Abstract
Tandem affinity purification (TAP) is a generic two-step affinity purification protocol for isolation of TAP-tagged proteins together with associated proteins. We used bacterial artificial chromosome to heterologously express TAP-tagged murine Sgo1 protein in human HeLa cells. This allowed us to test the functionality of the Sgo1-TAP protein by RNA interference-mediated depletion of the endogenous human Sgo1. Here, we present an optimized protocol for purification of TAP-tagged Sgo1 protein as well as KIAA1387 from HeLa cells with detailed instructions. The purification protocol can be completed in 1 day and it should be applicable to other proteins.
INTRODUCTION
Most cellular processes are carried out by multiprotein complexes. The identification of individual subunits is essential for understanding their function. To streamline the purification of protein complexes from cells, the TAP protocol was developed1,2. It has been successfully applied to purify protein complexes from various organisms including bacteria, yeasts, plants as well as mammalian cells3-14. We used the TAP protocol to purify proteins associated with murine Sgo1 protein15.
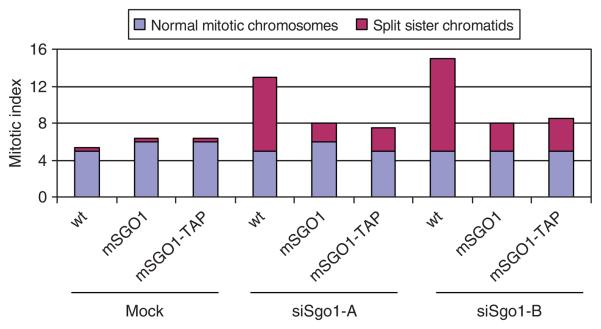
It is important to confirm that tag addition does not significantly affect the function of the tagged protein. In model organisms such as yeast, functionality of tagged proteins can be easily tested. In mammalian cells, the lack of efficient homologous recombination makes the functionality test difficult. Kittler et al.16 demonstrated that expression of murine bacterial artificial chromosomes (BAC) in human cells provides a reliable method to create RNA interference (RNAi)-resistant tagged transgenes. In such cells, the endogenous human gene can be knocked down by RNAi, while the corresponding murine gene expressed from the integrated BAC resists the RNAi treatment. We expressed murine Sgo1-TAP at physiological levels in HeLa cells from a BAC integrated into the HeLa genome15. This allowed us to determine whether the murine Sgo1-TAP was able to complement functionally the phenotype caused by depletion of the endogenous human Sgo1. As a control we used HeLa cells expressing untagged murine Sgo1 from a BAC integrated into the HeLa genome. Depletion of Sgo1 in HeLa cells by RNAi leads to precocious separation of sister chromatids17,18. In contrast, in RNAi-treated cells expressing the murine SGO1-TAP transgene, the levels of precocious sister chromatid separation dropped considerably toward wild-type levels (Fig. 1). These results demonstrate that the murine Sgo1-TAP is functional in HeLa cells as the SGO1-TAP transgene prevented precocious separation of sister chromatids caused by depleting endogenous human Sgo1 by RNAi. Notably, the rescue of the RNAi phenotype of SGO1 by BAC transgenesis also confirms the specificity of the RNAi experiment17. In addition to HeLa cells described here, we have succeeded in generating BAC-transgenic U2OS cells with the same protocol (data not shown). Given the fact that other BAC-transgenic cell lines, including mouse ES cells, have been successfully generated, it is likely that BAC-transgenic clones can also be generated from many different cells. However, it is probable that for other cell lines, different transfection protocols have to be used for the delivery of the BAC DNA.
Figure 1.
Rescue experiment. Wild-type HeLa cells (wt) and BAC-transgenic HeLa cells expressing either murine SGO1 (mSGO1) or murine SGO1-TAP (mSGO1-TAP) were transfected with two different Sgo1 siRNAs (siSgo1-A, siSgo1-B) or deionized H2O (mock). Cells were examined by chromosome spreading followed by Giemsa staining and mitotic cells were classified into two categories based on chromosome configuration.
As an example of possible applications for this system, we used murine Sgo1-TAP expressed from a BAC integrated into the HeLa genome to identify proteins that interact with Sgo1 in mitotic cells. Murine Sgo1-TAP, but not a control protein KIAA1387-TAP, associated with a specific form of protein phosphatase 2A (Tables 1 and 2). This finding helped us to elucidate the mechanism by which Sgo1 protects centromeric cohesion15,19. Notably, endogenous human Sgo1 also associated with murine Sgo1-TAP, suggesting that Sgo1 proteins form homo-oligomers (Table 1). Thus, our protocol also allows detection of oligomer formation of the tagged protein.
TABLE 1.
List of proteins identified by mass spectrometry co-purifying with murine Sgo1-TAP.
| Protein (GeneInfo identifier) | Description | Mascot score | |
|---|---|---|---|
| 1 | 32140473 | Protein kinase, catalytic polypeptide | 3087 |
| 2 | 56205916 | Retinoblastoma-associated factor 600 (RBAF600) | 2523 |
| 3 | 4506787 | IQ motif containing GTPase activating protein 1 | 2228 |
| 4 | 62485049 | Shugoshin A1/A2 protein (Mus musculus) | 1440 |
| 5 | 231443 | Alpha isoform of scaffold subunit PR65 of PP2A | 909 |
| 6 | 3603418 | Beta isoform of scaffold subunit of PP2A | 862 |
| 7 | 46015216 | Chain F of Ef3-Cam complexed With Pmeapp | 803 |
| 8 | 57165052 | Thyroid autoantigen 70 kDa (Ku antigen) | 772 |
| 9 | 16507237 | Heat shock 70 kDa protein 5 | 767 |
| 10 | 17512093 | ATP-dependent DNA helicase II | 725 |
| 11 | 54695922 | Beta isoform of catalytic subunit of PP2A | 699 |
| 12 | 16303631 | Gamma isoform of regulatory subunit B56 of PP2A | 694 |
| 13 | 47077243 | Unnamed protein product (similar to gamma isoform B56 of PP2A) | 668 |
| 14 | 60302875 | Shugoshin-like 1 isoform A1/A2 | 631 |
| 15 | 31657094 | Anillin, actin binding protein | 627 |
| 16 | 16974825 | Chain A of calcium-calmodulin N-terminal domain | 619 |
| 17 | 13623235 | Q5MIZ7 protein | 614 |
| 18 | 12654673 | Delta 2 isoform of regulatory subunit B56 of PP2A | 604 |
| 19 | 38014029 | Retinoblastoma-associated factor 600 (RBAF600) | 598 |
| 20 | 61680528 | Chain A, trapped intermediate of calmodulin | 575 |
| 21 | 24308448 | Shugoshin-like 1 isoform C1/C2 | 567 |
| 22 | 18490282 | Alpha isoform of regulatory subunit B56 of PP2A | 559 |
| 23 | 4529892 | HSP70-2 | 557 |
| 24 | 68533509 | Myosin IE | 554 |
| 25 | 66360504 | Chain T of Ef in complex with calmodulin | 540 |
| 26 | 33286088 | Cullin 3 isoform | 531 |
| 27 | 338695 | Beta-tubulin | 512 |
| 28 | 18088719 | Beta-tubulin | 504 |
| 29 | 37852 | Vimentin | 487 |
| 30 | 434753 | KIAA0030 | 478 |
| 31 | 468704 | Polypeptide BM28 | 472 |
| 32 | 1418763 | Beta 2 isoform of regulatory subunit B56 of PP2A | 451 |
| 33 | 54696358 | Protein phosphatase 4, catalytic subunit | 445 |
| 34 | 62531280 | Epsilon isoform of regulatory subunit B56 of PP2A | 444 |
| 35 | 4502563 | Calpain 2, large subunit | 438 |
| 36 | 47169206 | Chain A of the N-terminal domain of N60d calmodulin | 413 |
| 37 | 20809886 | Tubulin, beta, 2 | 396 |
| 38 | 188492 | Heat shock protein | 382 |
| 39 | 340021 | Alpha-tubulin | 380 |
Proteins associated specifically with Sgo1-TAP but not with KIAA1387-TAP are in bold.
TABLE 2.
List of proteins identified by mass spectrometry co-purifying with murine KIAA1387-TAP.
| Protein (GeneInfo identifier) |
Description | Mascot score |
|
|---|---|---|---|
| 1 | 61743954 | AHNAK nucleoprotein isoform 1 | 4657 |
| 2 | 627367 | Desmyokin—human | 2665 |
| 3 | 3337389 | Pre-mRNA splicing factor (PRP16) (KIAA0224) | 2433 |
| 4 | 4506787 | IQ motif containing GTPase activating protein 1 | 2219 |
| 5 | 13623235 | KIAA1387 protein (Mus musculus) | 2142 |
| 6 | 34536452 | Unnamed protein product | 1831 |
| 7 | 51493205 | PREDICTED: chromosome 14 open reading frame 78 | 1557 |
| 8 | 18028273 | Hypothetical protein SBBI57 | 1445 |
| 9 | 346323 | Phosphoprotein phosphatase (EC 3.1.3.16) X catalytic chain | 1162 |
| 10 | 20521049 | KIAA0432 | 1048 |
| 11 | 16507237 | Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 1027 |
| 12 | 56205916 | Retinoblastoma-associated factor 600 (RBA600) | 928 |
| 13 | 8250239 | Protein phosphatase 4 regulatory subunit 2 | 856 |
| 14 | 17391461 | PRP19/PSO4 pre-mRNA processing factor 19 homolog | 828 |
| 15 | 18655710 | Chain F, crystal structure of the adenyl cyclase domain of anthrax edema factor (Ef) in complex with calmodulin |
749 |
| 16 | 5123454 | Heat shock 70 kDa protein 1A | 701 |
| 17 | 46015216 | Chain F, crystal structure of the Ef3-Cam complex with Pmeapp | 655 |
| 18 | 46852390 | Coiled-coil domain containing 6 | 609 |
| 19 | 1346343 | Keratin, type II cytoskeletal 1 (cytokeratin 1) (K1) (CK 1) (67 kDa cytokeratin) (hair alpha protein) | 592 |
| 20 | 535177 | AHNAK-related protein | 587 |
| 21 | 292059 | MTHSP75 | 584 |
| 22 | 66360504 | Chain T, crystal structure of anthrax edema factor (Ef) in complex with calmodulin | 572 |
| 23 | 28317 | Unnamed protein product | 524 |
| 24 | 37852 | Vimentin | 513 |
| 25 | 24899184 | KIAA2010 protein | 503 |
| 26 | 181402 | Epidermal cytokeratin 2 | 491 |
| 27 | 61680528 | Chain A, trapped intermediate of calmodulin | 490 |
| 28 | 1381146 | Ha-CUL-3 | 384 |
| 29 | 48146983 | SNW1 | 377 |
| 30 | 188492 | Heat shock-induced protein | 363 |
| 31 | 34783647 | SKIIP protein | 363 |
| 32 | 4502563 | Calpain 2, large subunit | 359 |
| 33 | 468704 | Polypeptide BM28 | 341 |
| 34 | 550021 | Ribosomal protein S5 | 321 |
| 35 | 18645167 | Annexin A2, isoform | 320 |
| 36 | 55957452 | Tripartite motif-containing 14 | 316 |
| 37 | 435476 | Cytokeratin 9 | 299 |
| 38 | 13528987 | Breast carcinoma amplified sequence 2 | 299 |
| 39 | 16974825 | Chain A, solution structure of calcium-calmodulin N-terminal domain | 295 |
Proteins associated specifically with KIAA1387-TAP but not with Sgo1-TAP are in bold.
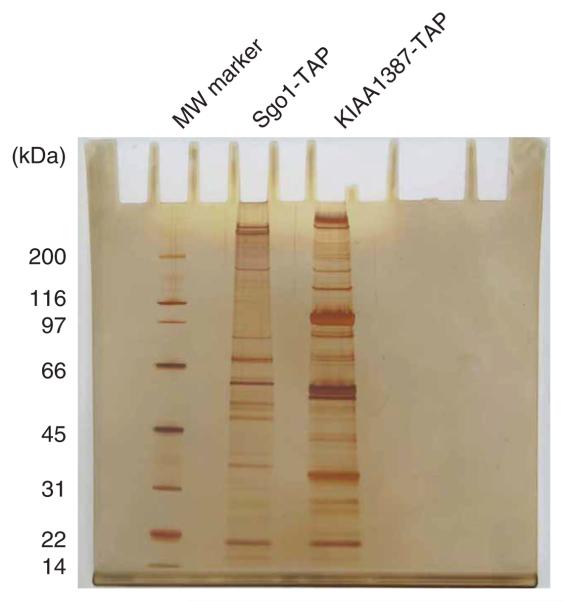
Here, we describe an optimized TAP purification protocol with detailed instructions. We also describe our mass spectrometry (MS) protocol (Step 20A) and our SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining protocol to visualize purified proteins (Step 20B; Fig. 2), which we have developed specifically for our target protein but could potentially be adapted for other proteins.
Figure 2.
Purified proteins visualized by silver staining. Protein complexes associated with murine Sgo1-TAP and murine KIAA1386-TAP were isolated by tandem affinity purification, separated by SDS-PAGE and visualized by silver staining. Molecular weight marker (MW marker) is indicated on the left.
MATERIALS
REAGENTS
Sterile Millex syringe-driven filters (pore size: 0.45 μm) (Millipore)
Sterile syringe (20 ml)
Capillary (Pico Tip, FS360-20-10, New Objective)
IgG sepharose beads (Amersham Biosciences)
Calmodulin sepharose beads (Amersham Biosciences)
Poly-prep chromatography columns (Bio-Rad)
Separation column: 0.075 mm ID × 150 mm length, 3 μm particle size (Dionex, packed both)
Protein molecular weight standard (e.g., Bio-Rad broad range)
4–12% (v/v) Bis-Tris gel (Invitrogen)
Phosphate-buffered saline (PBS; Sigma)
Calcium chloride (Sigma)
Methanol (p.a.) (Sigma)
Ethanol (p.a.) (Sigma)
Acetic acid (Sigma)
Formaldehyde (37%; Sigma)
NaCl (Sigma)
Na2S2O3 (Sigma)
Deionized water
Silver nitrate (Sigma)
Sodium deoxycholate (Sigma)
Trichloroacetic acid (Sigma)
Tris (Sigma)
Glycerol (Sigma)
Magnesium acetate (Sigma)
Imidazole (Sigma)
NP-40 (Sigma)
β-Mercaptoethanol (Sigma)
Dithiothreitol (DTT; Sigma)
AcTEV protease (Invitrogen)
Ammonium bicarbonate (Sigma)
Iodoacetamide (Sigma)
Trypsin (Sigma)
Acetonitrile (HPLC grade, Supra-Gradient, Biosolve B.V.)
Water (HPLC grade, Supra-Gradient, Biosolve B.V.)
300 μm ID × 5 mm length trap column cartridges (PepMap, C18, 5 μm particle size, 100 Å pore size, Dionex)
5% acetonitrile (HPLC grade, Supra-Gradient, Biosolve B.V.)
0.1% formic acid (Fluka)
0.1% trifluoroacetic acid (TFA; Pierce)
EQUIPMENT
Dounce homogenizers (Wheaton)
Xcell SureLock Mini-Cell apparatus for SDS-PAGE (Invitrogen)
Sterile roller bottles (BD Biosciences)
Ion trap mass spectrometer (LTQ, Thermo Finnigan)
Heated capillary (Pico Tip, FS360-20-10, New Objective)
UltiMate nano HPLC system (NAN-75 Flow Splitter) (Dionex) operated with Chromeleon chromatography software V 6.7 SP2 (Dionex)
75 μm ID × 150 mm length nano separation column (PepMap, C18, 3 μm particle size, 100 Å pore size, Dionex)
UltiMate separation system (NAN-75 Flow Splitter)
Mascot (Matrix Science; version 2.1.0)
Scaffold (version Scaffold-01_06_03, Proteome Software Inc.)
MS requires expensive equipment and highly skilled personnel supplemented by a well-established and integrated bioinformatic infrastructure
REAGENT SETUP
Media
Prepare standard DMEM medium supplemented with 10% fetal calf serum, 0.2 mM l-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin.
Extraction buffer
50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol (v/v), 0.2% NP-40 (v/v), Complete Mini Protease Inhibitor (Roche) (1 tablet per 10 ml) and 1 mM PMSF. ▲ CRITICAL Add inhibitors just before use.
Lysis buffer without inhibitors
50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol (v/v) and 0.1% NP-40 (v/v).
Lysis buffer
50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol (v/v), 0.1% NP-40 (v/v), Complete Mini Protease Inhibitor (Roche) (1 tablet per 10 ml) and 1 mM PMSF. ▲ CRITICAL Add inhibitors just before use.
TEV cleavage buffer
10 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol (v/v), 0.1% NP-40 (v/v), 0.5 mM EDTA and 1 mM DTT. ▲ CRITICAL Add DTT just before use.
Calmodulin binding buffer
10 mM β-mercaptoethanol, 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol (v/v), 0.1% NP-40 (v/v), 1 mM imidazole, 1 mM Mg-acetate and 2 mM CaCl2.
Calmodulin binding buffer-2
1 mM β-mercaptoethanol, 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM Mg-acetate and 2 mM CaCl2.
Fixing solution
50% methanol (v/v), 12% acetic acid (v/v) and 0.018% formaldehyde (v/v).
Silver nitrate solution
0.1% silver nitrate (w/v) and 0.027% formaldehyde (v/v).
Developing solution
6% (w/v) sodium carbonate, 0.00002% Na2S2O3 (w/v) and 0.018% formaldehyde (v/v).
EQUIPMENT SETUP
HPLC-MS
The UltiMate system consists of an UltiMate μHPLC pump and a UV detection unit with the nano UV-Z View flow cell with 3 nl cell volume, the Switchos μ-column-switching device with loading pump and two 10-port valves, and the FAMOS μ-autosampler equipped with the 250 μl sample loop, 15 μl injection needle and 250 μl syringe (Large Volume Injection Kit). For FAMOS, use the μL-Pickup injection method. As a transport liquid on FAMOS use 0.1% aqueous TFA (in Reagent Transport Vial 1). Use the 300 μm ID × 5 mm length trap column cartridges for sample trapping and cleaning before separation. Operate the trap column at 20 μl min−1 on Switchos loading pump. As a separation column, use the 75 μm ID × 150 mm length nano separation column. Operate the separation column at 275 nl min−1 on UltiMate separation system. Use the following tubing for separation: to deliver the flow from Switchos to the FAMOS autosampler and to transport the sample to the trap column, use 130 μm ID tubing. Keep the tubing length as short as possible. To connect the trap column to the Switchos valve, use the provided cartridge holder and two 30 μm ID fused silica connections, which are delivered with the trap column. To make all flow paths used for nano flow (275 nl min−1), use the 20 μm ID PEEKSil connection tubing. To connect the UV cell output with the MS inlet, use 20 μm ID fused silica and keep it as short as possible. Make all fluidic connections from Ultimate nano HPLC to the mass spectrometer using 20 μm ID fused silica capillaries, connected with low dead-volume micro tight connectors from Dionex.
Use the following mobile phases for the separation column:
HPLC mobile phase A: 95% water (HPLC grade), 5% acetonitrile (HPLC grade) and 0.1% formic acid;
HPLC mobile phase B: 30% water, 70% acetonitrile and 0.1% formic acid. Use water with loading mobile phase for the Switchos: 0.1% TFA is a loading mobile phase for the Switchos.
For separation, use the following HPLC gradient:
| Time (min) | % B |
|---|---|
| 0–30 | 0–50 |
| 30–31 | 50–100 |
| 31–35 | 100–0 |
| 36–50 | 0 |
Use the following tubing for separation: to deliver the flow from Switchos to the FAMOS autosampler and to transport the sample to the trap column, use 130 μm ID tubing. Keep the tubing length as short as possible. To connect the trap column to the Switchos valve, use the provided cartridge holder and two 30 μm ID PEEKSil connections delivered with the trap column. To make all flow paths used for nano flow (275 nl min−1), use the 20 μm ID PEEKSil connection tubing. To connect the UV cell output with the MS inlet, use 20 μm ID fused silica and keep it as short as possible.
Mass spectrometry
Transfer the eluting peptides online to a heated capillary of an ion trap mass spectrometer. Use the following ESI parameters:
| Spray voltage | 1.5 kV |
| Capillary temperature | 200 °C |
| Capillary voltage | 26 V |
| Tube lens offset voltage | 95 V |
| Electron multiplier | −800 V |
| Gain control | 20,000 |
The collision energy is set automatically depending on the mass of the parent ion.
PROCEDURE
Preparation of HeLa cell extract
1| Grow 8 liters of HeLa-S3 cells in roller bottles with DMEM medium supplemented with 10% fetal calf serum, 0.2 mM l-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin to a density of about 1×106 cells ml−1.
2| Pellet cells by centrifugation (500g for 5 min at 4 °C) and discard the supernatant.
3| Wash the pellet by resuspending the cells with 100 ml of ice-cold PBS gently by pipetting and repeating the centrifugation in Step 2. Repeat the wash once.
■ PAUSE POINT Cell pellets can be frozen in liquid nitrogen and stored at −80 °C for several days.
4| Resuspend the cells in 200 ml of ice-cold extraction buffer and transfer the cell suspension into the pre-chilled Dounce homogenizer.
5| Homogenize cells with 25 strokes in a Dounce homogenizer with a tight-fitting pestle and incubate on ice for 5 min; repeat the homogenization one more time.
6| Remove insoluble material by centrifugation at 13,000g for 10 min at 4 °C and keep the supernatant.
7| Centrifuge at 34,000g for 20 min at 4 °Cand keep the supernatant.
? TROUBLESHOOTING
8| Filter the supernatant through a sterile Millex syringe-driven filter (pore size: 0.45 μm) at 4 °C and proceed directly to protein complex purification.
? TROUBLESHOOTING
Protein complex purification
9| Wash 500 μl of IgG sepharose beads using a poly-prep chromatography column twice with 10 ml of lysis buffer at 4 °C.
10| Add the cell extract from step 8 to washed IgG beads from step 9 and incubate for 2 h at 4 °C on a rotating wheel.
? TROUBLESHOOTING
11| Wash beads twice with 10 ml of ice-cold lysis buffer at 4 °C (all washing steps are performed on poly-prep chromatography column).
12| Wash beads with 10 ml of ice-cold lysis buffer without inhibitors at 4 °C.
13| Wash beads with 10 ml of ice-cold TEV cleavage buffer at 4 °C.
14| Resuspend beads in 2 ml of ice-cold TEV cleavage buffer and add 50 μl of AcTEV protease (Invitrogen); incubate at 16 °C for 2 h (alternatively at 4 °C for 16 h) on a rotating wheel.
? TROUBLESHOOTING
15| Wash 70 μl of calmodulin sepharose beads (Amersham Biosciences), using a column (Bio-Rad), twice with 10 ml of calmodulin binding buffer at 4 °C.
16| Collect the TEV eluate (flow-through) from step 14 and adjust CaCl2 to 3 mM final concentration.
17| Add 6 ml of calmodulin binding buffer to the TEV eluate and transfer to the washed calmodulin beads from Step 15 and incubate for 2 h at 4 °C on a rotating wheel.
18| Wash beads with 10 ml of ice-cold calmodulin binding buffer at 4 °C.
19| Wash beads twice with 10 ml of ice-cold calmodulin binding buffer-2 at 4 °C.
20| Submit one half of the calmodulin beads for MS analysis (option A); use the remaining calmodulin beads for SDS-PAGE and silver staining (option B). If using option B, see also the protocol by Schagger20.
■ PAUSE POINT Calmodulin beads can be frozen in liquid nitrogen and stored at −80 °C for several days.
▲ CRITICAL STEP While the conventional TAP protocol uses EGTA to elute proteins from calmodulin beads, we were not able to efficiently elute proteins from calmodulin beads by EGTA. To overcome this limitation, we directly submitted calmodulin beads to tryptic digestion followed by MS analysis and we boiled calmodulin beads in SDS buffer for the SDS-PAGE and silver staining analysis.
(A) Tryptic digest, HPLC-MS/MS and database search
Tryptic digest: titrate the samples to pH 8.0 by addition of 1.0 M Tris-HCl, pH 8.5.
Wash calmodulin beads five times with 50 mM ammonium bicarbonate and 50 mM ammonium bicarbonate/30% acetonitrile by centrifugation (500g for 5 min at room temperature, i.e. 23 °C).
Reduce sample by incubation with 1 μg of DTT for 1 h at 56 °C.
Alkylate the sample by incubation with 5 μg of iodoacetamide for 30 min at room temperature in the dark.
Digest the proteins on beads with 200 ng trypsin for 4 h at 37 °C and then with an additional 200 ng trypsin overnight.
Stop the digest by adding 10 μl of 10% TFA. Use 2 μl of the supernatant for MS analysis.
Perform nano HPLC separations using an UltiMate nano HPLC system (see EQUIPMENT SETUP).
Collect the data in the centroid mode using an MS experiment (see EQUIPMENT SETUP) (Full-MS) followed by four MS/MS experiments of the four most intensive ions (intensity at least 10,000). Use dynamic exclusion for data acquisition with exclusion duration of 1 min and an exclusion mass width of ±3 Da. Make all fluidic connections from Ultimate nano HPLC to the mass spectrometer using 20 μm ID fused silica capillaries, connected with low dead-volume micro tight connectors from Dionex.
Database search: extract tandem mass spectra using extract-msn delivered with Bioworks 3.3. Do not perform charge state deconvolution and deisotoping. Analyze MS/MS samples using Mascot (Matrix Science; version 2.1.0). Set up the Mascot to search the human_KBMS_5.0.20050304.fa database (187752 entries) assuming the digestion enzyme trypsin. Search the Mascot with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 2.5 Da. Specify iodoacetamide derivative of cysteine in Mascot as a fixed modification. Specify oxidation of methionine in Mascot as a variable modification.
Criteria for protein identification: use Scaffold (version Scaffold-01_06_03, Proteome Software Inc.) to validate MS/MS-based peptide and protein identifications. Accept peptide identifications if they can be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm21. Accept protein identifications if they can be established at greater than 95.0% probability and contain at least two identified peptides. Assign protein probabilities by the Protein Prophet algorithm22. Group the proteins that contain similar peptides and cannot be differentiated based on MS/MS analysis alone to satisfy the principles of parsimony.
(B) SDS-PAGE and silver staining
Run the protein samples on a 4–12% Bis-Tris gel (Invitrogen) according to the manufacturer's instructions.
Fix the gel by incubating for 1 h in 100 ml of fixing solution.
Wash the gel for 10 min in 100 ml of 50% (v/v) ethanol.
Wash the gel for 10 min in 100 ml of 30% (v/v) ethanol.
Sensitize the gel by incubating for 1 min in 100 ml of 0.01% (w/v) Na2S2O3.
Wash the gel three times by incubating for 20 s in 100 ml of deionized water.
Incubate the gel for 20 min in 100 ml of silver nitrate solution.
Wash the gel three times by incubating for 20 s in 100 ml of deionized water.
Incubate the gel in developing solution until protein bands become visible (1–10 min).
Stop development by incubating the gel for 5 min in 100 ml of 15% (v/v) acetic acid.
Wash the gel twice with 100 ml of deionized water.
● TIMING
The protein complex purification protocol (Steps 2–20) can be completed in 1 day.
Step 1: growing HeLa cells, about 3 days
Steps 2 and 3: harvesting HeLa cells, 2 h
Steps 4–20: protein complex purification, 11 h
Step 20A(i)–(vi): tryptic digest, 1.5 days
Step 20A(vii) and (viii): nano HPLC and MS, 5 h
Step 20A(ix): database search, 4 h
Step 20B: SDS-PAGE and silver staining, 3 h
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
ANTICIPATED RESULTS
We developed this TAP protocol to optimize purification of proteins associated with mammalian Sgo115. Although each protein has unique properties, we believe that our TAP protocol should be applicable to other proteins. However, the functionality test of TAP-tagged murine transgenes is limited to those where human homologs can be identified. In some cases, artifactual binding partners may be identified owing to differences between human and mouse proteins. Although the conventional TAP tag has proven to be a very useful tool for protein complex purification, alternative affinity binding moieties are now available that may provide higher protein complex yield or allow for both protein isolation and live imaging of protein localization6,23.
TABLE 3.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| Tagged protein is not functional | Tag negatively interferes with protein's function |
Test different constructs with the tag at the N or C terminus. Insert a flexible oligopeptide linker in between the tag and protein |
|
| 7 | Tagged protein is not present in the supernatant |
Tagged protein is bound to membranes or insoluble particles |
Modify the preparation of cell extract |
| 8 | Tagged protein is lost after filtration |
Protein binds to the filter | Do not filter the protein extract (skip Step 8) |
| 10–14 | Degradation of proteins occurs | HeLa cells were grown in plates and harvested by trypsinization |
Avoid using trypsin, harvest cells by scraping |
| 14 | Degradation of proteins occurs | Presence of proteases in the sample | Add protease inhibitors into TEV cleavage buffer (1 tablet of Complete Mini Protease Inhibitor (Roche) per 20 ml, 0.5 mM PMSF) |
ACKNOWLEDGMENTS
This work was supported by Boehringer Ingelheim and Austrian Science Fund (P18955-B03). Work in the Mechtler lab was supported by the Austrian Proteomics Platform (APP) within the Austrian Genome Program (GEN-AU) and by the Mitocheck project within the Sixth Framework Program of the European Commission. J.G. was a recipient of the EMBO long-term fellowship. We thank Tony Hyman for the gift of the TAP tagging cassette, and Laurence Pelletier and Ralf Kittler for help with Sgo1 tagging. We also thank members of Karl Mechtler lab for help with MS analysis and Jan Michael Peters and members of his lab for helpful discussions.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
Rights and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 2.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 3.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 5.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 6.Burckstummer T, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 7.Rohila JS, Chen M, Cerny R, Fromm ME. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 2004;38:172–181. doi: 10.1111/j.1365-313X.2004.02031.x. [DOI] [PubMed] [Google Scholar]
- 8.Bouwmeester T, et al. A physical and functional map of the human TNF-alpha/ NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 9.Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Forler D, et al. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- 11.Chen CY, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 12.Gully D, Moinier D, Loiseau L, Bouveret E. New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett. 2003;548:90–96. doi: 10.1016/s0014-5793(03)00746-4. [DOI] [PubMed] [Google Scholar]
- 13.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 15.Riedel CG, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 16.Kittler R, et al. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture Cells. Proc. Natl. Acad. Sci. USA. 2005;102:2396–2401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate Cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 19.Kittler R, et al. An endoribonuclease-prepared siRNA screen in human Cells identifies genes essential for Cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 20.Schagger H. Tricine-SDS-PAGE. Nat. Protocols. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 22.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 23.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]