Abstract
Although the field of mass spectrometry-based proteomics is still in its infancy, recent developments in targeted proteomic techniques have left the field poised to impact the clinical protein biomarker pipeline now more than at any other time in history. For proteomics to meet its potential for finding biomarkers, clinicians, statisticians, epidemiologists and chemists must work together in an interdisciplinary approach. These interdisciplinary efforts will have the greatest chance for success if participants from each discipline have a basic working knowledge of the other disciplines. To that end, the purpose of this review is to provide a nontechnical overview of the emerging/evolving roles that mass spectrometry (especially targeted modes of mass spectrometry) can play in the biomarker pipeline, in hope of making the technology more accessible to the broader community for biomarker discovery efforts. Additionally, the technologies discussed are broadly applicable to proteomic studies, and are not restricted to biomarker discovery.
Keywords: targeted proteomics, multiple reaction monitoring, selected reaction monitoring, biomarker, mass spectrometry
Introduction
Biomarkers have proven to be invaluable in guiding our delivery of medical care to cancer patients.1 For example, biomarkers allow us to diagnose cancer early,2 to subtype within a disease category to prognosticate3 or to predict response to targeted therapies4,5 and to monitor patients for response to therapy or recurrent disease.6,7 Given the tremendous potential for protein biomarkers to improve our care of cancer patients, it comes as no surprise that there was tremendous hype in both the lay and scientific communities in 2002 when a landmark study was published that claimed to have discovered a novel approach using protein fingerprints in serum to diagnose ovarian cancer with a sensitivity of 100%, specificity of 95% and positive predictive value of 94%.8 Unfortunately, this initial hype was followed by a comparable level of disappointment when the results of this study were later determined to be due to an artifact,9–14 and the utility of the entire approach that had been described was called into serious question.15,16 This shaky beginning left the nascent field of mass spectrometry-based clinical proteomics reeling for the next few years, struggling to identify a productive application of its promising technologies to the biomarker field in the backdrop of a community left disgruntled and questioning whether there was any value of mass spectrometry to the biomarker field.17 This review article will focus on the evolving role of mass spectrometry in the development of novel protein biomarkers, highlighting tremendous progress in a field that is still in its infancy but that now appears to have righted itself and is on a path to make significant contributions to the clinical translation of novel biomarkers.
The Need for New Technologies in Cancer Biomarker Discovery
Historically, cancer protein biomarkers have been discovered in body fluids and tumor tissues (or cell lines) using 2d gel separations or by identifying immunogenic antigens on cancer cells.18 Conventional approaches have successfully produced nine FDA-approved, blood-based cancer biomarkers to date, most of which are used to monitor treatment.19 The number of new protein biomarkers achieving FDA approval has trended downwards for the past decade to a point where only 0–3 new markers are being approved per year (across all diseases).20,21 This disappointing downward trend suggests that conventional approaches have contributed what they can and that we now need to implement new approaches and new technologies to discover novel protein biomarkers of clinical relevance.18
Biomarkers in Tissue and Plasma
Protein biomarkers are measured either directly in tissues (by immunohistochemistry) or in plasma (by ELISA). Plasma biomarkers are highly desirable because they can be measured noninvasively and because they can be used to screen the general population for early disease detection. In contrast, measurement of tissue biomarkers is only useful once a potential cancer has been identified and a biopsy has been obtained. Both types of biomarkers have important roles to play during the natural history of the disease. Although this review will focus on plasma biomarkers, the emerging technologies described herein should apply equally to the development of novel tissue-based biomarkers.
The discovery of highly specific tumor-derived biomarkers by direct analysis of plasma presents an enormous analytical challenge: the 20 most abundant plasma proteins constitute 99% of the total protein mass in plasma,22 and the presence of these very high abundance proteins interferes with our ability to detect rare proteins that are shed or secreted into the circulation by tumor cells. For example, the most abundant plasma protein is albumin, which is present in plasma at a concentration of ~50 mg per milliliter. In contrast, known cancer-derived proteins in the circulation are present at a few nanograms per milliliter, 10 million times less abundant than albumin! Because of the very low abundance of cancer-derived proteins in the bloodstream, it is impossible to detect them using mass spectrometry unless the plasma sample is extensively fractionated, typically using biochemical methods. These extensive fractionation workflows severely reduce sample through put and introduce significant pre-analytical variation, severely crippling plasma-based biomarker discovery.
Using Tissues and Proximal Fluids to Discover Candidate Biomarkers
Despite these severe limitations, mass spectrometry can be used to identify circulating cancer biomarkers, albeit indirectly. This can be accomplished through the analysis of tumor tissues or proximal fluids (i.e., cerebrospinal fluid, urine, saliva, tumor interstitial fluid, etc.),23–25 from which protein biomarkers may be secreted, shed or leaked into the bloodstream. In tumor tissues and proximal fluids tumor-derived proteins are present at high enough local concentration that they can be detected by conventional mass spectrometers. Under this strategy, candidate biomarkers are first discovered in the tumors or proximal fluids and then subsequently measured out in the plasma using highly sensitive, targeted assay technologies (see below).26,27
A major advantage to using tumor tissue for the discovery of candidate circulating biomarkers is that genomic technologies such as gene expression profiling can also contribute candidates to the discovery process and may provide corroborating information with proteomic data to increase our confidence in a particular candidate. A second advantage is that tissue-based discovery datasets provide a rich source of tissue biomarker candidates in addition to potential circulating biomarker candidates. A disadvantage of this approach is that we currently do not know how to predict which proteins identified in tumor tissues or proximal fluids will ultimately access the plasma and have a long enough half-life to accumulate to measurable levels. Currently, it is assumed that proteins predicted to be either secreted or localized to the cell surface (and hence potentially shed) will have the highest probability of reaching the plasma;26 indeed there has been early success using this strategy in an animal model.27 Additional studies are currently testing the use of tumor cell line cultures23,28 or xenograft mouse models25 for the discovery of candidate plasma biomarkers, and these approaches may also have utility.23
Bottleneck in the Biomarker Pipeline
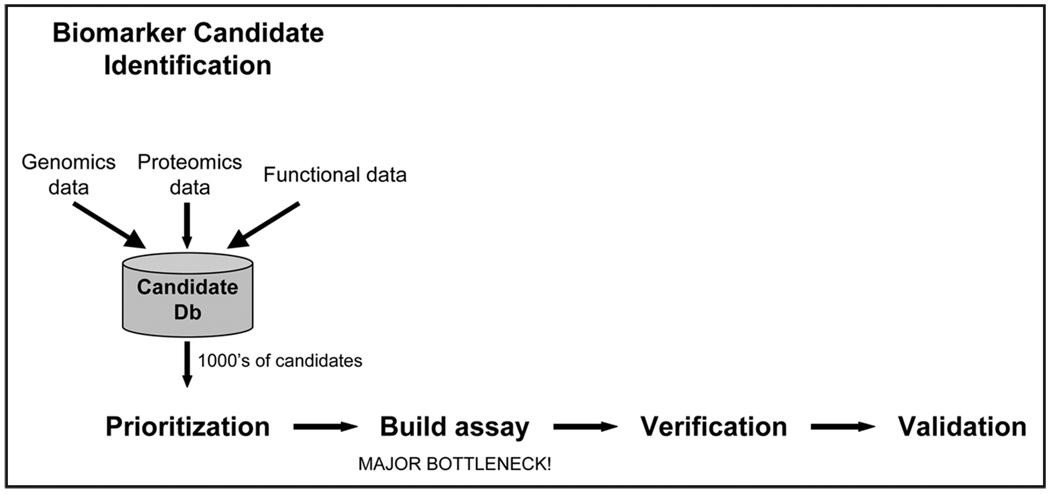
A typical protein biomarker pipeline is shown in Figure 1.29,30 As discussed above, application of genomic and proteomic technologies results in the identification of many hundreds to thousands of biomarker candidates for each disease. Each individual candidate must then be followed up in small-scale verification studies, followed by large-scale clinical validation trials. Verification and validation studies require a quantitative assay to measure the levels of each candidate biomarker in clinical plasma specimens. Only rarely is a quantitative assay available for a candidate of interest; more typically a novel quantitative assay must be developed de novo for every candidate biomarker that will be subjected to follow-up studies.
Figure 1.
Biomarker Pipeline. Candidate biomarkers are identified by compiling data from genomics, proteomics, and other sources. Because modern “-omics” experiments are capable of producing thousands of candidate biomarkers, the list must be prioritized. Furthermore, because these candidates come from a variety of experiments, they must be verified for their intended purpose (e.g., plasma-based diagnostic marker) and proven to show sufficient sensitivity and specificity in a large number of clinical samples (validation). Verification and validation efforts require a quantitative assay for each candidate biomarker, creating a large bottleneck in the pipeline (due to a lack of and high expense for developing such assays). Alleviating the bottleneck will allow evaluation of more candidates and thus inform the discovery and prioritization steps of those candidates most likely to reach success.
Traditionally, immunoassays (e.g., Enzyme-Linked Immuno-Sorbent Assay, also called ELISA) have been developed to measure the levels of each candidate protein in clinical samples (e.g., plasma, serum) from cases and controls. Because of the high expense ($100,000’s–$1,000,000’s) and long lead time (1–2 years) for ELISA development, candidates are prioritized for follow-up, and only a small fraction of the total candidates are actually pursued. Once assays are developed, early pilot studies (termed verification) are conducted to confirm that candidates are differentially present on average in cases vs. controls. Only the most promising few candidates are subsequently advanced into validation studies where the utility of the putative biomarker is tested in the targeted clinical application looking at thousands of individual patients.
Due to the high cost and long lead time associated with ELISA development, our ability to discover promising biomarker candidates far outstrips our ability to test each of those candidates for clinical utility, and this conundrum currently represents the most significant obstacle to translating novel protein biomarkers into clinical use.21,31 Although conventional proteomic technologies are far from being able to perform global protein biomarker discovery, they are brilliantly poised to relieve the most severe bottleneck in the biomarker pipeline: development of targeted assays to test individual candidate biomarkers.
The remainder of this review article will: (1) review the difference between commonly used untargeted mass spectrometry and emerging targeted mass spectrometry methods, (2) discuss current capabilities of targeted mass spectrometry methods and their potential in biomarker discovery, and (3) speculate regarding the use of integrative genomics to improve our ability to prioritize candidate biomarkers for testing.
Different Modes of Mass Spectrometry
The basics of mass spectrometry are presented in Technical Box 1. In this review we will largely focus on bottom up proteomics, in which proteins are digested to predictable peptide fragments using proteases such as trypsin. Tryptic digests of biological proteomes (e.g., tissue- or plasma-derived proteins) can be analyzed using different modes of mass spectrometry (Technical Box 1 and Table 1), depending on the desired application. For example, untargeted modes of mass spectrometry (i.e., MS fingerprinting, shotgun MS/MS) are used for de novo discovery of biomarker candidates such as from tumor tissues or proximal fluids. In contrast, targeted modes of mass spectrometry allow us to “tune” the instruments specifically to look for peptides (and hence proteins) of interest in clinical specimens; these targeted modes of mass spectrometry can be very useful for determining whether biomarker candidates discovered in tissues or proximal fluids are present (and elevated) in plasma from cancer patients compared to controls (Fig. 2). The remainder of this review will focus on the emerging use of targeted proteomic methods for testing candidate biomarkers in plasma.
Technical Box 1.
Table 1.
There are many types of mass spectrometer instruments as well as many modes of mass spectrometry
| LC-MS modes | Targeted | Masses programmed for detection |
Masses detected | Strength | Limitations | Comments |
|---|---|---|---|---|---|---|
| MS Fingerprinting |
No | None | parent ion m/z for all species that were successfullly ionized |
Provides richest, undirected coverage of ionizable analytes |
Not all analytes ionize efficiently. No sequence information is generated. |
Early clinical proteomic studies tested MS fingerprint “patterns” as diagnostics, but did not yield any viable biomarkers due to a variety of issues. Hence, although current studies may use MS fingerprint patterns to detect features that distinguish case from control, they always go on to identify (via MS/MS) the proteins that produce the features of interest. |
| ‘Shotgun’ MS/MS |
No | None | parent ion m/z and spectrum of fragment ions resulting from fragmentation of the most abundant detected ions |
MS/MS spectra allow absolute confirmation of target analyte |
Not all analytes ionize efficiently. Also, instrument speed is limiting, so only the most abundant ions are fragmented. |
This is a totally undirected mode in which the most abundant peptides are detected and identified by sequence. Low abundance analytes are generally not detected in complex biospecimens. This mode of LC-MS is useful for discovering candidate biomarkers in solid tissues and proximal fluids, where disease-specific proteins may be at high concentration. |
| Accurate Inclusion Mass Screening (AIMS) |
Yes | MS parent ion m/z | parent ion m/z and spectrum of fragment ions resulting from fragmentation of selected parent ions |
Improved sensitivity for detecting the target analyte; MS/MS spectra allow absolute confirmation of target analyte |
Not all analytes ionize efficiently. In complex mixtures, nontargeted analytes can interfere with detection of target analyte. |
This mode of LC-MS is useful when candidate protein biomarkers have been discovered in tissues or proximal fluids and it is unknown whether these candidate biomarkers can be detected in plasma. Representative (“proteotypic”) peptides from each candidate can be programmed into an inclusion list and used to analyze highly fractionated plasma to determine if the candidate biomarker can be detected. |
| Selected Reaction Monitoring (SRM) |
Yes | MS parent ion m/z and several MS/MS fragment ions (transitions) |
individual fragment ions resulting from fragmentation of programmed parent ion |
Highest sensitivity for detecting the target analyte; high duty cycle facilitates peak integration for quantification |
Not all analytes ionize efficiently. In complex mixtures, nontargeted analytes can interfere with detection of target analyte. |
This mode of LC-MS is useful when candidate protein biomarkers can be detected in plasma, and we want to determine if their concentration is elevated in plasma from disease cases compared with controls. |
Each mode has characteristic strengths and weaknesses, so it is essential to select the best instrument and mode of analysis for a given experimental goal.
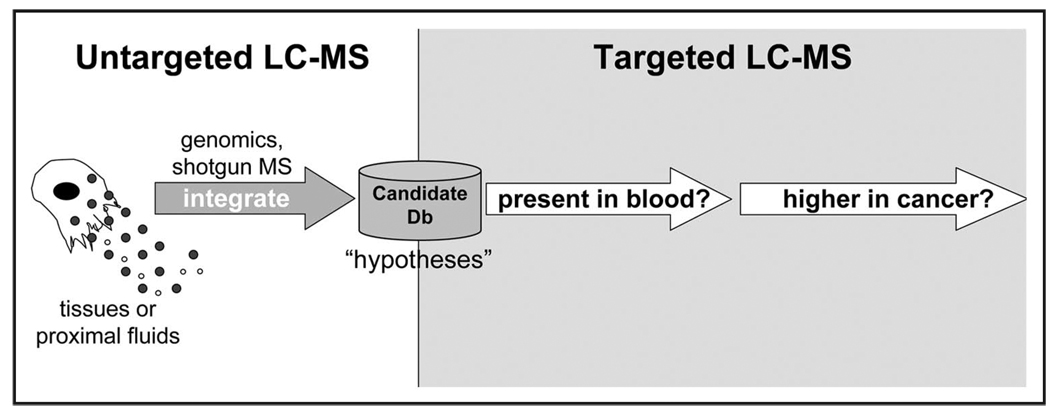
Figure 2.
Staged use of untargeted and targeted modes of mass spectrometry. Untargeted mass spectrometry is used in a discovery setting where identification of all potential biomarkers is desired. Untargeted discovery of biomarkers is conducted in a variety of biological specimens including tissues and proximal fluids. The resulting list of candidate biomarkers form a list of “hypotheses” which are interrogated using targeted mass spectrometry. Because candidates come from a variety of sources, initial targeted experiments are used to verify the presence of the candidate in plasma. Further studies are aimed at validating the effectiveness of the biomarker in discriminating cancer samples using a targeted mass spectrometry technique called selection reaction monitoring (SRM).
Emerging Use of Targeted Proteomic Methods for Testing Candidate Biomarkers in Plasma
Once candidate protein biomarkers have been discovered in tissues or proximal fluids, the next steps are to determine: (1) whether the candidate protein can be detected in the plasma (i.e., is it there?), and (2) whether the candidate protein is elevated in the plasma of cases compared to healthy controls. Two separate forms of targeted mass spectrometry can be used to answer these questions, as summarized below.
Can the Candidate Protein Biomarker be Detected in Plasma?
Recall that untargeted MS/MS analysis of plasma is extremely challenging, and the probability of identifying cancer-specific markers is low due to a cadre of high abundance proteins that interfere with detection of low abundance tumor-derived proteins. Hence, as discussed above, even if we are looking for a plasma-based biomarker, it makes the most sense to do our initial biomarker candidate discovery in tissues or proximal fluids where tumor-derived proteins can be detected using conventional mass spectrometers in an untargeted mode (and/or using genomics-based analyses). Once candidate biomarkers have been identified in tissues or proximal fluids, we must next determine whether each of the candidate proteins can be detected in plasma. In this situation, a targeted form of mass spectrometry called accurate inclusion mass screening (AIMS)32–34 is of great utility. In AIMS analysis (Techinical Box 1 and Table 1), the instrument is programmed to specifically “look at” peptides derived from candidate protein biomarkers; this is possible because if we know the candidate of interest, we can predict the mass to charge ratio (m/z) of each of the peptides the candidate will release upon digestion with trypsin. Each m/z of interest can be added to an inclusion list programmed into the instrument, which directs the instrument only to spend time analyzing peptides of interest while ignoring all other peptides. This effectively gives the instrument added sensitivity for detecting lower abundance proteins in plasma by reducing the undersampling effect in untargeted analyses (see Technical Box 1). To further facilitate our sensitivity for detecting low abundance proteins during AIMS analysis, a pool of plasma from cancer patients can be subjected to depletion of abundant proteins followed by trypsin digestion and strong cation exchange chromatography, producing 10–20 individual fractions that can be separately analyzed. Several thousand proteins can be comprehensively searched for in fractionated plasma within one month using a single dedicated instrument.
Is the Candidate Protein Biomarker Elevated in the Plasma of Cases Compared to Healthy Controls?
Once candidate protein biomarkers are confirmed to be detectable in plasma using AIMS, the next step is to determine whether the candidate is at a higher concentration in plasma from cases compared to healthy controls. A highly sensitive and specific quantitative assay is required for each candidate biomarker protein to determine its concentration in plasma from cancer patients and healthy controls. As discussed above, the immunoassay (e.g., ELISA) has been the mainstay for measuring candidate biomarkers. However, the high cost and long lead time for development of each immunoassay is prohibitive and presents a major bottleneck in the biomarker pipeline.
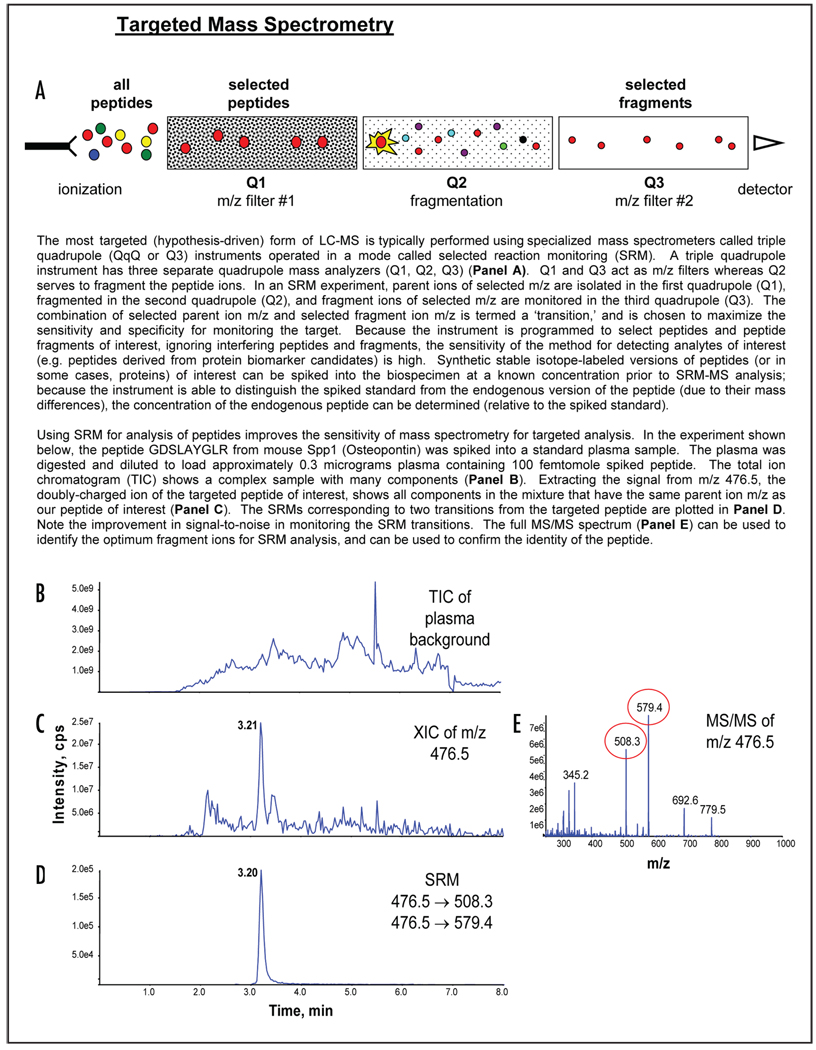
A second mode of targeted mass spectrometry, selected reaction monitoring (SRM), can be used to relieve this bottleneck. The sensitivity and specificity of SRM-MS are well-established in the measurement of small molecules; clinical reference laboratories employ this technique to measure drug metabolites and metabolites that accumulate in inborn errors of metabolism.35,36 The SRM-MS technology has recently been adapted to measure the concentration of candidate protein biomarkers, using proteotypic peptides as specific stoichiometric surrogates (Technical Box 2).27,37–40 Accurate calibration is achieved by spiking digested samples with known quantities of synthetic stable isotope-labeled peptides as internal standards. Without enrichment of the target peptides, SRM-MS alone is able to measure proteins present in the 100–1,000 nanograms per milliliter concentration range from small volumes (1–10 microliters) of plasma.41
Technical Box 2.
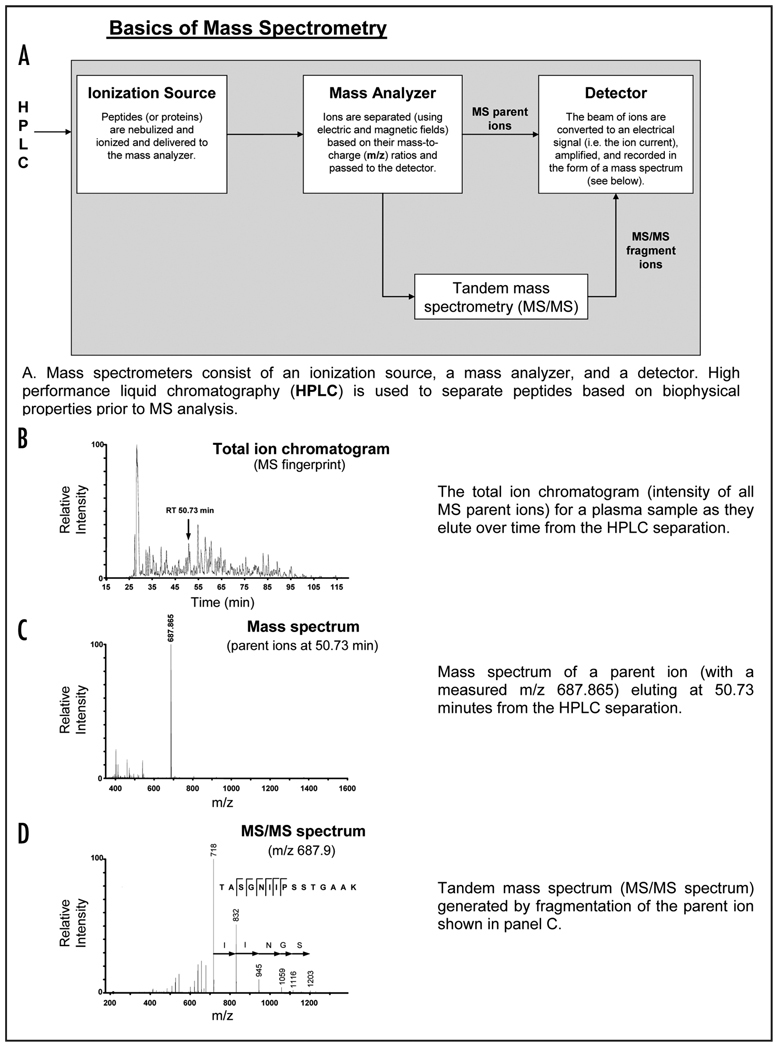
Technical Box 1.
Mass spectrometers consist of an ionization source, a mass analyzer, and a detector (panel A). Although there are a variety of ionization sources (e.g. electrospray and matrix assisted laser desorption ionization) and mass analyzers (e.g. quadrupoles, time-of-flight, quadrupole ion traps, and ion cyclotron resonance), all MS instruments have these basic features in common. In a typical analysis of a biological sample, proteins or peptides are introduced into the ionization source where they are converted to gas-phase charged particles (ionized) and passed to the mass analyzer. In the mass analyzer, the ions are separated (using electric and magnetic fields) based on their mass-to-charge (m/z) ratios. The detector electrically detects the beam of ions passing through the machine (i.e. the ion current) and amplifies the signal, which is recorded in the form of a mass spectrum
Typically, a mass spectrometer is coupled with a separation technique such as high performance liquid chromatography (HPLC). The sum of mass spectra accumulated over time as a sample is separated produces a total ion chromatogram (panel B). In this form of analysis, the sequences of peptides are not determined; rather the output consists of the ion currents at the m/z for each peptide or protein component detected in the specimen over the entire chromatographic elution period (i.e. a “fingerprint”). In an early application of mass spectrometry to cancer biomarker discovery, fingerprints generated from plasma samples from cancer patients and healthy controls were compared and differences were labeled as potential biomarkers. However, although there was a great deal of initial enthusiasm for the use of proteomic fingerprints to diagnose cancer, this approach has not produced any validated biomarkers due to a variety of issues, and this approach has been abandoned by the mainstream proteomic community.
Many instruments are capable of tandem mass spectrometry (i.e. MS/MS), which can be used to infer the sequence of the peptide being detected. During MS/MS, a desired analyte is isolated based on its m/z ratio (panel C) and fragmented (i.e. breaking peptide bonds within the peptides), producing a series of fragment ions that are detected as a MS/MS spectrum (panel D). The fragmentation pattern is compared to the theoretical fragmentation pattern for every peptide in the genome to find the closest match. In this way the sequence of the peptide ion is inferred from its fragmentation pattern.
A major limitation of conventional MS instruments is that they can neither detect nor fragment every peptide in a typical biospecimen; hence the proteome is significantly “undersampled.” There are two predominant reasons for undersampling. First, the ionization process is incomplete and many peptides do not become ionized and therefore cannot be detected by the instrument. Second, there is an impedance mismatch between the mass spectrometer and complex biological proteomes; the speed of the instrument is such that only a fraction of the peptide ions in the instrument can be selected for fragmentation. In a typical “data dependent acquisition” (i.e. “shotgun MS/MS” analysis), a MS spectrum is acquired and the five most abundant peptide ions are selected and sequentially fragmented by the instrument. This cycle is repeated for the duration of the sample analysis. Sampling a few of the most abundant ions in this way permits only a small sampling of the total diversity of the typical biospecimen. These issues are partially alleviated by increasing the separation of the complex peptide mixture. For instance, biological proteomes are frequently subjected to biochemical fractionation upstream of LC-MS to further reduce the complexity of the sample. Nonetheless, although these online and offline separations achieve some improvement in sampling, the vast majority of peptides (and hence proteins) in a typical complex proteome go undetected. Furthermore, there is a bias towards the detection of the most abundant peptides at the expense of lower abundance peptides, making direct discovery of low abundance cancer biomarkers directly in plasma an extremely challenging task. An alternative to data-dependent acquisition, targeted MS/MS can be used when one is asking if a protein of interest is present in the biospecimen. During targeted MS/MS, the instrument is programmed to fragment only peptides with a pre-selected m/z ratio. As a result, peptides of interest that might not otherwise have been targeted for fragmentation (due to more abundant interfering ions) can now be selected and identified.
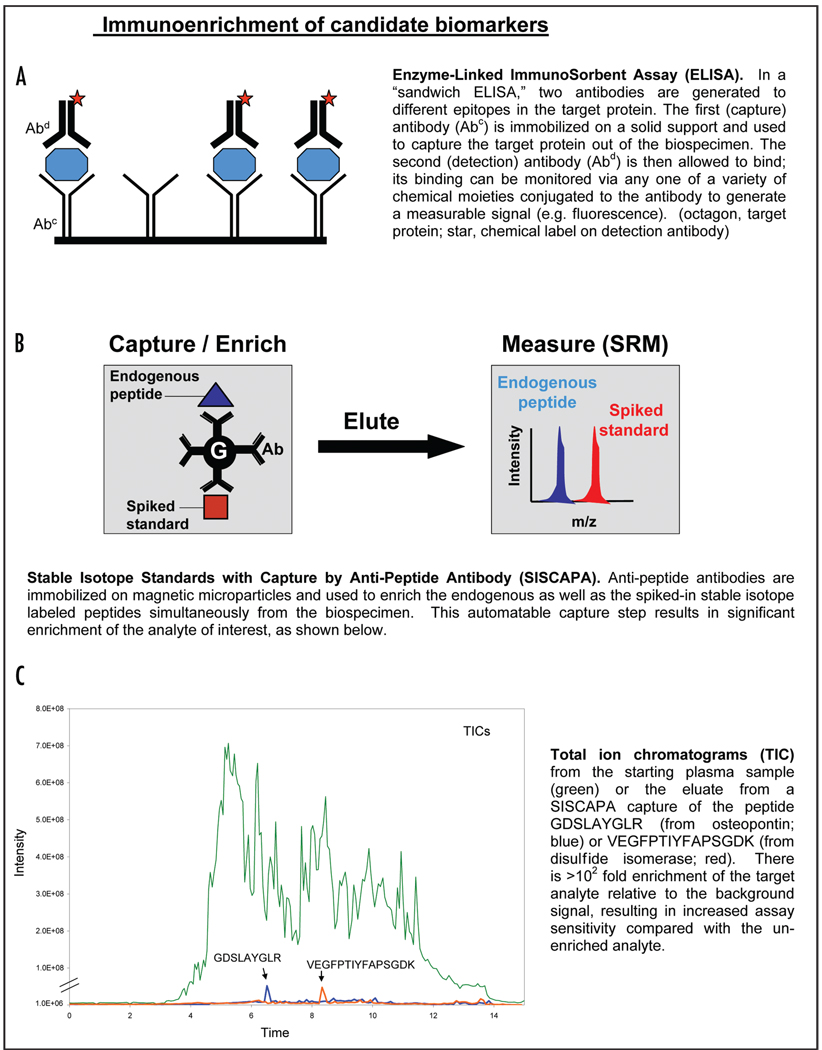
For candidates present at lower concentrations in plasma, an enrichment step is added. For example, previous studies have demonstrated the success of using limited SCX fractionation42 or glycopeptide enrichment43 to analyze low abundance analytes. Alternatively, targeted enrichment can be performed using a technology called SISCAPA (stable isotope standards and capture by anti-peptide antibodies).44–47 As shown in Technical Box 3, SISCAPA uses anti-peptide antibodies to enrich peptides of interest from plasma prior to SRM-MS analysis, increasing the sensitivity of the assay. Coupling SISCAPA to SRM-MS, it is possible to measure candidate protein biomarkers present in the plasma down to concentrations of 100 pg per milliliter (Whiteaker et al.).
Technical Box 3.
SISCAPA coupled to SRM-MS has great potential for relieving the assay bottleneck in the biomarker pipeline because protein assays can be generated relatively rapidly (it takes approximately 20 w to produce a new affinity-purified anti-peptide polyclonal antibody) and cheaper (<$5,000 in reagents costs per protein) compared to the traditional ELISA assay, thereby allowing a far higher number of candidates to undergo testing.29 Furthermore, SISCAPA-SRM assays also have several analytic advantages over traditional immunoassays. For example, while it is difficult to multiplex large numbers of ELISA assays, it should be possible to multiplex hundreds of SISCAPA assays using microliter quantities of plasma. Additionally, the presence of autoantibodies or anti-heterophile antibodies can interfere with ELISA assays.48–52 In contrast, interfering antibodies are digested for SISCAPA and the perfect specificity of the mass spectrometer as the detector circumvents these issues in SISCAPA-SRM assays.
Prioritizing Candidate Biomarkers for Assay Development
Until off-the-shelf assays are available for all candidate biomarkers,31 it will be necessary to prioritize for follow-up a subset of the several thousand candidate biomarkers that can be discovered for any given cancer. The AIMS analysis described above provides one level of prioritization by confirming which candidates are present in the circulation. We will then need a strategy for further prioritizing those markers detected in the plasma. Until sufficient flux has been established through the biomarker discovery pipeline to teach us paradigms about how to prioritize candidate biomarkers for ultimate clinical success, we are forced to rely upon clinical and theoretical biological considerations.
Clinical considerations for prioritizing candidate biomarkers
If we are to invest years of time and thousands of dollars in testing a given candidate biomarker, it is prudent to stack the deck in favor of choosing markers that will be of clinical utility. It is not sufficient that a candidate protein biomarker be at a higher average concentration in plasma of cancer patients compared to controls. In addition to being elevated in the plasma of cancer patients, the performance characteristics of candidate protein biomarkers (e.g., sensitivity, specificity, preclinical duration, etc.) must meet certain minimal requirements for clinical and economic acceptability in a given clinical application.29 Because the minimal requirements of acceptability will vary from one clinical setting to the next, it is essential that the desired clinical application be clearly defined at the outset of the biomarker discovery efforts so that every effort can be made to prioritize markers likely to have acceptable performance characteristics.
For example, if the goal is to identify a biomarker for screening for breast cancer, it is important to avoid overdiagnosis.2,53 Overdiagnosis occurs when we diagnose a patient with disease that in reality presents no risk to his or her life but in practice we are compelled to treat, thereby exposing patients to unnecessary emotional turmoil and treatment-related morbidity and costs. To avoid this, we might consider cross-referencing our candidates derived from the AIMS analysis with publicly available genomic data sets that identify genes whose expression correlates with clinical outcome.54 By prioritizing for follow-up our candidates that correlate with poor outcome we may avoid developing biomarkers that diagnose indolent disease.
Biological considerations for prioritizing candidate biomarkers: integrating genomic and proteomic data
In theory, proteomic biomarkers will provide more direct answers to clinical and pharmacological questions than genomic data, as the majority of known molecular markers and pharmaceutical targets are proteins,55 and the proteome provides a real time readout of physiology. On the other hand, because proteins are extremely dynamic, their changes are more difficult to fully monitor compared to genomic profiling. Despite rapid advances in the past decade, protein identification and quantification technologies still lag considerably behind those used to determine DNA sequence and mRNA expression levels on a genome-wide scale.56 Table 2 summarizes the strengths and weakness of different genomics/proteomics data types for protein biomarker discovery.
Table 2.
Multiple technologies can be used to discover candidate protein biomarkers
| Discovery technologies | Advantages | Disadvantages |
|---|---|---|
| RNA transcript (microarray) | global; quantitative; provides real time measurement of physiology |
no protein level information; applied to cells, not plasma |
| DNA copy number (CGH) | global; quantitative | no protein level information; reveals information about DNA blueprint but not real time information on physiology |
| DNA sequence | potentially global | no protein level information; reveals information about DNA blueprint but not real time information on physiology |
| Proteomic data (untargeted MS/MS) | protein level data; provides real time measurement of physiology |
not global; semi-quantitative at best |
Because these technologies have complementary strengths and weaknesses, it is beneficial to integrate data from a variety of experiments and approaches to prioritize the candidates most qualified for follow-up studies.
In human cancer studies, strong concordance between mRNA and protein expression levels is rarely observed or is only observed for a small subset of the proteins.55,57–59 This suggests that genomic and proteomic changes provide complementary information, as human tumors are complex and heterogeneous, and are caused by defects in numerous pathways and factors that operate at many levels.56 Thus, there is a pressing need to integrate data at multiple levels encompassing both proteomics and genomics in current biomarker studies.
As discussed above, targeted modes of MS can now be used to facilitate candidate biomarker testing. The simplest usage of genomic information is to provide corroborating information to help prioritize proteins implicated by proteomic studies. This helps to reduce the rate of false positive findings due to experimental errors (assuming independent errors in different high throughput experiments). Moreover, to date there have been more genomics experiments than proteomics experiments carried out in large scale clinical studies. Thus, including these genomics data sets in the candidate database helps us to better incorporate clinical information (e.g., disease outcomes) in the discovery stage. Many other benefits for carrying out an integrative-omics approach in biomarker studies stem from the more comprehensive genome coverage achieved by gene profiling than protein profiling (Table 2). Functional information learned from genomic studies provides a more global picture of the regulatory network, which helps to identify important protein groups and shed light on the “missing” measurements in proteomics experiments (see more discussion regarding the usage of networks in the next section). In summary, it is critical that we make use of every piece of information to improve biomarker identification, which can be best accomplished by an integrative approach using multiple “-omics” data sets.
Using interaction networks to refine biomarker candidate selection
As a “systems disease”, cancer cannot be understood by studying individual components only. This raises the challenge of reconciling the search for individual markers with a systems-level understanding, which may be especially useful to the rational design of putative biomarker panels. Protein-protein interaction networks and gene regulatory networks provide essential resources for characterizing the “system-level” behavior of genes and proteins. Some pioneering work has been done along this direction. For example, in a few recent breast cancer studies,60–62 expression profiles and protein interaction information were integrated and protein interaction subnetworks with coherent expression patters were identified. These subnetworks (interactome/modular) were then shown to be predictive of breast cancer prognosis. The merit of network-based analysis has also been recognized in a colon cancer study,63 in which the authors searched for protein interaction sub-networks enriched for proteins associated with colon cancer progression, and then successfully identified protein panels highly discriminative of stage D colon cancer versus control.
What are the basic approaches for us to use protein/gene interaction network information to further improve biomarker identification and prioritization? Intuitively, we can look for subnetworks (Fig. 3) enriched with nodes (molecules) and edges (molecular interactions) showing significant association with disease outcomes. Such subnetworks may represent functional molecular groups that play important roles in the disease process. This approach is in the same spirit as the “gene set” analysis, a common tool in the microarray community.64–70 Gene set analysis scores known pathways by the coherency of expression changes among their member genes regarding the association with disease phenotypes. By borrowing strength across the gene-set, there is potential for increased statistical power to identify disease association. In addition, signatures of gene-sets are more robust to biological and technical variability compared to the signatures of individual markers.
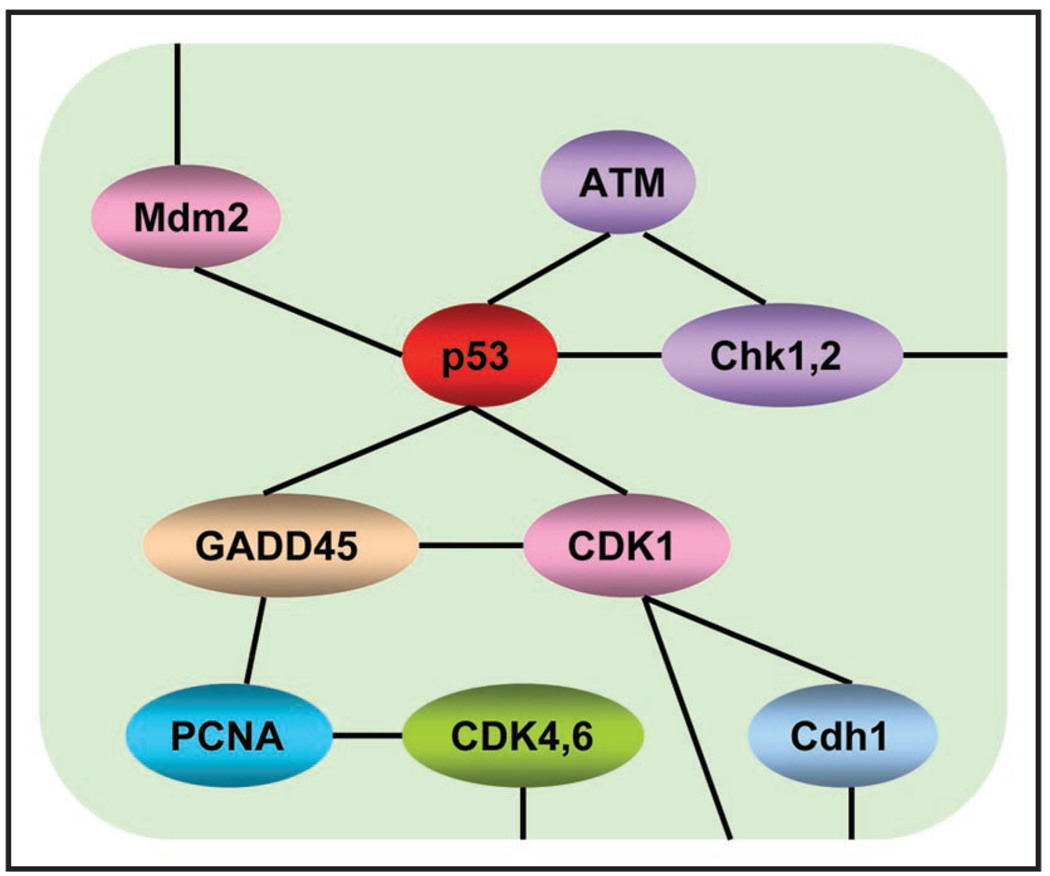
Figure 3.
Genes in one pathway have complex “net” interactions, which should be taken into consideration when analyzing proteomics/genomic data. Generally, in pathway analysis, each pathway is deemed as a set of genes, and the goal is often to detect associations between the overall activities of the genes in the pathway and a phenotype of interest (e.g., disease outcomes). However, simplifying “pathways” into “sets” of genes results in the loss of information—the interaction relationships among genes within the same pathway or across different pathways. This figure illustrates the gene-gene interaction patterns for part of the cell cycle pathway. The topology of the network is from the KEGG database; www.genome.ad.jp/kegg/pathway.html. Each gene in this network interacts with different neighbors and thus plays its unique role in maintaining the pathway function.
Although “gene set” analysis is believed to be a more effective means of marker identification, a remaining hurdle is that the majority of human genes have not yet been assigned to definitive pathways. Moreover, a biological “pathway” seldom holds a simple “chain” shape. Instead, components of the same “pathway” always have very complex “net” interactions (Fig. 3). Obviously, totally ignoring the topology of the interacting patterns among pathway components makes the analysis much less efficient. Thus, it is beneficial to use the interaction networks directly and seek subnetworks showing strong association with disease outcomes. The subnetworks enriched with both differentially expressed proteins and differentially expressed genes are very likely to contain good biomarkers to discriminate tumors from normals. In addition, this strategy helps us to recover viable candidates not identified in biomarker discovery experiments; a marker not observed in discovery experiments could be identified as a biomarker if it connects with many other proteins showing strong association with disease phenotypes. In the end, since interaction/regulatory networks usually contain rich information about markers’ functionality, they can also be used to facilitate the selection of a panel of markers covering a diverse set of biological pathways.
Future Directions and Unmet Needs
As described above, the role of mass spectrometry in the biomarker pipeline has evolved substantially since the first reports that claimed to identify proteomic fingerprints in serum that were diagnostic of cancer. Emerging technologies in mass spectrometry-based proteomics, especially targeted MS/MS, now offer extraordinary promise for hypothesis-driven testing of candidate protein biomarkers by relieving the severe bottleneck of assay generation in the biomarker pipeline.
A strength of using mass spectrometry for biomarker discovery is the ability to discern structural modifications to proteins (e.g., post-translational modifications). To date, mass spectrometry has been useful in profiling changes in the proteome such as phosphorylation and glycosylation.71–73 As these proteomic techniques improve there will likely be an increase in the flux of these modifications as biomarker candidates. Targeted mass spectrometry can be equally applied for testing these candidates, whereas it is difficult to build conventional assays to such targets.
Although targeted mass spectrometry methods in their current form will undoubtedly have a positive impact on biomarker testing, there is still tremendous room for improvement. For example, the sensitivity as well as the selectivity of SRM-based assays can be improved. Instrument vendors are actively working to build instruments with better ionization, enhanced ion transmission, and improved mass accuracy. Better ionization and ion transmission promises to lower our limits of quantitation and the hope is that someday we may achieve protein detection in plasma at comparable or better sensitivity than the immunoassay. Improved mass accuracy is also important because endogenous interferences emanating from analytes other than the peptide being targeted (i.e., matrix interference) can produce nonspecific signals in the channels being monitored during SRM-MS.74 Additionally, accurate quantitation by SRM-MS depends upon stoichiometric digestion of the plasma sample by the protease (e.g., trypsin). It is well-known that some proteins are resistant to proteolytic digestion and that the presence of post-translational modifications can also affect cleavage. Hence, there is a need for the development of standards and metrics for quality control of trypsin digestion of biospecimens prior to quantitative mass spectrometry.
Until mass spectrometers evolve to the point that they can detect very low abundance proteins in plasma (i.e., ≤ng/ml), it will be necessary to perform an upstream enrichment step for the protein of interest. As discussed above, the SISCAPA technology has shown great promise, with the best antibodies providing limits of quantitation in the hundreds of picograms of target protein per milliliter in plasma. Although these assays are far cheaper and faster to develop than the traditional ELISA, it still takes approximately 5 mo and a few thousands dollars per peptide to develop the required affinity purified polyclonal antibody. Hence, the assay bottleneck in the biomarker pipeline can be substantially relieved, but not yet eliminated. Biomarker testing could be greatly accelerated if there were more affordable affinity reagents as well as faster protocols for generating them. Alternatively, biomarker testing would be greatly facilitated if assays (and the required reagents) had already been assembled and validated and were available to any biomarker study.31
Acknowledgements
This work was funded by the National Cancer Institutes Clinical Proteomic Technology Assessment for Cancer (CPTAC) Program, the Paul G. Allen Family Foundation, and the Entertainment Industry Foundation.
Footnotes
The authors declare no competing interests.
References
- 1.Sturgeon CM, Duffy MJ, Stenman UH, Lilja H, Brunner N, Chan DW, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast and ovarian cancers. Clin Chem. 2008;54:11–79. doi: 10.1373/clinchem.2008.105601. [DOI] [PubMed] [Google Scholar]
- 2.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13:477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 4.Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27:1323–1333. doi: 10.1200/JCO.2007.14.8197. [DOI] [PubMed] [Google Scholar]
- 5.Fine B, Amler L. Predictive biomarkers in the development of oncology drugs: A therapeutic industry perspective. Clin Pharmacol Ther. 2009;85:535–538. doi: 10.1038/clpt.2009.9. [DOI] [PubMed] [Google Scholar]
- 6.Bacher U, Zander AR, Haferlach T, Schnittger S, Fehse B, Kroger N. Minimal residual disease diagnostics in myeloid malignancies in the post transplant period. Bone Marrow Transpl. 2008;42:145–157. doi: 10.1038/bmt.2008.185. [DOI] [PubMed] [Google Scholar]
- 7.Furihata T, Sawada T, Kita J, Iso Y, Kato M, Rokkaku K, et al. Serum alpha-fetoprotein level per tumor volume reflects prognosis in patients with hepatocellular carcinoma after curative hepatectomy. Hepatogastroenterology. 2008;55:1705–1709. [PubMed] [Google Scholar]
- 8.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 9.Wagner L. A test before its time? FDA stalls distribution process of proteomic test. J Natl Cancer Inst. 2004;96:500–501. doi: 10.1093/jnci/96.7.500. [DOI] [PubMed] [Google Scholar]
- 10.Garber K. Debate rages over proteomic patterns. J Natl Cancer Inst. 2004;96:816–818. doi: 10.1093/jnci/96.11.816. [DOI] [PubMed] [Google Scholar]
- 11.Check E. Proteomics and cancer: running before we can walk? Nature. 2004;429:496–497. doi: 10.1038/429496a. [DOI] [PubMed] [Google Scholar]
- 12.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 13.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 14.Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinformatics. 2003;4:24. doi: 10.1186/1471-2105-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin Chem. 2003;49:1272–1275. doi: 10.1373/49.8.1272. [DOI] [PubMed] [Google Scholar]
- 16.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 17.Hede K. $104 million proteomics initiative gets green light. J Natl Cancer Inst. 2005;97:1324–1325. doi: 10.1093/jnci/dji325. [DOI] [PubMed] [Google Scholar]
- 18.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 20.Anderson N, Anderson N. The human plasma proteome: history, character and diagnostic prospects. Mol Cell Prot. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.Carr SA, Anderson L. Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory? Clin Chem. 2008;54:1749–1752. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson NL, Anderson NG. The human plasma proteome: history, character and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Kulasingam V, Diamandis EP. Tissue culture-based breast cancer biomarker discovery platform. Int J Cancer. 2008;123:2007–2012. doi: 10.1002/ijc.23844. [DOI] [PubMed] [Google Scholar]
- 24.Celis JE, Gromov P, Cabezon T, Moreira JM, Ambartsumian N, Sandelin K, et al. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics. 2004;3:327–344. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Jansen FH, Krijgsveld J, van Rijswijk A, van den Bemd GJ, van den Berg MS, van Weerden WM, et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell Proteomics. 2009;8:1192–1205. doi: 10.1074/mcp.M800443-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Liu AY, Loriaux P, Wollscheid B, Zhou Y, Watts JD, Aebersold R. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 28.Sardana G, Jung K, Stephan C, Diamandis EP. Proteomic analysis of conditioned media from the PC3, LNCaP and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J Proteome Res. 2008;7:3329–3338. doi: 10.1021/pr8003216. [DOI] [PubMed] [Google Scholar]
- 29.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The Interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteom Clin Appl. 2008;2:1386–1402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 31.Anderson NL, Anderson NG, Pearson TW, Borchers CH, Paulovich AG, Patterson SD, et al. A human proteome detection and quantitation project: hPDQ. Mol Cell Proteomics. 2009;8:883–886. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinner O, Mueller LN, Hubalek M, Muller M, Gstaiger M, Aebersold R. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
- 34.Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 35.Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 36.Chace DH, Kalas TA. A biochemical perspective on the use of tandem mass spectrometry for newborn screening and clinical testing. Clin Biochem. 2005;38:296–309. doi: 10.1016/j.clinbiochem.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, et al. Isotope dilution—mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 38.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4:1175–1186. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 40.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3:644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 41.Anderson L, Hunter C. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 44.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 45.Whiteaker JL, Zhao L, Zhang HY, Feng LC, Piening BD, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54:1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berna M, Schmalz C, Duffin K, Mitchell P, Chambers M, Ackermann B. Online immunoaffinity liquid chromatography/tandem mass spectrometry determination of a type II collagen peptide biomarker in rat urine: investigation of the impact of collision-induced dissociation fluctuation on peptide quantitation. Anal Biochem. 2006;356:235–243. doi: 10.1016/j.ab.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 48.Spencer CA, Lopresti JS. Measuring thyroglobulin and thyroglobulin autoantibody in patients with differentiated thyroid cancer. Nat Clin Pract Endocrinol Metab. 2008;4:223–233. doi: 10.1038/ncpendmet0757. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe M, Uchida K, Nakagaki K, Kanazawa H, Trapnell BC, Hoshino Y, et al. Anti-cytokine autoantibodies are ubiquitous in healthy individuals. FEBS Lett. 2007;581:2017–2021. doi: 10.1016/j.febslet.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 50.Hennig C, Rink L, Fagin U, Jabs WJ, Kirchner H. The influence of naturally occurring heterophilic anti-immunoglobulin antibodies on direct measurement of serum proteins using sandwich ELISAs. J Immunol Meth. 2000;235:71–80. doi: 10.1016/s0022-1759(99)00206-9. [DOI] [PubMed] [Google Scholar]
- 51.Kricka LJ, Schmerfeld-Pruss D, Senior M, Goodman DB, Kaladas P. Interference by human anti-mouse antibody in two-site immunoassays. Clin Chem. 1990;36:892–894. [PubMed] [Google Scholar]
- 52.Nahm MH, Hoffmann JW. Heteroantibody: phantom of the immunoassay. Clin Chem. 1990;36:829. [PubMed] [Google Scholar]
- 53.Gotzsche PC, Jorgensen KJ, Maehlen J, Zahl PH. Estimation of lead time and overdiagnosis in breast cancer screening. Br J Cancer. 2009;100:219. doi: 10.1038/sj.bjc.6604762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 55.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lichtinghagen R, Musholt PB, Lein M, Romer A, Rudolph B, Kristiansen G, et al. Different mRNA and protein expression of matrix metalloproteinases 2 and 9 and tissue inhibitor of metalloproteinases 1 in benign and malignant prostate tissue. Eur Urol. 2002;42:398–406. doi: 10.1016/s0302-2838(02)00324-x. [DOI] [PubMed] [Google Scholar]
- 58.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 59.Orntoft TF, Thykjaer T, Waldman FM, Wolf H, Celis JE. Genome-wide study of gene copy numbers, transcripts and protein levels in pairs of non-invasive and invasive human transitional cell carcinomas. Mol Cell Proteomics. 2002;1:37–45. doi: 10.1074/mcp.m100019-mcp200. [DOI] [PubMed] [Google Scholar]
- 60.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Auffray C. Protein subnetwork markers improve prediction of cancer outcome. Mol Syst Biol. 2007;3:141. doi: 10.1038/msb4100183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 63.Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance M. Discovery and scoring of protein interaction sub-networks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 66.Pavlidis P, Lewis DP, Noble WS. Exploring gene expression data with class scores. Pac Symp Biocomput. 2002:474–485. [PubMed] [Google Scholar]
- 67.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23:1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- 70.Efron B, Tibshirani R. On testing the significance of sets of genes. Annals of Applied Statistics. 2007;1:107–129. [Google Scholar]
- 71.Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 72.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phospho-tyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 73.de Leoz ML, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, et al. Glycomic approach for potential biomarkers on prostate cancer: profiling of N-linked glycans in human sera and pRNS cell lines. Dis Markers. 2008;25:243–258. doi: 10.1155/2008/515318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sherman J, McKay MJ, Ashman K, Molloy MP. How specific is my SRM?: the issue of precursor and product ion redundancy. Proteomics. 2009;9:1120–1123. doi: 10.1002/pmic.200800577. [DOI] [PubMed] [Google Scholar]