth, and metastases of ESFT become more defined, potentials for new therapeutic targets will expand. This situation has created both a challenge and an opportunity to develop predictive biomarkers capable of selecting patients most likely to benefit from targeted therapy.
References
- 1.Ewing J. Diffuse endothelioma of bone. Proceedings of the New York Pathology Society. 1921;21:17–24. doi: 10.1097/01.blo.0000229311.36007.c7. [DOI] [PubMed] [Google Scholar]
- 2.Moll R, Gould VE, Berndt R. Immunocytochemical analysis of Ewing’s tumors. Patterns of expression of intermediate filaments and desmosomal proteins indicate cell type heterogeneity and pluripotential differentiation. American Journal of Pathology. 1987;127(2):288–304. [PMC free article] [PubMed] [Google Scholar]
- 3.Cavazzana AO, Miser JS, Jefferson J, Triche TJ. Experimental evidence for a neural origin of Ewing’s sarcoma of bone. American Journal of Pathology. 1987;127(3):507–518. [PMC free article] [PubMed] [Google Scholar]
- 4.Lipinski M, Braham K, Philip I. Neuroectoderm-associated antigens on Ewing’s sarcoma cell lines. Cancer Research. 1987;47(1):183–187. [PubMed] [Google Scholar]
- 5.Lipinski M, Hirsch M-R, Deagostini-Bazin H. Characterization of neural cell adhesion molecules (NCAM) expressed by ewing and neuroblastoma cell lines. International Journal of Cancer. 1987;40(1):81–86. doi: 10.1002/ijc.2910400115. [DOI] [PubMed] [Google Scholar]
- 6.Kawaguchi K, Koike M. Neuron-specific enolase and Leu-7 immunoreactive small round-cell neoplasm: the relationship to Ewing’s sarcoma in bone and soft tissue. American Journal of Clinical Pathology. 1986;86(1):79–83. doi: 10.1093/ajcp/86.1.79. [DOI] [PubMed] [Google Scholar]
- 7.Llombart-Bosch A, Lacombe MJ, Contesso G, Peydro-Olaya A. Small round blue cell sarcoma of bone mimicking atypical Ewing’s sarcoma with neuroectodermal features. An analysis of five cases with immunohistochemical and electron microscopic support. Cancer. 1987;60(7):1570–1582. doi: 10.1002/1097-0142(19871001)60:7<1570::aid-cncr2820600727>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 8.Stout AP. A tumor of the ulnar nerve. Proceedings of the New York Pathology Society. 1918;18:2–12. [Google Scholar]
- 9.Angervall L, Enzinger FM. Extraskeletal neoplasm resembling Ewing’s sarcoma. Cancer. 1975;36(1):240–251. doi: 10.1002/1097-0142(197507)36:1<240::aid-cncr2820360127>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 10.Soule EH, Newton W, Jr., Moon TE, Tefft M. Extraskeletal Ewing’s sarcoma. A preliminary review of 26 cases encountered in the intergroup rhabdomyosarcoma study. Cancer. 1978;42(1):259–264. doi: 10.1002/1097-0142(197807)42:1<259::aid-cncr2820420140>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 11.Nascimento AG, Cooper KL, Unni KK. A clinicopathologic study of 20 cases of large-cell (atypical) Ewing’s sarcoma of bone. American Journal of Surgical Pathology. 1980;4(1):29–36. [PubMed] [Google Scholar]
- 12.Askin FB, Rosal J, Sibley RK. Malignant small cell tumor of the thoracopulmonary region in childhood. A distinctive clinicopathologic entity of uncertain histogenesis. Cancer. 1979;43(6):2438–2451. doi: 10.1002/1097-0142(197906)43:6<2438::aid-cncr2820430640>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe R, Santamaria M, Yunis EJ. The neuroectodermal tumor of bone. American Journal of Surgical Pathology. 1984;8(12):885–898. doi: 10.1097/00000478-198412000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt D, Mackay B, Ayala AG. Ewing’s sarcoma with neuroblastoma-like features. Ultrastructural Pathology. 1982;3(2):143–151. doi: 10.3109/01913128209016638. [DOI] [PubMed] [Google Scholar]
- 15.Dehner LP. Peripheral and central primitive neuroectodermal tumors. A nosologic concept seeking a consensus. Archives of Pathology and Laboratory Medicine. 1986;110(11):997–1005. [PubMed] [Google Scholar]
- 16.Dehner LP. Primitive neuroectodermal tumor and Ewing’s sarcoma. American Journal of Surgical Pathology. 1993;17(1):1–13. doi: 10.1097/00000478-199301000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Yunis EJ. Ewing’s sarcoma and related small round cell neoplasms in children. American Journal of Surgical Pathology. 1986;10:54–62. [PubMed] [Google Scholar]
- 18.Navarro S, Cavazzana AO, Llombart-Bosch A, Triche TJ. Comparison of Ewing’s sarcoma of bone and peripheral neuroepithelioma: an immunocytochemical and ultrastructural analysis of two primitive neuroectodermal neoplasms. Archives of Pathology and Laboratory Medicine. 1994;118(6):608–615. [PubMed] [Google Scholar]
- 19.Le Deley MC, Delattre O, Schaefer KL, et al. Impact of EWS-ETS fusion type on disease progression in Ewing’s sarcoma/peripheral primitive neuroectodermal tumor: prospective results from the cooperative Euro-E.W.I.N.G. 99 trial. Journal of Clinical Oncology. 2010;28(12):1982–1988. doi: 10.1200/JCO.2009.23.3585. [DOI] [PubMed] [Google Scholar]
- 20.Riggi N, Cironi L, Provero P, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Research. 2005;65(24):11459–11468. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 21.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 22.Ordóñez JL, Osuna D, Herrero D, De Álava E, Madoz-Gúrpide J. Advances in Ewing’s sarcoma research: where are we now and what lies ahead? Cancer Research. 2009;69(18):7140–7150. doi: 10.1158/0008-5472.CAN-08-4041. [DOI] [PubMed] [Google Scholar]
- 23.Folpe AL, Goldblum JR, Rubin BP, et al. Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. American Journal of Surgical Pathology. 2005;29(8):1025–1033. [PubMed] [Google Scholar]
- 24.Llombart-Bosch A, Machado I, Navarro S, et al. Histological heterogeneity of Ewing’s sarcoma/PNET: an immunohistochemical analysis of 415 genetically confirmed cases with clinical support. Virchows Archiv. 2009;455(5):397–411. doi: 10.1007/s00428-009-0842-7. [DOI] [PubMed] [Google Scholar]
- 25.Dickman PS. Electron microscopy for diagnosis of tumors in children. Perspectives in pediatric pathology. 1987;9:171–213. [PubMed] [Google Scholar]
- 26.Kissane JM, Askin FB, Foulkes M. Ewing’s sarcoma of bone: clinicopathologic aspects of 303 cases from the Intergroup Ewing’s Sarcoma Study. Human Pathology. 1983;14(9):773–779. doi: 10.1016/s0046-8177(83)80300-1. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt D, Herrmann C, Jurgens H, Harms D. Malignant peripheral neuroectodermal tumor and its necessary distinction from Ewing’s sarcoma: a report from the Kiel Pediatric Tumor Registry. Cancer. 1991;68(10):2251–2259. doi: 10.1002/1097-0142(19911115)68:10<2251::aid-cncr2820681025>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 28.Pinto A, Grant LH, Hayes FA, Schell MJ, Parham DM. Immunohistochemical expression of neuron-specific enolase and Leu 7 in Ewing’s sarcoma of bone. Cancer. 1989;64(6):1266–1273. doi: 10.1002/1097-0142(19890915)64:6<1266::aid-cncr2820640617>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Ladanyi M, Heinemann FS, Huvos AG, Rao PH, Chen Q, Jhanwar SC. Neural differentiation in small round cell tumors of bone and soft tissue with the translocation t(11;22)(q24;q12): an immunohistochemical study of 11 cases. Human Pathology. 1990;21(12):1245–1251. doi: 10.1016/s0046-8177(06)80038-9. [DOI] [PubMed] [Google Scholar]
- 30.Bridge JA, Fidler ME, Neff JR, et al. Adamantinoma-like Ewing’s sarcoma: genomic confirmation, phenotypic drift. American Journal of Surgical Pathology. 1999;23(2):159–165. doi: 10.1097/00000478-199902000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Kanamori M, Antonescu CR, Scott M, et al. Extra copies of chromosomes 7, 8, 12, 19, and 21 are recurrent in Adamantinoma. Journal of Molecular Diagnostics. 2001;3(1):16–21. doi: 10.1016/S1525-1578(10)60644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enzinger FM, Weiss SW. Soft Tissue Tumors. 1st edition. St. Louis, Mo, USA: Mosby; 1983. [Google Scholar]
- 33.Cavazzana AO, Ninfo V, Roberts J, Triche TJ. Peripheral neuroepithelioma: a light microscopic, immunocytochemical, and ultrastructural study. Modern Pathology. 1992;5(1):71–78. [PubMed] [Google Scholar]
- 34.Katz RL, Quezado M, Senderowicz AM, Villalba L, Laskin WB, Tsokos M. An intra-abdominal small round cell neoplasm with features of primitive neuroectodermal and desmoplastic round cell tumor and a EWS/FLI-1 fusion transcript. Human Pathology. 1997;28(4):502–509. doi: 10.1016/s0046-8177(97)90042-3. [DOI] [PubMed] [Google Scholar]
- 35.Triche TJ, Askin FB, Kissane J. Neuroblastoma, Ewing’s sarcoma, and the differential diagnosis of small-, round-, blue-cell tumors. In: Finegold M, editor. Major Problems in Pathology: Pathology of Neoplasia in Children and Adolescents. Philadelphia, Pa, USA: WB Saunders; 1986. pp. 145–195. [Google Scholar]
- 36.Williams S, Parham DM, Jenkins JJ., III Peripheral neuroepithelioma with ganglion cells: report of two cases and review of the literature. Pediatric and Developmental Pathology. 1999;2(1):42–49. doi: 10.1007/s100249900088. [DOI] [PubMed] [Google Scholar]
- 37.Knezevich SR, Hendson G, Mathers JA, et al. Absence of detectable EWS/FLI1 expression after therapy-induced neural differentiation in Ewing sarcoma. Human Pathology. 1998;29(3):289–294. doi: 10.1016/s0046-8177(98)90049-1. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson AJ, Chatten J, Bertoni F, et al. CD99 (p30/32MIC2) neuroectodermal/Ewing's sarcoma antigen as an immunohistochemical marker: review of more than 600 tumors and the literature experience. Applied Immunohistochemistry. 1994;2(2):231–240. [Google Scholar]
- 39.Folpe AL, Hill CE, Parham DM, O’Shea PA, Weiss SW. Immunohistochemical detection of FLI-1 protein expression: a study of 132 round cell tumors with emphasis on CD99-positive mimics of Ewing’s sarcoma/primitive neuroectodermal tumor. American Journal of Surgical Pathology. 2000;24(12):1657–1662. doi: 10.1097/00000478-200012000-00010. [DOI] [PubMed] [Google Scholar]
- 40.Parham DM. Ewing's sarcoma, peripheral neuroepithelioma, and related tumors. In: Parham DM, editor. Pediatric Neoplasia: Morphology and Biology. New York, NY, USA: Raven Press; 1996. pp. 65–85. [Google Scholar]
- 41.Kovar H, Dworzak M, Strehl S, et al. Overexpression of the pseudoautosomal gene MIC2 in Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. Oncogene. 1990;5(7):1067–1070. [PubMed] [Google Scholar]
- 42.Cerisano V, Aalto Y, Perdichizzi S, et al. Molecular mechanisms of CD99-induced caspase-independent cell death and cell-cell adhesion in Ewing’s sarcoma cells: actin and zyxin as key intracellular mediators. Oncogene. 2004;23(33):5664–5674. doi: 10.1038/sj.onc.1207741. [DOI] [PubMed] [Google Scholar]
- 43.Schenkel AR, Dufour EM, Chew TW, Sorg E, Muller WA. The murine CD99-related molecule CD99-like 2 (CD99L2) is an adhesion molecule involved in the inflammatory response. Cell Communication and Adhesion. 2007;14(5):227–237. doi: 10.1080/15419060701755966. [DOI] [PubMed] [Google Scholar]
- 44.Rocchi A, Manara MC, Sciandra M, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. The Journal of Clinical Investigation. 2010;120(3):668–680. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 46.Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. International Journal of Biochemistry and Cell Biology. 1997;29(12):1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 47.Nilsson G, Wang M, Wejde J, Kreicbergs A, Larsson O. Detection of EWS/FLI-1 by immunostaining. An adjunctive tool in diagnosis of Ewing’s sarcoma and primitive neuroectodermal tumour on cytological samples and paraffin-embedded archival material. Sarcoma. 1999;3(1):25–32. doi: 10.1080/13577149977839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llombart-Bosch A, Navarro S. Immunohistochemical detection of EWS and FLI-1 proteins in ewing sarcoma and primitive neuroectodermal tumors: comparative analysis with CD99 (MIC-2) expression. Applied Immunohistochemistry and Molecular Morphology. 2001;9(3):255–260. doi: 10.1097/00129039-200109000-00010. [DOI] [PubMed] [Google Scholar]
- 49.Smart EJ, Graf GA, McNiven MA, et al. Caveolins, liquid-ordered domains, and signal transduction. Molecular and Cellular Biology. 1999;19(11):7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. American Journal of Physiology. 2005;288(3 57-3):C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- 51.Tirado OM, Mateo-Lozano S, Villar J, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Research. 2006;66(20):9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 52.Dickman PS, Triche TJ. Extraosseous Ewing’s sarcoma versus primitive rhabdomyosarcoma: diagnostic criteria and clinical correlation. Human Pathology. 1986;17(9):881–893. doi: 10.1016/s0046-8177(86)80638-4. [DOI] [PubMed] [Google Scholar]
- 53.Shimada H, Newton WA, Jr., Soule EH, Qualman SJ, Aoyama C, Maurer HM. Pathologic features of extraosseous Ewing’s sarcoma: a report from the intergroup rhabdomyosarcoma study. Human Pathology. 1988;19(4):442–453. doi: 10.1016/s0046-8177(88)80495-7. [DOI] [PubMed] [Google Scholar]
- 54.Parham DM, Hijazi Y, Steinberg SM, et al. Neuroectodermal differentiation in Ewing’s sarcoma family of tumors does not predict tumor behavior. Human Pathology. 1999;30(8):911–918. doi: 10.1016/s0046-8177(99)90244-7. [DOI] [PubMed] [Google Scholar]
- 55.Parham DM, Dias P, Kelly DR, Rutledge JC, Houghton P. Desmin positivity in primitive neuroectodermal tumors of childhood. American Journal of Surgical Pathology. 1992;16(5):483–492. doi: 10.1097/00000478-199205000-00007. [DOI] [PubMed] [Google Scholar]
- 56.Thorner P, Squire J, Chilton-MacNeill S, et al. Is the EWS/FLI-1 fusion transcript specific for Ewing sarcoma and peripheral primitive neuroectodermal tumor? A report of four cases showing this transcript in a wider range of tumor types. American Journal of Pathology. 1996;148(4):1125–1138. [PMC free article] [PubMed] [Google Scholar]
- 57.Sorensen PHB, Shimada H, Liu XF, Lim JF, Thomas G, Triche TJ. Biphenotypic sarcomas with myogenic and neural differentiation express the Ewing’s sarcoma EWS/FLI1 fusion gene. Cancer Research. 1995;55(6):1385–1392. [PubMed] [Google Scholar]
- 58.Boué DR, Parham DM, Webber B, Crist WM, Qualman SJ. Clinicopathologic study of ectomesenchymomas from intergroup rhabdomyosarcoma study groups III and IV. Pediatric and Developmental Pathology. 2000;3(3):290–300. doi: 10.1007/s100249910039. [DOI] [PubMed] [Google Scholar]
- 59.Huntly BJP, Gilliland DG. Cancer biology: summing up cancer stem cells. Nature. 2005;435(7046):1169–1170. doi: 10.1038/4351169a. [DOI] [PubMed] [Google Scholar]
- 60.Suvà M-L, Riggi N, Stehle J-C, et al. Identification of cancer stem cells in Ewing’s sarcoma. Cancer Research. 2009;69(5):1776–1781. doi: 10.1158/0008-5472.CAN-08-2242. [DOI] [PubMed] [Google Scholar]
- 61.Jiang X, Gwye Y, Russell D, et al. CD133 expression in chemo-resistant Ewing sarcoma cells. BMC Cancer. 2010;10, article 116 doi: 10.1186/1471-2407-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case 40-1981. The New England Journal of Medicine. 1981;305(14):814–823. doi: 10.1056/NEJM198110013051407. [DOI] [PubMed] [Google Scholar]
- 63.Spunt SL, Skapek SX, Coffin CM. Pediatric nonrhabdomyosarcoma soft tissue sarcomas. Oncologist. 2008;13(6):668–678. doi: 10.1634/theoncologist.2007-0182. [DOI] [PubMed] [Google Scholar]
- 64.De Alava E, Gerald WL. Molecular biology of the Ewing’s sarcoma/primitive neuroectodermal tumor family. Journal of Clinical Oncology. 2000;18(1):204–213. doi: 10.1200/JCO.2000.18.1.204. [DOI] [PubMed] [Google Scholar]
- 65.Ginsberg JP, De Alava E, Ladanyi M, et al. EWS-FLI1 and EWS-ERG gene fusions are associated with similar clinical phenotypes in Ewing’s sarcoma. Journal of Clinical Oncology. 1999;17(6):1809–1814. doi: 10.1200/JCO.1999.17.6.1809. [DOI] [PubMed] [Google Scholar]
- 66.Mertens F, Antonescu CR, Hohenberger P, et al. Translocation-related sarcomas. Seminars in Oncology. 2009;36(4):312–323. doi: 10.1053/j.seminoncol.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Romeo S, Dei Tos AP. Soft tissue tumors associated with EWSR1 translocation. Virchows Archiv. 2009;456(2):219–234. doi: 10.1007/s00428-009-0854-3. [DOI] [PubMed] [Google Scholar]
- 68.Ordóñez JL, Osuna D, García-Domínguez DJ, et al. The clinical relevance of molecular genetics in soft tissue sarcomas. Advances in Anatomic Pathology. 2010;17(3):162–181. doi: 10.1097/PAP.0b013e3181d98cbf. [DOI] [PubMed] [Google Scholar]
- 69.Zucman J, Melot T, Desmaze C, et al. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. The EMBO Journal. 1993;12(12):4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barr FG, Womer RB. Molecular diagnosis of Ewing family tumors too many fusions...? Journal of Molecular Diagnostics. 2007;9(4):437–440. doi: 10.2353/jmoldx.2007.070080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Alava E, Kawai A, Healey JH, et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. Journal of Clinical Oncology. 1998;16(4):1248–1255. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- 72.Zoubek A, Dockhorn-Dworniczak B, Delattre O, et al. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? Journal of Clinical Oncology. 1996;14(4):1245–1251. doi: 10.1200/JCO.1996.14.4.1245. [DOI] [PubMed] [Google Scholar]
- 73.De Alava E, Kawai A, Healey JH, et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. Journal of Clinical Oncology. 1998;16(4):1248–1255. doi: 10.1200/JCO.1998.16.4.1248. [DOI] [PubMed] [Google Scholar]
- 74.Codrington R, Pannell R, Forster A, et al. The Ews-ERG fusion protein can initiate neoplasia from lineage-committed haematopoietic cells. PLoS Biology. 2005;3(8, article e242) doi: 10.1371/journal.pbio.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Bhargava R, Zheng T, et al. Undifferentiated small roud cell sarcomas with rare EWS gene fusions: identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. Journal of Molecular Diagnostics. 2007;9(4):498–509. doi: 10.2353/jmoldx.2007.070053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ordi J, De Alava E, Torné A, et al. Intraabdominal desmoplastic small round cell tumor with EWS/ERG fusion transcript. American Journal of Surgical Pathology. 1998;22(8):1026–1032. doi: 10.1097/00000478-199808000-00014. [DOI] [PubMed] [Google Scholar]
- 77.Shing DC, McMullan DJ, Roberts P, et al. FUS/ERG gene fusions in Ewing’s tumors. Cancer Research. 2003;63(15):4568–4576. [PubMed] [Google Scholar]
- 78.Ng TL, O’Sullivan MJ, Pallen CJ, et al. Ewing sarcoma with novel translocation t(2;16) producing an in-flame fusion of FUS and FEV. Journal of Molecular Diagnostics. 2007;9(4):459–463. doi: 10.2353/jmoldx.2007.070009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Armengol G, Tarkkanen M, Virolainen M, et al. Recurrent gains of 1q, 8 and 12 in the Ewing family of tumours by comparative genomic hybridization. British Journal of Cancer. 1997;75(10):1403–1409. doi: 10.1038/bjc.1997.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tarkkanen M, Kiuru-Kuhlefelt S, Blomqvist C, et al. Clinical correlations of genetic changes by comparative genomic hybridization in Ewing sarcoma and related tumors. Cancer Genetics and Cytogenetics. 1999;114(1):35–41. doi: 10.1016/s0165-4608(99)00031-x. [DOI] [PubMed] [Google Scholar]
- 81.Nucci MR, Weremowicz S, Neskey DM, et al. Genetic imbalances revealed by comparative genomic hybridization in Ewing tumors. Genes Chromosomes and Cancer. 2001;32(2):164–171. doi: 10.1002/gcc.1178. [DOI] [PubMed] [Google Scholar]
- 82.Savola S, Klami A, Tripathi A, et al. Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer. 2009;9, article 17 doi: 10.1186/1471-2407-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohali A, Avigad S, Zaizov R, et al. Prediction of high risk Ewing's sarcoma by gene expression profiling. Oncogene. 2004;23(55):8997–9006. doi: 10.1038/sj.onc.1208060. [DOI] [PubMed] [Google Scholar]
- 84.Schaefer K-L, Eisenacher M, Braun Y, et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. European Journal of Cancer. 2008;44(5):699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 85.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9(5):405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 86.Owen LA, Lessnick SL. Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing’s sarcoma. Cell Cycle. 2006;5(18):2049–2053. doi: 10.4161/cc.5.18.3213. [DOI] [PubMed] [Google Scholar]
- 87.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7(2):250–256. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 88.Lessnick SL, Dei Tos AP, Sorensen PHB, et al. Small Round Cell Sarcomas. Seminars in Oncology. 2009;36(4):338–346. doi: 10.1053/j.seminoncol.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 89.Abudu A, Mangham DC, Reynolds GM, et al. Overexpression of p53 protein in primary Ewing’s sarcoma of bone: relationship to tumour stage, response and prognosis. British Journal of Cancer. 1999;79(7-8):1185–1189. doi: 10.1038/sj.bjc.6690190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of P53 status in Ewing sarcoma. Cancer. 2000;89(4):783–792. [PubMed] [Google Scholar]
- 91.Huang H-Y, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. Journal of Clinical Oncology. 2005;23(3):548–558. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 92.Brownhill SC, Taylor C, Burchill SA. Chromosome 9p21 gene copy number and prognostic significance of p16 in ESFT. British Journal of Cancer. 2007;96(12):1914–1923. doi: 10.1038/sj.bjc.6603819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lopez-Guerrero JA, Machado I, Scotlandi K, et al. Clinicopathological significance of cell cycle regulation markers in a large series of genetically confirmed Ewing's Sarcoma Family of Tumors. doi: 10.1002/ijc.25424. International Journal of Cancer. In press. [DOI] [PubMed] [Google Scholar]
- 94.Alldinger I, Schaefer KL, Goedde D, et al. Microsatellite instability in Ewing tumor is not associated with loss of mismatch repair protein expression. Journal of Cancer Research and Clinical Oncology. 2007;133(10):749–759. doi: 10.1007/s00432-007-0220-2. [DOI] [PubMed] [Google Scholar]
- 95.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(29):10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gangwal K, Lessnicke SL. Microsatellites are EWS/FLI response elements: genomic “junk“ is EWS/FLI’s treasure. Cell Cycle. 2008;7(20):3127–3132. doi: 10.4161/cc.7.20.6892. [DOI] [PubMed] [Google Scholar]
- 97.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Molecular Cancer Research. 2006;4(11):851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 98.Riggi N, Suvà M-L, Suvà D, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Research. 2008;68(7):2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 99.Torchia EC, Jaishankar S, Baker SJ. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Research. 2003;63(13):3464–3468. [PubMed] [Google Scholar]
- 100.Riggi N, Suvà M-L, De Vito C, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes and Development. 2010;24(9):916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu-Lieskovan S, Zhang J, Wu L, et al. EWS-FLI1 fusin protein up-regulates critical genes in neurtal crest development and is responsible for the observed phenotype of Ewing’s family of tumors. Cancer Research. 2005;65(11):4633–4644. doi: 10.1158/0008-5472.CAN-04-2857. [DOI] [PubMed] [Google Scholar]
- 102.Baliko F, Bright T, Poon R, Cohen B, Egan SE, Alman BA. Inhibition of notch signaling induces neural differentiation in Ewing sarcoma. American Journal of Pathology. 2007;170(5):1686–1694. doi: 10.2353/ajpath.2007.060971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shanfeld RL, Edelman J, Willis JE, Tuason L, Goldblum JR. Immunohistochemical analysis of neural markers in peripheral primitive neuroectodermal tumors (pPNET) without light microscopic evidence of neural differentiation. Applied Immunohistochemistry and Molecular Morphology. 1997;5(2):78–86. [Google Scholar]
- 104.Gascoyne DM, Dunne J, Behjati S, Sebire NJ, Anderson J, Latchman DS. EWS/ETS proteins promote expression and regulate function of the homeodomain transcription factor BRN3A. Oncogene. 2010;29(21):3134–3145. doi: 10.1038/onc.2010.72. [DOI] [PubMed] [Google Scholar]
- 105.van Doorninck JA, Ji L, Schaub B, et al. Current treatment protocols have eliminated the prognostic advantage of type 1 fusions in Ewing sarcoma: a report from the children’s Oncology Group. Journal of Clinical Oncology. 2010;28(12):1989–1994. doi: 10.1200/JCO.2009.24.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grier HE, Krailo MD, Tarbell NJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. The New England Journal of Medicine. 2003;348(8):694–701. doi: 10.1056/NEJMoa020890. [DOI] [PubMed] [Google Scholar]
- 107.Craft AW, Cotterill SJ, Bullimore JA, Pearson D. Long-term results from the first UKCCSG Ewing's tumour study (ET-1) European Journal of Cancer Part A. 1997;33(7):1061–1069. doi: 10.1016/s0959-8049(97)00043-9. [DOI] [PubMed] [Google Scholar]
- 108.Cotterill SJ, Ahrens S, Paulussen M, et al. Prognostic factors in Ewing's tumor of bone: analysis of 975 patients from the European Intergroup Cooperative Ewing's Sarcoma Study Group. Journal of Clinical Oncology. 2000;18(17):3108–3114. doi: 10.1200/JCO.2000.18.17.3108. [DOI] [PubMed] [Google Scholar]
- 109.Tageja N. Prognostic indicators for ewing's sarcoma. The Lancet. 2010;376(9737):p. 232. doi: 10.1016/S0140-6736(10)61141-5. [DOI] [PubMed] [Google Scholar]
- 110.Leavey PJ, Mascarenhas L, Marina N, et al. Prognostic factors for patients with Ewing sarcoma (EWS) at first recurrence following multi-modality therapy: a report from the children’s oncology group. Pediatric Blood and Cancer. 2008;51(3):334–338. doi: 10.1002/pbc.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ladenstein R, Pötschger U, Le Deley MC, et al. Primary disseminated multifocal Ewing sarcoma: results of the Euro-EWING 99 trial. Journal of Clinical Oncology. 2010;28(20):3284–3291. doi: 10.1200/JCO.2009.22.9864. [DOI] [PubMed] [Google Scholar]
- 112.Paulussen M, Ahrens S, Burdach S, et al. Primary metastatic (stage IV) Ewing tumor: survival analysis of 171 patients from the EICESS studies. Annals of Oncology. 1998;9(3):275–281. doi: 10.1023/a:1008208511815. [DOI] [PubMed] [Google Scholar]
- 113.Paulussen M, Ahrens S, Craft AW, et al. Ewing tumors with primary lungmetastases: survival analysis of 114 (European Intergroup) Ewing Sarcoma Studies patients. Journal of Clinical Oncology. 1998;16:3044–3052. doi: 10.1200/JCO.1998.16.9.3044. [DOI] [PubMed] [Google Scholar]
- 114.Rodríguez-Galindo C, Liu T, Krasin MJ, et al. Analysis of prognostic factors in Ewing sarcoma family of tumors. Cancer. 2007;110(2):375–384. doi: 10.1002/cncr.22821. [DOI] [PubMed] [Google Scholar]
- 115.Jurgens H, Exner U, Gardner H, et al. Multidisciplinary treatment of primary Ewing’s sarcoma of bone. Cancer. 1988;7:208–213. doi: 10.1002/1097-0142(19880101)61:1<23::aid-cncr2820610106>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 116.Miser JS, Kraito MD, Tarbell NJ. Treatment of metastatic Ewing’s sarcoma or primitive neuroectodermal tumor of bone: evaluation of combination ifosfamide and etoposide: a children’s Cancer group and Pediatric Oncology Group study. Journal of Clinical Oncology. 2004;22:2873–2876. doi: 10.1200/JCO.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 117.Rodríguez-Galindo C, Navid F, Liu T, Billups CA, Rao BN, Krasin MJ. Prognostic factors for local and distant control in Ewing sarcoma family of tumors. Annals of Oncology. 2008;19(4):814–820. doi: 10.1093/annonc/mdm521. [DOI] [PubMed] [Google Scholar]
- 118.Rodriguez-Galindo C, Spunt SL, Pappo AS. Review treatment of ewing sarcoma family of tumors: current status and outlook for the future. Medical Pediatric Oncology. 2003;40:276–287. doi: 10.1002/mpo.10240. [DOI] [PubMed] [Google Scholar]
- 119.Paulussen M, Ahrens S, Dunst J, et al. Localized Ewing tumor of bone: final results of the cooperative Ewing’s Sarcoma study CESS 86. Journal of Clinical Oncology. 2001;19(6):1818–1829. doi: 10.1200/JCO.2001.19.6.1818. [DOI] [PubMed] [Google Scholar]
- 120.Derek Jenkin R, Al-Fawaz I, Al-Shabanah M, et al. Localised Ewing sarcoma/PNET of bone—prognostic factors and international data comparison. Medical and Pediatric Oncology. 2002;39(6):586–593. doi: 10.1002/mpo.10212. [DOI] [PubMed] [Google Scholar]
- 121.Bacci G, Ferrari S, Longhi A, et al. Prognostic significance of serum LDH in Ewing’s sarcoma of bone. Oncology Reports. 1999;6(4):807–811. doi: 10.3892/or.6.4.807. [DOI] [PubMed] [Google Scholar]
- 122.Hattinger CM, Potsguer U, Tarkannen M, et al. Prognostic impact of chromosomal aberrations in Ewing tumors. British Journal of Cancer. 2002;86:1763–1769. doi: 10.1038/sj.bjc.6600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei G, Antonescu CR, De Alava E, et al. Prognostic impact of INK4A deletion in Ewing sarcoma. Cancer. 2000;89(4):793–799. doi: 10.1002/1097-0142(20000815)89:4<793::aid-cncr11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 124.Gupta AA, Pappo A, Saunders N. Clinical outcome of children and adults with localized Ewing sarcoma, impact of chemotherapy dose and timing of local therapy. Cancer. 2010;116:3189–3194. doi: 10.1002/cncr.25144. [DOI] [PubMed] [Google Scholar]
- 125.Ferrari S, Bertoni F, Palmerini E, et al. Predictive factors of histologic response to primary chemotherapy in patients with ewing sarcoma. Journal of Pediatric Hematology/Oncology. 2007;29(6):364–368. doi: 10.1097/MPH.0b013e3180640d08. [DOI] [PubMed] [Google Scholar]
- 126.Bacci G, Ferrari S, Bertoni F, et al. Prognostic factors in nonmetastatic Ewing’s sarcoma of bone treated with adjuvant chemotherapy: analysis of 359 patients at the Istituto Ortopedico Rizzoli. Journal of Clinical Oncology. 2000;18(1):4–11. doi: 10.1200/JCO.2000.18.1.4. [DOI] [PubMed] [Google Scholar]
- 127.Verrill MW, Judson IR, Harmer CL, Fisher C, Thomas JM, Wiltshaw E. Ewing’s sarcoma and primitive neuroectodermal tumor in adults: are they different from Ewing’s sarcoma and primitive neuroectodermal tumor in children? Journal of Clinical Oncology. 1997;15(7):2611–2621. doi: 10.1200/JCO.1997.15.7.2611. [DOI] [PubMed] [Google Scholar]
- 128.Fizazi K, Dohollou N, Blay J-Y, et al. Ewing’s family of tumors in adults: multivariate analysis of survival and long-term results of multimodality therapy in 182 patients. Journal of Clinical Oncology. 1998;16(12):3736–3743. doi: 10.1200/JCO.1998.16.12.3736. [DOI] [PubMed] [Google Scholar]
- 129.Lee J, Hoang BH, Ziogas A, Zell JA. Analysis of prognostic factors in Ewing sarcoma using a population-based cancer registry. Cancer. 2010;116(8):1964–1973. doi: 10.1002/cncr.24937. [DOI] [PubMed] [Google Scholar]
- 130.Hayes FA, Thompson EI, Meyer WH, et al. Therapy for localized Ewing’s sarcoma of bone. Journal of Clinical Oncology. 1989;7(2):208–213. doi: 10.1200/JCO.1989.7.2.208. [DOI] [PubMed] [Google Scholar]
- 131.Marina NM, Pappo AS, Parham DM, et al. Chemotherapy dose-intensification for pediatric patients with Ewing's family of tumors and desmoplastic small round-cell tumors: a feasibility study at St. Jude children’s research hospital. Journal of Clinical Oncology. 1999;17(1):180–190. doi: 10.1200/JCO.1999.17.1.180. [DOI] [PubMed] [Google Scholar]
- 132.Paulussen M, Craft AW, Lewis I, et al. Results of the EICESS-92 study: two randomized trials of Ewing’s sarcoma treatment—cyclophosphamide compared with ifosfamide in standard-risk patients and assessment of benefit of etoposide added to standard treatment in high-risk patients. Journal of Clinical Oncology. 2008;26(27):4385–4393. doi: 10.1200/JCO.2008.16.5720. [DOI] [PubMed] [Google Scholar]
- 133.DuBois SG, Grier HE. Chemotherapy: the role of ifosfamide and etoposide in Ewing sarcoma. Nature Reviews Clinical Oncology. 2009;6(5):251–253. doi: 10.1038/nrclinonc.2009.25. [DOI] [PubMed] [Google Scholar]
- 134.Evans R, Nesbit M, Askin F. Local recurrence, rate and sites of metastases, and time to relapse as a function of treatment regimen, size of primary and surgical history in 62 patients presenting with non-metastatic Ewing's sarcoma of the pelvic bones. International Journal of Radiation Oncology Biology Physics. 1985;11(1):129–136. doi: 10.1016/0360-3016(85)90371-2. [DOI] [PubMed] [Google Scholar]
- 135.Burgert EO, Jr., Nesbit ME, Garnsey LA, et al. Multimodal therapy for the management of nonpelvic, localized Ewing’s sarcoma of bone: intergroup study IESS-II. Journal of Clinical Oncology. 1990;8(9):1514–1524. doi: 10.1200/JCO.1990.8.9.1514. [DOI] [PubMed] [Google Scholar]
- 136.Craft A, Cotterill S, Malcolm A, et al. Ifosfamide-containing chemotherapy in Ewing's sarcoma: the second United Kingdom children’s Cancer Study Group and the Medical Research Council Ewing's Tumor Study. Journal of Clinical Oncology. 1998;16(11):3628–3633. doi: 10.1200/JCO.1998.16.11.3628. [DOI] [PubMed] [Google Scholar]
- 137.Grier H, Krailo M, Link M. Improved outcome in non metastatic Ewing’s Sarcoma (ES) and PPNET of bone with the addition of ifosfamide (I) and etoposide (E) to vincristine (V), adryamicin (Ad), cyclophosphamide (C), and actinomycin (A) : A Children’s Cancer group (CCG) and Pediatrin Oncology Group (POG) report. Procceedings Annual Meeting American Society Clinical Oncology. 1994;13 abstract no. 421. [Google Scholar]
- 138.Luksch R, Sampietro G, Collini P, et al. Prognostic value of clinicopathologic characteristics including neuroectodermal differentiation in osseous Ewing's sarcoma family of tumors in children. Tumori. 1999;85(2):101–107. [PubMed] [Google Scholar]
- 139.Granowetter L, Womer R, Devidas M, et al. Dose-intensified compared with standard chemotherapy for nonmetastatic ewing sarcoma family of tumors: a children’s oncology group study. Journal of Clinical Oncology. 2009;27(15):2536–2541. doi: 10.1200/JCO.2008.19.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dickman PS, Askin FB, Miser J, et al. Pathology and prognosis in osseous Ewing’s sarcoma-primitive neuroectodermal tumor (ES/PNET): preliminary data analysis from CCG 7881/POG 8850. Modern Pathology. 1999;12:p. 2. [Google Scholar]
- 141.Bacci G, Forni C, Longhi A, et al. Long-term outcome for patients with non-metastatic Ewing's sarcoma treated with adjuvant and neoadjuvant chemotherapies. 402 patients treated at Rizzoli between 1972 and 1992. European Journal of Cancer. 2004;40(1):73–83. doi: 10.1016/j.ejca.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 142.Bacci G, Longhi A, Ferrari S, Mercuri M, Versari M, Bertoni F. Prognostic factors in non-metastatic Ewing's sarcoma tumor of bone: an analysis of 579 patients treated at a single institution with adjuvant or neoadjuvant chemotherapy between 1972 and 1998. Acta Oncologica. 2006;45(4):469–475. doi: 10.1080/02841860500519760. [DOI] [PubMed] [Google Scholar]
- 143.Marec-Bérard P, Chotel F, Claude L. Tumeurs PNET/Ewing: prise en charge actuelle et perspectives. Bulletin du Cancer. 2010;97(6):707–713. doi: 10.1684/bdc.2010.1120. [DOI] [PubMed] [Google Scholar]
- 144.Balamuth NJ, Womer RB. Ewing's sarcoma. The Lancet Oncology. 2010;11(2):184–192. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 145.Chen C-S, Lin JT, Goss KA, He Y-A, Halpert JR, Waxman DJ. Activation of the anticancer prodrugs cyclophosphamide and ifosfamide: identification of cytochrome P450 2B enzymes and site-specific mutants with improved enzyme kinetics. Molecular Pharmacology. 2004;65(5):1278–1285. doi: 10.1124/mol.65.5.1278. [DOI] [PubMed] [Google Scholar]
- 146.Scripture CD, Sparreboom A, Figg WD. Modulation of cytochrome P450 activity: implications for cancer therapy. The Lancet Oncology. 2005;6(10):780–789. doi: 10.1016/S1470-2045(05)70388-0. [DOI] [PubMed] [Google Scholar]
- 147.Fujita K-I. Cytochrome P450 and anticancer drugs. Current Drug Metabolism. 2006;7(1):23–37. doi: 10.2174/138920006774832587. [DOI] [PubMed] [Google Scholar]
- 148.Furlanut M, Franceschi L. Phramacology of Ifosfamide. Oncology. 2003;65(supplement 2):2–6. doi: 10.1159/000073350. [DOI] [PubMed] [Google Scholar]
- 149.Scotlandi K, Remondini D, Castellani G, et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. Journal of Clinical Oncology. 2009;27(13):2209–2216. doi: 10.1200/JCO.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- 150.Townsend DM, Tew KD. The role of gluthatione-S-transferasse in anticancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Salzer-Kuntschik M, Delling G, Beron G, et al. Morphologicla grades of regression in osteosarcoma after polychemotherapy COSS 80. Journal of Cancer Research Clinical Oncology. 1983;106(supplement 1):21–24. doi: 10.1007/BF00625047. [DOI] [PubMed] [Google Scholar]
- 152.Womer RB, West DC, Krailo MD, et al. Randomized comparison of every-two-week v. every-three-week chemotherapy in Ewing sarcoma family of tumors (ESFT) Journal of Clinical Oncology. 2008;26(supplement 15) abstract no. 10504. [Google Scholar]
- 153.Dirksen U, Jürgens H. Approaching Ewing sarcoma. Future Oncology. 2010;6(7):1155–1162. doi: 10.2217/fon.10.76. [DOI] [PubMed] [Google Scholar]
- 154.Ricci G, De Maria F, Antonini G, et al. -7,nitro-2,1,3 benzoxadiazole derivatives a new class 0f suicide inhibitors for gluthatione S-transferases . Mechanism of action of potential anticancer drugs. The Journal of Biological Chemistry. 2005;280:26397–26405. doi: 10.1074/jbc.M503295200. [DOI] [PubMed] [Google Scholar]
- 155.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing's sarcoma. Oncogene. 2010;29(32):4504–4516. doi: 10.1038/onc.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Maire G, Brown CW, Bayani J, et al. Complex rearrangement of chromosomes 19, 21, and 22 in Ewing sarcoma involving a novel reciprocal inversion-insertion mechanism of EWS-ERG fusion gene formation: a case analysis and literature review. Cancer Genetics and Cytogenetics. 2008;181(2):81–92. doi: 10.1016/j.cancergencyto.2007.11.002. [DOI] [PubMed] [Google Scholar]