Abstract
Reversible protein phosphorylation is a major regulatory mechanism in a cell. A chemical-genetic strategy to conditionally inactivate protein kinases has been developed recently. Mutating a single residue in the ATP-binding pocket confers sensitivity to small-molecule inhibitors. The inhibitor can only bind to the mutant kinase and not to any other wild-type kinase, allowing specific inactivation of the modified kinase. Here, we describe a protocol to construct conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. This protocol can be completed in about 3 weeks and should be applicable to other organisms as well.
INTRODUCTION
To inactivate essential proteins conditionally, temperature-sensitive alleles, regulatable promoters, degron alleles, conditionally active inteins as well as other strategies have been used1-7. However, these alleles are often leaky, require analysis under non-physiological conditions (high temperature) or their inactivation requires a long period of time. In addition, the molecular mechanism of temperature-sensitive protein inactivation is rarely understood. Small-molecule inhibitors selective for a target protein provide a valuable tool for conditional inactivation of proteins and provide several advantages over genetic approaches. Total as well as partial inhibition of the target protein can be achieved, depending on the amount of inhibitor added. The cell-permeable nature of these inhibitors allows reversible inhibition of the target protein both in vitro and in vivo. Rapid inactivation of the target protein by addition of the inhibitor leaves the cell with little time to adapt to or compensate for the missing protein activity. Small-molecule inhibitors do not typically alter the expression level of the target protein, nor do they disrupt protein complexes. In many cases, a small-molecule inhibitor and a genetic mutation can perturb a protein's activity in different ways. This may result in different phenotypes, often providing complementary information about the cellular function of a given kinase8. Small-molecule inhibitors can therefore reveal new biological functions of proteins that have already been studied genetically9.
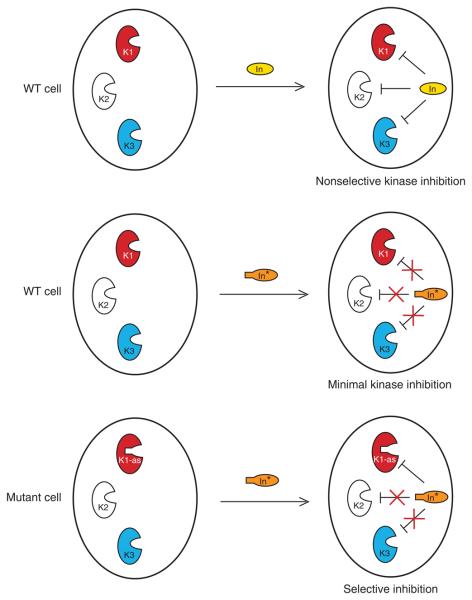
Numerous small-molecule inhibitors have been discovered and employed in the study of various protein kinases. However, due to the large number of kinases in the genome and their highly conserved active sites, which these inhibitors target, many inhibitors suffer from poor selectivity10. Moreover, little is known about the specificity of these inhibitors against yeast kinases, as most of these inhibitors were developed and characterized against mammalian kinases. A chemical-genetic strategy for sensitizing protein kinases to small-molecule inhibitors has been developed recently11. A single residue in the ATP-binding pocket, termed the gate-keeper residue, controls sensitivity of protein kinases to designed small-molecule inhibitors. This gate-keeper residue is conserved as a bulky hydrophobic amino acid in the protein kinase superfamily. Mutation of the gate-keeper to a small residue (alanine or glycine) creates a novel pocket that can be uniquely targeted by inhibitors with properly enlarged substituents. The mutation thus confers inhibitor sensitivity but does not interfere with kinase function in the absence of inhibitor (Fig. 1). The ATP-binding pocket of kinases is so conserved that the inhibitor can only bind to the mutant kinase and not to any wild-type kinases due to steric clash of the enlarged substituent and the bulky wild-type gate-keeper residue, hence allowing specific inactivation of the mutant kinase. Importantly, the fact that only sensitized protein kinase is inhibited allows a critical control experiment to be performed in which wild-type cells are treated with the inhibitor, thus revealing any possible off-target effects of the inhibitor. Such a control experiment should be performed in parallel with every experiment involving sensitized protein kinase and the inhibitor. The gate-keeper residue can be easily identified from primary sequence alignments in many protein kinases from various kinase subfamilies (http://sequoia.ucsf.edu/ksd/) (see ref. 12). Thus, the same strategy can be used to construct conditional analog-sensitive alleles of a wide variety of protein kinases11,13-18. This technology can also be used for identification of kinase substrates and has multiple applications in the drug discovery process19-21. Moreover, this methodology of generating inhibitors that selectively target a sensitized enzyme is not limited to protein kinases. A similar approach has been successfully used to inhibit proteins from other families, including phosphatases, motor proteins and GTPases22-26.
Figure 1.
Strategy for sensitizing protein kinases to small-molecule inhibitors. A conventional small-molecule inhibitor (In) typically blocks multiple kinases in the cell. Mutation of the gate-keeper residue in a protein kinase (K1) creates a new pocket where an enlarged inhibitor (In*) can bind. The inhibitor (In*) cannot bind to wild-type (WT) kinases, allowing a specific inactivation of the mutant kinase.
Despite many advantages, the chemical-genetic strategy of protein kinase sensitization contains a number of limitations. The catalytic activity of the protein kinase may be diminished by mutating the gate-keeper residue. In such cases, a second-site suppressor mutation can be introduced to rescue the kinase activity27. Another limitation is that this approach is only applicable to organisms in which genetic techniques are available to knock out or inactivate the target kinase and introduce the sensitized kinase allele. Exquisite skills and lengthy procedures are necessitated to carry out gene replacement in high eukaryotes such as Caenorhabditis elegans, Drosophila melanogaster and Mus musculus, impeding chemical-genetic analysis of protein kinases from these organisms. However, such limitation does not apply to the fission yeast S. pombe, a single-celled fungus, which has been extensively studied and has served as an excellent model organism. Importantly, the genome of S. pombe has been sequenced28.
Here, we describe a detailed protocol for the construction of conditional analog-sensitive kinase alleles in S. pombe (Fig. 2). Hhp1 and Hhp2 are members of a highly conserved casein kinase 1 family involved in DNA repair in S. pombe. Mutants lacking both Hhp1 and Hhp2 proteins grow extremely slowly, which precludes careful analysis of the double mutant phenotype29,30. We therefore constructed an analog-sensitive hhp1 mutant and combined it with an hhp2 deletion mutant. This allowed us to grow sufficient amounts of cells in the absence of inhibitor and to harvest them after adding the inhibitor. In this way, we were able to analyze the mutant phenotype in cells with inactivated Hhp1 kinase and lacking the Hhp2 protein31. We mutated methionine 84 of hhp1 to a glycine residue (hhp1-as) and expressed it in an S. pombe strain lacking wild-type hhp1. The mutated gene hhp1-as complemented the phenotype caused by deleting the hhp1 gene and resulted in only a minor growth defect. Importantly, the hhp1-as mutant, but not a wild-type strain, was sensitive to the inhibitor 1-NM-PP1 (Figs. 3 and 4) (see ref. 31).
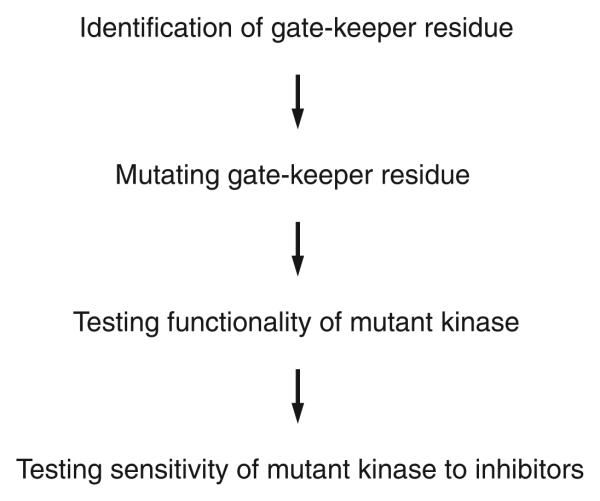
Figure 2.
Flowchart of the protocol.
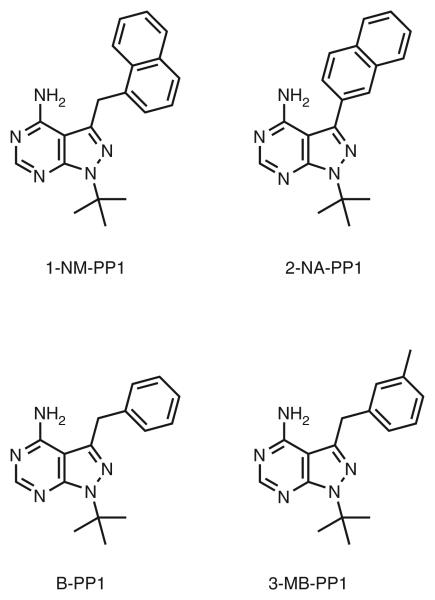
Figure 3.
Chemical structures of commonly used inhibitors for analog-sensitized kinases.
Figure 4.
Sensitivity of cells expressing Hhp1-as to various inhibitors. (a) Serial dilutions of hhp1Δ hhp2Δ cells expressing either wild-type Hhp1 (K13619) or the analog-sensitive Hhp1-as (K14637) protein were spotted on YES plates containing or lacking inhibitors and grown for 2 d at 32 °C. We noticed that in the presence of 2-NA-PP1, crystals formed on YES plates. (b) DIC (differential interference contrast) images of cells grown on YES plates containing or lacking inhibitors as described in (a).
MATERIALS
REAGENTS
Inhibitors 1-NA-PP1 and 1-NM-PP1 (Calbiochem; http://www.emdbiosciences.com/product/529579, http://www.emdbiosciences.com/product/529581); other inhibitors are available from Kevan Shokat on request
PCR kit (Takara; PCR kits from other suppliers should also work); PCR premix (see REAGENT SETUP)
High-fidelity DNA polymerase (e.g., PfuTurbo DNA polymerase; Stratagene)
Restriction enzymes (Roche; restriction enzymes from other suppliers should also work)
pCloneHyg1 (EF101286) is the vector used for cloning. It can be requested from Kim Nasmyth (Kim Nasmyth DNA collection number: 4802) or Juro Gregan lab
S. pombe genomic DNA from which the gene can be amplified
Anion exchange columns (Qiagen) for DNA purification from PCR, restriction digest or agarose gel; products from other suppliers should also work
Salmon sperm DNA (Stratagene) for use as a carrier for yeast transformation
Dimethyl sulfoxide (Sigma)
Agarose (Sigma)
EQUIPMENT
Benchtop centrifuge
Incubators
DNA gel electrophoresis apparatus
REAGENT SETUP
Plasmid DNA
Prepare a midi-prep (Qiagen) of the cloning vector (pCloneHyg1).
Lithium acetate solution
Prepare 0.1 M lithium acetate in 1× TE buffer, pH 7.5.
PEG solution
Prepare 40% (wt/vol) PEG 3350 in 1× TE buffer, pH 7.5.
Competent Escherichia coli
Make transformation-competent E. coli DH5alpha using CaCl2 method32. Alternatively, use commercially available competent E. coli cells.
Media
Prepare standard 2× bacto-tryptone yeast (TY) medium for E. coli. For selection, add ampicillin (100 μg ml−1). For yeast cultivation, prepare standard YES medium supplemented with 0.15g liter−1 adenine and 0.1g liter−1 of each of uracil, l-histidine, l-lysine and l-leucine. For selection, add hygromycin B at 200 μg ml−1.
Primers
Dissolve oligonucleotide primers in TE buffer to 100 μM concentration. The oligonucleotides required to amplify the kinase gene are hhp1BHIterm2, ATATGGATCCGTATTATTAGCAAATGTACTAATAT and hhp1XhoIprom2, ATATCTCGAGAATATTATTAGATTTTGTATATAG. The mutagenic oligonucleotides required are h1AntisM84G, GGACCCAATAAATCCCCCACCAT AGCGTTG and h1M84G, CAACGCTATGGTGGGGGATTTATTGGGTCC. The oligonucleotides required to check correct integration are hhp1ATG-check2, ATCCACTGCCAATTTTACGACC and hhp1pchk2, ACAGCTTTTATTTTCGTCTGAG.
PCR premix
Contains 1× PCR buffer (containing 1.5 mM MgCl2, 0.2 mM of each dNTP mixture, 0.5 μM of each primer, 2 U per 100 μl Taq DNA polymerase and template DNA).
PROCEDURE
Identification of gate-keeper residue
1| Search the kinase sequence database (http://sequoia.ucsf.edu/ksd/) to identify the gate-keeper residue. The gate-keeper position, which is used to introduce space-creating mutations, is highlighted in red in the sequence. Other residues contacting ATP in the active site are marked green. Supplementary Table 1 shows S. pombe protein kinases whose gate-keeper residue can be identified. The gate-keeper residue can be easily identified from primary sequence alignments.
? TROUBLESHOOTING
Mutating the gate-keeper residue
2| Amplify the hhp1 gene together with its promoter region using oligonucleotides (hhp1BHIterm2, ATATGGATCCGTATTATTAGCAAATGTACTAATAT and hhp1XhoIprom2, ATATCTCGAGAATATTATTAGATTTTGTATATAG) and high-fidelity DNA polymerase (e.g., PfuTurbo DNA polymerase; Stratagene). Use S. pombe genomic DNA as a template. Prepare 50 μl of reaction mixture for PCR containing 0.2 μM oligonucleotides, 0.2 mM dNTP mix, about 100 ng template DNA, 1× polymerase reaction buffer and 2.5 U DNA polymerase. Run a PCR: 3 min at 94 °C; 33 cycles of 50 s at 94 °C, 50 s at 50 °C, 90 s at 72 °C; 5 min at 72 °C.
▲ CRITICAL STEP The promoter region must contain a unique restriction site that will be used to linearize the construct for integrating it to the genome (Step 9).
3| Clone the hhp1 into plasmid pCloneHyg1 (see ref. 33) using XhoI and BamHI restriction sites. The resulting plasmid is pCloneHyg1-hhp1. It is possible to use other integrative vectors carrying a suitable selection marker.
4| Verify the cloned hhp1 gene by sequencing.
5| Design mutagenic oligonucleotide primers according to instructions given in the QuikChange II site-directed mutagenesis kit (Stratagene). It is possible to use a QuikChange primer design program (www.stratagene.com). To introduce mutation met84gly (hhp1-as) use oligonucleotides (h1AntisM84G, GGACCCAATAAATCCCCCACCATAGCGTTG and h1M84G, CAACGCTATGGTGGGGGATTTATTGGGTCC).
6| Use a QuikChange II site-directed mutagenesis kit (Stratagene) and the plasmid pCloneHyg1-hhp1 to mutate the gate-keeper residue according to the manufacturer's instructions. The QuikChange II site-directed mutagenesis kit allows site-specific mutation in virtually any double-stranded plasmid. The expected colony number is between 10 and 1,000 colonies. More than 80% of the E. coli colonies should contain the desired mutation. It is also possible to use mutagenesis kits from other manufacturers.
▲ CRITICAL STEP Ensure that the plasmid DNA template is isolated from a dam+E. coli strain (the majority of the commonly used E. coli strains are dam+).
? TROUBLESHOOTING
7| Isolate plasmid DNA using QIAprep spin miniprep kit (Qiagen) according to the manufacturer's instructions. It is possible to use mini-prep kits from other manufacturers (e.g., GeneJET plasmid miniprep kit; Fermentas).
8| Sequence the insert of interest (hhp1 gene) to verify that selected clones contain the desired mutation and do not contain unwanted second-site mutations. The resulting plasmid is pCloneHyg1-hhp1-as.
Testing functionality of mutant kinase
9| Linearize the plasmid pCloneHyg1-hhp1-as with the restriction enzyme SfuI and integrate into the hhp1 promoter of a strain where hhp1 open reading frame has been deleted. Use the yeast transformation protocol described by Gregan et al.33. As a control, integrate the plasmid pCloneHyg1-hhp1 containing wild-type hhp1.
10| Confirm the integration by colony PCR33 using oligonucleotides (hhp1ATGcheck2, ATCCACTGCCAATTTTACGACC and hhp1pchk2, ACAGCTTTTATTTTCGTCTGAG).
11| Analyze the functionality of the hhp1-as mutant by comparing it with hhp1 knockout strain and strain carrying a wild-type hhp1 allele (use a strain containing hhp2Δ mutation). hhp1Δ hhp2Δ double knockout mutant grows extremely slowly, forms very small colonies and cells display abnormal morphology (elongated cells). This phenotype can be rescued by introducing a wild-type hhp1 allele as well as hhp1-as mutant (Fig. 4).
? TROUBLESHOOTING
Testing sensitivity of mutant kinase to inhibitors
12| Dissolve each candidate inhibitor in dimethyl sulfoxide to 5 mM final concentration.
13| Add each candidate inhibitor to autoclaved YES medium containing 2% agar (wt/vol) to 25 μM final concentration.
14| Prepare serial dilutions of freshly grown cells expressing hhp1-as, wild-type hhp1 and hhp1 knockout cells. Grow the cells on YES plates containing 25 μM of each candidate inhibitor for 2 to 3 d at 32 °C. Choose effective and mutant-specific inhibitors for further analysis.
● TIMING
The entire protocol can be completed in about 3 weeks.
Steps 1–4, cloning of the gene: 3 d
Steps 5–8, mutagenesis: 5 d
Steps 9-10, yeast transformation: 5 d
Step 11, testing functionality: 4 d
Steps 12–14, testing sensitivity: 4 d
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1 | The kinase of interest could not be found in the kinase sequence database |
The database is incomplete and does not contain this kinase |
Align the primary sequence of the kinase to kinases with known gate-keeper, such as PKA, CDK2 or c-Src. Identify its gatekeeper from the alignment |
| It is not possible to identify the gate-keeper residue |
Atypical kinase, which does not align to typical kinases well |
Use multiple sequence alignments of closely related kinases. Take a candidate approach and mutate several amino acid candidates for the gate-keeper residue. Alternatively, use a different approach to inactivate the kinase |
|
| 6 | Low transformation efficiency or low mutagenesis efficiency |
Amount of DNA template in the mutagenesis reaction not sufficient |
Visualize the DNA template on a gel to verify its quantity and quality. Ensure that sufficient DNA template is used in the mutagenesis reaction. See also the QuikChange II site-directed mutagenesis kit (Stratagene) manual |
| Low transformation efficiency or low mutagenesis efficiency |
Formation of secondary structures may be inhibiting the mutagenesis reaction |
Increase the annealing temperature (up to 68 °C). See also the QuikChange II site-directed mutagenesis kit (Stratagene) manual |
|
| 11 | Mutating the gate-keeper to glycine residue results in non-functional protein |
A bulky residue is required at the gate-keeper position for the kinase to be functional |
Mutate the gate-keeper to some other small residue (e.g., alanine). Alternatively, introduce a second-site suppressor mutation to rescue the kinase activity27 |
ANTICIPATED RESULTS
This protocol can be used to engineer protein kinases sensitive to cell-permeable inhibitors. Here we describe the construction of the first analog-sensitive allele of an S. pombe kinase (Fig. 4). As the gate-keeper residue can be identified in most protein kinases, we expect that our protocol will also work for other S. pombe protein kinases as well as other organisms. Analog-sensitive kinase alleles have been successfully created and studied in various organisms including the budding yeast Saccharomyces cerevisiae, mouse and human cells11,34,35.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Austrian Science Fund (P18955-B03). L.C. was a recipient of EMBO and FEBS short-term fellowships. We thank Mark Petronczki and Maria Siomos for helpful discussions and comments on the manuscript.
Footnotes
Note: Supplementary information is available via the HTML version of this article.
Rights and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Zeidler MP, et al. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 2004;22:871–876. doi: 10.1038/nbt979. [DOI] [PubMed] [Google Scholar]
- 2.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 3.Gregan J, et al. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell. 2003;14:3876–3887. doi: 10.1091/mbc.E03-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 5.Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 6.Lindner K, Gregan J, Montgomery S, Kearsey SE. Essential role of MCM proteins in premeiotic DNA replication. Mol. Biol. Cell. 2002;13:435–444. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregan J, et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 8.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Carroll AS, Bishop AC, DeRisi JL, Shokat KM, O'Shea EK. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA. 2001;98:12578–12583. doi: 10.1073/pnas.211195798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 12.Buzko O, Shokat KM. A kinase sequence database: sequence alignments and family assignment. Bioinformatics. 2002;18:1274–1275. doi: 10.1093/bioinformatics/18.9.1274. [DOI] [PubMed] [Google Scholar]
- 13.Burkard ME, et al. Chemical genetics reveals the requirement for polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan L, Zhang C, Shokat KM, Hollingsworth NM. Chemical inactivation of cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–1774. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura JJ, et al. Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 17.D'Aquino KE, et al. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Jones MH, et al. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Habelhah H, et al. Identification of new JNK substrate using ATP pocket mutant JNK and a corresponding ATP analogue. J. Biol. Chem. 2001;276:18090–18095. doi: 10.1074/jbc.M011396200. [DOI] [PubMed] [Google Scholar]
- 20.Shokat K, Velleca M. Novel chemical genetic approaches to the discovery of signal transduction inhibitors. Drug Discov. Today. 2002;7:872–879. doi: 10.1016/s1359-6446(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 21.Shah K, Shokat KM. A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 2002;9:35–47. doi: 10.1016/s1074-5521(02)00086-8. [DOI] [PubMed] [Google Scholar]
- 22.Hwang YW, Miller DL. A mutation that alters the nucleotide specificity of elongation factor Tu, a GTP regulatory protein. J. Biol. Chem. 1987;262:13081–13085. [PubMed] [Google Scholar]
- 23.Hoffman HE, Blair ER, Johndrow JE, Bishop AC. Allele-specific inhibitors of protein tyrosine phosphatases. J. Am. Chem. Soc. 2005;127:2824–2825. doi: 10.1021/ja043378w. [DOI] [PubMed] [Google Scholar]
- 24.Holt JR, et al. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell. 2002;108:371–381. doi: 10.1016/s0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 25.Gillespie PG, Gillespie SK, Mercer JA, Shah K, Shokat KM. Engineering of the myosin-ibeta nucleotide-binding pocket to create selective sensitivity to N(6)-modified ADP analogs. J. Biol. Chem. 1999;274:31373–31381. doi: 10.1074/jbc.274.44.31373. [DOI] [PubMed] [Google Scholar]
- 26.Kapoor TM, Mitchison TJ. Allele-specific activators and inhibitors for kinesin. Proc. Natl. Acad. Sci. USA. 1999;96:9106–9111. doi: 10.1073/pnas.96.16.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 28.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 29.Kearney PH, Ebert M, Kuret J. Molecular cloning and sequence analysis of two novel fission yeast casein kinase-1 isoforms. Biochem. Biophys. Res. Commun. 1994;203:231–236. doi: 10.1006/bbrc.1994.2172. [DOI] [PubMed] [Google Scholar]
- 30.Dhillon N, Hoekstra MF. Characterization of two protein kinases from Schizosaccharomyces pombe involved in the regulation of DNA repair. EMBO J. 1994;13:2777–2788. doi: 10.1002/j.1460-2075.1994.tb06571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petronczki M, et al. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 32.Cohen SN, Chang AC, Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA. 1972;69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregan J, et al. High-throughput knockout screen in fission yeast. Nat. Protoc. 2006;1:2457–2464. doi: 10.1038/nprot.2006.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larochelle S, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol. Cell. 2007;25:839–850. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaeschke A, et al. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.