Abstract
Interleukin-12 (IL-12) has potent antitumor activities via natural killer cells and cytotoxic T lymphocytes. However, the molecular mechanisms whereby IL-12 induces tumoricidal activities are poorly understood. Here, we report the genome-wide analysis of gene expression in a primary murine mammary carcinoma model that resembles human breast cancer, following the therapeutic application of recombinant IL-12, which restricted tumor growth and metastasis. IL-12 was able to curtail neovascularization in the tumor as well as enhance the number of tumor-infiltrating lymphocytes. Comprehensive examination of global gene expression revealed IL-12-induced molecular changes associated with tumor regression and reduced lung metastasis, thus providing a high-resolution snapshot of a host response against a developing malignancy and a rich source of potential targets for therapeutic intervention of breast cancer.
Keywords: Interleukin-12, Interferon-γ, mammary carcinoma, tumor-infiltrating lymphocytes, metastasis, angiogenesis
Tumors often possess a number of potential recognition sites for immunologic effector cells, which, in theory, could make them susceptible to immune surveillance. Nevertheless, most of such tumors grow progressively in their natural hosts or syngeneic recipients, without being controlled effectively by the immune system. The lack of apparent immunogenicity of tumors in situ might be due to special properties of the tumor cells, e.g., lack of costimulatory molecules, downregulation of MHC molecules, or production of immunosuppressive factors1,2 or due to intrinsic tolerance mechanisms of the immune system.3
Two principal types of cells are immunologically potent tumor-killing effectors: NK and CTL. A major activator of both cell types is Interleukin-12 (IL-12). NK cells kill tumors by intrinsic and nonspecific mechanisms hinged on a lack of MHC molecules on the surface of the tumor. CTL, on the other hand, recognizes specific antigenic peptides that are derived from the tumor and presented to them by antigen-presenting cells such as dendritic cells (DCs). In recent years, immunotherapy has been rekindled that attempts to either mark the tumor by upregulating the surface antigens for enhanced interaction with immune effector cells2,4 or by directly activating DC, T and NK cells for their heightened “scouting” capacity and increased cytolytic potency. IL-12 is a factor belonging to the latter class.5 IL-12 has powerful anti-tumor and anti-metastatic activities against many murine tumors as well as human tumors.6 Recent encouraging developments in clinical applications of IL-12 for human T cell lymphoma,7,8 B cell non-Hodgkin lymphoma,9 melanoma10–14 and renal carcinoma15 and SIV-infection model in rhesus macaques16 strongly underscore the importance of understanding the cellular and molecular mechanisms of IL-12mediated anti-tumor responses. The potent anti-malignancy activities of IL-12 are thought to be mediated through similar mechanisms that are used by IL-12 against infectious agents, i.e., via the activation of NK cells for the bulk nonantigen specific clearance, and activation of CTL and CD4 for tumor-specific elimination and long-term immunity; 4T1 is a tumor cell line isolated from a single spontaneously arising mammary tumor from a BALB/BfC3H mouse (MMTV+).17 It is an excellent model system for breast cancer research because its tumor development is well characterized both oncologically and immunologically. The 4T1 tumor closely mimics human breast cancer in its anatomical site (mammary gland), immunogenicity, growth characteristics and metastatic properties.18 4T1 tumor spontaneously metastasizes to a variety of target organs including the lung, heart, bone, brain and liver through primarily a hematogenous route.19 4T1 is also poorly immunogenic in that immunization with irradiated 4T1 cells provides only slight delays in tumor growth against wild-type tumor, not sufficient to protect the animal.20 IL-12 administration to 4T1-bearing mice resulted in a substantial reduction in tumor size and in spontaneous metastases in the lungs of 4T1 tumor-bearing mice and significantly prolonged their survival time.21 Tumor-draining lymph node cells obtained from 4T1 tumor-bearing mice treated with IL-12 cDNA exhibit increased natural killer (NK) activity and produced enhanced levels of IFN-γ, suggesting the involvement of both NK cells and IFN-γ in this effect.21 These properties make this mammary tumor model an excellent system in which to investigate the cellular and molecular changes throughout the malignant development.
To investigate the molecular mechanisms whereby IL-12 exerts its potent anti-tumor activities, we carried out a comprehensive analysis of gene expression in the 4T1 primary tumor following IL-12 treatment in vivo.
MATERIAL AND METHODS
Mice
Female BALB/c mice (6–8 week old) were purchased from the Jackson Laboratory (Bar Harbor, Maine). All mice were housed at the Weill Medical College of Cornell University Animal Facilities in accordance with the Principles of Animal Care (NIH publication number 85–23, revised 1985). Mice bearing 4T1 tumors were all sacrificed no later than day 28 due to the morbidity caused by large tumor size and strong metastasis.
Tumor implantation, size measurement, lung metastasis assay
4T1 mammary carcinoma cells (1×105) were injected subcutaneously into the abdominal mammary gland area of recipient mice in 0.1 ml of a single-cell suspension in phosphate-buffered saline (PBS) on day 0. The dose of tumor implantation was empirically determined to give rise to tumors of ~10 mm in diameter in untreated wild-type mice in 28 days. Primary tumors were measured using electronic calipers every other day. Reported measurements are the square root of the product of 2 perpendicular diameters. Numbers of metastatic cells in lung were determined by the clonogenic assay.18 In brief, lungs were removed from each mouse, finely minced and digested in 5 ml of enzyme cocktail containing 1× PBS and 1 mg/ml collagenase type IV for 2 hr at 37°C on a platform rocker. After incubation, samples were filtered through 70 μm nylon cell strainers and washed twice with PBS. Resulting cells were resuspended and plated serially diluted in 10 cm tissue culture dishes in medium RPMI1640 containing 60 μM thioguanine for clonogenic growth. 6-Thioguanine-resistant tumor cells formed foci within 10–14 days, at which time they were fixed with methanol and stained with 0.03% methylene blue for counting.
IL-12 treatment
Recombinant murine IL-12 was provided by Genetics Institute (Cambridge, MA). IL-12 treatment was given by intraperitoneal injection at 1 μg per mouse every other day starting on day 7 until the end of each experiment unless otherwise described. This regimen of IL-12 was well tolerated with no signs of overt toxicity.
Histopathology
On the day of sacrifice, primary tumors were removed, fixed, sectioned and stained with H&E, and examined by light microscopy for their architecture, evidence of lymphocyte infiltrating and angiogenesis.
Microarray experiment
The high-density oligonucleotide microarray system of Affymetrix (Santa Clara, CA), murine Genome U74A Array version 2 containing 12,488 genes was used. Total RNA was isolated from freshly isolated 4T1 tumors of all surviving mice on day 23. RNA samples of each mouse within each experiment group were pooled from 7–10 mice. Ten micrograms of total RNA was used to synthesize cDNA using Superscript cDNA synthesis kit (Invitrogen, Carlsbad, CA) with a primer containing oligo (dT) and T7 RNA polymerase promoter sequences. Double-stranded cDNA was then purified by phase lock gel (Eppendorf, Westbury, NY) with phenol/chloroform extraction. The purified cDNA was used as a template to generate biotinylated cRNA using the Bioarray High Yield RNA Transcript Labeling Kit (Enzo Biochem, Farmingdale, NY) and then biotinylated cRNAs were fragmented and hybridized to Affymetrix Test 3 chips (Affymetrix, Inc., Santa Clara, CA). All RNA samples passed quality control (ratio of 3′ to 5′ < 3) and then the samples were hybridized to the Murine Genome Array U74Av2 array, which contains 12,488 well-substantiated mouse genes. After overnight hybridization, the arrays were washed, stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) on the GeneChip Fluidics Station (Affymetrix) and scanned according to the standard Affymetrix protocol.
Microarray data collection and analysis
Affymetrix GeneChip 5.0 was used as the image acquisition software for the U74Av2 chips. The signal, which represents the intensity of each gene, was extracted from the image. The target intensity value from each chip was scaled to 250. Data normalization, log transformation, and statistical analysis were performed with GeneSpring software (Silicon Genetics, Redwood City, CA).
Reverse transcription-polymerase chain reaction (RT-PCR)
Primers used for PCR were 1. ASM-like phosphodiesterase 3a (PDE3a): sense- TGGCTGGGTGAAGAAGCTAT, anti-sense- GGAGATAGCCCACTGGAACA; 2. Serum amyloid A 2 (SAA2): sense- GCGAGCCTACACTGACATGA, anti-sense- GGCAGTCCAGGAGGTCTGTA; 3. spi2 proteinase inhibitor (spi2/eb1): sense- ACATTGATGGTGCTGGTGAA, anti-sense- AGTGCAGGACAGCTCCTCAT; 4. Immunoresponsive gene 1 (Irg1): sense- CGTGAGAAAGCACCTTGTGA, anti-sense- CTGTGGAAGGATGGGACAGT; 5. sex-limited protein alpha-gamma chain Slp(w7) : sense- TGCCTTCCGTCTCTTTGAGT; antisense, ACATTTGTTCCGAAGGCATC; 6. IL-12Rβ2: sense- AATTCAGTACCGACGCTCTCA, anti-sense- ATCAGGGGCTCAGGCTCTTCA; 7. hypoxanthineguanine phosphoribosyl transferase (HPRT): sense- GTTGGATACAGGCCAGACTTTGTTG, anti-sense- GAGGGTAGGCTGGCCTATGGCT; 8. RANTES: sense- CCCTCACCATCATCCTCACT, anti-sense- CTTCTTCTCTGGGTTGGCAC; 9. MIG: sense- AGAACTCAGCTCTGCCATGAAGTC, anti-sense-CTAGGCAGGTTTGATCTCCGTTCT; 10. MCP2: sense-GATCTACGCAGTGCTTCTTTGCCT, anti-sense GACATACCCTGCTTGGTCTGGAAA and 11. SDF-1α: sense- GCTCTGCATCAGTGACGGTA, anti-sense-TGGGCTGTTGTGCTTACTTG.
Statistical tests
Tumor growth and metastasis data to be compared were first subjected to normality test. Where the samples studied were normally distributed, statistical comparisons were performed using the Student’s t-test. Where the samples deviated from normality, a nonparametric, Mann Whitney Rank-Sum test was used for comparisons. Statistical analyses were performed using SigmaStat software. For all experiments, the mean and the SD are depicted.
RESULTS
IL-12 induces anti-tumor activities in vivo
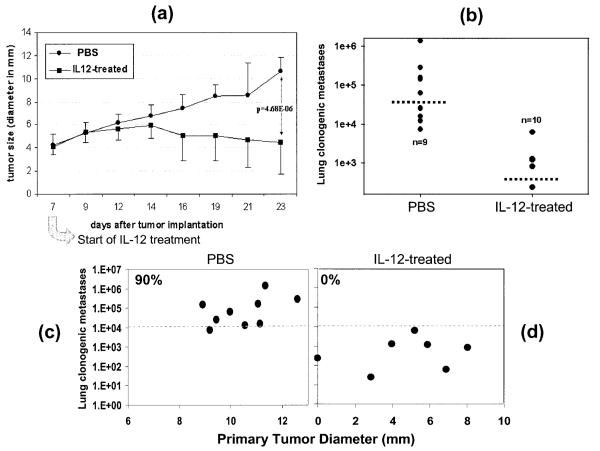
To assess the effects of IL-12 in tumor regression and metastasis, 4T1 mammary carcinoma was initiated by s.c. injection of 4T1 cells into mice on the syngeneic BALB/c background. Recombinant IL-12 was given i.p. starting on day 7 post tumor injection to both types of mice when the primary tumor had grown to ~ 4 mm in diameter. The timing of IL-12 administration was based on potential therapeutic considerations to mimic clinical situations in which breast cancer patients do not get therapy until the presence of malignant growth in the breast has been identified by mammogram or other means. As shown in Figure 1a, by day 23, there were significant differences in tumor growth in mice with or without IL-12 treatment. The pattern of lung metastasis in these mice in response to IL-12 treatment was very similar to that of the primary tumor growth (Fig. 1b). There was a positive correlation between primary tumor size and the number of metastatic cells in the lung, i.e., the larger the tumor, the greater the lung metastases (Fig. 1c,d). IL-12 treatment prevented 4T1 lung metastasis from crossing the lethal threshold of 104 metastatic tumor cells per lung22 in all 10 mice while in nontreated mice (1d), 90% of the mice bore lung metastases in excess of 104 tumor cells (Fig. 1c). IL-12 treatment also reduced the death rate resulting from the tumor from 47% in nontreated mice to 13% in 6 separate studies over the 28-day experimental period.
Figure 1.
IL-12 induces anti-tumor activities in vivo. (a) 4T1 tumor cells were injected s.c. in the abdominal mammary gland with 0.1 ml of a single-cell suspension containing 105 tumor cells. Tumor growth was monitored every 2–3 days and tumor size in diameter (mm) was measured with an electronic caliper. Each data point is comprised of 10 mice. Error bars represent standard deviation. IL-12 treatment started on Day 7 and was injected i.p. every 2 days until day 21. (b) Lung metastasis was measured at the end of this experiment (day 23) by the 6-thiogaunine clonogenicity assay. The dashed horizontal lines represent the mean of each group. (c,d) The same data shown in (a,b) are plotted on X and Y axis to show the correlation between the size of the primary tumors and their ability to metastasize to the lung.
IL-12 enhances lymphocyte infiltration and reduces neovascularization in the tumor
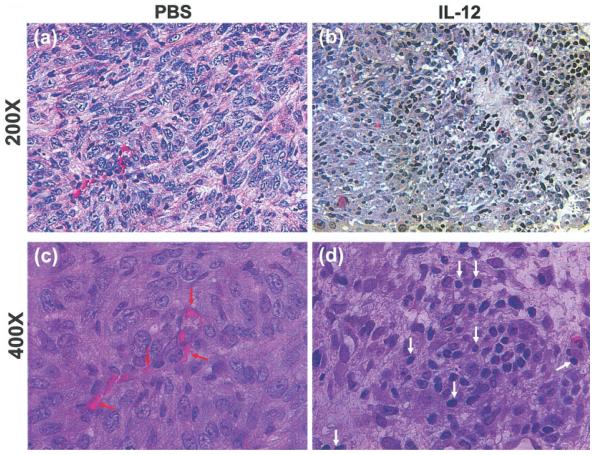
We examined microscopically tumor sections with or without IL-12 treatment (Fig. 2). One of the readily recognizable features of these tumor sections is the large amount of neovascularization in PBS-treated mice (Fig. 2a,c, indicated by red arrows). IL-12 treatment strongly blocked the formation of these vessels (Fig. 2b,d), confirming the well-established role of IL-12 in inhibition of neoangiogenesis.23 In addition, PBS-treated tumors had few infiltrating T lymphocytes (Fig 2a,c). IL-12 treatment dramatically increased the number of tumor-infiltrating lymphocytes (TILs) (Fig. 2b,d, indicated by white arrows). These results are confirmatory for the anti-angiogenic activity of IL-12 and its ability to induce TILs.24
Figure 2.
Angiogenesis and lymphocyte infiltration in primary 4T1 tumors. Parafin-embedded tumor sections were stained with H&E and microscopically examined for blood vessels and tumor infiltrating lymphocytes at 200× (a,b), and 400× (c,d) magnification. Shown for each group is 1 representative slide of 3 tumor samples randomly picked from the 2 experimental groups. Red arrows, blood vessels; white arrows, TILs.
Genome-wide analysis of gene expression in primary tumors
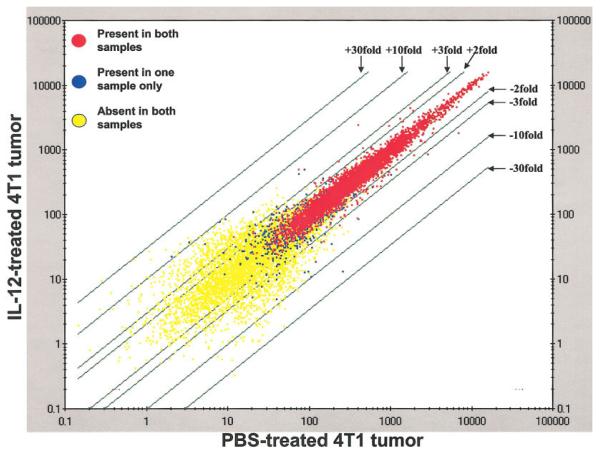
To obtain a detailed appreciation of the molecular events taking place in 4T1 tumors exposed in vivo to IL-12, we performed global gene expression analysis of the 2 groups of tumors on day 23 using the Affymetrix oligonucleotide microarray system (Murine Genome U74A Array version 2 containing 12,488 genes). To reduce variations between individual mice, RNA samples were pooled from all mice within each group for the microarray analysis. We applied this technology to the search of genes that undergo altered expression in 4T1 tumor cells in vivo following IL-12 therapy in an attempt to identify the downstream targets of IL-12. RNA samples were prepared from the 2 experimental groups shown in Figure 1 for comparison of differential gene expression. This microarray experiment (10 mice from each group) yielded a large amount of data, which was processed through the GeneSpring software for i) data normalization (bias correction), ii) data transformation (to ensure normal distribution) and (iii) gene filtering to identify specific genes that are expressed differentially by using appropriate statistic tools. The group of PBS-treated wild-type mice was set as the baseline (with an expression value of 1.0) to which the IL-12-treated group was compared (Fig. 3). Most of the genes exhibit no altered expression. The vast majority of the genes were either present equally (red dots) or absent (yellow dots) in both samples. A small number of genes manifested changes in expression to varying degrees (blue dots) as indicated by the distances they move away from the central line. This microarray experiment was repeated once with high similar results.
Figure 3.
Affymetrix microarray analysis Total RNA samples were extracted from 4T1 tumors excised out of all mice treated or not with rmIL-12 as shown in Figures 1 and 2, and mixed equally within each group to eliminate bias in sampling. The RNA was converted into biotinylated cRNA by in vitro transcription. Microarray hybridization was performed and data graphically represented by scatter plot of the data points. The baseline data points (PBS-treated tumor) are plotted along the X-axis (horizontal), and those of the experimental group (IL-12-treated tumor) along the Y-axis (vertical). The green lines indicate signal intensities above (+) or below (−) the center line, which is set as 1. This microarray experiment was independently repeated with similar results.
Genes that exhibit altered expression following IL-12 in vivo treatment
To further examine the details of the genes differentially expressed in the 2 groups of 4T1 tumor in an effort to understand the molecular mechanisms involved in IL-12-mediated anti-tumor activities, we categorized these genes using additional stringencies, i.e., by choosing only those genes that were expressed 2-fold or higher in IL-12-treated tumors with statistically significant change p values (p < 0.05) and present calls. By these criteria, 52 genes were found to be induced in IL-12-treated tumor by at least 2 fold to a detectable level (called present), compared to PBS-treated tumor (Table Ia), and 17 genes were inhibited by at least 2-fold (Table Ib).
TABLE I.
GENES IN 4T1 TUMOR1
| Genbank Accession |
Signal ratio (log of 2) |
Change p-value | Gene description | Gene function |
|---|---|---|---|---|
| a. Genes that show enhanced expression in 4T1 tumor following IL-12 treatment in vivo | ||||
| AF0659473 | 3.20 | 0 | NOD/LtJ small inducible cytokine A5 (ScyA5) RANTES |
Chemoattractant for Mϕ/DC, T/NK/basophil/ eosinophil |
| AF061569 | 2.60 | 0.00001 | c-myc coding region determinant binding protein |
Stabilizes c-myc mRNA |
| L12029 | 2.30 | 0.000001 | Stromal cell derived factor 1-α | Chemoattractant |
| X064543 | 2.20 | 0 | Sex-limited protein Slp(w7) alpha-gamma chain |
Complement-like |
| U348813 | 2.10 | 0.000649 | CD8 antigen, alpha chain | Cytotoxic and DC marker |
| K027823 | 1.80 | 0.000008 | Complement component C3 mRNA, alpha and beta subunits |
Complement |
| X04673 | 1.80 | 0.000408 | Adipsin | Complement D activity, activates alternative pathway |
| M348153 | 1.80 | 0 | Monokine induced by gamma interferon (MIG); CXCL9 |
Chemoattractant for activated T cells |
| AB0234183 | 1.80 | 0 | monocyte chemoattractant protein-2 (MCP-2), CCL8 |
Chemoattractant for T/monocyte/basophil/ eosinophil |
| AL0786302 | 1.80 | 0.000388 | Mouse genomic DNA sequence from clone 573K1 (EST) |
Unknown |
| M63335 | 1.80 | 0.004863 | lipoprotein lipase | Hydrolyzes triglycerides in lipid metablism |
| X046533 | 1.70 | 0 | Ly-6 alloantigen (Ly-6E.1) | T-lymphocyte activation |
| AF0102543 | 1.50 | 0.000001 | Complement component 1 inhibitor | Inhibitor of C1 and several plasma serine proteases |
| X03505 | 1.50 | 0 | Serum amyloid A 3 | Acute stress response |
| M64085 | 1.50 | 0 | spi2 proteinase inhibitor (spi2/eb1) | Inhibitor of serine proteases |
| AW1243372 | 1.40 | 0.000002 | UI-M-BH2.1-apq-f-08-0-UI.s1 Mus musculus cDNA, 3 end |
Homologous to microsomal glutathione S- transferase 1 |
| AF0612603 | 1.40 | 0.000031 | Immunosuperfamily protein BI2, nectin-like protein 2 (Necl2) |
Cell-cell adhesion and transmembrane protein localization |
| U353303 | 1.40 | 0 | Histocompatibility 2, class II, locus Mb1 | Antigen presentation to CD4+/T cells |
| U069243 | 1.40 | 0 | Signal transducer and activator of transcription 1 (Stat1) |
IFN-γ signaling |
| AF071180 | 1.40 | 0.000001 | N-formylpeptide receptor-like 2 gene | Chemotactic for phagocytes (antibacterial and tissue repair) |
| X009583 | 1.30 | 0 | I-E (beta-b) gene | Antigen presentation to CD4+ T cells |
| Y08135 | 1.30 | 0 | Acid Sphingomyelinase-like phosphodiesterase 3a |
Novel binding protein of tumor suppressor DBCCR1 (deleted in bladder cancer chromosome region 1) |
| AA7903072 | 1.30 | 0 | vw17a10.r1 Mus musculus cDNA, 5′ end (Onzin) |
Unknown |
| U53219 | 1.30 | 0 | GTPase IGTP | Interferon induced GTP-binding protein |
| X78445 | 1.30 | 0.000001 | Cytochrome P450 | Detoxification by oxidation |
| X023393 | 1.30 | 0.000115 | CD3 antigen, delta polypeptide | T-lymphocyte marker |
| AI3860933 | 1.30 | 0.000008 | Similar to protein tyrosine kinase ZAP-70 | T-lymphocyte activation signaling |
| U88566 | 1.20 | 0.000001 | Secreted frizzled related protein sFRP-1 (Sfrp1) |
Inhibitor of Wnt, induces apoptosis |
| V008333 | 1.20 | 0 | E-alpha gene for immune response gene | Antigen presentation to CD4+/T cells |
| U353233 | 1.10 | 0 | H2-M chain, H2-M2, H2-M1, low molecular weight protein 2 (Lmp2) |
Antigen presentation to CD4+ T cells |
| M219323 | 1.10 | 0 | MHC class II H2-I-A-beta gene (k haplotype) | Antigen presentation to CD4+ T cells |
| M16238 | 1.10 | 0 | Fibrinogen-like protein 2 | Thrombosis, cytotoxic T-specific protein |
| D44464 | 1.10 | 0 | Uridine phosphorylase | Catalyzes the reversible phosphorolysis of uridine to uracil |
| M689033 | 1.10 | 0.000001 | Integrin beta 7, Ly69 | Lymphocyte homing and retention |
| AA914345 | 1.10 | 0.000001 | Interferon-inducible GTPase | Interferon induced GTP-binding protein |
| U434283 | 1.10 | 0.000018 | Nitric oxide synthase 2, inducible, macrophage | Synthesis of nitric oxide |
| M55544 | 1.10 | 0 | Guanylate nucleotide binding protein 1 | GTPase |
| AF084524 | 1.10 | 0.000018 | Cellular repressor of E1A-stimulated genes CREG |
Inhibits cell growth and/or promotes cell differentiation |
| AW049768 | 1.10 | 0 | Tubulointersititial nephritis antigen-related protein |
Unknown |
| U12884 | 1.00 | 0 | Vascular cell adhesion molecule-1 truncated form T-VCAM-1 |
Unknown |
| X526433 | 1.00 | 0 | Histocompatibility 2, class II antigen A, alpha | Antigen presentation to CD4+ T cells |
| AI8428282 | 1.00 | 0.000002 | EST UI-M-AP1-agi-h-10-0-UI.s1 Mus musculus cDNA (EST) |
Unknown |
| M55181 | 1.00 | 0.000311 | Preproenkephalin 2 | Five-amino-acid neuropeptide with morphin- like activity |
| AJ0079703 | 1.00 | 0 | GBP-2 protein | GTPase inducible by IFN-γ in macrophages |
| AJ007971 | 1.00 | 0 | IIGP protein | IFN-γ-inducible GTPase |
| M161183 | 1.00 | 0.000009 | T-cell receptor insulin (A-chain) reactive alpha chain VJC |
T-cell receptor |
| X75129 | 1.00 | 0 | Xanthine dehydrogenase | Catalyzes oxidation of hypoxanthine, xanthine, and purine. |
| AA9589032 | 1.00 | 0.000001 | EST: ua19f08.r1 Mus musculus cDNA (EST) | Unknown |
| AW0474762 | 1.00 | 0 | EST: UI-M-BH1-all-g-04-0-UI.s1 Mus musculus cDNA (EST) |
Unknown |
| D317883 | 1.00 | 0.000001 | BP-3 alloantigen, bone marrow stromal cell antigen 1 (BST-1) |
CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities |
| AJ006474 | 1.00 | 0.000004 | Carbonic anhydrase III | Protects proteins from oxidation catalyzed by iron-containing degradation products of haemoglobin and myoglobin |
| AV2396532 | 1.00 | 0.000006 | EST: Mus musculus cDNA, 3′ end/clone = 4732435F04 (EST) |
Unknown |
| b. Genes that show reduced expression in 4T1 tumor following IL-12 treatment | ||||
| AA592182 | −1.8 | 0 | Nephronectin | Extracellular matrix protein associated with integrin α8β1 |
| AW0478752 | −1.8 | 0 | EST, UI-M-BH1-als-g-03-0-UI.s1 | Unknown |
| Y13185 | −1.7 | 0.000002 | Matrix metalloproteinase 10 | Modulator of extracellular matrix composition and structure |
| M76124 | −1.6 | 0 | EGP314, epithelial glycoprotein | Involved in metastasis |
| Y17793 | −1.6 | 0.000063 | Roundabout homolog 1, mouse homologue of human DUTT1 |
Lung development |
| M83219 | −1.5 | 0 | MRP14, myeloid related protein | Neutrophil and monocyte activation |
| U43327 | −1.4 | 0 | Lamc2, Laminin, gamma 2 | Constituent of lamin-5, a major adhesion ligand of epithelial cells |
| AJ242625 | −1.4 | 0.000001 | Dmp-1, dentin matrix protein-1 | Involved in matrix mineralization |
| M83218 | −1.3 | 0 | MRP8, intracellular calcium-binding protein | Neutrophil and monocyte activation |
| U88064 | −1.3 | 0 | Basonuclin | Increases transcription of the ribosomal RNA genes |
| AF087825 | −1.3 | 0.000011 | Claudin 7 | A transmembrane protein that seals tight junctions |
| AA7169632 | −1.2 | 0 | EST, vu69f10.r1 | Unknown |
| AF017994 | −1.2 | 0.000008 | Peg1/MEST | Imprinted gene of unknown function expressed in mouse embryo |
| C766433 | −1.2 | 0.000002 | EST, D15Ertd30e | Unknown |
| X537983 | −1.1 | 0.000002 | MIP2, macrophage inflammatory protein-2 | Chemotactic for monocytes/macrophages |
| AJ2429123 | −1.1 | 0.000003 | Decysin, disintegrin metalloprotease | Expressed in macrophages and dendritic cells |
| AV112006 | −1.1 | 0.000025 | Procollagen, type XV | Precursor of constituent of skin and bone |
The signal ratios given (in log of 2) have been normalized as follows the 50th percentile of all measurements was used as a positive control for each sample; each measurement for each gene was divided by this synthetic positive control, assuming that this was at least 10. The bottom tenth percentile was used as a test for correct background subtraction. This was never less than the negative of the synthetic positive control. Each gene was normalized to itself by making a synthetic positive control for that gene and dividing all measurements for that gene by this positive control, assuming it was at least 0.01. This synthetic control was the median of the gene’s expression values over all the samples. Lastly, normalized values below 0 were set to 0. Change p values reflect statistical significance in differential expression between the 2 groups (IL-12-treated vs. PBS-treated).
EST clones.
Genes with primary immune cell origins.
The gene lists include only those that were reproduced in 2 separate experiments (72% reproducibility rate).
It is clear from a glance at the gene list in Table Ia that IL-12 in vivo treatment induced a whole host of genes involved in many aspects of its antitumor activities: metabolism, cell cycle, proliferation, apoptosis, metastasis, antigen presentation, chemotaxis, immune response etc. in the form of the altered expression of genes that encode surface, intracellular and extracellular molecules. Many of the genes have well-defined functions; others are poorly or not at all characterized (EST clones). Of the 45 induced genes with known functions, 23 are likely derived primarily frominfiltrating lymphocytes and phagocytes such as IFN-γ-induced proteins, chemokines, complement factors, Fas ligand, CD8, CD3, Ly-6E.1, MHC class II antigens, iNOS etc., likely representing an IL-12-induced, concerted response against the developing mammary tumor. The remaining 22 genes are probably expressed by the tumor cells or the stroma.
In contrast to IL-12-induced genes, the list of IL-12-inhibited genes (Table Ib) were predominantly expressed in cells of nonhematopoietic origin.
Verification of differential gene expression in vitro and in vivo
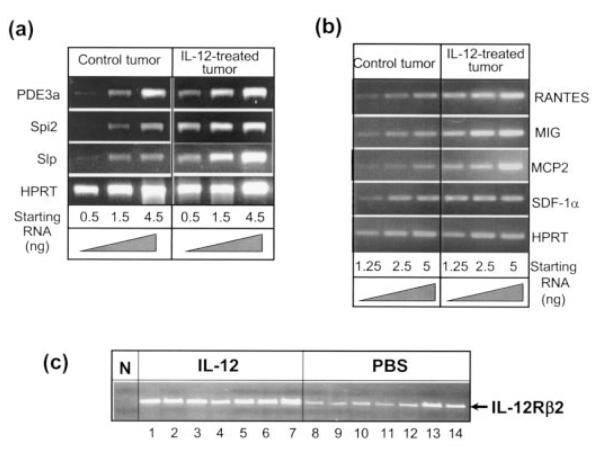
We randomly selected 3 genes from this list [ASM-like phosphodiesterase 3a (PDE3a), spi2 proteinase inhibitor (spi2/eb1) and sex-limited protein alpha-gamma chain (Slpw7)] and performed RT-PCR to verify their status of differential mRNA expression between PBS-treated and IL-12-treated tumors (Fig. 4a). All 3 genes exhibited significantly differential expression between PBS and IL-12 treatments. The expression pattern was then compared to 4T1 cells treated in vitro with IL-12 for 24, 48 and 72 hr. In contrast to 4T1 tumor cells treated in vivo with IL-12, the expression of PDE3a, Spi2 and Slp was not detectable in vitro in IL-12-treated or PBS-treated samples (data not shown). These results suggest that the induction of many genes shown in Table I may require additional factors other than IL-12. These factors are available in vivo presumably provided by other cells but not in vitro in isolated 4T1 cells.
Figure 4.
Confirmation of differential mRNA expression by RT-PCR. (a) Reverse transcription (RT) was performed with the same total RNA samples used for the microarray experiment followed by PCR amplification using appropriately designed primers for each cDNA. Thirty-four cycles of amplification were carried out with serially diluted cDNA samples. The equivalent corresponding starting amount of total RNA each serial dilution represents is indicated at the bottom. (b) RT-PCR was performed as described in (a) with RNA samples taken from 4T1 cells treated or not in vivo with rIL-12 for RANTES, SDF-1α, MIG and MCP-2. (c) RT-PCR was performed to examine the mRNA expression of IL-12Rβ2 in 4T1 tumors exposed to IL-12 or not in vivo. Each lane represents an individual tumor tissue isolated from one mouse. Lanes 1–7, tumors from IL-12-treated mice; lanes 8 –14, tumors from PBS-treated mice. N, negative control (PCR without cDNA added).
Several chemokines were notably stimulated by the IL-12 treatment. This microarray data was also confirmed by RT-PCR for regulated upon activation of normal T cells expressed and secreted (RANTES), stroma cell-derived factor-1α (SDF-1α), monokine induced by γ interferon (MIG, CXCL9) and monocyte chemoattractant protein-2 (MCP-2, CCL8) (Fig. 4b).
We also examined the expression of IL-12Rβ2 mRNA expression by RT-PCR in the primary 4T1 tumors with or without the IL-12 treatment (Fig. 4c). It appears that IL-12Rβ2 was expressed in tumors tissues without exogenous IL-12, and the IL-12 treatment significantly enhanced its expression. These results suggest that 4T1 tumors are able to respond to IL-12 directly because they express the critical IL-12Rβ2 chain.25 However, presently it’s not clear whether it is the 4T1 tumor cells themselves or the TILs that express IL-12R.
DISCUSSION
IL-12 has potent antitumor activities with strong clinical relevance to cancer therapy. However, the molecular mechanisms that underline these activities of IL-12 are poorly understood, presenting an obstacle to its clinical applications. The current work constitutes an important step towards comprehensively identifying the key factors involved in host response induced by IL-12 against a developing malignancy.
The approach that we took to examine the global gene expression profile in the whole tumor with its stroma and lymphocytic infiltrates, as opposed to looking at certain purified populations of cells within the tumor, provides us with the ability to view all of the transcriptional changes (limited by the number of genes on the microarray and by the detection limit) effected by IL-12, thus permitting a more comprehensive analysis of the interactions between malignant cells and its microenvironment without potential perturbations to the various cell populations embodied in the tumor due to in vitro manipulation. The counter argument is that this “wholesome” approach would not allow the identification of specific cell types that may play a more disproportionate role in IL-12-mediated anti-tumor responses than other cells. One of the ways to circumvent this dilemma is to analyze whole tumor mass in mice that are genetically deficient in producing certain immune cell types.
A prominent feature among the genes induced by IL-12 in vivo is the production of chemokines: RANTES, SDF-1α, MIG, MCP-2, N-formylpeptide receptor-like 2. The production of these chemotactic factors may be responsible for the influx of TILs observed in 4T1 tumors treated with IL-12 (Fig. 2d).
Some of the 22 IL-12-induced genes apparently of nonhematopoietic origins are recognized to be associated with tumor growth and/or metastasis. Take adipsin and lipoprotein ligase for example. Induction of adipocyte-specific gene expression including these 2 genes in the rat mammary carcinoma is correlated with tumor regression by the retinoid X receptor-ligand LGD1069 (targretin).26 Phosphodiesterase (PDE) 3a and 3b were detected in human neoplastic submandibular gland intercalated duct HSG cells. The PDE3-specific inhibitor, cilostamide, inhibited the growth of HSG cells. PDE3 thus appears to be a potential target for antiproliferative therapies.27 Cellular repressor of E1A-stimulated gene(CREG) is a secreted glycoprotein during differentiation of embryonic stem cells and is able to enhance differentiation of the human embryonal carcinoma cell line NTERA-2 characterized by changes in morphology, altered patterns of gene expression, reduced proliferative potential and a loss of tumorigenicity.28 Therefore, induction of the expression of these growth-inhibiting genes by IL-12 may impede the succession of the mammary tumor.
One of the interesting novel genes that were induced by IL-12 treatment is onzin whose function is currently unknown. Human onzin was identified as a differentially expressed gene in a subtractive hybridization screening between IL-3/CD40L-activated plasmacytoid DCs (tester) and CD40-activated monocyte-derived DCs (driver). This novel gene, also called C-15, encodes a putative cytokine because its deduced polypeptide is composed of 112 aa (115 aa for mouse C-15), including a 23-aa signal peptide which allows its secretion. Onzin/C-15 is expressed in different hematopoietic cells of T, B, monocyte, granulocyte lineages.29 Human C-15 contains 16 cysteines, 15 of which are conserved in the mouse homologue. So it could also be a member of the defensin-like molecules given that defensins are a family of small peptides with 3 or 4 intramolecular cysteine disulfide bonds.30 Onzin/C-15 was also identified as the most strongly stimulated gene in a microarray analysis of draining auricular lymph node tissue sampled at 48 hr following exposure to the potent contact allergen 2,4-dinitrofluorobezene (DNFB).31 Expression of onzin/C-15 has been reported to be inhibited by c-myc in the myeloid cell line 32D,32 suggestive of a role in cell cycling. Taken together, onzin/C-15 could be an important player in IL-12-mediated activation and migration of DCs and lymphocytes to confer its anti-tumor activity.
The most remarkable feature among the 17 genes whose mRNA expression was inhibited by the IL-12 treatment in vivo is the dominance of genes involved in extracellular matrix (ECM) remodeling and cell adhesion functions. Structural changes in ECM are necessary for cell migration during normal and pathologic tissue remodeling and neoplastic cell invasion. The matrix metalloproteinases (MMPs) and their inhibitors are critical modulators of ECM composition and are thus important in neoplastic cell progression, invasion and metastasis.33–36 Strong expression of MMP-3 and -10 was found in childhood astrocytomas,37 in lung adenocarcinomas,38 especially in the ECM adjacent to blood vessels. Expression of MMPs and their inhibitors correlates with invasion and metastasis in squamous cell carcinoma of the head and neck.39 EGP314 is a panepithelial glycoprotein.40 Transfection of a low metastasizing fibrosarcoma, pheochromoblastoma and adenocarcinoma with the rat ortholog of human and mouse EGP314, D5.7A facilitated tumor metastasis via the lymphatic system or hematogeneously, depending on the origin of the tumor.41 Particularly after proteolytic cleavage, D5.7A exhibited enhanced cell-cell adhesion and provided a proliferative signal upon crosslinking.41 PEG1/MEST is an imprinted gene involved in the growth of the fetus and/or placenta.42 Pedersen et al.43 reported that monoallelic PEG1 expression was observed in normal breast tissues, indicating the presence of a functional imprint. However, they noted a loss of imprinting in all informative invasive breast carcinomas examined, possibly involving a novel mechanism of promoter switch that causes biallelic PEG1expression.44 Thus, inhibition of the expression of these tumor/metastasis-promoting genes by IL-12 may directly blunt the progression of the malignancy.
In summary, the comprehensive analysis of gene expression in the 4T1 tumor model following IL-12 treatment uncovers many candidates that may be involved in IL-12-regulated immune responses against tumor progression, or in direct modification of the cancer cell, its stroma, and the microenvironment such that the growth and metastasis of the tumor is retarded. It also provides a rich pool of prospective targets desired for the development of therapeutic agents to control breast cancer.
REFERENCES
- 1.Browning MJ, Bodmer WF. MHC antigens and cancer: implications for T-cell surveillance. Curr Opin Immunol. 1992;4:613–8. doi: 10.1016/0952-7915(92)90036-e. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Ashe S, Brady WA, Hellstrom I, Hellstrom KE, Ledbetter JA, McGowan P, Linsley PS. Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell. 1992;71:1093–102. doi: 10.1016/s0092-8674(05)80059-5. [DOI] [PubMed] [Google Scholar]
- 3.Naor D. Suppressor cells: permitters and promoters of malignancy? Adv Cancer Res. 1979;29:45–125. doi: 10.1016/s0065-230x(08)60846-5. [DOI] [PubMed] [Google Scholar]
- 4.Townsend SE, Allison JP. Tumor rejection after direct costimulation of CD8+ T cells by B7- transfected melanoma cells. Science. 1993;259:368–70. doi: 10.1126/science.7678351. [DOI] [PubMed] [Google Scholar]
- 5.Coughlin CM, Salhany KE, Gee MS, LaTemple DC, Kotenko S, Ma X, Gri G, Wysocka M, Kim JE, Liu L, Liao F, Farber JM, et al. Tumor cell responses to IFNgamma affect tumorigenicity and response to IL-12 therapy and antiangiogenesis. Immunity. 1998;9:25–34. doi: 10.1016/s1074-7613(00)80585-3. [DOI] [PubMed] [Google Scholar]
- 6.Trinchieri G, Scott P. Interleukin-12: basic principles and clinical applications. Curr Top Microbiol Immunol. 1999;238:57–78. doi: 10.1007/978-3-662-09709-0_4. [DOI] [PubMed] [Google Scholar]
- 7.Rook AH, Wood GS, Yoo EK, Elenitsas R, Kao DM, Sherman ML, Witmer WK, Rockwell KA, Shane RB, Lessin SR, Vonderheid EC. Interleukin-12 therapy of cutaneous T-cell lymphoma induces lesion regression and cytotoxic T-cell responses. Blood. 1999;94:902–8. [PubMed] [Google Scholar]
- 8.Rook AH, Zaki MH, Wysocka M, Wood GS, Duvic M, Showe LC, Foss F, Shapiro M, Kuzel TM, Olsen EA, Vonderheid EC, Laliberte R, et al. The role for interleukin-12 therapy of cutaneous T cell lymphoma. Ann N Y Acad Sci. 2001;941:177–84. doi: 10.1111/j.1749-6632.2001.tb03721.x. [DOI] [PubMed] [Google Scholar]
- 9.Ansell SM, Witzig TE, Kurtin PJ, Sloan JA, Jelinek DF, Howell KG, Markovic SN, Habermann TM, Klee GG, Atherton PJ, Erlichman C. Phase 1 study of interleukin-12 in combination with rituximab in patients with B-cell non-Hodgkin lymphoma. Blood. 2002;99:67–74. doi: 10.1182/blood.v99.1.67. [DOI] [PubMed] [Google Scholar]
- 10.Mortarini R, Borri A, Tragni G, Bersani I, Vegetti C, Bajetta E, Pilotti S, Cerundolo V, Anichini A. Peripheral burst of tumor-specific cytotoxic T lymphocytes and infiltration of metastatic lesions by memory CD8+ T cells in melanoma patients receiving interleukin 12. Cancer Res. 2000;60:3559–68. [PubMed] [Google Scholar]
- 11.Gollob JA, Mier JW, Veenstra K, McDermott DF, Clancy D, Clancy M, Atkins MB. Phase I trial of twice-weekly intravenous interleukin 12 in patients with metastatic renal cell cancer or malignant melanoma: ability to maintain IFN-gamma induction is associated with clinical response. Clin Cancer Res. 2000;6:1678–92. [PubMed] [Google Scholar]
- 12.Lee P, Wang F, Kuniyoshi J, Rubio V, Stuges T, Groshen S, Gee C, Lau R, Jeffery G, Margolin K, Marty V, Weber J. Effects of interleukin-12 on the immune response to a multipeptide vaccine for resected metastatic melanoma. J Clin Oncol. 2001;19:3836–47. doi: 10.1200/JCO.2001.19.18.3836. [DOI] [PubMed] [Google Scholar]
- 13.Kang WK, Park C, Yoon HL, Kim WS, Yoon SS, Lee MH, Park K, Kim K, Jeong HS, Kim JA, Nam SJ, Yang JH, et al. Interleukin 12 gene therapy of cancer by peritumoral injection of transduced autologous fibroblasts: outcome of a phase I study. Hum Gene Ther. 2001;12:671–84. doi: 10.1089/104303401300057388. [DOI] [PubMed] [Google Scholar]
- 14.Gajewski TF, Fallarino F, Ashikari A, Sherman M. Immunization of HLA-A2+ melanoma patients with MAGE-3 or MelanA peptide-pulsed autologous peripheral blood mononuclear cells plus recombinant human interleukin 12. Clin Cancer Res. 2001;7:895s–901s. [PubMed] [Google Scholar]
- 15.Portielje JE, Kruit WH, Schuler M, Beck J, Lamers CH, Stoter G, Huber C, de Boer-Dennert M, Rakhit A, Bolhuis RL, Aulitzky WE. Phase I study of subcutaneously administered recombinant human interleukin 12 in patients with advanced renal cell cancer. Clin Cancer Res. 1999;5:3983–9. [PubMed] [Google Scholar]
- 16.Ansari AA, Mayne AE, Sundstrom JB, Bostik P, Grimm B, Altman JD, Villinger F. Administration of recombinant rhesus interleukin-12 during acute simian immunodeficiency virus (SIV) infection leads to decreased viral loads associated with prolonged survival in SIV-mac251-infected rhesus macaques. J Virol. 2002;76:1731–43. doi: 10.1128/JVI.76.4.1731-1743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller FR, Miller BE, Heppner GH. Characterization of metastatic heterogeneity among subpopulations of a single mouse mammary tumor: heterogeneity in phenotypic stability. Invasion Metastasis. 1983;33:22–31. [PubMed] [Google Scholar]
- 18.Pulaski BA, Ostrand-Rosenberg S. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res. 1998;58:1486–93. [PubMed] [Google Scholar]
- 19.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Research. 1992;52:1399–405. [PubMed] [Google Scholar]
- 20.Morecki S, Yacovlev L, Slavin S. Effect of indomethacin on tumor-igenicity and immunity induction in a murine model of mammary carcinoma. Int J Cancer. 1998;75:894–9. doi: 10.1002/(sici)1097-0215(19980316)75:6<894::aid-ijc12>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Rakhmilevich AL, Janssen K, Hao Z, Sondel PM, Yang NS. Interleukin-12 gene therapy of a weakly immunogenic mouse mammary carcinoma results in reduction of spontaneous lung metastases via a T-cell-independent mechanism. Cancer Gene Ther. 2000;7:826–38. doi: 10.1038/sj.cgt.7700176. [DOI] [PubMed] [Google Scholar]
- 22.Pulaski BA, Clements VK, Pipeling MR, Ostrand-Rosenberg S. Immunotherapy with vaccines combining MHC class II/CD80+ tumor cells with interleukin-12 reduces established metastatic disease and stimulates immune effectors and monokine induced by interferon gamma. Cancer Immunol Immunother. 2000;49:34–45. doi: 10.1007/s002620050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerbel RS, Hawley RG. Interleukin 12: newest member of the anti-angiogenesis club. J Natl Cancer Inst. 1995;87:557–9. doi: 10.1093/jnci/87.8.557. [DOI] [PubMed] [Google Scholar]
- 24.Mortarini R, Borri A, Tragni G, Bersani I, Vegetti C, Bajetta E, Pilotti S, Cerundolo V, Anichini A. Peripheral burst of tumor-specific cytotoxic T lymphocytes and infiltration of metastatic lesions by memory CD8+ T cells in melanoma patients receiving interleukin 12. Cancer Res. 2000;60:3559–68. [PubMed] [Google Scholar]
- 25.Szabo SJ, Dighe AS, Gubler U, Murphy KM. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med. 1997;185:817–24. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal VR, Bischoff ED, Hermann T, Lamph WW. Induction of adipocyte-specific gene expression is correlated with mammary tumor regression by the retinoid X receptor-ligand LGD1069 (targretin) Cancer Res. 2000;60:6033–8. [PubMed] [Google Scholar]
- 27.Murata T, Sugatani T, Shimizu K, Manganiello VC, Tagawa T. Phosphodiesterase 3 as a potential target for therapy of malignant tumors in the submandibular gland. Anticancer Drugs. 2001;12:79–83. doi: 10.1097/00001813-200101000-00011. [DOI] [PubMed] [Google Scholar]
- 28.Veal E, Groisman R, Eisenstein M, Gill G. The secreted glycoprotein CREG enhances differentiation of NTERA-2 human embryonal carcinoma cells. Oncogene. 2000;19:2120–8. doi: 10.1038/sj.onc.1203529. [DOI] [PubMed] [Google Scholar]
- 29.Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint Vis B, Briere F, Bates EE. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 30.Hoover DM, Chertov O, Lubkowski J. The structure of human beta-defensin-1: new insights into structural properties of beta-defensins. J Biol Chem. 2001;276:39021–6. doi: 10.1074/jbc.M103830200. [DOI] [PubMed] [Google Scholar]
- 31.Betts CJ, Moggs JG, Caddick HT, Cumberbatch M, Orphanides G, Dearman RJ, Ryan CA, Hulette BC, Gerberick G Frank, Kimber I. Assessment of glycosylation-dependent cell adhesion molecule 1 as a correlate of allergen-stimulated lymph node activation. Toxicology. 2003;185:103–17. doi: 10.1016/s0300-483x(02)00598-x. [DOI] [PubMed] [Google Scholar]
- 32.Nesbit CE, Tersak JM, Grove LE, Drzal A, Choi H, Prochownik EV. Genetic dissection of c-myc apoptotic pathways. Oncogene. 2000;19:3200–12. doi: 10.1038/sj.onc.1203636. [DOI] [PubMed] [Google Scholar]
- 33.Liotta LA, Rao CN, Barsky SH. Tumor invasion and the extracellular matrix. Lab Invest. 1983;49:636–49. [PubMed] [Google Scholar]
- 34.Mignatti P, Robbins E, Rifkin DB. Tumor invasion through the human amniotic membrane: requirement for a proteinase cascade. Cell. 1986;47:487–98. doi: 10.1016/0092-8674(86)90613-6. [DOI] [PubMed] [Google Scholar]
- 35.Matrisian LM. The matrix-degrading metalloproteinases. Bioessays. 1992;14:455–63. doi: 10.1002/bies.950140705. [DOI] [PubMed] [Google Scholar]
- 36.Ossowski L. Invasion of connective tissue by human carcinoma cell lines: requirement for urokinase, urokinase receptor, and interstitial collagenase. Cancer Res. 1992;52:6754–60. [PubMed] [Google Scholar]
- 37.Bodey B, Bodey B, Jr., Siegel SE, Kaiser HE. Matrix metalloproteinase expression in childhood astrocytomas. Anticancer Res. 2000;20:3287–92. [PubMed] [Google Scholar]
- 38.Bodey B, Bodey B, Jr., Groger AM, Siegel SE, Kaiser HE. Invasion and metastasis: the expression and significance of matrix metalloproteinases in carcinomas of the lung. In Vivo. 2001;15:175–80. [PubMed] [Google Scholar]
- 39.P OC, Rhys-Evans PH, Eccles SA. Expression of matrix metalloproteinases and their inhibitors correlates with invasion and metastasis in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2001;127:813–20. [PubMed] [Google Scholar]
- 40.Claas C, Herrmann K, Matzku S, Moller P, Zoller M. Developmentally regulated expression of metastasis-associated antigens in the rat. Cell Growth Diff. 1996;7:663–78. [PubMed] [Google Scholar]
- 41.Wurfel J, Rosel M, Seiter S, Claas C, Herlevsen M, Weth R, Zoller M. Metastasis-association of the rat ortholog of the human epithelial glycoprotein antigen EGP314. Oncogene. 1999;18:2323–34. doi: 10.1038/sj.onc.1202542. [DOI] [PubMed] [Google Scholar]
- 42.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–9. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen IS, Dervan PA, Broderick D, Harrison M, Miller N, Delany E, O’Shea D, Costello P, McGoldrick A, Keating G, Tobin B, Gorey T, et al. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59:5449–51. [PubMed] [Google Scholar]
- 44.Pedersen IS, Dervan P, McGoldrick A, Harrison M, Ponchel F, Speirs V, Isaacs JD, Gorey T, McCann A. Promoter switch: a novel mechanism causing biallelic PEG1/MEST expression in invasive breast cancer. Hum Mol Genet. 2002;11:1449–53. doi: 10.1093/hmg/11.12.1449. [DOI] [PubMed] [Google Scholar]