Abstract
Induced pluripotent stem (iPS) cells are derived by epigenetic reprogramming, but their DNA methylation patterns have not yet been analyzed on a genome-wide scale. Here, we find substantial hypermethylation and hypomethylation of cytosine-phosphate-guanine (CpG) island shores in nine human iPS cell lines as compared to their parental fibroblasts. The differentially methylated regions (DMRs) in the reprogrammed cells (denoted R-DMRs) were significantly enriched in tissue-specific (T-DMRs; 2.6-fold, P < 10−4) and cancer-specific DMRs (C-DMRs; 3.6-fold, P < 10−4). Notably, even though the iPS cells are derived from fibroblasts, their R-DMRs can distinguish between normal brain, liver and spleen cells and between colon cancer and normal colon cells. Thus, many DMRs are broadly involved in tissue differentiation, epigenetic reprogramming and cancer. We observed colocalization of hypomethylated R-DMRs with hypermethylated C-DMRs and bivalent chromatin marks, and colocalization of hypermethylated R-DMRs with hypomethylated C-DMRs and the absence of bivalent marks, suggesting two mechanisms for epigenetic reprogramming in iPS cells and cancer.
Induced pluripotent stem (iPS) cells can be derived from somatic cells by introduction of a small number of genes: for example, POU5F1, MYC, KLF4 and SOX21–4. As direct derivatives of an individual’s own tissue, iPS cells offer considerable therapeutic promise5, avoiding both immunologic and ethical barriers to their use. iPS cells differ from their somatic parental cells epigenetically, and thus a comprehensive comparison of the epigenome in iPS and somatic cells would provide insight into the mechanism of tissue reprogramming. Although two recent targeted studies6,7 examined a subset of the genome—7,000 (ref. 6) and 66,000 (ref. 7) CpG sites—in a small cohort of three iPS-fibroblast pairs, a global assessment of genome-wide methylation has not yet been performed.
Recently, we described differential methylation patterns that distinguish among normal tissue types (T-DMRs) and patterns that can segregate colorectal cancer tissue from matched normal tissues (C-DMRs)8. Unexpectedly, these two DMRs occur 13-fold more frequently at CpG island ‘shores’, regions of comparatively low CpG density that are located near traditional CpG islands, than at the CpG islands themselves. Cancers showed approximately equal numbers of hypomethylated and hypermethylated regions, and 45% of C-DMRs overlapped T-DMRs, suggesting that epigenetic changes in cancer involve reprogramming of the normal pattern of tissue-specific differentiation8.
Here we used a similar approach to the question of iPS cell reprogramming, first comparing six human iPS cell lines to the fibroblasts from which they were derived using comprehensive high-throughput array-based relative methylation (CHARM) analysis9. This approach allows the interrogation of ~4.6 million CpG sites genome-wide using a custom designed NimbleGen HD2 microarray, including almost all CpG islands and shores in the human genome. Genomic DNA from iPS cells3,5, their parental fibroblasts and human embryonic stem (hES) cells (Online Methods) was digested with the enzyme McrBC, fractionated, labeled and hybridized to a CHARM array.
A total of 4,401 regions (including 96,404 CpG sites) were found to differ in iPS cell lines from the fibroblasts of origin (Table 1, Supplementary Table 1) at a false discovery rate (FDR) of 5%; we term these regions R-DMRs. Of these R-DMRs, DMRs that were hypermethylated in iPS cells compared to fibroblasts predominated over hypomethylated DMRs (60%:40%). Of the 4,401 DMRs, 1,969 were within 2 kb of the transcriptional start site of a gene.
Table 1.
Differentially methylated regions (DMRs) found by CHARM that overlap with tissue-specific differentially methylated regions (T-DMRs)
| DMRs | Total no. | Meth. | Total DMRs | DMRs within 2 kb of a gene TSS | Overlap with T-DMRs |
|||
|---|---|---|---|---|---|---|---|---|
| No. | Fold enrich. | P value | ||||||
| iPS-fib. R-DMRsa (n = 6) | iPS > Fib. | 2,663 | 1,278 | Observed | 1,425 | 2.51 | <0.0001 | |
| 4,401 | Random | 568 | ||||||
| Fib > iPS | 1,738 | 691 | Observed | 1,038 | 2.67 | <0.0001 | ||
| Random | 389 | |||||||
| iPS-ES DMRsb (n = 3) | 71 | iPS > ES | 51 | 23 | Observed | 37 | 3.32 | <0.0001 |
| Random | 11 | |||||||
| ES > iPS | 20 | 9 | Observed | 9 | 2.00 | 0.020 | ||
| Random | 4 | |||||||
| iPS-fib. R-DMRsb (n = 3) | 2,179 | iPS > Fib. | 988 | 497 | Observed | 630 | 3.30 | <0.0001 |
| Random | 191 | |||||||
| Fib > iPS | 1,191 | 384 | Observed | 679 | 2.86 | <0.0001 | ||
| Random | 237 | |||||||
Fib., fibroblast; meth., methylation; enrich., enrichment.
FDR cutoff of 0.05.
Absolute area cutoff of 10.0.
P value based on 10,000 permutations. Random values are averages over all 10,000 iterations.
The genes that were associated with these R-DMRs showed functionally important features based on bioinformatic analyses. First, gene ontology (GO) annotation analysis of these genes revealed significant enrichment for genes involved in developmental and regulatory processes (Supplementary Table 2). For example, 38% of the genes that were hypomethylated in iPS compared to fibroblasts (P = 3.56 × 10−60) and 22% of the genes that were hypermethylated in iPS compared to fibroblasts (P = 1.73 × 10−12) were involved in developmental processes. To further elucidate the functional significance of these R-DMRs, we looked at their overlap with bivalent domains, which mark developmental genes in embryonic stem (ES) cells10,11. Notably, 65% of the R-DMRs that were hypomethylated in iPS cells compared to fibroblasts showed significant association with bivalent domain marks (P < 0.0001 by 10,000 permutations), whereas only 18.6% of hypermethylated R-DMRs overlapped with these domains (P = 0.5699 by 10,000 permutations) (Supplementary Table 3). Furthermore, when we observed the overlap of the R-DMRs with known binding sites for pluripotency markers such as POU5F1, NANOG and SOX2 (ref. 12), we saw a similar relationship, in which the hypomethylated R-DMRs showed significant overlap (P < 0.0001 by 10,000 permutations) whereas the hypermethylated DMRs did not (P = 1 by 10,000 permutations; Supplementary Table 4). These observations indicate that the sites of demethylation during reprogramming of fibroblasts to iPS cells are tightly linked to genes that are functionally important for pluripotency.
The R-DMRs showed several noteworthy features. First, over 70% of the R-DMRs were associated with CpG island shores rather than with the associated CpG islands (Fig. 1a), regardless of whether the R-DMRs were hypermethylated or hypomethylated in iPS cells relative to fibroblasts (Supplementary Fig. 1a). Second, 56% of R-DMRs overlapped T-DMRs previously identified as distinguishing tissues representing the three germ cell lineages, namely, brain, liver and spleen8 (Table 1). This overlap was statistically significant (P < 0.0001 by 10,000 permutations). Furthermore, both hypermethylated and hypomethylated R-DMRs in iPS cells showed similar overlap with known T-DMRs, overlapping at 54% and 60%, respectively (Table 1). Thus, R-DMRs are heavily enriched in CpG island shores and largely overlap T-DMRs that are involved in normal development. There was also a 61% overlap of the gene-proximal R-DMRs with the T-DMRs.
Figure 1.
Reprogramming differentially methylated regions (R-DMRs). (a) Enrichment of R-DMRs at CpG island shores. The CHARM array (left, labeled CpG regions) is enriched in CpG islands, and the R-DMRs (right, labeled R-DMR) show marked enrichment at CpG island shores. Islands are denoted as regions that include >50% of a CpG island or are wholly contained in an island, and overlap regions are denoted as regions that include 0.1–50% of a CpG island. Specific base intervals of regions not overlapping islands are indicated; (0–500) means from 1 to 500 bases. Percentage of the distribution (y axis) is given for the CpG regions (CHARM array, null hypothesis) and reprogramming differentially methylated regions (R-DMRs). (b,c) Examples of DMRs. The gene encoding bone morphogenetic protein 7 (BMP7) is indicated in b, and the gene encoding goosecoid (GSC) is indicated in c. In each case, the upper panels show a plot of methylation (M value; see Online Methods) versus genomic location, where the curve represents averaged smoothed M values; the location of CpG dinucleotides (black tick marks), CpG density, location of CpG islands (orange line), as well as the gene annotation are shown. The bottom panels show validation by bisulfite pyrosequencing (mapping to red box in upper panel). Bars represent the mean methylation (triplicate measurement) ± s.d. of iPS cells (pink), fibroblasts (gray) and ES cells (blue) as well as the generally highly methylated HCT116 colon cancer cell line and a generally hypomethylated double DNA methyltransferase 1/3B double knockout line (DKO) derived from it. In each case, five separate CpG sites were assayed quantitatively, shown as differing shades.
We then repeated the CHARM analysis on a separate set of three iPS cell lines and the fibroblasts from which they were derived, as well as three human ES cell lines. We could not perform an FDR statistical test on this smaller number of lines, so we used a similar area cutoff in the curves that corresponded in magnitude to the 5% FDR cutoff of the previous experiment. In this second analysis, 2,179 R-DMRs were identified, with a slight excess of hypomethylated versus hypermethylated DMRs (55% compared to 45%) in iPS cells. Notably, 80% of the DMRs overlapped those found in the first experiment (see Supplementary Table 5 for full list). As in the first analysis, there was a substantial enrichment for CpG island shores (78%, Supplementary Fig. 1b), and 60% of the R-DMRs overlapped T-DMRs (Table 1).
This second analysis provided insight into the methylome of iPS cells as compared to ES cells. Although the two cell types had very similar DNA methylation, 71 DMRs distinguished them, with 51 showing hypermethylation and 20 showing hypomethylation in iPS cells (Supplementary Table 6). GO annotation of these DMRs showed significant enrichment of developmental processes in the genes that were hypermethylated in iPS cells as compared to ES cells (Supplementary Table 7). In 32 of the DMRs that distinguish iPS cells from ES cells, the DMRs were near genes of interest, including HOXA9 and two genes that encode the zinc finger proteins ZNF568 and ZFP112. In some cases, the methylation in iPS cells was intermediate between differentiated fibroblasts and ES cells; this was true, for example, of TBX5, which encodes a transcription factor that is involved in cardiac and limb development. In other cases, methylation in iPS cells differed from both fibroblasts and ES cells, suggesting that the iPS cells occupy a distinct and possibly aberrant epigenetic state. An example was PTPRT, encoding a protein tyrosine phosphatase involved in many cellular processes including differentiation. For some ES-iPS differences, the methylation levels changed in the same direction as for ES cells compared to fibroblasts, but to a greater degree; for example, methylation of the homeobox gene HOXA9 was greater in iPS compared to ES, whose methylation at this gene was greater than in fibroblasts.
We validated these data in two ways. First we verified the methylation results from CHARM by bisulfite pyrosequencing of nine DMRs, examining 2–6 CpGs within each DMR. For all of these genes, the bisulfite pyrosequencing data confirmed the differential methylation data from CHARM (Fig. 1b,c, Supplementary Fig. 2).
We also performed global gene expression analysis using the Affymetrix HGU133 Plus 2.0 microarray. There was a strong inverse correlation between differential gene expression and differential DNA methylation at R-DMRs that are within 500 bp of the transcriptional start site (TSS) of a gene: P < 10−3 for both hypermethylation and hypomethylation (Supplementary Fig. 3a, Supplementary Table 8). The significant association held true even when the R-DMR was within 1 kb of a TSS (P = 0.01 and P < 10−3 for hypermethylated and hypomethylated R-DMRs, respectively, Supplementary Fig. 3b). Moreover, this correlation was enhanced in DMRs that were in CpG island shores.
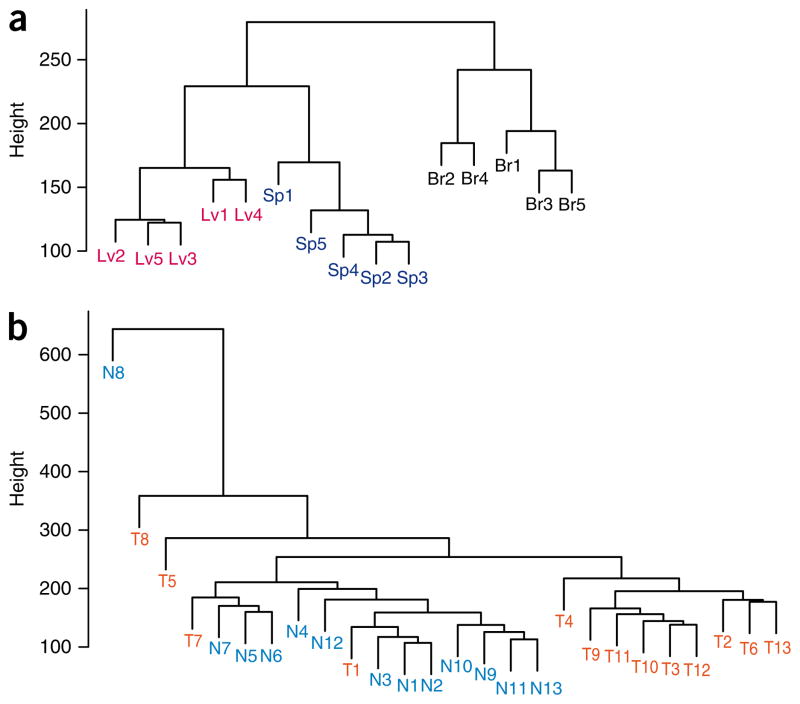
Furthermore, we performed an unsupervised cluster analysis using the R-DMRs to determine to what degree the methylation at these locations distinguished normal brain, liver and spleen from each other. Notably, there was complete separation of these three tissues, indicating that the sites of the methylation changes that occur during reprogramming normally distinguish these disparate tissues (Fig. 2a). In addition, the R-DMRs could largely distinguish normal colonic mucosa from colorectal cancer, indicating that the R-DMRs are also involved in abnormal reprogramming in cancer (Fig. 2b). As a test of significance, none of 1,000 randomly generated lists of the CHARM array regions of equal length and number clustered the tissues as well, as assessed either by whether they yielded a median euclidean distance among samples of a given tissue type at least as low as that found when using the R-DMRs, or yielded a median euclidean distance among samples of different tissue types at least as great as that found when using the R-DMRs. This was true both for the comparison between normal tissues and for the cancer-to-normal-tissue comparison.
Figure 2.
DNA methylation at R-DMRs distinguishes normal tissues from each other and colon cancer from normal colon. (a,b) The M values of all tissues from the 4,401 regions (FDR < 0.05) corresponding to R-DMRs (iPS cells compared to parental fibroblasts) were used for unsupervised hierarchical clustering comparing (a) normal brain, spleen and liver (denoted as Br, Sp and Lv, respectively) and (b) colorectal cancer and matched normal colonic mucosa (denoted as T and N, respectively). Notably, all of the normal brain, spleen and liver tissues are completely discriminated by the regions that differ between iPS cells and fibroblasts (R-DMRs). The major branches in the dendrograms correspond perfectly to tissue type. Furthermore, most of the colorectal cancer samples are discriminated from matched normal colonic mucosa by R-DMRs.
We compared the R-DMRs to those obtained in a genome-scale comparison of DNA methylation in colorectal cancer and matched normal colonic mucosa from the same individuals (C-DMRs)8. We had previously found a much smaller number of C-DMRs than T-DMRs (2,707 compared to 16,379), and 45% of the C-DMRs overlapped T-DMRs. Approximately 16% of the R-DMRs in the present study overlapped the C-DMRs of the previous study, whereas only 4.5% on average would be predicted by permutation analysis to overlap (P < 0.0001 based on 10,000 permutations) (Supplementary Table 9). Notably, hypomethylated R-DMRs (iPS compared to fibroblasts) were associated with hypermethylated C-DMRs (cancer compared to normal, P < 0.0001 based on 10,000 permutations) (Supplementary Table 9). Of the 294 DMRs found to overlap between hypomethylated R-DMRs and hypermethylated C-DMRs, 251 (85%) also overlapped bivalent chromatin marks. In contrast, hypermethylated R-DMRs were associated with hypomethylated C-DMRs (P < 0.0001 based on 10,000 permutations) (Supplementary Table 9). Of the 293 DMRs found to overlap between hypermethylated R-DMRs and hypomethylated C-DMRs, only 37 (13%) also overlapped bivalent chromatin marks. Because bivalent chromatin marks are associated with recruitment of Polycomb group proteins, these data suggest that there are two independent epigenetic mechanisms for cell reprogramming and tumorigenesis. One mechanism involves decreased DNA methylation and chromatin modifications at bivalent sites during reprogramming and increased methylation in cancer. The other mechanism involves increased methylation during reprogramming and loss of methylation in cancer.
In summary, we have found that epigenetic reprogramming of human fibroblasts to iPS cells involves substantial changes in DNA methylation largely affecting the same CpG island shores in T-DMRs that mark normal differentiation. It is notable that the R-DMRs completely distinguish brain from liver from spleen tissues and largely distinguish colon cancer from normal colon tissue. These results provide compelling evidence of the importance of CpG island shores and T-DMRs in both normal development and somatic cell reprogramming. Indeed, the target loci for normal tissue programming, epigenetic reprogramming to pluripotency and aberrant programming of cancers largely overlap. A secondary finding is that certain loci in iPS cells remain incompletely reprogrammed, whereas others are aberrantly reprogrammed, thus establishing that the methylation pattern of iPS cells differs both from those of the parent somatic cells and from those of human ES cells.
Our results contrast with prior studies that were primarily directed toward developing powerful new tools to analyze DNA methylation of targeted genomic regions rather than genome-scale studies of iPS cell methylation. Our more extensive genome-scale analysis of nine paired sets of iPS cells and parental fibroblasts detected roughly equal levels of hypo- and hypermethylation and revealed the predominant involvement of CpG island shores over islands themselves. Limitations of our study include the still-incomplete genome coverage of the CHARM array, which although including islands and shores, still does not examine single or very low density CpG methylation; the use of iPS cells derived from a single cell type; and the still relatively limited database for comparison of T-DMRs, involving only three normal tissue types, and C-DMRs, involving only one cancer type. Nevertheless, the present study reveals a host of loci that represent targets of epigenetic remodeling that are central to somatic cell reprogramming. These R-DMRs include both hypomethylated and hypermethylated regions and are a subset of the previously described T-DMRs and C-DMRs, indicating that these R-DMRs at CpG island shores are critical epigenetic targets for defining cell fate.
Finally, the colocalization of hypomethylated R-DMRs in iPS cells with hypermethylated C-DMRs in cancer and bivalent chromatin marks, and hypermethylated R-DMRs with hypomethylated C-DMRs and the absence of these marks, suggest two parallel mechanisms for epigenetic reprogramming in iPS cells and in cancer, one involving a loss of DNA methylation in iPS and a chromatin-dependent gain of DNA methylation in cancer and the other involving a gain of methylation in iPS and a chromatin-independent loss of DNA methylation in cancer.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgments
This research was supported by the US National Institutes of Health (A.P.F. and G.Q.D.). G.Q.D affiliations include the Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School; the Division of Hematology, Brigham and Women’s Hospital; Harvard Stem Cell Institute; and the Manton Center for Orphan Disease Research.
Footnotes
Accession codes. NCBI GEO: Gene expression microarray data and CHARM microarray data have been submitted under accession number GSE18111.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
A.D. performed CHARM, bisulfite pyrosequencing and data analysis; B.W. performed initial experiments and helped design and analyze the study; I.-H.P., J.R., S.L., J.M. and T.S. performed cell culture and prepared nucleic acids; P.M., M.J.A., R.I., B.H. and C.L.-A. performed statistical analysis; G.Q.D. and A.P.F. designed the study, supervised the experiments and wrote the paper with A.D. and B.W.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:485. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 11.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.